Abstract
Mice lacking the interleukin 7 receptor (IL-7R) generate α/β T cells at a detectable but greatly reduced rate, but γ/δ T cells are completely absent. The special role of IL-7R signaling in γ/δ T cell development has remained unclear. IL-7Rα−/− mice exhibit a paucity of γ gene rearrangements. This striking observation can be explained by a defect in T cell receptor (TCR)-γ gene rearrangement, a defect in TCR-γ gene transcription leading to death of γ/δ lineage cells, and/or a requirement for IL-7R in commitment of cells to the γ/δ lineage. To determine the role of IL-7R signaling in γ/δ T cell development, we examined transcription of a prerearranged TCR-γ transgene in IL-7Rα−/− mice, as well as the effects of IL-7 on transcription of endogenous, rearranged TCR-γ genes in α/β lineage cells. The results demonstrate that IL-7R–mediated signals are necessary for the normal expression of rearranged TCR-γ genes. Equally significant, the results show that the poor expression of TCR-γ genes in IL-7Rα−/− mice is responsible for the selective deficit in γ/δ cells in these mice, since a high copy TCR-γ transgene exhibited sufficient residual expression in IL-7Rα−/− mice to drive γ/δ cell development. The results indicate that the absence of γ/δ T cells in IL-7Rα−/− mice is due to insufficient TCR-γ gene expression.
Keywords: T cell development, interleukin 7, lineage commitment, T cell receptor gene rearrangement, transcription
Interleukin (IL)-7 is crucial for the survival and normal differentiation of lymphocytes. It is produced by bone marrow and thymic stromal cells from embryonic day 12 onward 1. IL-7R expression is first detected on the lymphoid lineage-restricted progenitor in the bone marrow 2. Subsequent immature intermediates in T cell development express it at varying levels. The IL-7R complex consists of at least two subunits, the unique IL-7Rα chain and the common γ (γc) chain that is also a component of the high-affinity receptors for IL-2, -4, -9, and -15 3.
IL-7 appears to play a role at multiple stages of T and B lymphocyte development. Analysis of mice deficient for IL-7 4 or its receptor 5 6 7 demonstrates that the cytokine is indispensable for the normal maintenance and proliferation of early thymic progenitor cells, before expression of the TCR. For example, IL-7−/− mice exhibit a 20-fold reduction in the number of CD25+CD44+CD4−CD8− pro-T cells 8, and a corresponding reduction in thymus cellularity 4. Nevertheless, some development of mature T cells occurs in IL-7−/− mice, such that the percentages of α/β lineage thymic subsets, including CD4+CD8+ double positive (DP)1 cells and mature CD4+ and CD8+ single positive (SP) cells, are near normal 4 8. In contrast, the proportion of TCR-γ/δ+ thymocytes is strongly reduced in IL-7−/− mice 8, consistent with previous in vitro experiments that suggested a specific function of IL-7 in γ/δ T lineage development separate from its function in early T cell progenitors 9 10.
Like IL-7−/− mice, induced mutant mice with defective genes encoding for IL-7Rα 5, γc 6 7, or Janus kinase (JAK)3 11 12 13, a kinase involved in IL-7R signal transduction, exhibit substantially reduced numbers of mature lymphocytes. However, the phenotypes of the IL-7Rα−/− and γc−/− mice are more extreme. The number of α/β lineage T cells is more noticeably decreased (30–40-fold), and γ/δ T cells are completely absent 14 15. The more severe effects of mutations in the receptor compared with mutations in the cytokine have led to the proposal that another cytokine can also interact with the IL-7R 5.
The finding that mutations in IL-7 and its receptor have a more profound effect on γ/δ T cell development than α/β T cell development has led to the suggestion that the role of IL-7 is not limited to supporting the survival and proliferation of uncommitted T cell progenitor cells, and that it might play a unique role in γ/δ T cell development. This conclusion was bolstered by the finding that α/β T cell development was restored in IL-7R−/− mice by constitutive expression of the antiapoptotic gene bcl-2, whereas γ/δ T cell development was not 16 17. This result suggested that the IL-7R provides a survival signal during early α/β T cell development. Importantly, the failure of Bcl-2 expression to restore γ/δ T cell development suggests that the IL-7R provides a necessary and distinct type of signal in γ/δ T cell development.
The specific role of the IL-7R in γ/δ T cell development is controversial 18. It has been suggested that IL-7R–mediated signals facilitate TCR-γ gene rearrangement and/or transcription 10 19 20 21, possibly by inducing and maintaining open chromatin structure at the locus. In vitro experiments using fetal or adult lymphocyte precursors have shown that the addition of IL-7 in culture leads to increased levels of TCR-β and -γ gene rearrangements 10 22. In young adult IL-7Rα−/− mice, TCR-δ, -β, and -α genes were rearranged to a similar or a marginally reduced level to that seen in normal mice, but TCR-γ gene rearrangements were reported to be absent or selectively reduced for some Vγ genes but not others 19 21. In young adult γc−/− mice, TCR-γ, but not -δ, gene rearrangements were decreased significantly, but not all Vγ genes were affected to the same degree 20.
It has been reported that TCR-γ gene rearrangements may not be completely abrogated in IL-7Rα−/− mice, but rather that the rearranged TCR-γ genes in these mice are not transcribed 21. The functional relevance of the low levels of TCR-γ gene rearrangement in IL-7Rα−/− mice in this study is not known. Hence, the relative contributions of IL-7R–mediated signals to TCR-γ gene rearrangement, as opposed to transcription of the rearranged TCR-γ genes, are not clear. Since TCR-γ/δ+ thymocytes are not present in IL-7Rα−/− mice, it is difficult to directly assess the role of IL-7R–mediated signals in TCR-γ gene transcription in γ/δ lineage thymocytes. As a whole, although the data suggest that the IL-7R plays an important role in inducing TCR-γ gene rearrangements and/or transcription, it remains possible that the observed effects are partly or even wholly due to the preferential survival or proliferation of a thymic subset in which rearrangement/transcription occurs.
Several nonexclusive mechanisms can be considered to account for the selective absence of γ/δ T cells in IL-7Rα−/− mice. First, lack of IL-7R–mediated signaling results in a selective loss of γ/δ lineage–committed pro-T cells. Second, TCR-γ and/or -δ gene rearrangements are impaired in IL-7R−/− mice, leading to a block in γ/δ lineage commitment and/or development. Third, the TCR-γ/δ genes rearrange normally, but transcription of the rearranged genes requires IL-7R–mediated signaling, in the absence of which γ/δ thymocytes fail to differentiate. Finally, mature γ/δ thymocytes, or at least certain subsets of mature γ/δ thymocytes, absolutely require IL-7R–mediated signaling to survive and/or proliferate. As one means to distinguish among these possibilities, we have introduced a prerearranged TCR-γ transgene into IL-7Rα−/− mice. By providing a prerearranged gene, we can circumvent any effects of IL-7R–mediated signals on TCR-γ gene rearrangement. The results clearly demonstrate that IL-7R–mediated signaling is necessary for optimal transcription of a rearranged TCR-γ gene. Nevertheless, significant transgene transcription occurred in a high copy transgenic IL-7Rα−/− line, resulting in a rescue of γ/δ T cell development. Hence, the selective defect in γ/δ cell development in IL-7R−/− mice is due to effects on TCR-γ gene transcription and attendant effects on recombination.
Materials and Methods
Mice.
G8 TCR-γ transgenic mice have been characterized previously 23. C57BL/6 (B6), B6 IL-7Rα−/−, and B6 recombination activating gene (RAG)-1−/− mice were purchased from The Jackson Laboratory. G8 TCR-γ transgenic mice that had been backcrossed to B6 mice at least three times were crossed to IL-7Rα−/− mice to generate TCR-γ transgenic+ or transgenic− IL-7R−/− or IL-7R+/− littermates. Mice were typed by PCR using published primers and by flow cytometric analysis of peripheral blood lymphocytes. Mice were bred and maintained in specific pathogen-free facilities at the University of California, Berkeley.
Antibodies, Flow Cytometry, and Cell Culture.
Anti-Vγ2 TCR (UC3-10A6), anti–TCR-δ (GL3), anti–TCR-β (H57), anti-CD5 (53-7.3), anti-NK1.1 (PK136), and anti–heat-stable antigen (anti-HSA; J11d) mAbs were purified and conjugated with FITC or biotin according to standard protocols. Anti-CD8α–Tricolor (53-6.7), anti-CD44–FITC (IM7.8.1), and anti-CD25–PE (PC61.5.3) mAbs were purchased from Caltag; anti-CD4–Red613 was from GIBCO BRL; and streptavidin-PE was from Molecular Probes. Red blood cell–lysed single cell suspensions of thymocytes and splenocytes were stained with the relevant antibodies at saturating concentrations, and 1–2 × 105 labeled cells (side and forward gated) were analyzed by four-color cytometry using an EPICS® XL-MCL flow cytometer (Beckman Coulter). In some cases, splenocytes were passed through nylon wool to enrich for T cells before antibody staining. Cells were sorted using an ELITE® cell sorter (Beckman Coulter). Flow cytometric profiles were analyzed using the WinMDI program (John Trotter, Salk Institute, San Diego, CA.).
Induction of TCR-γ gene expression in mature α/β thymocytes was carried out by culturing sorted mature (HSAlo) CD4+ or CD8+ α/β (TCR-βhi) thymocytes along with γ/δ thymocytes in complete medium (RPMI 1640 supplemented with 10% FCS, 50 μM 2-ME, 2 mM l-glutamine, 20 mM Hepes, and antibiotics) containing recombinant IL-2 (5 ng/ml; Chiron) with or without IL-7 (2–100 ng/ml recombinant murine IL-7 [Genzyme] or 1% IL-7 supernatant from J558 plasmacytoma transfected with an expression vector containing IL-7 cDNA 24) for 5 h to 4 d. Each sorting experiment used three to six B6 mice (4–8-wk-old) and involved depletion of HSAhi cells using a magnetic-activated cell sorter (MACS) before cell sorting.
Southern Blot Analysis.
Genomic DNA (gDNA) isolation and Southern blotting were carried out as described 25. 15 μg of gDNA was digested (except for the sample from RAG-1−/− mice, which represents 2 μg of gDNA) with EcoRI, electrophoresed on 0.7% agarose gels, and blotted. The blot was hybridized with a Cγ1 probe that cross-hybridizes with Cγ2 and Cγ3 genes 26.
RNase Protection Assay.
[α-32P]rUTP (Amersham Pharmacia Biotech)–labeled riboprobe specific for the TCR-γ transgene transcripts was generated from a linearized pKS Bluescript vector (Stratagene) containing the 273-bp KpnI-BsrI fragment that includes the V-J junction sequence of G8 transgene under the control of T7 promoter. Control riboprobe specific for γ-actin mRNA was generated using SP6 RNA polymerase. RNase protection assay was carried out essentially as described 25 using 5 μg of total RNA purified from sorted thymocyte populations (a pool of 9–15 IL-7Rα−/− mice per transgenic line was used in two independent sorting experiments). RNA was isolated using Ultraspec RNA solution (Biotecx). Each sample contained both probes to eliminate quantitative variations in input RNA. Densitometric analysis was performed using a PhosphorImager (Molecular Dynamics).
Reverse Transcription PCR.
Reverse transcription (RT) of 1–2 μg of total RNA from sorted (and cultured) thymocytes was performed using oligo-dT primers and avian reverse transcriptase (both from Boehringer Mannheim). Each sorting experiment used pooled thymocytes from four to eight IL-7Rα−/− mice for the analysis of transgene expression and pooled thymocytes from three to six B6 mice for the analysis of endogenous TCR-γ gene expression in α/β cells. cDNA was serially diluted 3- or 4-fold, and PCR was performed for 28 cycles with the addition of 1.0 μCi of [α-32P]dCTP (Amersham Pharmacia Biotech) per sample in a total reaction volume of 50 μl. Samples without the RT step were subjected to the same PCR protocol. Starting sample concentration of tubulin PCR reactions was three- or fourfold lower than the corresponding target gene PCRs. Products were resolved by gel electrophoresis on a 5% polyacrylamide gel and visualized by autoradiography. Densitometric analysis was performed using a PhosphorImager (Molecular Dynamics). For nonradioactive PCR, 35–39 cycles of amplification were performed. Products were subjected to gel electrophoresis on a 1.5% agarose gel and visualized by ethidium bromide staining. The following PCR primers were used: 5′ Vγ2, CTGGGAATTCAACCTGGCAGATGA; 3′ Jγ1, GGGAAGCTTACCAGAGGGAATTACTATGAG; 3′ G8 junction, GTGAAAACCTGAG-CTCCCCTCCC; 5′ tubulin, CAGGCTGGTCAATGTGGC-AACCAGATCGGT; 3′ tubulin, GGCGCCCTCTGTGTAG-TGGCCTTTGGCCCA.
Results
Impaired TCR-γ Transgene Expression in IL-7R–deficient Mice.
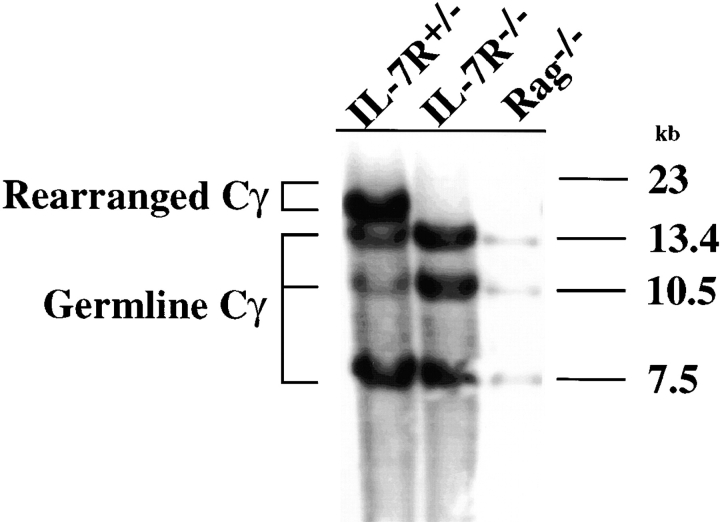
The TCR-γ gene rearrangement status in IL-7Rα−/− mice has been somewhat controversial. Southern blot analysis of TCR-γ gene rearrangement in thymocytes demonstrated that the rearranged TCR-γ genes are virtually undetectable in IL-7Rα−/− mice and that the pattern is very similar to that observed in RAG-1−/− mice (Fig. 1). This result supports previous Southern analysis 19 but is in contrast to the recent PCR data that indicated a significant level of TCR-γ gene rearrangement in IL-7Rα−/− mice 21. This deficiency in TCR-γ gene rearrangement can be caused by a specific defect in TCR-γ gene rearrangement/transcription and/or a selective loss of cells that rearrange/transcribe TCR-γ genes.
Figure 1.
Lack of significant TCR-γ gene rearrangement in thymocytes of IL-7Rα−/− mice. Southern blot analysis of EcoR1-digested genomic DNA probed with a Cγ-specific probe reveals a virtually undetectable level of TCR-γ gene rearrangement in thymocytes of IL-7Rα−/− mice. Three germline bands at 13.4, 10.5, and 7.5 kb detected in the RAG-1−/− lane represent unrearranged J-C gene segments of Cγ1, Cγ2, and Cγ3 loci, respectively. Cγ3 gene segment is a pseudogene and is found mostly in the germline configuration in normal thymocytes. Rearranged Cγ genes migrate at 16–20 kb, representing two predominant gene rearrangements, Vγ2-Jγ1-Cγ1 and Vγ1.2-Jγ2-Cγ2, found in adult thymocytes. The RAG-1−/− lane contains approximately eightfold less digested genomic DNA than the other two lanes. Numbers on the right represent DNA sizes (in kb).
To investigate the role of IL-7R signaling in TCR-γ gene expression, we crossed TCR-γ transgenes onto an IL-7Rα−/− background. The use of a prerearranged transgene allowed us to examine TCR-γ gene expression independent of TCR-γ gene recombination. Two distinct α/β lineage developmental phenotypes of IL-7Rα−/− mice have been documented: in type I IL-7Rα−/− mice the thymus is composed exclusively of immature CD4−CD8− (double negative [DN]) thymocytes 5, whereas in type II mice they exhibit near normal CD4/CD8 thymic composition 14. The basis for the phenotypic variation is unknown. All IL-7Rα−/− mice lack γ/δ T cells and show a drastically decreased thymic cellularity. In our colony, the majority of the B6 IL-7Rα−/− mice (>80%) exhibit the type II phenotype.
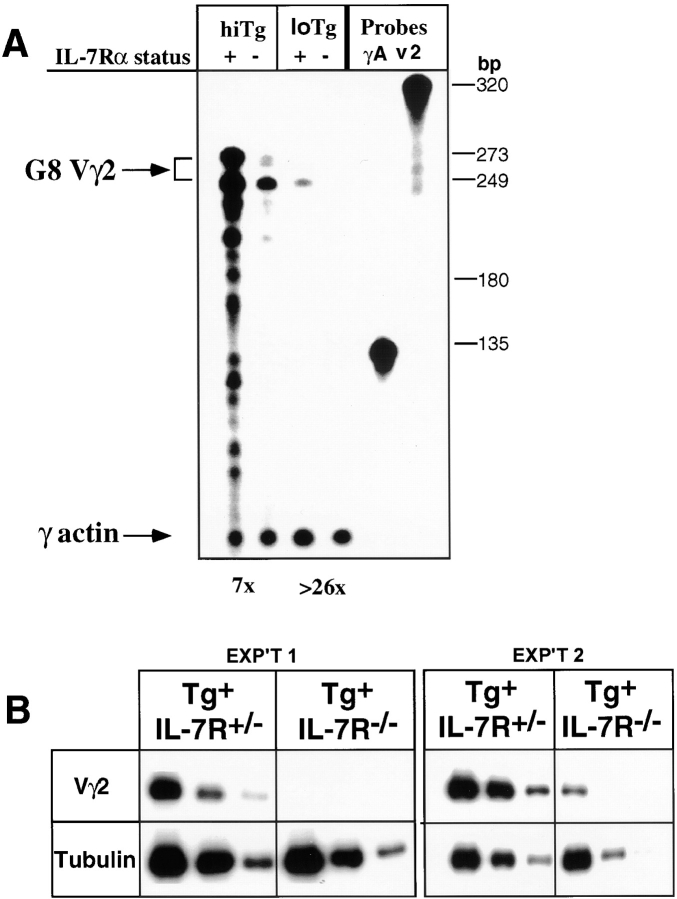
Mice harboring a productively rearranged TCR-γ (Vγ2-Jγ1-Cγ1) transgene 27 from the G8 cell line have been described previously 23. It has been demonstrated that expression of this transgene correlates well with transgene copy number and is not subject to transgene position effects 23 28. We compared 2 TCR-γ transgenic lines containing the identical transgene, but with different numbers of transgene copies (30 vs. 2 copies). Expression levels were determined in RNA samples from sorted (>99% pure) CD4−CD8−TCR-β− thymocytes, with an RNase protection assay using a V-J junction spanning probe specific for the transgene (Fig. 2 A). The results were normalized with reference to an internal γ-actin control. In addition, semiquantitative RT-PCR was performed using transgene-specific primers (Fig. 2 B).
Figure 2.
Expression of rearranged TCR-γ transgene in thymocyte precursors is dependent on IL-7R signaling. (A) A representative RNase protection assay of transgene-specific RNA in purified CD4−CD8−TCR-β− thymocytes from high (>30) and low (<2) copy transgenic IL-7Rα+/− (designated +) and IL-7Rα−/− (−) mice using the transgene V-J junction–specific riboprobe. Labeled, undigested probes are shown on the right (γA, γ-actin; v2, G8 VJ junction–specific probe). Identities of the protected bands are indicated beside the arrows. The two protected bands marked as originating from G8 TCR-γ gene template (G8 Vγ2) at 273 and 249 bp are so designated based on the predicted sizes of the protected VJ region of the G8 mRNA and are only detected in transgenic RNA samples. Size markers were included in all of the gels. The 249-bp fragment corresponds to correctly spliced mRNA, and the 273-bp fragment represents an mRNA in which the correct J-C splice has not occurred. Numbers below the panel represent the degrees of reduction in transgene-specific RNA level (combined quantities of two bands marked as G8γ) in transgenic IL-7Rα−/− mice compared with IL-7Rα+/− mice as determined by the PhosphorImager densitometric analysis with γ-actin RNA level as a loading control. Numbers on the right represent RNA sizes (in bp). Sub-bands present in the high copy IL-7R+ lane are transgene transcription specific and may originate from incorrect splicing, aberrant transcription, and/or unstable mRNA, and are possibly enhanced by an abnormally elevated expression of the transgene. (B) Semiquantitative RT-PCR assay for transgene-specific RNA in purified CD4−CD8−TCR-β− thymocytes from low copy transgenic IL-7Rα+/− and IL-7Rα−/− mice. Results from fourfold serial dilutions of the cDNA samples used for PCR are shown. Tubulin RT-PCR was used as a control. The highest initial amount used for tubulin PCR was fourfold diluted compared with the highest amount used for Vγ2-G8 junction–specific PCR. No significant level of product was detected in PCR reactions without the RT step, and no RT-PCR product was detectable in nontransgenic samples (data not shown). Results from two of three independent sorting experiments are shown.
The low copy TCR-γ transgene is expressed in the normal thymus at similar levels as the endogenous TCR-γ genes in a γ/δ T cell hybridoma (29; Kang, J., and D.H. Raulet, unpublished results). Transgene transcript levels were reduced by >26-fold in CD4−CD8−TCR-β− thymocytes from the low copy transgenic IL-7Rα−/− mice compared with control transgenic IL-7Rα+/− thymocytes (Fig. 2 A). The dramatic reduction in transgene transcription was confirmed in highly sensitive RT-PCR experiments. In two out of three such experiments, no transgene transcripts were detected in sorted CD4−CD8−TCR-β− thymocytes from transgenic IL-7Rα−/− mice, whereas abundant transcripts were detected in transgenic IL-7Rα+/− mice (for example, see experiment 1 [EXP'T 1] in Fig. 2 B). In one experiment, transcripts were detected in sorted thymocytes from IL-7Rα−/− mice, but the levels were reduced by more than eightfold compared with IL-7Rα+/− mice (Fig. 2 B, experiment 2). This level of transgene transcription was only detectable in one experiment, and only in the PCR assay.
The high 30 copy transgene was expressed at much higher levels in transgenic IL-7Rα+/− thymocytes than the low copy transgene, as expected from the greater number of transgene copies (Fig. 2 A). Nevertheless, transgene mRNA levels were substantially reduced in sorted thymocytes from IL-7Rα−/− transgenic mice, by a factor of at least five- to sevenfold (Fig. 2 A). No significant difference was seen between samples from transgenic IL-7R+/+ and IL-7R+/− mice (data not shown). The less dramatic effects of IL-7R deficiency on transgene transcription in the high copy line may be due to cooperative effects of multiple tandem transgene copies, which may partly overcome the requirement for IL-7R–dependent factors. Taken together, the results demonstrate that the transcription of both high and low copy TCR-γ transgenes is highly dependent on IL-7Rα expression.
The High Copy TCR-γ Transgene Rescues γ/δ T Cell Development in IL-7R−/− Mice.
Although optimal TCR-γ gene transcription depended on IL-7R signaling, the high copy TCR-γ transgene supported significant levels of transgene transcription. This persistent but reduced transgene transcription was likely due in part to the higher number of templates for transcription as well as to a partial reduction in IL-7R dependence due to tandem multimerization of the transgene. The significant expression of the transgene in the high copy line allowed us to address whether the absence of γ/δ cells in IL-7Rα−/− mice is due to the absence of TCR-γ chain synthesis.
It was shown previously that in otherwise normal mice, TCR-γ transgene expression resulted in a two- to threefold enhancement in the development of γ/δ thymocytes. The resulting γ/δ cell population was >90% Vγ2+ compared with ∼40% Vγ2+ thymocytes in nontransgenic littermates (23; Fig. 3 A). The increase in γ/δ cell numbers in the transgenic mice was independent of transgene copy number, but was dependent on the production of functional endogenous TCR-δ chains.
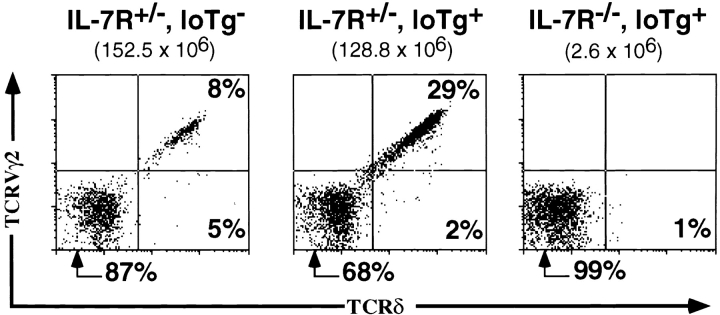
Figure 3.
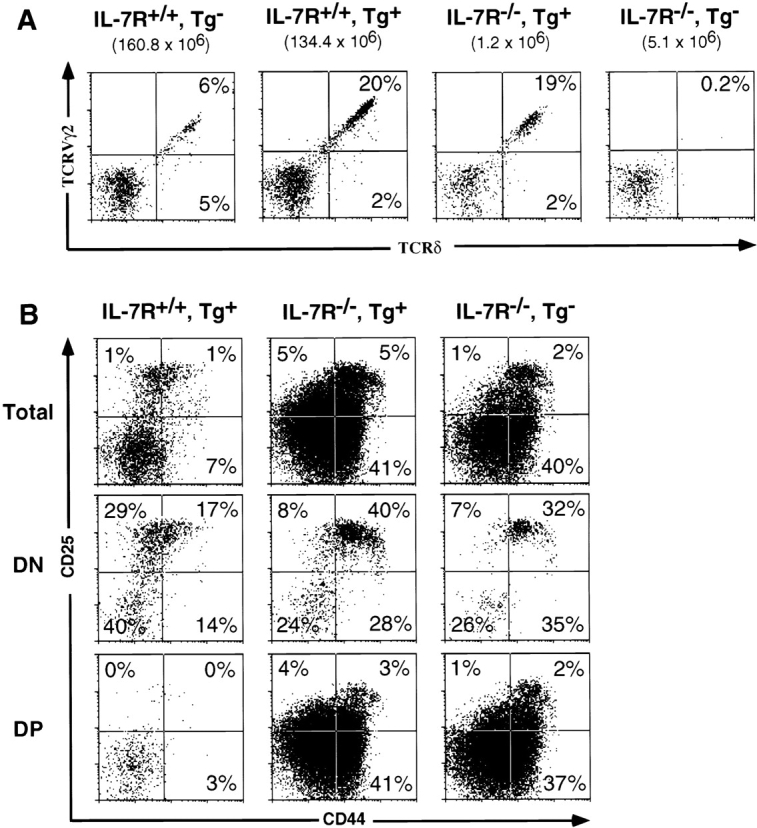
Expression of a rearranged TCR-γ gene results in γ/δ T cell development in IL-7Rα−/− mice. (A) Proportions of gated CD4−CD8− DN thymocytes expressing TCR-γ/δ in representative nontransgenic B6, high copy transgenic B6, high copy transgenic B6 IL-7Rα−/−, and nontransgenic B6 IL-7Rα−/− littermate mice. Transgenic mice predominantly express TCR-γ/δ composed of G8 Vγ2 chain. Numbers in brackets represent total thymocyte number. (B) CD25/CD44 expression profiles of total and gated DN and DP thymocytes in high copy transgenic IL-7Rα+/+ or IL-7Rα−/− mice. No significant difference is seen in the CD25/CD44 subset distribution between the nontransgenic and transgenic IL-7R+/+ mice. Tg, transgenic.
γ/δ T cells were not detectable in IL-7Rα−/− mice 14 15. The rearranged TCR-γ transgene, when present at 30 transgene copies, rescued γ/δ T cell (CD4−, CD8−, CD5med, TCR-β−, NK1.1−, HSAlo to hi; data not shown) development in the thymus of IL-7Rα−/− mice (Fig. 3 A). The proportion of DN thymocytes that expressed TCR-γ/δ in transgenic IL-7Rα−/− mice (20.6 ± 1.8%) was similar to that of transgenic littermates that were IL-7Rα+/− or IL-7Rα+/+ (19.3 ± 2.4%, Fig. 3 A). Moreover, the proportion of the DN thymocytes that were HSAloTCR-γ/δ+, a provisional phenotype for mature T cells, did not differ significantly between transgenic IL-7Rα−/− mice and wild-type mice (data not shown). The cells were stained with an anti–TCR-δ mAb, indicating that δ chain expression is independent of IL-7R signaling.
Although the percentage of γ/δ thymocytes in transgenic IL-7Rα−/− mice was normal, the absolute number of these cells was reduced ∼14-fold at 4–5 wk of age compared with transgenic IL-7Rα+/− mice. This is to be expected, given the >20-fold reduction in the number of T lineage–committed progenitor cells (CD25+CD44+ c-kit+CD4−CD8−CD3−) in these mice (17; Fig. 3 B, and data not shown). Hence, the absolute number of γ/δ thymocytes generated per progenitor T cell in the transgenic IL-7Rα−/− mice was not substantially different from that in normal mice. Examination of DN developmental intermediates using CD25/CD44 expression revealed no significant alterations in the subset distribution between the transgenic and nontransgenic IL-7Rα−/− littermates, and a distinct CD25+CD44− DN pre-T cell subset was not evident even when the transgene was expressed (Fig. 3 B, middle panel).
Peripheral γ/δ T cells were essentially undetectable in nontransgenic IL-7Rα−/− mice (14 15; Table ). Significant numbers of peripheral γ/δ T cells were detected in transgenic IL-7Rα−/− mice older than 3 wk, although they were reduced in absolute cell number by ∼13-fold compared with transgenic IL-7Rα+/− mice (Table ), as might be expected considering the reduction in thymic progenitor cells in these mice. These observations suggest that IL-7R signaling is not absolutely required for γ/δ T cell maintenance in the periphery.
Table 1.
γ/δ T Cell Development in High Copy TCR-γ Transgenic IL-7Rα2/− Mice
| Genotype | n | (age, wk) | Cell no. (×105) | ||
|---|---|---|---|---|---|
| All thymocytes | Thymic γ/δ | Splenic γ/δ | |||
| average ± SD | |||||
| IL-7Rα1/−, Tg− | 4 | (3–5) | 2,158 ± 420 | 4.3 ± 1.2 | 7.2 ± 1.4 |
| IL-7Rα1/−, Tg+ | 11 | (3–5) | 1,723 ± 294 | 10.2 ± 2.8 | 21.5 ± 3.2 |
| IL-7Rα2/−, Tg− | 10 | (2–3) | 38 ± 17 | <0.003 | <0.01 |
| IL-7Rα2/−, Tg+ | 8 | (2–3) | 14 ± 5 | 0.22 ± 0.10 | 0.07 ± 0.06 |
| IL-7Rα2/−, Tg− | 16 | (4–5) | 49 ± 22 | <0.01 | <0.01 |
| IL-7Rα2/−, Tg+ | 12 | (4–5) | 35 ± 26 | 0.69 ± 0.36 | 1.6 ± 0.5 |
Collectively, the results indicate that sufficient expression of a rearranged TCR-γ gene alone can lead to the development of γ/δ thymocytes in the absence of IL-7R–mediated signals. Production of endogenous TCR-δ chains was not dependent on IL-7R. However, it cannot be ruled out that a subtle difference in endogenous TCR-δ gene transcription exists in IL-7R−/− mice, although such a difference is not expected to affect functional TCR-δ chain generation. Importantly, the level of TCR-γ/δ receptors on the surface of the transgenic thymocytes was no higher than the receptor levels on γ/δ cells in normal mice (23; and Fig. 3 A), suggesting that γ/δ T cell development in the transgenic IL-7Rα−/− mice is not due to aberrantly enhanced TCR-γ/δ signaling. These results argue against the possibility that IL-7R–mediated signals are essential for the survival and/or maintenance of γ/δ T cells. Instead, the data suggest that an IL-7R-mediated signal(s) is necessary for the synthesis of TCR-γ chains.
The Low Copy Transgene Fails to Rescue γ/δ T Cell Development in IL-7Rα−/− Mice.
To determine whether the rescue of γ/δ T cells in IL-7Rα−/− mice is related to transgene copy number, the low 2 copy transgenic IL-7Rα−/− mice were analyzed. Flow cytometric analysis of DN thymocytes confirmed that the low copy transgene expression enhanced γ/δ T cell development in IL-7Rα+/− mice (Fig. 4), similar to what was observed with the high copy line (Fig. 3). However, unlike the high copy line, provision of the low copy transgene failed to rescue γ/δ cell development in IL-7Rα−/− mice (Fig. 4). These data indicate that the impaired transcription of the rearranged transgene in low copy IL-7Rα−/− mice results in levels of TCR-γ chain synthesis that are too low to stimulate γ/δ T cell development.
Figure 4.
γ/δ T cells are not generated in low copy TCR-γ transgenic (loTg) IL-7Rα−/− mice. Proportions of gated DN thymocytes expressing TCR-γ/δ in representative nontransgenic IL-7Rα+/−, low copy TCR-γ transgenic IL-7Rα+/−, and low copy TCR-γ transgenic IL-7Rα−/− littermates. Fractions of TCR-γ/δ cells that express Vγ2 chain are also compared. Numbers in brackets represent total thymocyte number.
IL-7 Can Reverse Transcriptional Suppression of the TCR-γ Gene in α/β Lineage Thymocytes.
TCR-γ genes are rearranged in the majority of CD4+ and CD8+ α/β lineage cells, but they are normally maintained in a transcriptionally inactive state 26 29 30. Rearranged Vγ2-Jγ1Cγ1 genes, in particular, are present in most α/β lineage cells but are completely repressed. To determine if the transcription of endogenous rearranged TCR-γ genes can also be influenced by IL-7R–mediated signals, we tested whether IL-7 can activate expression of endogenous TCR-γ genes in α/β lineage cells from normal mice. In contrast to immature DP thymocytes, the majority of CD4+ SP mature thymocytes from B6 mice express the IL-7Rα chain (31; and data not shown). A significant fraction of CD8+ SP mature thymocytes also express the IL-7Rα chain, but at a reduced level per cell compared with CD4+ thymocytes (31; and data not shown).
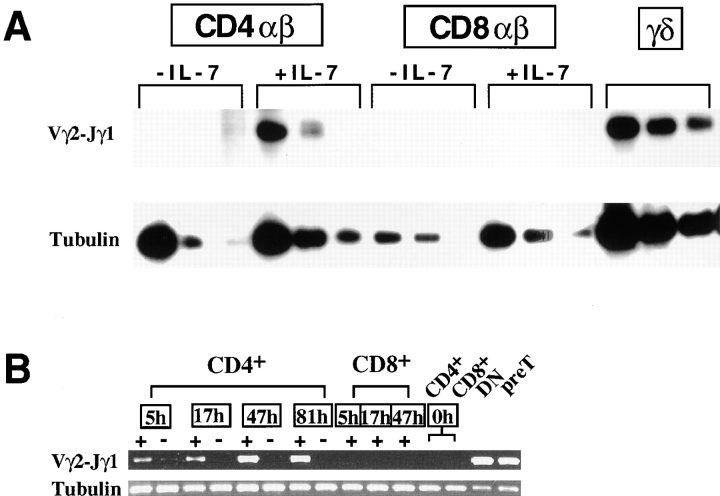
Sorted CD4+ or CD8+ SP HSAloTCR-α/β+ as well as CD4−CD8−TCR-γ/δ+ thymocytes (all >97% pure) from B6 mice were cultured in IL-2 for 2 d in the presence or absence of IL-7, and the level of TCR-γ gene transcription was measured by semiquantitative RT-PCR. The proportion of cells surviving after culture for all different conditions was similar in five independent experiments (data not shown). Fig. 5 A shows results from one representative RT-PCR experiment of four assays with identical results. Consistent with previous results, α/β T cells cultured without IL-7 failed to express the Vγ2-Jγ1 gene, despite the fact that a large fraction of α/β T cells harbor Vγ2-Jγ1 rearrangements 26. Remarkably, however, IL-7 induced the expression of the endogenous Vγ2-Jγ1 TCR-γ gene in mature CD4+ SP thymocytes, to a level that was 1/3 to 1/2 the level seen in γ/δ thymocytes cultured with or without IL-7 (Fig. 5, data not shown). The induction of transcription occurred rapidly after only 5 h in culture, and persisted until the end of a 4-d culture (Fig. 5 B). Treatment with IL-7 did not result in a significant induction of rag1/2 gene expression in these cultures (data not shown). These results demonstrate that IL-7R signals can induce transcription of the endogenous rearranged TCR-γ gene in CD4+ α/β lineage cells in vitro, and suggest an active role of the downstream mediators of IL-7R signaling in promoting transcriptional activities at the TCR-γ locus. In contrast to its effects on CD4+ SP thymocytes, IL-7 failed to induce Vγ2-Jγ1 TCR-γ gene transcription in mature CD8 SP thymocytes (Fig. 5), perhaps due in part to the lower levels of IL-7R expression by these cells. The reason for the lack of transcription of rearranged endogenous TCR-γ gene in CD4+ SP thymocytes in vivo is unclear, since IL-7 is produced by some thymic stromal cells. It appears that CD4+ SP thymocytes do not receive sufficient IL-7R signaling in situ, possibly as a result of limiting local IL-7 concentration or antagonistic effects of other thymic inductive signals.
Figure 5.
IL-7 induces transcription of endogenous, rearranged TCR-γ genes in purified CD4+ α/β thymocytes. (A) Semiquantitative RT-PCR assay of endogenous, rearranged Vγ2-Jγ1–specific RNA from sorted TCR-γ/δ+ or CD4+ or CD8+HSAloTCR-β+ thymocytes cultured for 2 d in IL-2 alone (designated as –IL-7) or in medium supplemented with IL-2 and IL-7 (1% supernatant of transfected IL-7 cDNA–expressing J558 cells, designated as +IL-7). Addition of IL-7 did not significantly alter the level of TCR-γ gene expression in cultures of γ/δ thymocytes (data not shown). Results from threefold serial dilutions of the cDNA samples used for PCR are shown. The highest amount used for control tubulin PCR was threefold diluted compared with the highest amount used for Vγ2-Jγ1–specific PCR. No product was detected in PCR reactions without the RT step (data not shown). (B) Kinetics of endogenous Vγ2-Jγ1 gene expression in sorted mature α/β thymocytes cultured with 2 ng/ml rIL-7. RT-PCR products specific for rearranged Vγ2-Jγ1 and tubulin at various times after culture (numbers in the boxes refer to hours in culture) were resolved by gel electrophoresis and visualized by ethidium bromide staining. Sorted HSAloTCR-β+CD4+ or CD8+ thymocytes (designated as CD4+ or CD8+) were cultured in IL-2 medium with (+) or without (−) IL-7. No significant TCR-γ gene expression was detectable in cultured CD8+ thymocytes or sorted CD4+ or CD8+ thymocytes at the beginning of culture (0h). Abundant TCR-γ transcripts were detected in DN (purified by complement kill of CD4+ and/or CD8+ thymocytes) and sorted CD25+CD44− triple negative cells (pre-T) containing γ/δ lineage cells. No product was detected in PCR reactions without the RT step (data not shown).
Discussion
At least two aspects of T cell precursor differentiation are regulated by IL-7R–mediated signals. IL-7R signals ensure survival by preventing programmed cell death of developing T cells 16 17, and they directly and specifically regulate the production of TCR-γ chains. In IL-7Rα−/− mice, greatly reduced numbers of α/β thymocytes develop, but γ/δ thymocytes are completely absent. Based on the dramatically decreased levels of TCR-γ, but not -δ, -β, and -α, gene rearrangement observed in thymocytes of IL-7Rα−/− mice, it has been suggested that the IL-7R signals are necessary for activating the TCR-γ gene locus 19. We show here that transcripts emanating from a prerearranged TCR-γ transgene are markedly reduced in IL-7Rα−/− mice, indicating that IL-7Rα signaling directly affects TCR-γ gene expression, independent of rearrangement. This conclusion is strikingly supported by the studies showing that IL-7 induces transcription of the endogenous rearranged Vγ2-Jγ1Cγ1 gene in isolated mature CD4+ SP thymocytes from normal mice. The downstream mediators of IL-7R have been previously shown to activate germline, sterile TCR-γ transcription in nonlymphocytes 32, consistent with the proposed role of IL-7R in regulating TCR-γ gene transcription.
Equally important, the present data indicate that the poor expression of TCR-γ genes in IL-7Rα−/− mice is mainly responsible for the selective deficit of γ/δ cells in these mice, since γ/δ cell development could be restored with a high copy rearranged TCR-γ transgene. The number of γ/δ T cells generated per pro-T cell was similar in the wild-type and high copy transgenic IL-7Rα−/− mice. Hence, it appears that the γ/δ lineage precursor cell generation, γ/δ lineage development, and maintenance are not strictly dependent on IL-7R signals as long as functional TCR-γ chains are present. Since no TCR-δ transgene was provided, these data also suggest that IL-7Rα signaling is not crucial for TCR-δ gene rearrangement and TCR-δ chain production, consistent with previous reports 19 20.
The residual level of transgene transcription in the high copy transgenic IL-7Rα−/− line was probably due to two factors. First, the high gene dosage provided a greater number of templates for γ gene transcription. Second, adjusting for gene dosage, it appeared that IL-7Rα deficiency had somewhat less effect on transcript levels in the high copy line compared with the low copy line, possibly reflecting a dysregulation that can occur in multiple tandemly integrated transgenes 33. Nevertheless, transgene transcription was substantially reduced in thymocytes of these mice compared with IL-7Rα+ mice.
In the IL-7Rα−/− low copy γ transgenic mice, no restoration of γ/δ cell development was observed. Previous studies showed that the level of transgene-directed γ mRNA in IL-7Rα+ thymocytes harboring the low copy γ transgene is comparable to the level of TCR-γ transcripts in a normal cloned γ/δ T cell line 29. This level of expression is sufficient to increase the number of γ/δ cells in the thymus of IL-7R+ mice to the same extent as that seen in the high copy IL-7R+ transgenic mice 23. The IL-7R mutation is not permissive for this normal level of expression, and the sharp reduction in expression is sufficient to completely prevent γ/δ cell development. This finding suggests that the deficiency in TCR-γ transcription in IL-7Rα−/− mice could account for the lack of γ/δ cells in these mice even if the locus underwent rearrangement normally.
However, it is likely that TCR-γ gene rearrangement is also affected in IL-7Rα−/− mice. Although it might be proposed that the low levels of TCR-γ gene rearrangements observed in IL-7Rα−/− thymi are due to the rapid turnover of cells that have rearranged TCR-γ genes but do not survive as a result of the lack of IL-7R signaling, this is unlikely to be the complete explanation. Previous studies have shown that the majority of TCR-γ gene rearrangements in the normal thymus are nonproductive ones in α/β lineage CD4+CD8+ DP thymocytes 34. The IL-7Rα−/− mice we examined contained appreciable percentages of DP and SP αβ thymocytes, yet TCR-γ gene rearrangement was nevertheless sharply reduced. In IL-7Rα−/− mice, TCR-γ gene rearrangements are rare even in α/β lineage cells, which do not depend on TCR-γ/δ expression for survival. This consideration suggests that T cells of IL-7Rα−/− mice exhibit a specific deficiency in TCR-γ gene rearrangement. Many previous studies have correlated transcription of B and T cell receptor genes with rearrangement 35. For example, enhancer and promoter elements that support transcription have been shown to play critical roles in rearrangement of the genes as well. Hence, the defective γ gene transcription in IL-7Rα−/− mice probably also results in reduced levels of γ gene rearrangement.
Two previous reports have provided indirect evidence for a possible role of IL-7R signals in controlling rearranged TCR-γ gene transcription. First, during fetal ontogeny in γc−/− mice, rearrangement of some TCR-γ genes was evident, but fetal γ/δ thymocytes still did not develop 20. It was not established that this absence of γ/δ cells was due to a defect in TCR-γ gene transcription. Second, in one study of fetal and adult IL-7Rα−/− mice, it appeared that TCR-γ gene rearrangements were not completely absent as assayed by PCR, but the corresponding transcripts were undetectable 21. In our hands, IL-7Rα deficiency reduces the level of thymic γ gene rearrangements by at least 40-fold, as detected by genomic Southern blotting (Fig. 1). In the aforementioned study, it was not clear whether the level of gene rearrangement detected was sufficient to result in a measurable quantity of mRNA. Furthermore, it was not established that the rearrangements that were detected were present in cells that are normally capable of expressing TCR-γ genes. It is known that γ gene expression is extinguished in certain thymic lineages, such as in CD4+CD8+ DP cells 29 30. By using a prerearranged transgene present in all thymocyte lineages, we were able to demonstrate directly that TCR-γ gene expression is impaired in the absence of IL-7Rα signaling.
One other aspect of the data is notable. At 2–3 wk of age, the total thymocyte number in the high copy TCR-γ transgenic IL-7Rα−/− mice was only 36% of the number observed in nontransgenic IL-7Rα−/− counterparts (Table ). Thus, the TCR-γ transgene expression depressed the number of thymocytes in IL-7Rα−/− mice. One possible explanation for this observation is that TCR-γ transgene expression in young mice enhances the rate of generation of thymocytes expressing TCR-γ/δ, resulting in reduced numbers of α/β lineage cells. Since thymic proliferative expansion occurs mainly in α/β but not γ/δ lineage cells, the reduction in α/β lineage cells would lead to a significant reduction in thymic cellularity. A similar effect of the TCR-γ transgene on thymic cellularity has been described previously in mice that generate α/β lineage cells suboptimally 23.
Our results with the G8 (Vγ2) TCR-γ transgenic IL-7Rα−/− mice differ somewhat from a study that employed a different TCR-γ transgene (T3.13.3, Vγ1.1) that was crossed into γc−/− mice 20. The latter mice contained only 10% the number of splenic γ/δ T cells that we observed, although the difference in thymic γ/δ cell number was limited to two- to threefold. A plausible explanation for this difference in peripheral γ/δ T cell numbers is that γc-dependent cytokines other than IL-7 may be involved in the maintenance of mature γ/δ T cells.
How does the IL-7R regulate TCR-γ gene expression? The TCR Cγ1 locus contains a downstream enhancer element, 3′ECγ1 36 37. We have recently identified another regulatory element, called 5′HsA, that cooperates with the 3′ECγ1 to stimulate optimal TCR-γ gene rearrangement and transcription 28. 5′HsA is a locus control region (LCR)-like element that does not exhibit enhancer activity as measured with transient transfection assays, but is active when integrated in the genome. It appears to play a role in rendering the TCR-γ locus accessible for gene rearrangement and expression in vivo. 3′ECγ1 contains a consensus binding site for signal transducer and activator of transcription 5 (STAT5 38), a transcription factor that is activated by JAK1 and JAK3, which are in turn activated by the IL-7R. 5′HsA contains binding sites for other STATs. Reportedly, thymocytes of IL-7Rα−/− mice are devoid of nuclear STAT5 21. Further studies will be necessary to assess the role, if any, that STATs play in TCR-γ gene transcription.
Acknowledgments
We thank Dr. C. Chambers for helpful discussions and review of the manuscript, P. Schow for expert technical assistance with flow cytometric cell sorting, and Russell Vance and Dr. A. Volkman for comments on the paper.
This work was supported by a grant from the National Institutes of Health (RO1-AI30171). J. Kang is a research fellow of the National Cancer Institute of Canada supported by the Canadian Cancer Society.
Footnotes
1used in this paper: B6, C57BL/6; DN, CD4−CD8− double negative; DP, CD4+CD8+ double positive; HSA, heat-stable antigen; JAK, Janus kinase; RAG, recombination activating gene; RT, reverse transcription; SP, CD4+ or CD8+ single positive; STAT, signal transducer and activator of transcription
M. Coles's present address is Department of Molecular Immunology, National Institute for Medical Research, Mill Hill, London, NW7 1AA, UK.
References
- Wiles M.V., Ruiz P., Imhof B.A. Interleukin-7 expression during mouse thymus development. Eur. J. Immunol. 1992;22:1037–1042. doi: 10.1002/eji.1830220424. [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lin J.X., Migone T.S., Tsang M., Friedmann M., Weatherbee J.A., Zhou L., Yamauchi A., Bloom E.T., Mietz J., John S. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- von Freeden-Jeffry U., Vieira P., Lucian L.A., McNeil T., Burdach S.E., Murray R. Lymphopenia in interleukin (IL)-7 gene–deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon J.J., Morrissey P.J., Grabstein K.H., Ramsdell F.J., Maraskovsky E., Gliniak B.C., Park L.S., Ziegler S.F., Williams D.E., Ware C.B. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J. Exp. Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSanto J.P., Müller W., Guy-Grand D., Fischer A., Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Shores E.W., Hu-Li J., Anver M.R., Kelsall B.L., Russell S.M., Drago J., Noguchi M., Grinberg A., Bloom E.T. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- Moore T.A., von Freeden-Jeffry U., Murray R., Zlotnik A. Inhibition of γδ T cell development and early thymocyte maturation in IL-7−/− mice. J. Immunol. 1996;157:2366–2373. [PubMed] [Google Scholar]
- Plum J., De Smedt M., Leclercq G. Exogenous IL-7 promotes the growth of CD3−CD4−CD8− CD44+CD25+/− precursor cells and blocks the differentiation pathway of TCR-αβ cells in fetal thymus organ culture. J. Immunol. 1993;150:2706–2716. [PubMed] [Google Scholar]
- Appasamy P.M., Kenniston T.W., Jr., Weng Y., Holt E.C., Kost J., Chambers W.H. Interleukin 7–induced expression of specific T cell receptor γ variable region genes in murine fetal liver cultures. J. Exp. Med. 1993;178:2201–2206. doi: 10.1084/jem.178.6.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomis D.C., Gurniak C.B., Tivol E., Sharpe A.H., Berg L.J. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- Nosaka T., van Deursen J.M., Tripp R.A., Thierfelder W.E., Witthuhn B.A., McMickle A.P., Doherty P.C., Grosveld G.C., Ihle J.N. Defective lymphoid development in mice lacking Jak3 [published erratum appears in Science. 1996. 271:17] Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Saijo K., Takahashi T., Osawa M., Arase H., Hirayama N., Miyake K., Nakauchi H., Shirasawa T., Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- Maki K., Sunaga S., Komagata Y., Kodaira Y., Mabuchi A., Karasuyama H., Yokomuro K., Miyazaki J.I., Ikuta K. Interleukin 7 receptor-deficient mice lack gammadelta T cells. Proc. Natl. Acad. Sci. USA. 1996;93:7172–7177. doi: 10.1073/pnas.93.14.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.W., Malek T.R. Interleukin-7 receptor α is essential for the development of γδ+ T cells, but not natural killer cells. J. Exp. Med. 1996;184:289–293. doi: 10.1084/jem.184.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi K., Kondo M., von Freeden-Jeffry U., Murray R., Weissman I.L. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- Maraskovsky E., O'Reilly L.A., Teepe M., Corcoran L.M., Peschon J.J., Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1 −/− mice. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- Malek T.R., Porter B.O., He Y.W. Multiple γc-dependent cytokines regulate T cell development. Immunol. Today. 1999;20:71–76. doi: 10.1016/s0167-5699(98)01391-7. [DOI] [PubMed] [Google Scholar]
- Maki K., Sunaga S., Ikuta K. The V-J recombination of T cell receptor-γ genes is blocked in interleukin-7 receptor–deficient mice. J. Exp. Med. 1996;184:2423–2427. doi: 10.1084/jem.184.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Pereira P., Gerber D.J., Malissen B., DiSanto J.P. The common cytokine receptor γ chain controls survival of γ/δ T cells. J. Exp. Med. 1997;186:1277–1285. doi: 10.1084/jem.186.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal N.B., Kenniston T.W., Jr., Tweardy D.J., Dyer K.F., Hoffman R., Peschon J., Appasamy P.M. TCR-γ genes are rearranged but not transcribed in IL-7Rα-deficient mice. J. Immunol. 1997;158:5744–5750. [PubMed] [Google Scholar]
- Muegge K., Vila M.P., Durum S.K. Interleukin-7a cofactor for V(D)J rearrangement of the T cell receptor beta gene. Science. 1993;261:93–95. doi: 10.1126/science.7686307. [DOI] [PubMed] [Google Scholar]
- Kang J., Coles M., Cado D., Raulet D.H. The developmental fate of T cells is critically influenced by TCRγδ expression. Immunity. 1998;8:427–438. doi: 10.1016/s1074-7613(00)80548-8. [DOI] [PubMed] [Google Scholar]
- Karasuyama H., Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur. J. Immunol. 1988;18:97–104. doi: 10.1002/eji.1830180115. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. Molecular CloningA Laboratory Manual 1989. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: pp. 545 [Google Scholar]
- Garman R.D., Doherty P.J., Raulet D.H. Diversity, rearrangement and expression of murine T cell gamma genes. Cell. 1986;45:733–742. doi: 10.1016/0092-8674(86)90787-7. [DOI] [PubMed] [Google Scholar]
- Dent A.L., Matis L.A., Hooshmand F., Widacki S.M., Bluestone J.A., Hedrick S.M. Self-reactive γδ T cells are eliminated in the thymus. Nature. 1990;343:714–719. doi: 10.1038/343714a0. [DOI] [PubMed] [Google Scholar]
- Baker J.E., Kang J., Xiong N., Chen T., Cado D., Raulet D.H. A novel element upstream of the Vγ2 gene in the murine T cell receptor γ locus cooperates with the 3′ enhancer to act as a locus control region. J. Exp. Med. 1999;190:669–680. doi: 10.1084/jem.190.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Fehling H.J., Laplace C., Malissen M., Cado D., Raulet D.H. T cell receptor γ gene regulatory sequences prevent the function of a novel TCRγ/pTα pre-T cell receptor. Immunity. 1998;8:713–721. doi: 10.1016/s1074-7613(00)80576-2. [DOI] [PubMed] [Google Scholar]
- Ishida I., Verbeek S., Bonneville M., Itohara S., Berns A., Tonegawa S. T-cell receptor γδ and γ transgenic mice suggest a role of a γ gene silencer in the generation of αβ T cells. Proc. Natl. Acad. Sci. USA. 1990;87:3067–3071. doi: 10.1073/pnas.87.8.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo T., Nishikawa S., Ohno N., Akiyama N., Tamakoshi M., Yoshida H. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle F.W., Wang D., Nosaka T., Thierfelder W.E., Stravopodis D., Weinstein Y., Ihle J.N. Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol. Cell. Biol. 1996;16:1622–1631. doi: 10.1128/mcb.16.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Tan–Un K.C., Harper A., Michalovich D., Yannoutsos N., Philipsen S., Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- Kang J., Baker J., Raulet D. Evidence that productive rearrangements of TCRγ genes influence the commitment of progenitor cells to differentiate into αβ or γδ T cells. Eur. J. Immunol. 1995;25:2706–2709. doi: 10.1002/eji.1830250946. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Gorman J.R., Alt F.W. Accessibility control of antigen-receptor variable-region gene assemblyrole of cis-acting elements. Annu. Rev. Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- Spencer D.M., Hsiang Y.-H., Goldman J.P., Raulet D.H. Identification of a T-cell-specific transcriptional enhancer located 3′ of Cγ1 in the murine T-cell receptor γ locus. Proc. Natl. Acad. Sci. USA. 1991;88:800–804. doi: 10.1073/pnas.88.3.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes D., Browne C.P., Tonegawa S. Identification of a T-cell-specific enhancer at the locus encoding T-cell antigen receptor γ chain. Proc. Natl. Acad. Sci. USA. 1991;88:2204–2208. doi: 10.1073/pnas.88.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle J.N. STATssignal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]