Abstract
Salmonella enterica serovar Typhimurium strain 798 is a clinical isolate from a pig and is known to be able to cause persistent, asymptomatic infections. This strain also is known to exist in two phenotypes (adhesive and nonadhesive to enterocytes) and can switch between the two phenotypes at a rate consistent with phase variation. Cells in the adhesive phenotype are more readily phagocytosed by leukocytes than nonadhesive cells. Once in a leukocyte, adhesive-phase cells survive while nonadhesive-phase cells die. In the present study, nonadhesive mutants were obtained with the transposon TnphoA. A nonadhesive mutant was selected for study and was shown by electron microscopy not to produce fimbriae. The gene encoding the adhesin was cloned and sequenced. Based on its sequence, the adhesin was shown to be FimA, the major subunit of type 1 fimbriae. The nonadhesive mutant was attenuated in its ability to colonize both mouse and pig intestines, but remained capable of systemic spread in mice. The nonadhesive mutant was phagocytosed to the same extent as parental cells in the adhesive phase and then survived intracellularly. These results demonstrated that type 1 fimbriae were important for attachment to enterocytes and promoted intestinal colonization. However, they were not important in promoting phagocytosis or intracellular survival.
Salmonella enterica is a ubiquitous, gram-negative enteric pathogen found in humans, livestock, wild mammals, reptiles, birds, and even insects. S. enterica produces a wide spectrum of disease ranging from diarrhea, to septicemia, meningitis, and osteomyelitis (1, 27). S. enterica also causes asymptomatic persistent infections (28). Adhesion to the intestinal epithelium is generally considered to be the first step in pathogenesis preceding invasion (10-12). Fimbriae (pili) are believed to mediate this adhesive process. It has been reported that among the vast array of different serotypes, S. enterica can produce at least nine fimbrial types, only some of which have been well characterized (3-5, 23). A single bacterial isolate often can express more than one type of fimbria. Following adhesion, S. enterica organisms invade and subsequently cross the mucosal epithelium. Early studies indicated that brush border epithelial cells were an important route of invasion for S. enterica (24). However, later studies suggested that the primary site of invasion and spread is the ileal Peyer's patch and possibly the cecal lymphoid patches (6, 14). These lymphoid areas are covered by the follicle-associated epithelium on the luminal surface. M cells, regarded as antigen sampling cells, are an important cellular component of this luminal covering and are a target of invasion. S. enterica serovar Typhimurium utilizes different types of fimbriae to target attachment to different cell types within the intestinal epithelium (4). Long polar fimbriae (Lpf) confer attachment to M cells (5), while plasmid-encoded fimbriae (Pef) confer attachment to murine small intestines and to certain tissue culture cells (3). Type 1 fimbriae are produced by S. enterica serovar Typhimurium and confer attachment to a wide variety of cells (13, 18, 25). Once salmonellae have invaded, they may remain locally within the intestinal tissues or may spread systemically within macrophages and neutrophils. Systemic spread occurs via the lymphatics and the bloodstream with seeding of the reticuloendothelial system and replication in the spleen and liver. In order to cause systemic disease, the organism must survive and replicate within macrophages.
S. enterica is a commonly recognized pathogen of swine. The two serovars isolated most frequently from swine are Choleraesuis and Typhimurium. Infection with S. enterica serovar Choleraesuis usually results in death due to septicemia, diarrhea, and/or pneumonia (26). In contrast, S. enterica serovar Typhimurium in pigs generally causes a self-limiting diarrhea that rarely results in mortality. A characteristic of S. enterica serovar Typhimurium, which is a cause for concern, is its ability to persistently colonize swine without producing clinical signs (28). Persistently infected pigs may shed the organism in their feces for anywhere from a few weeks to several months following exposure (28). The stresses associated with transportation and feed withdrawal prior to slaughter cause pigs to regain shedder status (15). This shedding can result in contamination of food products during the slaughter and processing stage. When pigs are persistently infected, organisms can be isolated most frequently from tonsils, caudal jejunum, ileum, cecum, colon, and mandibular and ileocolic lymph nodes but are difficult to find in feces (15, 28). Currently, little is known about the mechanism of persistent infection by S. enterica serovar Typhimurium. It is our hypothesis that persistence is the result of prolonged colonization of the mucosal surface of the intestines, through fecal-oral reinoculation, or both. Identification of carrier animals is complicated by the fact that the organism is shed intermittently, most often during periods of stress such as food deprivation and transport to the abattoir (15). Furthermore, detection of Salmonella from animals requires lengthy enrichment techniques, and the techniques probably are not particularly sensitive.
In a previous publication from this laboratory, a clinical isolate of S. enterica serovar Typhimurium strain 798 was shown to undergo phase variation with cells switching between an adhesive and nonadhesive phenotype (16). Cells in the adhesive phenotype, designated ι519, adhered to porcine intestinal epithelial cells in vitro, while cells in the nonadhesive phenotype, designated ι518, did not adhere to epithelial cells. The estimated rate of phase variation was 10−2 to 10−5. In addition, it was shown that cells in the adhesive phase produced 10 to 15 unique envelope-associated proteins, were more readily phagocytosed by porcine leukocytes in vitro, and once phagocytosed were capable of intracellular survival. Cells of the nonadhesive phenotype did not survive within phagocytes. Using electron microscopy, the presence of fimbriae on cells of the adhesive phenotype was observed. Fimbriae were not observed on cells of the nonadhesive phenotype. Earlier studies stressed the importance of adhesion for virulence by demonstrating that the loss of adhesiveness correlated with decreases in virulence (8, 10). However, subsequent publications indicated that knocking out of a single fimbrial type was not sufficient to attenuate S. enterica serovar Typhimurium (4). In the present study, transposon mutagenesis was utilized to create mutants defective for attachment to porcine intestinal brush borders in vitro. Based on the analysis of the nonadhesive mutants, the adhesin was identified as type 1 fimbriae. Furthermore, type 1 fimbriae were shown to promote intestinal colonization in vivo.
MATERIALS AND METHODS
Bacterial strains and media
The strains used in this study are described in Table 1. Trypticase soy agar (TSA; Baltimore Biological Laboratories) was used as a semisolid medium and was supplemented with the appropriate antibiotics when required (tetracycline at 12.5 μg/ml, kanamycin at 50 μg/ml, and ampicillin at 50 μg/ml). 5-Bromo-4-chloro-3-indolyl phosphate [XP] (40 μg/ml) was added to TSA to screen for alkaline phosphatase activity. Alkaline phosphatase-producing cells appear as blue colonies on this medium. Terrific Broth (TB) and Luria-Bertani (LB) broth were prepared as previously described (21) and used to grow bacteria for the isolation of plasmids or genomic DNA. Tryptone phosphate broth (Difco) (TPB) was used to grow cells when enhanced adhesiveness was desired (16). P22 broth was prepared as previously described to prepare bacteriophage P22HT int lysates (20). Green plates or EBU plates were prepared as previously described and used to screen for pseudo-lysogens and bacteriophage-free colonies (20).
TABLE 1.
S. enterica serotype Typhimurium strains used in this study
| Strain no. | Origin | Reference |
|---|---|---|
| 798 | Pig | 27 |
| ι518 | Nonadhesive phenotype of 798 | 15 |
| ι519 | Adhesive phenotype of 798 | 15 |
| ι519′ | phoN52::Tn10(Tet) in ι519 | This study |
| Mutant 14 | fimA::TnphoA in ι519′ | This study |
| ι939 | Restored fimA in mutant 14 | This study |
| LT2 | S. Maloy | |
| MS308 | LT2/phoN::Tn10 (Tet) | S. Maloy |
| MS1621 | LT2, TnphoA | S. Maloy |
| ISF145 | pISF101 | S. Clegg |
| CC118 (E. coli) | λpir, pRE107 | 8 |
Transduction and TnphoA mutagenesis.
A high-titer (1.3 × 1012) lysate of bacteriophage P22 HT int was initially prepared with S. enterica serovar Typhimurium strain LT2. Bacteriophage P22 was grown on S. enterica serovar Typhimurium strain MS308 containing phoN52::Tn10(Tet) and the phoN mutation in MS308 was transduced into strain ι519. This step was employed to reduce nonspecific acid phosphatase activity prior to TnphoA mutagenesis. The phoN mutant was designated ι519′. The bacteriophage P22 lysate prepared on strain MS1621 was used to transduce TnphoA into strain ι519′ as described previously, except the initial incubation was at 30°C to enhance transduction efficiency. Transductants were plated onto TSA containing kanamycin and XP to identify TnphoA insertions (Kanr) that generated exported fusion proteins (blue color indicating expression of phoA). Five hundred-twenty colonies that were blue on XP and were Kanr were selected and cleaned up from residual phage by using green plates or EBU (20).
Restoration of adhesiveness.
A 4.7-kb SstI-SphI fragment containing fimA from the type 1 fimbrial gene cluster encoded on pISF101 (obtained from S. Clegg, University of Iowa) was cloned into the suicide vector pRE107 (9). The fragment was then introduced into the fimA::TnphoA mutant 14 by allelic exchange. Restoration of fimbriation was demonstrated by agglutination of yeast cells. The agglutination was inhibited by d-mannose.
Absorbed antiserum and pellicle selection assay.
Anti-ι519 antiserum was prepared previously by immunizing rabbits with envelope extracts from ι519. Cross-reacting material was removed from the serum by absorption with ι518 as described previously (16). The antiserum was used at a 1:10 dilution, which caused agglutination of ι519′ in less than 20 s, but not of ι518. For the pellicle selection assay, cells were inoculated into LB broth and incubated without shaking at 37°C for 7 to 10 days. If a pellicle formed, it was found at the meniscus of the tube at the liquid-air interface.
In vitro adhesion assays using porcine enterocytes.
The assay employed was described previously (16). Briefly, enterocytes were harvested from 0- to 1-day-old pigs by scraping the mucosal surface of the small intestine. The enterocyte concentration was adjusted to 105 to 106 cells per ml in phosphate-buffered saline (PBS). Equal volumes of enterocytes (∼105) and test organisms (∼2 × 108) were mixed and incubated for 1 h with gentle shaking. Free bacteria were removed by centrifugation and washed with PBS. Adherent bacteria were counted by visualization with a phase-contrast microscope. The number of adherent bacteria from a minimum of 20 intact epithelial cells with brush borders was determined. Mutants considered nonadhesive had an average of 0 to 4 bacteria per enterocyte (i.e., the same as nonadhesive ι518 cells). The mutants characterized as adhesive had an average of >10 bacteria per enterocyte (same as strains ι519 and ι519′). Strains ι518, ι519, and ι519′ were included as controls in each experiment.
Mouse infections.
Groups of six ICR mice (Harlan) were inoculated orally with a mixture containing equal numbers of ι519′ cells and the nonadhesive mutant 14 or ι939, the strain in which type 1 fimbrial production was restored to wild type. The final inoculum contained ∼5 × 107 cells of each strain. The actual concentration of each cell type was determined by plate count prior to oral inoculation of mice. Two days postchallenge, mice were euthanatized by exposure to carbon dioxide, and the following tissues were aseptically harvested: thymus, lung, liver, spleen, cecum, ileum, and colon. The tissues were weighed, homogenized, serially diluted in sterile saline, plated on TSA containing tetracycline and kanamycin, and incubated overnight at 37°C. The following day, colonies were counted. Kanr colonies were the nonadhesive mutant containing the TnphoA insertion, while tetracycline-resistant colonies represented both the parent and mutant [due to the phoN::Tn10(Tet) insertion]. The number of parent colonies was calculated by subtracting the number of mutants (kanamycin resistant) from the total (tetracycline resistant). A competition index was calculated by dividing the number of wild-type cells by the number of mutant cells. For mortality time course studies, groups of BALB/c mice (five per group) were orally inoculated with either S. enterica serotype Typhimurium strain ι519′ or the nonadhesive mutant 14. BALB/c mice were used because, unlike for ICR mice, the result of oral challenge with S. enterica serovar Typhimurium is systemic disease and death. The challenge doses were 108, 109, or 1010. The time to death after challenge was recorded.
Swine colonization studies.
Two groups of eight pigs each were inoculated orally with either the wild-type parental strain 798 or the fimA mutant 14. The inoculum was in a gelatin capsule filled with ∼0.75 g of pelleted feed and ∼0.8 ml of culture (∼2 × 108 CFU/ml). The capsule was administered orally. Four animals from each group were euthanatized at 1 or 2 weeks postchallenge. Tissues at the ileocecal junction and cecum were harvested, and the concentration of challenge organism was determined by most-probable-number analysis after tissue homogenization, enrichment, and plating (29).
Molecular cloning of fimA::TnphoA.
Total chromosomal DNA was extracted from cells grown in TB by a procedure employing hexadecyltrimethyl ammonium bromide (CTAB) (2). Total DNA was digested with the enzyme SacI (GIBCO) at 37°C for 2 h. RNase A (final concentration, 100 μg/ml) was added to the chromosomal digests during the final 30 min of the restriction enzyme digestion. The DNA subsequently was subjected to phenol extraction. The digested chromosomal DNA was ligated into the dephosphorylated SacI site of the vector pGEM4Z by using T4 DNA ligase (GIBCO) with overnight incubation at room temperature. Ligated DNA was introduced into Escherichia coli strain DH5α by electroporation with an InVitrogen electroporation system (Carlsbad, Calif.). Cells were plated onto TSA with kanamycin, ampicillin, and XP as previously described. Blue colonies were picked. The site of the TnphoA insertion was determined by DNA sequencing with a Sequenase version 2.0 DNA sequencing kit (U.S. Biochemicals, Cleveland, Ohio). The following sequencing primers were used: PhoA primer (5′ AGTAATATCGCCCTGAGCAG 3′), FimA forward (5′ GGTTACCGTAATCCCTCGTC 3′), FimA reverse (5′ TTCGAAGGTAACTGGTTAAC 3′), FimH forward (5′ AATATACTCAGCGCTATTGC 3′), and FimH reverse (5′ GCGCAGTAATCGGCCCTTCC 3′).
Phagocyte uptake and killing assays.
Porcine leukocytes were isolated from freshly collected blood and purified as described previously (16). Blood was collected from healthy adult male pigs (15 to 20 months old). Solutions were adjusted to a final concentration of 107 cells per ml. Phagocyte uptake studies were performed by mixing leukocytes (106) with S. enterica serotype Typhimurium (108) in PBS and incubating this mixture at 37°C as described previously (17). Excess bacteria were removed by centrifugation at 800 × g for 10 min and washed once with PBS. Any remaining extracellular bacteria were killed by the addition of gentamicin (100 μg/ml) and incubation at 37°C for 30 min. Leukocytes were collected and washed with PBS and resuspended in PBS containing sodium dodecyl sulfate (SDS [0.05%]) and gentamicin (2 μg/ml). The samples were diluted serially in sterile saline and plated in duplicate on TSA. For phagocyte killing studies, after the first addition of gentamicin, the cells were incubated at 37°C. Samples were removed at 0, 30, 60, 90, 120, and 240 min, and the number of viable organisms was determined as described above. All strain comparisons used leukocytes from the same pig.
Electron microscopy.
S. enterica serovar Typhimurium cultures were mixed with an equal volume of sodium phosphotungstate (1%). A drop was placed on a Formvar-coated grid, and excess liquid was removed with a piece of filter paper. The grids were air dried and observed in a JOEL transmission electron microscope.
Statistical test.
Statistical significance was evaluated by Wilcoxon paired-samples analysis.
RESULTS
Generation of a nonadhesive mutant.
Because the enterocyte-specific adhesin was expected to be located on the outer surface of S. enterica serovar Typhimurium, a mutagenesis strategy using TnphoA was employed to create nonadhesive mutants. In this mutagenesis scheme, selection for an active fusion between the genes encoding the adhesin and alkaline phosphatase (phoA) could be detected by blue coloration of colonies when grown on XP. TnphoA was transduced into a phoN-deficient mutant (ι519′) and 520 mutants that were blue on XP and were resistant to kanamycin were selected for further screening.
The 520 mutants were screened with the in vitro enterocyte binding assay. Four mutants were found to be nonadhesive. These mutants were then screened for the loss of agglutination by using the adsorbed antiserum specific for the adhesive parent. This antiserum was previously shown to agglutinate cells of the adhesive phenotype but not of the nonadhesive phenotype. Therefore, use of this antiserum helped to determine the phenotypic phase of the cells. Colonies containing cells that agglutinated were considered to be in the adhesive phase (i.e., ι519), while nonagglutinating cells were in the nonadhesive phase (i.e., ι518). Mutant 14 agglutinated with the antiserum, although it did not adhere to enterocytes in vitro. Mutants 398, 430, and 457 were not agglutinated by the antiserum and did not adhere to enterocytes in vitro. The loss of adhesion could result from phase variation, rather than a mutation in the adhesin gene.
Thus, mutants 398, 430, and 457 could simply be nonadhesive phase variants. Therefore, mutant 14 was selected for further study, since it retained the phenotype of the adhesive parent as measured by serum agglutination while selectively losing adhesiveness.
Characterization of the nonadhesive mutant.
To determine the location of the TnphoA insertion and to determine which gene was mutated as a result of the transposon insertion, the TnphoA element and DNA flanking the transposon from mutant 14 were cloned into pGEM4Z. This piece of DNA was sequenced with a primer specific for the 5′ end of TnphoA. The DNA sequence was then subjected to a homology search with BLASTN and the database maintained by the National Center for Biotechnology Information (GenBank). The sequence flanking TnphoA was found to be identical to that of fimA, the gene encoding the major structural subunit (pilin) of type 1 fimbriae. Based on the sequence of fimA, the entire gene was PCR amplified, and using a primer specific to the 5′ end of fimA, the entire gene was sequenced. Again it was identical to the published sequence for fimA. To determine if the type 1 adhesin gene (fimH) was the same as what has been published for other S. enterica serotype Typhimurium strains, a PCR product containing the entire fimH sequence was prepared and sequenced. The sequence of fimH from ι519 was identical to the sequence stored in GenBank. Out of curiosity, the three nonadhesive, nonagglutinating mutants were analyzed for the location of the TnphoA insertions. Interestingly, in each case, the transposon was located in fimA.
Mutant 14 was examined by electron microscopy to determine if it produced fimbriae. As shown in Fig. 1A, mutant 14 did not produce fimbriae. In comparison, the parent ι519′ did produce fimbriae that looked like type 1 fimbriae (Fig. 1B). Cells that produce type 1 fimbriae produce a pellicle at the meniscus of a broth culture if incubated for 7 to 10 days without shaking. As another means of testing whether mutant 14 was a phase variant, it was subjected to the pellicle selection assay. No pellicle formed, consistent with the conclusion that this was a nonadhesive mutant that was otherwise in the adhesive phase.
FIG. 1.
Electron micrographs of mutant 14 (A) and the parent strain, ι519′ (B). The cells were negatively stained with phosphotungstate. Fimbriae can be seen on ι519′ but not on mutant 14.
Mouse challenge model.
To examine the in vivo effects of the mutation in the adhesin of mutant 14, oral challenges of mice were performed. Competition studies between mutant 14 and the parental strain ι519′ were performed to determine if type 1 fimbriae provided a competitive advantage in the colonization of and dissemination to various tissues. The two strains were mixed together in equal concentrations and administered orally. Two days postchallenge, the mice were sacrificed, and the concentrations of the two strains were determined by plating on media containing selective antibiotics (kanamycin or tetracycline). The results are shown in Table 2. A competition index value of less than 1 meant that the mutant was able to outcompete the parent, while a competition index of greater than 1 meant that the mutant was less competitive in the site than the parent. Compared to the wild-type parent, mutant 14 was less able to colonize the three mouse intestinal tissues examined: ileum, cecum, and colon. The calculated ratios for the intestinal sites ranged from 8 to 20, and each was statistically significant (P < 0.05). Peripheral tissues (lung, liver, and spleen) also were analyzed for the presence of each challenge strain. In these tissues, there were no significant differences in the concentrations of the parent and the mutant.
TABLE 2.
Results from oral challenges of mice with the parent S. enterica serovar Typhimurium strain, the fimA mutant, and the fimA-repaired strain
| Tissue | Competition index
|
|
|---|---|---|
| ι519′/mutant 14 | ι519′/ι939 | |
| Lung | 2.57 | NDb |
| Liver | 1.25 | ND |
| Spleen | 1.0 | ND |
| Cecum | 8.0a | 0.75 |
| Ileum | 7.1a | 0.29 |
| Colon | 20a | 0.35 |
Significant at P ≤ 0.05 by Wilcoxon paired-sample analysis.
ND, not done.
To determine if the defect in intestinal colonization was the result of the fimA mutation, the mutated fimA gene was replaced with the wild-type gene by allelic exchange (strain ι939) and used in the mouse challenge along with the wild-type parent. For each intestinal tissue examined, the reversion to wild type restored the ability of the parent to colonize the site (Table 2).
Both S. enterica serotype Typhimurium strain ι519 and mutant 14 were used individually to orally challenge BALB/c mice. Both strains had very high 50% lethal dose (LD50) values (between 1 × 109 and 2 × 1010). It was noted, however, that the mean time to death at all challenge doses was delayed approximately 4 days for mutant 14 (Table 3). The mean times to death were 11 days for ι519 and 15 days for mutant 14 when the challenge dose was 109. The difference in the time to death was not statistically significant (Wilcoxon paired-samples analysis), even though it was a reproducible effect in three replicates and at three doses.
TABLE 3.
Time to death of BALB/c mice after challenge with ι519 or mutant 14
| S. enterica serovar Typhimurium dose | Time to death (days)a
|
|
|---|---|---|
| Strain ι519 | Mutant 14 | |
| 109 | 12.5 | 18 |
| 1010 | 11 | 14 |
| 1011 | 11.5 | 15 |
At the 109 dose, only 40% mortality was achieved. Therefore, all values are stated for 40% mortality.
Pig model.
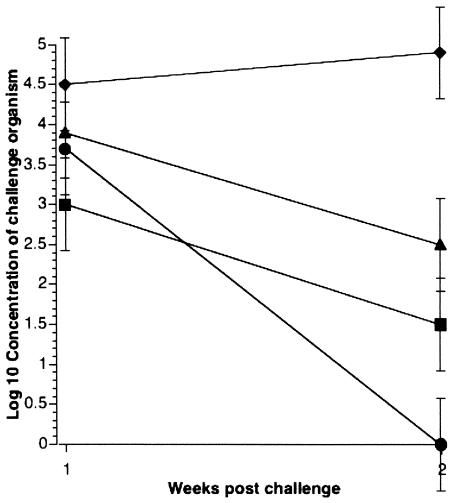
The mouse model provided evidence that the fimA mutant was less capable than its parent in colonizing intestines. Since this isolate of S. enterica serovar Typhimurium was originally isolated from a pig, we wanted to know if the fimA mutant was less capable of colonizing pig intestines as well. Therefore, postweaned pigs were challenged orally with strain 798 or mutant 14. Strain 798, being the parent of ι518 and ι519, contains a mixture of both. Samples were collected at 1 or 2 weeks postchallenge. As indicated in Fig. 2, the mutant strain was cleared more rapidly from the ileocecal junction and mid-ileum than the parental strain. At the ileocecal junction 2 weeks postinfection, the parental strain was recovered in increasing numbers, while the mutant had decreased by 1.5 log10. In the mid-ileum, both strains were being cleared; however, while strain 798 was still recovered 2 weeks postchallenge, the mutant could no longer be detected.
FIG. 2.
Colonization of pig intestines by S. enterica serovar Typhimurium 1 and 2 weeks postchallenge. Symbols show results for strain 798 in the mid-ileum (▪) and ileocecal junction (♦) and mutant 14 in the mid-ileum (•) and ileocecal junction (▴). Results are the mean of four pigs per time point.
Phagocyte uptake and intracellular survival.
Phagocyte uptake and intracellular survival studies were performed to determine if type 1 fimbriae were important in either step in pathogenesis. It was demonstrated previously that ι518 was less readily phagocytosed but then more rapidly killed than ι519 (16). In the present experiments, the fimA mutant retained characteristics of the adhesive phase. The mutant 14 cells were phagocytosed as readily as ι519 and then survived intracellularly (Fig. 3). Thus, type 1 fimbriae did not appear to enhance phagocyte uptake or intracellular survival.
FIG. 3.
Uptake (A) and survival (B) of S. enterica serovar Typhimurium strains ι518 (▪), ι519′ (•), and mutant 14 (▴) in porcine leukocytes. Representative results from three replicates are presented. The differences in the uptake at 30 and 60 min between ι518 and ι519 and ι518 and mutant 14 were statistically significant (P = 0.0001). Comparison of survival at 120 and 240 min between ι518 and ι519 was statistically significant (P = 0.0004), as was that between ι518 and mutant 14 after adjusting for the differences in the initial numbers of cells in the leukocytes.
DISCUSSION
Previously, two distinct phenotypes of S. enterica serovar Typhimurium strain 798 were described (16). This strain was originally isolated from a pig and is able to cause asymptomatic persistent infections of pigs (28). The two phenotypes were distinguishable based on the expression of several different traits, including the ability to adhere to porcine enterocytes, the expression of fimbriae, the production of unique outer membrane proteins, agglutination by phenotype-specific antiserum, and the ability to colonize phagocytes (invasion and survival). It was demonstrated that cells switched between the two phenotypes at a rate of 10−2 to 10−5 per generation and that each of the traits was simultaneously turned on or off in response to the phenotype switch. Because the frequency of switching between phenotypes was high, it was assumed that this change represented phase variation. Phase variation is a common phenomenon in bacteria, and several mechanisms have been identified that regulate this process. Clegg et al. have studied phase variation of type 1 fimbriae in S. enterica serovar Typhimurium, and unlike the process in Escherichia coli, phase variation is not mediated by the inversion of a DNA element containing a promoter (7).
In the studies described here, transposon mutagenesis was employed to knock out the enterocyte-specific adhesin produced by cells of the adhesive phenotype of strain 798. Using TnphoA mutagenesis, a mutant was created that lost the ability to adhere to enterocytes in vitro. This mutant retained the other properties of the adhesive phenotype, including agglutination by adhesive phenotype-specific antiserum, enhanced uptake into leukocytes, and intracellular survival in leukocytes. In addition, we have found that the two phenotypes produce different colored (white and blue) colonies when plated on EBU agar (S. Patterson and R. E. Isaacson, unpublished result). The fimA mutant 14 retains the adhesive phenotype's color (white) when plated on EBU. Thus, it was concluded that the mutant lost the ability to attach to enterocytes as a result of the mutation and was not a simple nonadhesive-phase variant.
To identify the site of the mutation in the nonadhesive mutant 14, a DNA fragment containing the transposon insertion (TnphoA) was cloned and sequenced. The site of the TnphoA insertion in mutant 14 was shown to be in fimA, the major fimbrial subunit of type 1 fimbriae. Our original studies on this strain suggested that the enterocyte-specific adhesin was not type 1 fimbriae, because adhesion was not inhibited by d-mannose. Therefore, the entire fimA gene and the functional adhesin gene, fimH, were completely sequenced. No differences in the sequences were detected compared to the previously published sequences for fimA and fimH. Thus, we conclude that type 1 fimbriae are an important adhesin mediating attachment to enterocytes. Other studies previously have shown that type 1 fimbriae possess adhesive characteristics that may be important in intestinal colonization (13, 18, 25). However, while previous studies were targeted toward the study of type 1 fimbriae, we employed a genetic approach to knock out the enterocyte-specific adhesin without regard to a priori assumptions about specific adhesins. The fact that type 1 fimbriae were selected corroborates the other studies and strongly suggests an important role for this adhesin at the epithelial cell level. Another important difference between this study and previous studies is that we used an S. enterica serovar Typhimurium strain that is not highly virulent (strain 798) in the mouse model but is capable of causing long-term (chronic) asymptomatic infections of pigs.
S. enterica serovar Typhimurium is an invasive organism, and adhesion is thought to be a prerequisite for invasion (10-12). M cells of the ileal Peyer's patches are currently considered the major route of invasion and systemic spread (6, 14, 19). Invasion across enterocytes is known to occur as well (24). Once S. enterica serovar Typhimurium crosses the intestinal epithelium, it is phagocytosed and carried throughout the reticuloendothelial system by macrophages residing in the gut-associated lymphoid tissues underlying the M cells. In order to produce systemic disease, therefore, the organism must survive within phagocytes. Previous studies demonstrated that cells of the nonadhesive strain were phagocytosed less readily and killed more rapidly by porcine phagocytes (16). Because fimbriae are both adhesive and highly antigenic, it was hypothesized that these structures could be involved in entry into leukocytes. However, uptake and killing studies performed with mutant 14 showed no difference in the entry into leukocytes when compared with the adhesive parent. Likewise, once phagocytosed, the fimA mutant was equally insensitive to intracellular killing as its parent (ι519′). Thus, we conclude that type 1 fimbriae are not required for phagocyte uptake and intracellular survival. Therefore, other virulence-related genes expressed by cells of the adhesive phase are involved in these virulence-related activities.
To determine if type 1 fimbriae were important in vivo, pigs and mice were challenged with the wild-type strain and mutant 14. In pigs, mutant 14 was cleared more rapidly from the mid-ileum and the ileocecal junction than its wild-type parent. In mice, the ability to colonize the intestinal tract and peripheral sites was assessed by a competition between the parent and fimA mutant. At all of the intestinal sites sampled, the mutant was less able to colonize than its parent. These differences were statistically significant. However, it was very interesting that this advantage did not extend to peripheral sites (lung, liver, and spleen). We did note that with mutant 14, there was a reproducible increase in time to death of BALB/c mice. Murray and Lee identified a similar response in hilA mutants (22). These results lead us to hypothesize that type 1 fimbriae are important in vivo and mediate localized colonization of intestines. The reason that the mutant still is able to distribute systemically is because it is not deficient in its ability to interact with and enter M cells. The M cell-specific adhesin is Lpf (long polar fimbriae) (5), and since mutant 14 contains a single insertion in fimA, we would expect that this mutant still produces Lpf. Since mutant 14 retains other traits of the adhesive phenotype, it still enters and survives in leukocytes and therefore retains the ability to spread systemically. The reason why there should be a delay to death in BALB/c mice is not clear even though the mutant appears to spread systemically in ICR mice as well as the wild-type parent. This could result if tissue distribution and death over time are also dependent on enterocyte-specific adhesion. It also could be a trait difference between ICR mice and BALB/c mice.
While strain 798 produces transient diarrhea in pigs, its real significance is its ability to cause persistent infections. Carrier animals create a potential source of contaminated pork products. The results of these studies are consistent with the hypothesis that the ability to persistently colonize a host is mediated by attachment to enterocytes rather than to M cells, while systemic spread is dependent upon the interaction with M cells. Studies by Bäumler et al. showed that mutants lacking the ability to produce Lpf failed to attach to M cells and did not spread systemically in mice (5). On the other hand, the fimA mutant described in these studies failed to attach to enterocytes and poorly colonized both mouse and pig small intestines, but still disseminated to peripheral sites as well as the parental strain. These results are consistent with type 1 fimbriae being an enterocyte-specific adhesin and a major contributor to persistent, asymptomatic infections. As a result, this strain may be an important source of contamination of meats at slaughter plants.
Acknowledgments
We thank R. Weigel for assistance with the statistical analyses.
This work was supported by a grant from the United States Department of Agriculture, National Research Initiatives Competitive Grants Program (no. 98-02811).
Editor: A. D. O'Brien
REFERENCES
- 1.Acha, P. N., and B. Szyfres. 1987. Zoonoses and communicable diseases common to man and animals, 2nd ed. Pan American Health Organization, Washington, D.C.
- 2.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology, vol. 1. John Wiley and Sons, New York, N.Y.
- 3.Bäumler, A. J., R. M. Tsolis, F. A. Bowe, J. G. Kusters, S. Hoffmann, and F. Heffron. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clegg, S., L. S. Hancox, and K.-S. Yeh. 1996. Salmonella typhimurium fimbrial phase variation and FimA expression. J. Bacteriol. 178:542-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duguid, J. P., M. R. Darekar, and D. W. Wheater. 1976. Fimbriae and infectivity in Salmonella typhimurium. J. Med. Microbiol. 9:459-473. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 10.Gahring, L. C., F. Heffron, B. B. Finlay, and S. Falkow. 1990. Invasion and replication of Salmonella typhimurium in animal cells. Infect. Immun. 58:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan, J. E., and R. Curtiss. 1989. Cloning and molecular charcterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginocchio, C., J. Pace, and J. E. Galan. 1992. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of Salmonellae into cultured epithelial cells. Proc. Natl. Acad. Sci. USA 89:5976-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancox, L. S., K. S. Yeh, and S. Clegg. 1997. Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 19:289-296. [DOI] [PubMed] [Google Scholar]
- 14.Hohmann, A. W., G. Schmidt, and D. Rowley. 1978. Intestinal colonization and virulence of Salmonella in mice. Infect. Immun. 22:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isaacson, R. E., L. D. Firkins, R. M. Weigel, F. A. Zuckermann, and J. A. DiPietro. 1999. Effect of transportation and feed withdrawal on shedding of Salmonella typhimurium among experimentally infected pigs. Am. J. Vet Res. 60:1155-1158. [PubMed] [Google Scholar]
- 16.Isaacson, R. E., and M. Kinsel. 1992. Adhesion of Salmonella typhimurium to porcine intestinal epithelial surfaces: identification and characterization of two phenotypes. Infect. Immun. 60:3193-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindquist, B. L., E. Lebenthal, P.-C. Lee, M. W. Stinson, and J. M. Merrick. 1987. Adherence of Salmonella typhimurium to small-intestinal enterocytes of the rat. Infect. Immun. 55:3044-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucas, R. L., and C. A. Lee. 2000. Unraveling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 20.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Miller, J. H. 1971. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Murray, R. A., and C. A. Lee. 2000. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 68:5050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swenson, D. L., S. Clegg, and D. C. Old. 1991. The frequency of fim genes among Salmonella serovars. Microb. Pathog. 10:487-492. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi, A. 1967. Electron microscopic studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am. J. Pathol. 50:109-136. [PMC free article] [PubMed] [Google Scholar]
- 25.Thankavel, K., A. H. Shah, M. S. Cohen, T. Ikeda, R. G. Lorenz, R. Curtiss, and S. N. Abraham. 1999. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J. Biol. Chem. 274:5797-5809. [DOI] [PubMed] [Google Scholar]
- 26.Turk, J. R., W. H. Fales, C. Maddox, M. Miller, L. Pace, J. Fischer, J. Kreeger, G. Johnson, S. Turnquist, J. A. Ramos, and H. S. Gosser. 1992. Pneumonia associated with Salmonella choleraesuis infection in swine: 99 cases (1987-1990). J. Am. Vet. Med. Assoc. 201:1615-1616. [PubMed] [Google Scholar]
- 27.Williams, L. P. 1980. Salmonellosis, p. 11-34. In J. H. Steel (ed.), CRC handbook in zoonosis. CRC Press, Inc., Boca Raton, Fla.
- 28.Wood, R. L., A. Pospischil, and R. Rose. 1989. Distribution of persistent Salmonella typhimurium infection in internal organs of swine. Am. J. Vet. Res. 50:1015-1021. [PubMed] [Google Scholar]
- 29.Wood, R. L., and R. Rose. 1992. Populations of Salmonella typhimurium in internal organs of experimentally infected carrier swine. Am. J. Vet. Res. 53:653-658. [PubMed] [Google Scholar]