Abstract
Bacillus anthracis is nonhemolytic, even though it is closely related to the highly hemolytic Bacillus cereus. Hemolysis by B. cereus results largely from the action of phosphatidylcholine-specific phospholipase C (PC-PLC) and sphingomyelinase (SPH), encoded by the plc and sph genes, respectively. In B. cereus, these genes are organized in an operon regulated by the global regulator PlcR. B. anthracis contains a highly similar cereolysin operon, but it is transcriptionally silent because the B. anthracis PlcR is truncated at the C terminus. Here we report the cloning, expression, purification, and enzymatic characterization of PC-PLC and SPH from B. cereus and B. anthracis. We also investigated the effects of expressing PlcR on the expression of plc and sph. In B. cereus, PlcR was found to be a positive regulator of plc but a negative regulator of sph. Replacement of the B. cereus plcR gene by its truncated orthologue from B. anthracis eliminated the activities of both PC-PLC and SPH, whereas introduction into B. anthracis of the B. cereus plcR gene with its own promoter did not activate cereolysin expression. Hemolytic activity was detected in B. anthracis strains containing the B. cereus plcR gene on a multicopy plasmid under control of the strong B. anthracis protective antigen gene promoter or in a strain carrying a multicopy plasmid containing the entire B. cereus plc-sph operon. Slight hemolysis and PC-PLC activation were found when PlcR-producing B. anthracis strains were grown under anaerobic-plus-CO2 or especially under aerobic-plus-CO2 conditions. Unmodified parental B. anthracis strains did not demonstrate obvious hemolysis under the same conditions.
In the Bacillus genus, the Bacillus cereus group of spore-forming soil bacteria (Bacillus cereus, Bacillus thuringiensis, Bacillus anthracis, and Bacillus mycoides) is one of the most taxonomically ambiguous groups. All four species have been placed in Bacillus subgroup 1 based on their large cell widths and certain characteristics of their spores, which do not distend the sporangium (30). DNA studies also grouped these Bacillus species because they all have AT-rich genomes. B. cereus and B. thuringiensis are highly polymorphic, whereas B. anthracis is viewed as monomorphic (14, 19). Phylogenetic analyses based on sequence and enzyme electrophoresis data also revealed that while B. cereus and B. thuringiensis are very similar, B. anthracis could be considered systematically rather distinct (40). In contrast to B. cereus and B. thuringiensis, B. anthracis is penicillin sensitive, produces a polypeptide capsule, is nonhemolytic, and does not produce phospholipase C. In addition, it produces the lethal and edema toxins. However, analysis of the B. anthracis genome (with the TIGR database at http://www.tigr.org) reveals the presence of structural genes for penicillin resistance and hemolytic activities: two β-lactamase genes corresponding to the type I and type II β-lactamases of B. cereus and orthologues of the B. cereus hemolytic genes producing phosphatidylcholine-specific phospholipase C (PC-PLC), phosphatidylinositol-specific phospholipase, sphingomyelinase (SPH), and cereolysin O (25; Y. Chen, J. Succi, and T. M. Koehler, abstr. Proc. 4th Int.Workshop Anthrax, Annapolis, Md., 2001). All of these genes are silent in B. anthracis. Cloning of the type I β-lactamase gene into Escherichia coli and Bacillus subtilis conferred penicillin resistance to both recipient bacteria (Chen et al., Proc. 4th Int. Workshop Anthrax). This suggested that some additional regulatory factor required for β-lactamase production in B. cereus is not present in B. anthracis.
An interesting observation has been the fact that the 16-bp palindrome known to be the target of the positive transcriptional regulator PlcR (1) is located upstream of every B. anthracis hemolysis-related gene (25). PlcR, the pleiotropic regulator of extracellular virulence factors, is active both in B. cereus and in B. thuringiensis. It activates transcription of at least 15 genes encoding secreted proteins, including phospholipases, proteases, and two enterotoxin complexes (10). Expression of the plcR gene is autoregulated and activated at the onset of the stationary phase. The putative PapR protein produced from the short open reading frame (orf2) located downstream of the plcR gene probably activates PlcR expression (1, 22, 27, 39). Expression of PlcR at the onset of the stationary phase is also dependent on the growth medium and is controlled by the transition state regulator Spo0A (23). Thus, hemolytic activity is greatly reduced in strains of B. cereus and B. thuringiensis with mutations in plcR (ΔplcR) (34). Recent evidence shows that an oligopeptide permease is also required for expression of the plcR regulon (11). The plcR gene is present in, and probably restricted to, all members of the B. cereus group. However, while the B. cereus and B. thuringiensis PlcR proteins appear to be functionally equivalent, the B. anthracis PlcR protein is truncated and does not operate as a transcriptional activator (1). Expression of the B. thuringiensis PlcR in B. anthracis organisms resulted in the transcriptional activation of genes that are only weakly expressed in the absence of PlcR. The transcriptional activation was also evident from increased enzyme activity, including that of PC-PLC (25). It has been reported recently (20) that B. anthracis hemolytic genes, including plc and sph, are induced by strictly anaerobic conditions, suggesting that alternative regulatory mechanisms come into play under such conditions.
Here we compare the activities of the PC-PLC and SPH enzymes of B. anthracis and B. cereus and some aspects of their regulation. For this purpose, the four structural genes were cloned and purified as His-tagged derivatives from an E. coli T7 expression system. We also investigated the kinetics of PlcR synthesis in B. cereus strain 569 and in a PlcR-deficient derivative. The latter was obtained through a single crossover between the chromosomal plcR gene and a plasmid-borne gene encoding the truncated PlcR protein from B. anthracis. Consistent with previous observations, inactivation of PlcR greatly decreased PC-PLC and SPH expression in B. cereus. The functional B. cereus plcR gene was introduced into B. anthracis by means of several different plasmids in which plcR was under control of its own promoter or a strong, constitutive promoter. This technique allowed us to study the dynamics of recombinant PlcR synthesis in B. anthracis and its influence on PC-PLC and SPH activities. We also compared PC-PLC and hemolytic activities of several B. anthracis and B. cereus strains grown on agar containing lecithin or sheep or human blood under aerobic, aerobic-plus-CO2, and anaerobic-plus-CO2 conditions.
MATERIALS AND METHODS
Growth conditions.
E. coli strains were grown in Luria-Bertani (LB) broth (35) and used as hosts for cloning and protein production. Media were supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich, St. Louis, Mo.) for induction of expression of T7-lac promoter plasmids. L agar was used for the selection of transformants and for the estimation of the hemolytic properties of isolated enzymes. B. anthracis and B. cereus strains were grown in brain heart infusion (BHI) medium and in LB medium. Solid media were supplemented with 5% fresh sheep or human blood for determinations of hemolytic activity or with 0.02% l-α-phosphatidylcholine (lecithin; Sigma-Aldrich) for determinations of PC-PLC activity. The following antibiotics were purchased from Sigma-Aldrich and added to media when appropriate to give the indicated final concentrations: ampicillin (100 μg/ml), erythromycin (5 μg/ml), kanamycin (10 μg/ml), and tetracycline (5 μg/ml). SOC medium (Quality Biologicals, Inc., Gaithersburg, Md.) was used to grow cells during transformation.
Anaerobic and/or CO2-enriched conditions were produced in a jar system using BBL GasPak Plus Anaerobic System Envelopes with palladium catalyst or BBL GasPak CO2 System Envelopes (Becton Dickinson Microbiology Systems, Sparks, Md.) These produce gaseous environments containing 4 to 10% CO2 and either anaerobic or aerobic conditions. The GasPak dry methylene blue anaerobic indicator was used to confirm establishment of an anaerobic environment. BHI agar supplemented with 5% sheep or human blood was used in these experiments for estimating hemolytic activity. Specific medium containing 37 g of BHI medium, 0.01 g of resazurin (sodium salt; Sigma-Aldrich, St. Louis, Mo.) 50 ml of egg yolk suspension (Sigma-Aldrich), and 14 g of agar per liter (7) was used in these experiments for estimating PC-PLC activity. Suspensions of the B. anthracis Sterne, Sterne 34F2deltaT (SdT), and SdT2 strains and the B. cereus 569 and 541 strains were prepared with an A600 close to 0.25, and 3 μl of each suspension was inoculated on plates which were then incubated at 37°C for 48 h under aerobic or anaerobic (with or without CO2) conditions.
DNA isolation and manipulation.
The preparation of plasmid DNA from E. coli, the transformation of E. coli, and recombinant DNA techniques were carried out by standard procedures (35). E. coli XL2-Blue and SCS110 competent cells were purchased from Stratagene, Inc., La Jolla, Calif. Recombinant plasmid construction was carried out in E. coli XL2-Blue.
Plasmid DNA from B. anthracis and B. cereus organisms was isolated according to the Plasmid Protocol: Purification of Plasmid DNA from Bacillus subtilis (QIAGEN Inc., Valencia, Calif.). Chromosomal DNA from B. anthracis and B. cereus organisms was isolated with a Wizard Genomic Purification Kit (Promega, Madison, Wis.) in accordance with the protocol for isolation of genomic DNA from gram-positive bacteria (Promega). B. cereus cells were electroporated with plasmid DNA from E. coli as described elsewhere (13). B. anthracis cells were electroporated with unmethylated plasmid DNA isolated from E. coli SCS110. Electroporation-competent cells were prepared as previously described (28). Restriction enzymes, T4 ligase, Klenow fragment, and alkaline phosphatase were purchased from MBI Fermentas (Vilnius, Lithuania) or New England Biolabs (Beverly, Mass.). Taq polymerase kits were purchased from TaKaRa Shuzo Co., Ltd. (Otsu, Japan) or Invitrogen/Life Technologies (Rockville, Md.). The GeneRuler DNA Ladder Mix from MBI Fermentas was used for determination of DNA fragment length. All constructs were verified by DNA sequencing.
Strain construction.
Strains, plasmids, and their relevant characteristics are listed in Table 1. Oligonucleotide primers are listed in Table 2. The B. cereus plcR mutant was constructed by replacement of the plcR coding sequence with the B. anthracis truncated plcR (ΔplcR) coding sequence by means of single-crossover technology (42). Briefly, the antisense B. anthracis ΔplcR flanked by two SmaI restriction sites was cloned into the EcoRV site of vector pYJ335, which contains a hybrid xylose-tetracycline-controlled promoter (17). The resulting plasmid pYJ335-anti-plcR was electroporated into B. cereus cells with selection for erythromycin resistance. Transformants were selected at 37°C on BHI agar containing erythromycin and lecithin. Bacteria from the edges of the colonies were repeatedly passed on new plates and screened for disappearance of the halo around the growing cells. Halo-negative colonies were analyzed by PCR to verify that single-crossover recombination events had occurred. Separately, clones containing the extrachromosomal plasmid pYJ335-anti-plcR were examined for the ability of the tetracycline-controlled antisense transcript to down regulate expression (17). For this purpose, B. cereus 569(pYJ335-anti-plcR) was grown with shaking in BHI medium and erythromycin at 37°C to an A600 of 0.2 to 0.3. Cultures were divided, and different concentrations of tetracycline (0 to 1,000 ng/ml) were added. Each culture was plated after 3 h onto BHI agar plates containing lecithin or sheep blood, erythromycin, and corresponding concentrations of tetracycline. Colonies were examined for halos or zones of hemolysis.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Plasmid | ||
| pCR 2.1-TOPO | Cloning vector; Apr Kmr | Invitrogen |
| TOPO-antAB | 2.1-kb PCR fragment (A1-A2 primers) containing plc and sph genes of B. anthracis cloned into pCR2.1-TOPO | This work |
| TOPO-cerAB | 2.1-kb PCR fragment (A1-A2 primers) containing plc and sph genes of B. cereus cloned into pCR2.1-TOPO | This work |
| TOPO-ant(plcR-papR) | 1.37-kb PCR fragment (P1-P2 primers) containing ΔplcR and papR genes of B. anthracis cloned into pCR2.1-TOPO | This work |
| TOPO-cer(plcR-papR) | 1.37-kb PCR fragment (P3-P4 primers) containing plcR and papR genes of B. cereus cloned into pCR2.1-TOPO | This work |
| pET-15b | Expression vector for E. coli | Novagen |
| pET-15b-antA | 740-bp PCR fragment (AP1-AP2 primers) encoding mature B. anthracis PC-PLC | This work |
| pET-15b-cerA | 740-bp PCR fragment (AP1-AP2 primers) encoding mature form of B. cereus PC-PLC | This work |
| pET-15b-antB | 920-bp PCR fragment (AS1-AS2 primers) encoding mature form of B. anthracis SPH | This work |
| pET-15b-cerB | 920-bp PCR fragment (CS1-AS2 primers) encoding mature form of B. cereus SPH | This work |
| pUTE29 | Vector for gene replacement in B. anthracis; Apr in E. coli; Tcr in B. anthracis | 21 |
| pUTE29-plcR-papR | 1.38-kb BamHI-PstI B. cereus DNA fragment from TOPO-cer (plcR-papR) | This work |
| pSJ115 | Encodes anthrax toxin lethal factor (lef) behind signal sequence of anthrax protective antigen (pag), in shuttle plasmid; Apr in E. coli; Kmr in B. anthracis | 28 |
| pSJ115a | pSJ115 having NdeI site at bp 212 eliminated | This work |
| pSW4 | pSJ115a with both lef gene and signal sequence of pag deleted | This work |
| pAE5 | pSW4 containing 858-bp PCR fragment (R1-R2 primers) encoding B. cereus plcR gene | This work |
| pAE5::IS10 | pAE5 with IS10 inserted into plcR gene | This work |
| pOB12 | pE194 vector containing plc-sph operon of B. cereus BKM-B164; Emr in B. anthracis | 29 |
| pYJ335 | Vector expressing antisense from Tet-inducible promoter; Apr in E. coli; Emr in B. cereus | 17 |
| pYJ335-anti-plcR | pYJ335 containing 691-bp PCR fragment (BAP1-BAP2) of B. anthracis ΔplcR cloned in antisense orientation | This work |
| pYJ335-plcR-papR | pYJ335 containing 1.38-kb fragment of B. cereus DNA from pUTE29-plcR-papR cloned in sense orientation | This work |
| Strain | ||
| B. anthracis | ||
| Ames 34 | pXO1− pXO2+ strain similar to ΔAmes-1 | 12 |
| Ames 33 | pXO1− pXO2− Ames 34 derivative strain | This work |
| UM44-1C9 | pXO1− pXO2− Plasmid-cured UM44-1 strain | 2 |
| Sterne 34F2 | pXO1+ pXO2− | 16 |
| Sterne 34F2 DeltaT (SdT) | Sterne 34F2 cured of pXO1; therefore pXO1− pXO2− | 16 |
| SdT1 | SdT(pUTE29-plcR-papR); Tcr; does not produce intracellular PlcR, nonhemolytic strain | This work |
| SdT2 | SdT(pAE5); Kmr; produces intracellular PlcR; weakly hemolytic | This work |
| SdT3 | SdT(pAE5::IS10); Kmr; does not produce intracellular PlcR; nonhemolytic | This work |
| SdT4 | SdT electroporated with pOB12; Emr; hemolytic | This work |
| B. cereus | ||
| 569 | Wild strain; produces extracellular PC-PLC | 32 |
| 540 | 569 electroporated with plasmid pYJ335-anti-plcR; produces extracellular PC-PLC | This work |
| 541 | 540 with integrated plasmid pYJ335-anti-plcR; ΔPlcR; does not produce extracellular PC-PLC | This work |
| 6A3 | B. cereus Frankland and Frankland NRRL-569 | BGSCb |
| 6A5 | B. cereus Frankland and Frankland ATCC 14579 | BGSC |
| B. thuringiensis | ||
| 4A2 | Wild-type isolate, serotype 1 | BGSC |
| 4B1 | Wild-type isolate, serotype 2 | BGSC |
| E. coli | ||
| XL2-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr) Amy Cmr] | Stratagene |
| SCS110 | rpsL (Smr) thr leu endA thi-l lacY galK galT ara tonA tsx dam dcm supE44D (lac-proAB) [F′ traD36 proAB lacIqZDM15] | Stratagene |
| BL21 (DE3) | E. coli B; F−ompT hsdS(rB− mB−) dcm+ Tetrgal (DE3) endA Hte | Stratagene |
Apr, ampicillin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; Emr, erythromycin resistant; NRRL, Agricultural Research Service Culture Collection; ATTC, American Type Culture Collection.
BGSC, Bacillus Genetic Stock Center.
TABLE 2.
Primers used in this study
| Primer | 5′-3′ Sequencea (location) | Relevant property | Restriction sites |
|---|---|---|---|
| A1 | GTATTCATTCATTATATTCACTGTG (180 bp before plc start codon) | Used to amplify plc and sph from 5′ | |
| A2 | CTACTTCATAGAAATAGTCGCCT (C-end of sph) | Used to amplify plc and sph from 3′ | |
| P1 | GGATAAAAAAGACCGAGTGTAATG | Used to amplify plcR and papR from 5′ | |
| P2 | CGTTTGGAGGTTACTCCAC | Used to amplify plcR and papR from 3′ | |
| P3 | GCTAGGATCCGGGCAAAGAAGACCGAATGTA | Used to amplify ΔplcR and papR from 5′ | BamHI |
| P4 | GGGCTGCAGGACGTTTGGATGTTACTCCAT | Used to amplify ΔplcR and papR from 3′ | PstI |
| AP1 | GGGCATATGTCTGCTGAAGATAAACATAAA | Used to amplify B. anthracis and B. cereus Pc-Plc genes (mature forms) from 5′ | NdeI |
| AP2 | GCTACTCGAGTTAACGATCTCCGTACGTATCAAA | Used to amplify B. anthracis and B. cereus Pc-Plc genes (mature forms) from 3′ | XhoI |
| AS1 | GGGCATATGGCAGATACGTCTACAGATCAA | Used to amplify B. anthracis Sph gene (mature form) from 5′ | NdeI |
| AS2 | GCTACTCGAGCTACTTCATAGAAATAGTCGCCT | Used to amplify B. anthracis and B. cereus Sph genes (mature forms) from 3′ | XhoI |
| CS1 | GGGCATATGGCAGAAGCATCTACAAATCAA | Used to amplify B. cereus Sph gene (mature form) from 5′ | NdeI |
| B1 | GCAATCAGATCTTCCTTCAGGT | Used for pSW4 construction | BglII |
| B2 | GGGGGATCCCATATGCGTTCTCCTTTTTGTAT | Used for pSW4 construction | BamHI, NdeI |
| R1 | GATCCATATGCACGCAGAGAAATTAGGAAGTG | Used to amplify B. cereus plcR gene from 5′ | NdeI |
| R2 | CCCGGGATCCTTATTTCTTCATTTTTTTCATAAA | Used to amplify B. cereus plcR gene from 3′ | BamHI |
| BAP1 | GGGGGGCCCGGGTATAGTGGGATGGTGAGTAAG | Used to amplify B. anthracis ΔplcR from 5′ | SmaI |
| BAP2 | GGGGGGCCCGGGAATAGCTTTATTTGCATGACA | Used to amplify B. anthracis ΔplcR from 3′ | SmaI |
| ISp1 | CTGATGAATCCCCTAATGAT | Inner primer used to amplify IS10 5′ region | |
| ISp2 | TTTTAGGTGACGGGTGGTGAC | Inner primer used to amplify IS10 3′ region |
Restriction enzyme recognition sites are underlined.
Plasmid-free B. anthracis strain Ames 33 was selected as a spontaneous rough variant of the pXO2-containing B. anthracis Ames 34 strain as described elsewhere (12). The strain was confirmed to be free of pXO2 by PCR.
DNA cloning and sequencing.
The plc-sph regions from B. cereus and B. anthracis were obtained as 2.1-kb DNA fragments by PCR using primers A1 and A2 (Table 2). Similarly, the plcR-papR regions of both species were obtained as 1.37-kb DNA fragments by PCR using primers P1 and P2. The fragments isolated from SeaKem agarose gels (Rockland, Maine) were cloned into the vector pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) Nucleotide sequencing of the cloned fragments was performed by the dideoxy chain termination technique with a Taq dye primer cycle sequencing kit. M13 reverse and forward primers were used initially; primers complementary to the determined sequences were subsequently used.
The NCBI BLAST and FASTA programs (http://www.ncbi.nlm.nih.gov/) were used for homology searches in the GenBank and Swiss-Prot databases. B. anthracis genome nucleotide sequence data from the Institute for Genomic Research (http://www.tigr.org/) were used for comparison.
A DNA fragment encoding the entire plcR-papR region of B. cereus 569 was PCR amplified with primers P3 and P4, cut with BamHI and PstI, and inserted into the same sites of plasmid pUTE29, provided by Theresa Koehler (21). The same fragment was blunted with Klenow fragment and cloned into the EcoRV site of vector pYJ335 (17). The resulting pUTE29-plcR-papR and pYJ335-plcR-papR plasmids were transformed into several B. anthracis strains (Table 1).
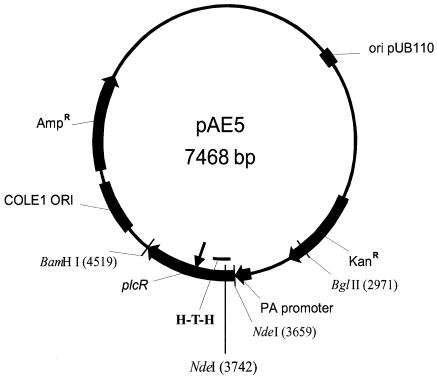
Plasmid pSJ115, a shuttle vector that expresses anthrax toxin lethal factor from the anthrax toxin protective antigen (PA) promoter (28), was used for construction of plasmids pSW4 and pAE5. For this purpose, the NdeI site at position 212 of plasmid pSJ115 was eliminated, and a PCR fragment was amplified from plasmid pSJ115 with primers B1 and B2, restricted with BglII and BamHI, and inserted into the same sites of pSJ115. The resulting plasmid pSW4 contained the B. anthracis PA gene promoter (without the PA signal peptide gene sequence) immediately preceding the unique NdeI site. The unique BamHI and NdeI sites of plasmid pSW4 allowed directional cloning of a PCR fragment containing the B. cereus plcR gene amplified with the primer R1-R2. The resulting plasmid pAE5 was introduced into several different B. anthracis strains (Table 1).
Expression and purification of His-tagged enzymes.
For the expression of mature forms of B. anthracis and B. cereus PC-PLC and SPH as their His6-tagged derivatives, DNA fragments containing added NdeI and XhoI sites were amplified with primers AP1-AP2 (for both B. anthracis and B. cereus PC-PLC), AS1-AS2 (B. anthracis SPH), and CS1-AS2 (B. cereus SPH) by Platinum TaqDNA Polymerase High Fidelity (Invitrogen/Life Technologies). The fragments were isolated from agarose gels and inserted into the corresponding restriction sites of vector pET-15b (Novagen, Madison, Wis.) The resulting plasmids (Table 1) were introduced into E. coli BL21(DE3). The proteins with His6 tags at the N termini were purified from 1 liter of IPTG-induced cultures grown in LB medium at 37°C. Protein purification by elution with imidazole from nickel-nitrilotriacetic acid His-Bind resin was performed essentially as recommended by the manufacturer (QIAGEN Inc.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to analyze the purity of the proteins.
PC-PLC and SPH enzymatic assays.
Bacteria were grown in BHI broth at 37°C. Aliquots of growing cultures were centrifuged for 15 min at 6,500 × g. The supernatants and cell pellets were frozen in dry ice. The proteins from selected supernatants were concentrated with Centriprep YM-10 units (Amicon, Inc., Beverly, Mass.) and stored at −20°C until used. Concentrations of the proteins in the samples were determined with BCA protein assay reagent (Pierce Biotechnology, Rockford, Ill.).
The activities of the PC-PLC and SPH enzymes of the samples were determined with, respectively, the Amplex Red phosphatidylcholine-specific phospholipase C assay kit and the Amplex Red sphingomyelinase assay kit (Molecular Probes, Eugene, Oreg.). For measurement of PC-PLC, each reaction mixture contained 200 μM Amplex Red reagent (10-acetyl-3,7-dihydroxyphenoxazine), 1 U of horseradish peroxidase/ml, 4 U of alkaline phosphatase/ml, 0.1 U of choline oxidase/ml, 0.5 mM lecithin, and 20 to 100 mU of PC-PLC/ml in 50 mM Tris-HCl (pH 7.4)-140 mM NaCl-10 mM dimethylglutarate-2 mM CaCl2. (Enzyme units are defined by the kit manufacturer, and except for horseradish peroxidase, 1 U of enzyme produces 1 μmol of product per min under optimal conditions.) For measurement of SPH, each reaction mixture contained 50 μM Amplex Red reagent, 1 U of horseradish peroxidase/ml, 4 U of alkaline phosphatase/ml, 0.1 U of choline oxidase/ml, 0.25 mM sphingomyelin, and 0.2 to 1.0 mU of SPH/ml in 0.1 M Tris-HCl-10 mM MgCl2 (pH 7.4). Reaction mixtures were incubated in the dark at 37°C for 30 to 60 min. Fluorescence was measured with a Wallac 1420 VICTOR 96-well plate reader (Perkin Elmer, Boston, Mass.) with excitation at 530 nm and emission at 590 nm. The activity of unknowns was compared to that of the standard enzyme supplied with the kit to calculate milliunits per microgram of total protein.
PlcR antisera preparation.
The multiple antigen peptides (MAPs) technique (4) was used for the production of antibodies to PlcR. Sequences of linear peptides corresponding to the carboxyl and amino termini of PlcR and having favorable antigenic indices were selected with the JaMBW computer program (41) as LEKLGYDETESEEAY (C-PlcR) and EIYNKVWNELKKEEY (N-PlcR), respectively. The peptides were synthesized with their N termini attached to a branched poly-l-lysine core sequence, with eight peptide chains incorporated in each molecule. Synthesis was performed by the Center for Biologics Evaluation and Research core facility, Food and Drug Administration, Bethesda, Md.
Rabbits were immunized with a total of 10 mg of each MAP, administered in four equal doses over a 75-day period, with a final bleeding at day 89 (performed by Covance Research Products Inc., Denver, Penn.). The antisera were tested by enzyme-linked immunosorbent assay (ELISA). Briefly, 96-well plates (Greiner, Monroe, N.C.) were coated overnight at 4°C with 100 μl of MAP at 10 μg/ml in 10 mM potassium phosphate buffer, pH 9.0. All steps thereafter were performed with 100 μl per well. After the samples were washed with buffer (0.1% gelatin in 10 mM Tris-HCl [pH 8.0]-50 mM NaCl), serially diluted rabbit sera were added and incubated overnight at 4°C. After repeated washings of the samples, goat anti-rabbit immunoglobulin G-horeseradish peroxidase (sc-2054; Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) was added at a final concentration of 0.2 μg/ml. After incubation for 1.5 h at room temperature (20 to 25°C), plates were washed thoroughly. Finally, the wells were incubated with 1 mg of ABTS substrate [2,2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid; Sigma-Aldrich] per ml in 100 mM potassium phosphate (pH 5.0)-0.003% H2O2 and the absorbance at 405 nm was measured.
Western immunoblotting of PlcR.
The frozen bacterial cell pellets were thawed, suspended in ice-cold phosphate-buffered saline (PBS; 0.15 M NaCl-10 mM sodium phosphate [pH 7.5]), added to prechilled FastPROTEIN BLUE tubes and homogenized with glass beads by using a FastPrep FP120 instrument (Qbiogene; BIO 101 Systems, Carlsbad, Calif.) at a speed of 6.0 for two 30-s periods. After homogenization, the tubes were centrifuged for 15 min at 20,000 × g, and the supernatants were transferred to new tubes for protein determination. Equal amounts of each sample (50 to 100 μg of protein) were separated on SDS-polyacrylamide (10 to 20%) gels (Novex precast gels; Invitrogen). The MultiMark multicolored standard (Invitrogen) was used as a molecular weight marker. The separated proteins were transferred to nitrocellulose membrane (PROTRAN B85; Schleicher & Schuell) in a Novex transfer unit (Invitrogen). PlcR was detected in most cases with a 1:3,000 dilution of rabbit serum 1451, directed to the C-terminal peptide LEKLGYDETESEEAY, because this serum was more specific (see Fig. 5). The blot was developed with a 0.2-μg/ml concentration of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (sc-2054; Santa Cruz Biotechnology) and an enhanced chemiluminescence substrate (SuperSignal; Pierce Biotechnology).
FIG. 5.
PlcR antiserum characterization. (A) B. cereus 569 PlcR amino acid sequence. The N-PlcR peptide immunogen corresponds to aa 93 to 107, and the C-PlcR peptide corresponds to aa 246 to 260. The vertical arrow indicates the site at which the B. anthracis PlcR protein is truncated. (B) ELISA reactivity of N-PlcR and C-PlcR antiserum at 1:3,000 dilutions on plates coated with either the N-PlcR peptide (left graph) or the C-PlcR peptide (right graph). (C) Western blot of whole-cell proteins from B. anthracis SdT (lane 1) and B. anthracis SdT2 (lane 2). Membranes were treated with N-PlcR antiserum (serum 1447) or C-PlcR antiserum (serum 1451) at a 1:2,000 dilution. Mr, position of external molecular mass marker.
Nucleotide sequence accession numbers.
The nucleotide sequences comprising the B. cereus 569 genes were submitted to GenBank, with plc and sph submitted under accession number AY195600 and plcR and papR submitted under accession number AY195601.
RESULTS
Nucleotide sequence comparisons of plc, sph, plcR, and papR genes.
We cloned and sequenced the plc and sph genes from B. anthracis strain UM44-1C9 and B. cereus 569 and compared these sequences with previously published sequences of B. anthracis and B. cereus. The plc-sph region sequence determined for B. anthracis UM44-1C9 exactly matched that of the B. anthracis Ames strain, available from TIGR at http://www.tigr.org, and was nearly identical to that of B. anthracis strain A2012 (31) (GenBank accession number AAAC01000001). However, the B. anthracis plc and sph genes differ slightly from those of B. cereus, especially in the promoter regions (Fig. 1). The sequence of the putative 16-bp PlcR-binding site upstream of the B. anthracis plc start codon contains T instead of G at the fourth position, and therefore it deviates from the perfect palindrome (TATGnAnnnnTnCATA, where n is any of the four nucleotides, A, G, C, or T) found in most B. cereus and B. thuringiensis plc genes (1). A possible PlcR-binding site was found just 5 bp upstream of the start codon of the B. anthracis sph gene, overlapping the ribosome binding site. Although this sequence also differs from the canonical PlcR binding site sequence (G instead of A at the sixth position), the same sequence is found in the sph genes of B. cereus strain SE-1 (18) and strain BKM-B164 (8). A very similar 19-bp TATGTTATGTACCTCCATA sequence was found at the same distance (5 bp) upstream of the sph start codons of B. cereus strains 569, GP-4 (9), and IAM 1208 (45). However, in all these B. cereus strains, this sequence is located on the cDNA strand.
FIG. 1.
Sequence alignment of the promoter regions for plc and sph genes of B. anthracis strain UM44-1C9 (B.a.) and B. cereus (B.c.) strains SE-1, BKM-B164, GP-4, and 569. Putative PlcR binding sites are located upstream of σA −35 and −10 sequences for the plc gene and downstream of the sph σA −35 and −10 sequences. Consensus sequences of regulatory elements are indicated in bold type under the corresponding aligned sequences. Gray areas indicate nucleotide sequence differences.
The B. anthracis plc gene −35 and −10 promoter elements (TAGAAA and TATTCT, respectively) were found to be identical to those of all reported B. cereus plc genes. The B. anthracis sph gene −35 and −10 promoter sequences (TTAAAA and TAGAGT, respectively) were identical to the corresponding sequences of sph genes from B. cereus SE-1 (18) and BKM-B164 (8). However, the B. anthracis −35 sequence differed from the TTGAAA sequence reported for B. cereus 569, B. cereus IAM 1208 (45), and B. cereus GP-4 (9).
The PC-PLC and SPH proteins of B. anthracis are predicted to be very similar in sequence and structure to those of B. cereus. The genes encode signal peptides (24 amino acids [aa] for PC-PLC and 27 aa for SPH) and a 14-aa propeptide for PC-PLC. A conserved ribosome binding site, GGAGG, is located upstream of the initiation codons for PC-PLC (ATG) and SPH (GTG). The primary structure of the mature B. anthracis PC-PLC protein (245 aa) is almost identical to the structures of the proteins of B. cereus SE-1 (18) and B. cereus BKM-B164 (8), differing by only two substitutions: H156Y and D174E. On the other hand, the B. cereus 569 mature PC-PLC protein also differed from that of B. cereus GP-4 (9) by two substitutions, T29K and D244N. The mature B. anthracis SPH protein (306 aa) differs from the B. cereus SE-1 (18) and B. cereus BKM-B164 (8) proteins by six substitutions. The SPH protein of B. cereus 569 differs from that of B. cereus GP-4 (9) by only 5 aa. The SPH proteins of B. cereus 569 and B. cereus GP-4 have more than 30 aa differences from the very similar SPH proteins of B. anthracis and B. cereus strains SE-1, BKM-B164, and IAM 1208. In particular, the NIRIMITLIIIQ sequence at the C-terminal end (from position 277 to 299) of the SPH protein is specific for only the B. cereus 569 and GP-4 strains.
The nucleotide sequences of the plcR and papR genes from B. anthracis UM44-1C9 and B. cereus 569 were aligned with published sequences of these genes from B. thuringiensis 407 (22), B. cereus ATCC 14579 (27), B. anthracis 9131 (1), and B. anthracis Ames (TIGR database). No differences were found among the nucleotide sequences of this region in the B. anthracis Ames, 9131, and UM44-1C9 strains. The PlcR operon structures of B. cereus 569, B. cereus 14579, and B. thuringiensis 407 are the same, consisting of the two genes, plcR and papR, which encode polypeptides of 285 aa (complete PlcR of B. cereus) or 212 aa (truncated PlcR of B. anthracis) and 48 aa (PapR), respectively. Both the plcR and papR genes contain perfect 16-bp PlcR palindromes upstream of the ATG initiation codons. The amino acid sequences of the various PlcR and PapR polypeptides are rather varied. For example, in B. cereus 569, 16 unusual amino acids were found for PlcR and 5 were found for PapR compared to the other Bacillus strains discussed here. Interestingly, the unique C-terminal KFMKKMKK sequence of B. cereus 569 PlcR contains two MKK repeats, and this tripeptide sequence is also at the N terminus of PapR.
B. anthracis and B. cereus PC-PLC and SPH enzymes have similar activities.
The enzymes expressed in E. coli were purified as His6-tagged proteins and analyzed by SDS-PAGE. The electrophoretic mobilities of the purified proteins corresponded well to the calculated molecular mass values of 28 kDa for PC-PLC and 34 kDa for SPH (Fig. 2A). The catalytic activity of the enzymes was largely as expected, with both SPH proteins having sphingomyelinase activity (Fig. 2B) and both PC-PLC enzymes hydrolyzing phosphatidylcholine (Fig. 2C). There were no significant differences between the enzymes from B. anthracis and B. cereus. However, it was notable that the SPH enzymes were also able to cleave phosphatidylcholine (Fig. 2C). The recombinant enzymes exhibited hemolytic properties like those of the extracellular enzymes from B. cereus (3). Thus, sheep erythrocytes were lysed by SPH but not by PC-PLC (Fig. 2D). Combining the enzymes did not appreciably enhance lysis over that caused by SPH alone.
FIG. 2.
Molecular and functional properties of recombinant B. anthracis and B. cereus PC-PLC and SPH. (A) SDS-PAGE of the proteins purified from E. coli, stained with Coomassie blue R-250. Mr, molecular mass marker. (B and C) SPH and PC-PLC activities of the four recombinant phospholipases. The order of the samples is the same as in panel A. (D) Ability of PC-PLC, SPH, and a 1:1 mixture of these proteins to lyse sheep red blood cells. Each well was filled with 20 μl of a 0.5-mg/ml solution of enzyme. Wells with both phospholipases were filled with 20 μl of each enzyme.
Role of PlcR in control of PC-PLC activity.
To analyze the role of PlcR in controlling the expression of PC-PLC, we constructed a vector expressing tetracycline-inducible antisense to plcR (Fig. 3A). Introduction of this plasmid, pYJ335-anti-plcR, into B. cereus 569 and induction on agar plates containing tetracycline, lecithin, and erythromycin gave colonies of the resulting B. cereus strain 540 that had normal halos due to PC-PLC hydrolysis of lecithin. Thus, the antisense appeared unable to effectively repress PlcR action. Plasmid pYJ335-anti-plcR could be recovered from this Ermr, PC-PLC-positive bacterium, demonstrating that it was extrachromosomal (Fig. 3B, panel 2).
FIG. 3.
Inactivation of B. cereus 569 plcR gene. (A) Scheme of plasmid pYJ335-anti-plcR integration into plcR gene of B. cereus 569 chromosome. The truncated antisense-oriented B. anthracis plcR gene (ΔplcR) was recombined with the B. cereus 569 chromosomal plcR gene so that the only intact plcR, at the right end, lacks a promoter. PCR primers used in analysis include P1 and P2 (external primers matching chromosomal DNA), M13f and M13r (internal primers that match only the pYJ335-antisense plasmid), and BAP1 and BAP2 (primers that belong to chromosomal and plasmid DNAs). (B) DNA analysis of recombinant strains. Panel 1, PCR fragments from the left end of the inserted plasmid; panel 2, plasmid content of B. cereus 569 derivatives having integrated (B. cereus 541) and extrachromosomal (B. cereus 540) pYJ335-anti-plcR plasmids. The arrow (panel 2) indicates a band of plasmid pYJ335-anti-plcR. Other bands are endogenous B. cereus 569 plasmids. Panel 3, PCR fragments from the right end of the inserted plasmid. Mr, molecular mass marker. (C) Hemolysis of B. cereus 541 is weaker than that of B. cereus 569 (panel I). B. cereus 541 does not hydrolyze lecithin (panel II).
However, repeated passages of B. cereus strain 540 on BHI agar with lecithin and erythromycin allowed selection of a variant strain, B. cereus 541, containing a single crossover event that introduced the plasmid into the chromosome (Fig. 3A). Four passages on BHI agar, with samples taken each time from the edge of the growing area, were needed to obtain this halo- and plasmid-negative B. cereus 541 clone (Fig. 3C). This strain also grew faster on solid medium than either of the parental strains, 569 and 540. PCR analysis of DNA from this clone confirmed introduction of plasmid pYJ335-anti-plcR into the plcR gene (Fig. 3B). Introduction of plasmid pUTE29-plcR-papR (described in the next section) into B. cereus 541 restored the hemolytic properties of B. cereus (data not shown). These data confirm that PlcR is required to activate PC-PLC expression in B. cereus.
Presence of B. cereus plcR-papR operon in B. anthracis is not sufficient to induce PC-PLC activity.
To test the hypothesis that lack of hemolysis by B. anthracis is due to the C-terminal truncation of the PlcR protein, we sought to complement this defect. Plasmid pYJ335 was used again but this time to express the B. cereus plcR-papR operon as an inducible sense transcript. Plasmid pYJ335-plcR-papR was introduced into B. anthracis Ames 33 and SdT with erythromycin selection. The resulting transformants were grown and passed repeatedly on plates containing sheep blood or lecithin and erythromycin and in the presence and absence of tetracycline. The presence of tetracycline did not cause the colonies to produce amounts of PC-PCL and SPH that were sufficient to generate hemolytic zones or halos. Even after four passages, colonies with hemolytic zones or halos did not appear.
The fragment containing the complete B. cereus plcR operon was also inserted into plasmid pUTE29, which has been used previously in B. anthracis (21, 33). The resulting pUTE29-plcR-papR plasmid was electroporated into B. anthracis strains SdT and UM44-1C9. Transformants were selected on tetracycline-containing medium supplemented with sheep blood or lecithin. The plasmid was reisolated from the B. anthracis transformants, and the restriction digest patterns of the isolated plasmid were compared with those of the plasmid used for the electroporation. No differences were found in the patterns or in the nucleotide sequences of the fragment containing the plcR and papR genes. However, even in this case, colonies with hemolytic zones or halos did not appear (Fig. 3C). The results with the pYJ335 and PUTE29 constructs suggest that regulated artificial promoters and the B. cereus endogenous promoters do not produce sufficient amounts of the PlcR and PapR proteins or that other regulatory factors are needed for PlcR transcriptional activation in B. anthracis.
Expression of PlcR from a strong promoter in B. anthracis induces weak hemolysis of sheep blood.
As an alternative way to complement the defective plcR gene in B. anthracis, we constructed plasmid pAE5 (Fig. 4) containing B. cereus plcR under control of the B. anthracis PA gene promoter (without the PA signal sequence). This plasmid was transformed into B. anthracis SdT, and following plating on BHI plates containing kanamycin and sheep blood, only four slightly hemolytic colonies (designated SdT2) were found among hundreds of nonhemolytic transformants (designated SdT1). The same isolates were not hemolytic when plated on LB agar containing sheep blood. Restriction analysis showed that plasmids isolated from the hemolytic SdT2 colonies were identical to the original pAE5. However, plasmids isolated from the nonhemolytic SdT1 colonies demonstrated insertion of additional DNA within the plcR gene. Determination of the nucleotide sequence of the inserted DNA showed that it belonged to the 3′ terminal IS10 element of Tn10 (5). Repeated transformations of B. anthracis by the two plasmids showed that the efficiency of transformation by pAE5 was much lower than that of plasmid pAE5::IS10 carrying the inactivated plcR gene. PCR analysis of E. coli XL2-Blue and SCS110 chromosomal DNAs and of chromosomal DNAs from the B. anthracis Ames 33, UM44-1C9, and SdT strains demonstrated that only E. coli XL2-Blue chromosomal DNA contains an IS10 element. (Primers ISp1 and ISp2 were used for PCR detection of IS10 DNA.) This result clearly indicated that strong expression of an active PlcR is toxic to B. anthracis. Consistent with this interpretation, we noted that the hemolytic activity of two of the four B. anthracis hemolytic clones containing plasmid pAE5 disappeared during repeated passages on BHI agar containing sheep blood and antibiotic, whereas the other two appeared stable.
FIG. 4.
Genetic and restriction map of plasmid pAE5. The plcR gene of B. cereus 569 is under control of the B. anthracis protective antigen gene promoter. The signal peptide of the protective antigen gene was eliminated in order to retain PlcR inside the cell. The location of the inserted IS10 found in pAE5::IS10 is indicated by the arrow. H-T-H, helix-turn-helix motif of PlcR.
Characterization of rabbit polyclonal antisera to PlcR.
To allow detection and quantitation of PlcR expression, we prepared antisera by immunization with synthetic peptides matching N- and C-terminal regions of the protein. The N-terminal sequence selected is present in both B. anthracis and B. cereus PlcR proteins, whereas the C-terminal sequence would recognize the B. cereus protein but not the truncated B. anthracis protein (Fig. 5A). An ELISA showed that the resulting sera react strongly to the corresponding peptides (A405, >2 at a 1:3,000 dilution) (Fig. 5B) and do not interact with the heterologous antigen (A405, <0.2 at a 1:3,000 dilution). Both antisera reacted with a band of the size expected for B. cereus PlcR (34 kDa) in extracts from B. anthracis SdT2 (SdT with pAE5). The C-PlcR antisera reacted only with this band, whereas the N-PlcR antiserum also reacted with additional proteins of higher molecular weight from both SdT2 and SdT (Fig. 5C). Neither antibody detected a band corresponding to ΔPlcR of B. anthracis (calculated molecular mass of 25.4 kDa), suggesting that the truncated B. anthracis PlcR polypeptide either is not expressed or is unstable. The demonstrated specificity of the C-PlcR antiserum for the B. cereus plcR gene allowed its use in the characterization of PlcR expression presented below.
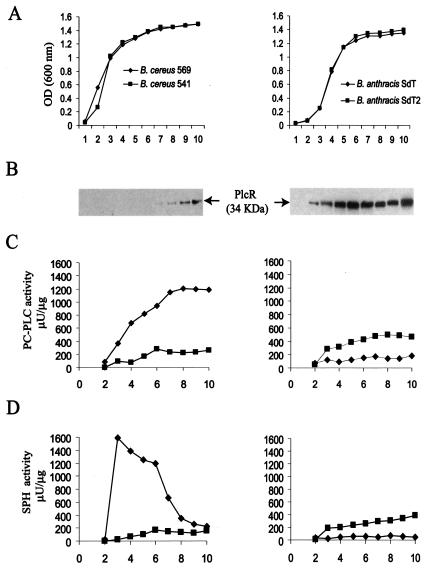
PlcR expression in B. cereus 569 at onset of stationary phase correlates with induction of PC-PLC activity and repression of SPH activity.
Growth curves demonstrated that B. cereus 569 grew in BHI broth without a lag phase whereas B. cereus 541 (PlcR negative) experienced a short lag (Fig. 6A, left panel). This lag was small but was observed in repeated experiments and is consistent with the report that a B. cereus ATCC 14579 ΔplcR strain grew more slowly than its wild-type parent (10). Western blotting showed that the PlcR protein appeared in B. cereus 569 grown in BHI medium only after 6 to 7 h, reaching a maximum at 9 h (Fig. 6B, left panel), a time that probably corresponds to the onset of the stationary phase (23). Similarly, PC-PLC activity reached a maximum at 7 to 8 h (Fig. 6C, left panel). Interestingly, PC-PLC activity increased gradually in the period from 3 to 7 h, in contrast to the sharp increase described for plcA transcriptional activation in B. thuringiensis (22). On the other hand, SPH activity increased at the beginning of the exponential phase of growth (Fig. 6D, left panel). This activity decreased later and disappeared completely by 9 h when the stationary phase was reached. B. cereus strain 541, in which plcR was disrupted, showed no production of PlcR protein by Western blot analysis (data not shown) and no induction of PC-PLC or SPH activity (Fig. 6C and D, left panels).
FIG. 6.
Growth, PlcR production, and enzyme activities for B. cereus and B. anthracis strains. (A) Growth curves for strains incubated at 37°C in BHI broth. OD, optical density. (B) Western blot analyses of whole-cell proteins from B. cereus 569 (left panel) and B. anthracis strain SdT2 (right panel) with C-PlcR antiserum 1451 as described above. No PlcR production was found for B. cereus 541 or B. anthracis SdT (not shown). (C and D) PC-PLC (C) and SPH (D) activities of extracellular proteins. Equal amounts of extracellular protein (2 to 10 μg for PC-PLC and 0.1 to 0.5 μg for SPH) were assayed by the Red Amplex reagents. Maximum root-mean-square deviations did not exceed 10% for the PC-PLC and SPH determinations. Filled square, B. cereus 541; filled diamond, B. cereus 569.
PlcR expression in B. anthracis causes only weak expression of PC-PLC and SPH.
The growth rates of the parental B. anthracis SdT and of the weakly hemolytic pAE5 transformant, SdT2, were indistinguishable (Fig. 6A, right panel). Both strains grew with a lag phase of about 2 h. Expression of the full size B. cereus PlcR protein from the PA promoter of plasmid pAE5 in B. anthracis SdT2 took place from the beginning of the exponential phase of growth in BHI medium (Fig. 6B, right panel). The production of the protein in B. anthracis SdT2 was more extensive than that in B. cereus 569 and in comparison almost independent of time. On the other hand, although the activities of the PlcR-regulated enzymes PC-PLC and SPH in SdT2 increased with time, the peak activity levels (Fig. 6C and D, right panels) never reached values equal to those of B. cereus 569. These data indicate that the molecular mechanisms of PlcR action are different for these two bacteria. The parental B. anthracis SdT strain produced no PlcR protein (data not shown), and the PC-PLC or SPH activities were negligible (Fig. 6C and D, right panels).
Growth of B. anthracis producing PlcR under aerobic-plus-CO2 or under anaerobic-plus-CO2 conditions slightly induces both its PC-PLC and hemolytic activities.
Recent studies employing reverse transcription-PCR reported that B. anthracis Sterne 34F2 hemolysin-related genes including plc and sph were induced under strictly anaerobic conditions (20). We examined whether the same was true for B. anthracis SdT2 and included experiments in which plates were grown in aerobic and CO2-enriched environments. The parental B. anthracis Sterne and SdT strains and B. cereus strains 569 and 541 were used for comparison. Under aerobic conditions (Fig. 7, Aerobic), only B. cereus strain 569 demonstrated high PC-PLC and hemolytic activities for both sheep and human blood. B. anthracis SdT2 and B. cereus 541 slightly lysed sheep blood but did not hydrolyze lecithin or lyse human blood. The remaining B. anthracis strains were inactive even for sheep blood (Fig. 7, Aerobic). Growth in an aerobic-plus-CO2 environment appeared to increase PC-PLC and hemolytic activities in B. anthracis SdT2, and some activity was seen in B. cereus 541. The other strains did not demonstrate PC-PLC activity in the aerobic-plus-CO2 atmosphere. Under anaerobic-plus-CO2 conditions, we found that B. cereus strain 569 retained PC-PLC and hemolytic activities, although the levels of both activities were less than those under aerobic-plus-CO2 conditions. However, comparisons are difficult because all strains grew less vigorously in the anaerobic atmosphere, as Klichko et al. (20) also observed. Very low PC-PLC and hemolytic activities were found for the PlcR-producing SdT2 strain, and no activities were found for the other B. anthracis strains. B. cereus 541 induced some hemolytic activity for sheep blood (Fig. 7, Anaerobic + CO2). We repeated all the assays shown in Fig. 7 with separate plates for every strain and growth condition and obtained equivalent results.
FIG. 7.
Hemolysis and lecithinase production by B. anthracis and B. cereus strains grown for 48 h at 37°C in aerobic, aerobic-plus-CO2, or anaerobic-plus-CO2 atmospheres. Lecithin (left) column shows agar plates containing egg yolk suspension (full recipe given in Materials and Methods), and sheep blood (center) and human blood (right) columns show agar plates with 5% sheep and human blood, respectively. On each of the nine plates, B. anthracis strains SdT2, SdT, and Sterne were spotted in the top horizontal row and B. cereus strains 541 and 569 were spotted in the vertical column. The arrangement of strains on all plates is that shown in the upper left panel.
Extracellular factors increase PlcR activation in B. anthracis.
Studies of B. thuringiensis and B. cereus showed that peptides derived from PapR promote gene activation by PlcR (39). We examined whether the same was true of B. anthracis by testing whether extracellular materials from several B. cereus and B. thuringiensis strains could activate the PC-PLC activity of B. anthracis SdT2. Growing B. cereus 6A5 and B. thuringiensis 4A2 (serotype 1) and B. thuringiensis 4B1 (serotype 2) next to SdT2 did confer on it the ability to hydrolyze lecithin (Fig. 8). Presumably, B. cereus strain 6A5 and B. thuringiensis strains of serotype 1 and 2 produce extracellular peptides derived from PapR that can be processed to pentapeptides having a Leu in the N terminus, whereas other strains have a Met or Val in this position and therefore yield inactive peptides (39). However, in our experiments, B. cereus strain 6A3 also activated the PC-PLC activity of B. anthracis SdT2, even though its pentapeptide has a Met in the N terminus (Fig. 8).
FIG. 8.
Spot tests showing that both B. cereus and B. thuringiensis release activators of B. anthracis SdT2 PC-PLC production. For each test, 5 μl of B. cereus or B. thuringiensis overnight culture was spotted on LB agar containing lecithin between spots of B. anthracis SdT and SdT2. The top illustration demonstrates growth of B. anthracis strains SdT2 and SdT on LB agar with 5% sheep blood. Bacteria were grown on the plates for 24 h at 37°C.
Expression of hemolytic enzymes by B. anthracis containing plc and sph on a multicopy plasmid.
In previous work, it was shown that introduction of the multicopy plasmid pOB12 carrying B. cereus plc and sph genes into B. anthracis led to production of PC-PLC and SPH (29). It is now recognized that B. anthracis ΔPlcR is not active (1), and our experiments confirmed that B. anthracis has no detectable PlcR (Fig. 6). That fact is of interest because hemolysis and PC-PLC production in B. anthracis strains carrying pOB12 did not require specific media and were detected on LB medium, whereas B. anthracis strain SdT2 requires BHI broth for the expression of cereolysins. To further examine this observation, enzyme activity and PlcR production were determined for B. cereus 569 and B. anthracis strains SdT2 and SdT4 (SdT with pOB12) after growth for 16 h in LB broth (Fig. 9, bottom). Although a high level of PlcR production was found in the SdT2 sample, no PC-PLC or SPH activity was detected. PC-PLC activity was detected in both the SdT4 and B. cereus 569 samples, whereas SPH activity was found only in the SdT4 sample. Obviously, no production of full-length PlcR was found in the sample of strain SdT4 (Fig. 9, top). PC-PLC activation in B. cereus 569 at the stationary phase of growth, along with the loss of SPH activity at the onset of the stationary phase, confirmed our hypothesis about the dual role of PlcR as an activator of PC-PLC expression and as an inhibitor of SPH expression. However, it is not yet clear what regulates PC-PLC and SPH production in the SdT4 strain.
FIG. 9.
PlcR production and PC-PLC and SPH activities of B. anthracis SdT2, B. cereus 569, and B. anthracis SdT4. The strains were grown for 16 h in LB broth at 37°C. Production of PlcR was determined by Western blot analysis of whole-cell proteins with C-PlcR antiserum as described above. The PC-PLC and SPH activities of culture supernatants of the same three strains were determined as described in the legend to Fig. 6.
DISCUSSION
In this study, we have shown that the chromosomally located B. anthracis plc and sph genes are almost identical in sequence to their B. cereus orthologues and encode functionally active proteins with activities equal to the proteins from B. cereus. Sheep erythrocytes were lysed by both B. anthracis and B. cereus SPH but not by PC-PLC, probably owing to their high content of sphingomyelin (45 to 53%) and low content of phosphatidylcholine (3 to 4%). The auxiliary phosphatidylcholine hydrolysis activity found for SPH may contribute to hemolysis. However, it was found that the cooperative actions of B. cereus PC-PLC and SPH were needed to lyse human erythrocytes (3), which contain almost ten times more phosphatidylcholine (31%) and two times less sphingomyelin (25%). It has also been shown that the synergistic action of B. cereus PC-PLC and SPH is required to produce the maximum rate of lysis for human but not for ruminant or swine erythrocytes (3).
The presence of almost identical plc and sph structural genes encoding the same functional proteins in B. anthracis and B. cereus highlights the differences in gene regulation that must account for their contrasting hemolytic abilities. In a recent study of the role of PlcR, the B. thuringiensis plcR-papR operon was introduced into B. anthracis and found to induce expression of genes containing PlcR binding sites (25), which include plc and sph. Unfortunately, no information was presented about the genetic structure of the genes introduced into B. anthracis. In contrast, the introduction that we describe here of the B. cereus plcR-papR genes into B. anthracis did not activate expression of hemolytic genes, in particular, plc, even though we confirmed that the plasmid was intact in the recipient strain. Perhaps the greater activation seen in the previous work is due to differences in plasmid copy number or in the ability of various culture media to support PlcR action.
Because the B. cereus plcR gene is positively self-regulated and may require some additional host factor for regulation (22), we decided to produce this protein in B. anthracis under the control of a strong constitutive promoter. High intracellular expression of PlcR was achieved in B. anthracis by using the PA promoter (28). However, overexpression of the protein appeared to be deleterious to B. anthracis, because nearly all transformants contained an expression plasmid in which an IS10 element had disrupted the plcR gene. Less than 1% of the pAE5 transformants were hemolytic. Direct comparison showed that a plasmid containing the disrupted plcR transformed B. anthracis at a much higher transformation efficiency than a plasmid with intact plcR. It is probably not surprising that overexpression of a broadly acting transcriptional regulator would be deleterious.
It is well known that a significant number of clones in the public databases are contaminated by the mobile genetic element IS10, owing to the use of Tn10 in the construction of bacterial strains (15). We used the E. coli XL2-Blue strain, which apparently was the source for the IS10 inserted into plasmid pAE5. Although the frequency of insertions was probably low in E. coli, around 10−4 per cell per bacterial generation (37), this was easily detected because the B. anthracis recipient selected for plasmid with inactivated plcR.
A BLAST search of the PlcR sequence showed that the N-terminal portion of the protein is homologous with more than 30 bacterial transcriptional regulators belonging to the PBSX family (44), including the 112-aa transcriptional repressor RtsR of Vibrio cholerae (43) and the 111-aa SinR protein of the B. subtilis repressor-antirepressor complex (24). PlcR also contains an approximately 70-aa conservative helix-turn-helix motif. This DNA-binding property of PlcR may explain its toxicity when overexpressed in B. anthracis. It is important to mention that the uncharacterized 65-aa pXO1-40 polypeptide of B. anthracis contains the same helix-turn-helix motif (26). The data presented previously showed an incompatibility between the PlcR- and AtxA-controlled regulons (25). Perhaps polypeptide pXO1-40 may also be involved in incompatibility with PlcR.
Although a high level of recombinant B. cereus PlcR production was achieved in the B. anthracis SdT2 strain, only weak PC-PLC and SPH activities were found in this recombinant strain. This suggests that expression of PlcR-dependent genes may require some additional factor. Expression of this factor may be induced by growth in an aerobic-plus-CO2 atmosphere, because both the PC-PLC and hemolytic activities of B. anthracis SdT2 were increased by CO2 compared to the levels under simple aerobic conditions. Growth under anaerobic-plus-CO2 conditions did not cause any of the B. anthracis strains to induce hemolysis of sheep or human blood cells, whereas B. cereus strain 569 retained activity for sheep or human cells under the same conditions. It is also possible that an activating factor is provided by BHI medium, explaining the hemolytic activity observed for the SdT2 strain in the BHI medium (Fig. 7) that is not observed in LB medium (Fig. 8). A similar effect of BHI medium on the hemolytic activity of B. anthracis was found recently (36).
A relatively small amount of PlcR is synthesized in B. cereus, but this amount nevertheless initiates strong expression of PC-PLC. Maximal expression of this enzyme occurred at the onset of the stationary phase, when production of PlcR was still low (Fig. 6). For the plc gene, the PlcR binding site is located upstream of the promoter (Fig. 1), suggesting that PlcR can serve as an activator. The increase in PC-PLC activity paralleling PlcR intracellular content demonstrates that PlcR serves as authentic activator for B. cereus 569 plc gene expression. The gradual increase of PC-PLC activity in comparison with the sudden rise of activity found previously for phospholipase A of B. thuringiensis (22) could be explained by the dual PC-PLC and SPH activities detected for the mature form of the SPH protein of B. cereus (Fig. 2).
There are some data indicating that B. cereus SPH is able to hydrolyze not only phosphatidylcholine (38) but also phosphatidylethanolamine and phosphatidylserine (6). It has been shown also that the PC-PLC proteins of both native and recombinant B. cereus are capable of hydrolyzing sphingomyelin but at rates that are 200-fold lower than for SPH (3). Apparently, this low activity is not sufficient to induce hemolysis of sheep erythrocytes, in which the major phospholipid is sphingomyelin (Fig. 2).
An especially intriguing finding in our work was the evidence that PlcR may be acting to inhibit SPH expression in B. cereus 569. Thus, SPH activity completely disappeared when production of PlcR was most evident and when the activity of PC-PLC reached its maximal value (Fig. 6). It is possible that an extracellular protease, possibly one whose expression is activated by PlcR, acts to destroy SPH or damage its activity. The latter would be consistent with the detection by gel electrophoresis of the SPH protein in the supernatant of B. cereus ATCC 14579 at the onset of the stationary phase (10). Another possible explanation for the absence of SPH activity at the onset of the stationary phase of growth is that PlcR may act as a repressor of sph. Because a convincing PlcR binding site is located on the complementary strand between the sph start codon and promoter, and overlapping the ribosome binding site, it is possible that PlcR could block sph transcription in B. cereus 569. However, the absence of SPH expression in B. cereus 541, the PlcR knockout strain, clearly shows that this activator also plays a positive role in stimulating sph transcription. The positive role of PlcR in SPH expression may be indirect, because no consensus PlcR binding site was found upstream of sph. Thus, PlcR may have both positive and negative roles in SPH expression.
The PlcR-independent expression of the plasmid-encoded B. cereus plc and sph genes in B. anthracis strain SdT4 suggests that several factors could be involved in the regulation of plc and sph. First, the copy number of plc and sph would be higher for the plasmid-encoded genes. Insertion of the plasmid into the chromosome and subsequent amplification of these genes could be activating transcription of the plc and sph without PlcR, as has been previously reported (29). DNA topology is also important because the binding of many proteins to DNA is profoundly affected by DNA bending, twisting, and supercoiling. Because these parameters are different for plasmid and chromosomal DNAs, some steric factor could be involved in the presentation of plc and sph promoters for interaction with transcriptional complexes. It is not clear now what kind of protein or cofactors might be part of this complex. Clearly, more work is needed to elucidate the exact mechanism of plc and sph regulation in the B. anthracis and the B. cereus and B. thuringiensis strains.
Acknowledgments
We thank Theresa Koehler for providing plasmid pUTE29, Yinduo Ji for plasmid pYJ335, Robert Boykins for assistance with MAP peptides, MJ Rosovitz for advice, and Violetta Kivovich and Dana Hsu for assistance with some experiments.
Editor: J. T. Barbieri
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Battisti, L., B. D. Green, and C. B. Thorne. 1985. Mating system for transfer of plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. J. Bacteriol. 162:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beecher, D. J., and A. C. Wong. 2000. Cooperative, synergistic and antagonistic haemolytic interactions between haemolysin BL, phosphatidylcholine phospholipase C and sphingomyelinase from Bacillus cereus. Microbiology 146:3033-3039. [DOI] [PubMed] [Google Scholar]
- 4.Briand, J. P., C. Barin, M. H. Van Regenmortel, and S. Muller. 1992. Application and limitations of the multiple antigen peptide (MAP) system in the production and evaluation of anti-peptide and anti-protein antibodies. J. Immunol. Methods. 156:255-265. [DOI] [PubMed] [Google Scholar]
- 5.Chalmers, R., S. Sewitz, K. Lipkow, and P. Crellin. 2000. Complete nucleotide sequence of Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, E. C., C. C. Chang, Y. S. Li, C. A. Chang, C. C. Chiou, and T. Z. Wu. 2000. Purification and characterization of neutral sphingomyelinase from Helicobacter pylori. Biochemistry 39:4838-4845. [DOI] [PubMed] [Google Scholar]
- 7.de Vasconcellos, F. J., and L. Rabinovitch. 1995. A new formula for an alternative culture medium, without antibiotics, for isolation and presumptive quantification of Bacillus cereus in foods. J. Food Protection 58:235-238. [DOI] [PubMed] [Google Scholar]
- 8.Gavrilenko, I. V., G. E. Baida, A. V. Karpov, and N. P. Kuz'min. 1993. Nucleotide sequence of phospholipase C and sphingomyelinase genes from Bacillus cereus BKM-B164. Bioorg. Khim. 19:133-138. [PubMed] [Google Scholar]
- 9.Gilmore, M. S., A. L. Cruz-Rodz, M. Leimeister-Wächter, J. Kreft, and W. Goebel. 1989. A Bacillus cereus cytolytic determinant, cereolysin AB, which comprises the phospholipase C and sphingomyelinase genes: nucleotide sequence and genetic linkage. J. Bacteriol. 171:744-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gohar, M., O. A. Økstad, N. Gilois, V. Sanchis, A.-B. Kolstø, and D. Lereclus. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2:784-791. [DOI] [PubMed] [Google Scholar]
- 11.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 12.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grones, J., and J. Turna. 1995. Transformation of microorganisms with the plasmid vector with the replicon from pAC1 from Acetobacter pasteurianus. Biochem. Biophys. Res. Commun. 206:942-947. [DOI] [PubMed] [Google Scholar]
- 14.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, F., C. Gemund, V. Benes, W. Ansorge, and T. J. Gibson. 2000. An estimate of large-scale sequencing accuracy. EMBO Rep. 1:29-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivins, B. E., J. W. Ezzell, Jr., J. Jemski, K. W. Hedlund, J. D. Ristroph, and S. H. Leppla. 1986. Immunization studies with attenuated strains of Bacillus anthracis. Infect. Immun. 52:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansen, T., T. Holm, P. H. Guddal, K. Sletten, F. B. Haugli, and C. Little. 1988. Cloning and sequencing of the gene encoding the phosphatidylcholine-preferring phospholipase C of Bacillus cereus. Gene 65:293-304. [DOI] [PubMed] [Google Scholar]
- 19.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klichko, V. I., J. Miller, A. Wu, S. G. Popov, and K. Alibek. 2003. Anaerobic induction of Bacillus anthracis hemolytic activity. Biochem. Biophys. Res. Commun. 303:855-862. [DOI] [PubMed] [Google Scholar]
- 21.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2749-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lereclus, D., H. Agaisse, C. Grandvalet, S. Salamitou, and M. Gominet. 2000. Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int. J. Med. Microbiol. 290:295-299. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, R. J., J. A. Brannigan, W. A. Offen, I. Smith, and A. J. Wilkinson. 1998. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J. Mol. Biol. 283:907-912. [DOI] [PubMed] [Google Scholar]
- 25.Mignot, T., M. Mock, D. Robichon, A. Landier, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 26.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okstad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A. B. Kolsto. 1999. Sequence analysis of three Bacillus cereus loci carrying PIcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology 145:3129-3138. [DOI] [PubMed] [Google Scholar]
- 28.Park, S., and S. H. Leppla. 2000. Optimized production and purification of Bacillus anthracis lethal factor. Protein Expr. Purif. 18:293-302. [DOI] [PubMed] [Google Scholar]
- 29.Pomerantsev, A. P., N. A. Staritsin, Y. Mockov, and L. I. Marinin. 1997. Expression of cereolysine AB genes in Bacillus anthracis vaccine strain ensures protection against experimental hemolytic anthrax infection. Vaccine 15:1846-1850. [DOI] [PubMed] [Google Scholar]
- 30.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 3-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D. C.
- 31.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2034. [DOI] [PubMed] [Google Scholar]
- 32.Reddy, A., L. Battisti, and C. B. Thorne. 1987. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J. Bacteriol. 169:5263-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salamitou, S., F. Ramisse, M. Brehelin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Shannon, J. G., C. L. Ross, T. M. Koehler, and R. F. Rest. 2003. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect. Immun. 71:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen, M. M., E. A. Raleigh, and N. Kleckner. 1987. Physical analysis of Tn10- and IS10-promoted transpositions and rearrangements. Genetics 116:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sillence, D. J., and D. Allan. 1998. Utilization of phosphatidylcholine and production of diradylglycerol as a consequence of sphingomyelin synthesis. Biochem. J. 331:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ticknor, L. O., A.-B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil Isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toldo, L. I. 1997. JaMBW 1.1: Java-based Molecular Biologists' Workbench. Comput. Appl. Biosci. 13:475-476. [DOI] [PubMed] [Google Scholar]
- 42.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 43.Waldor, M. K., E. J. Rubin, G. D. Pearson, H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXphi are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 44.Wood, H. E., K. M. Devine, and D. J. McConnell. 1990. Characterisation of a repressor gene (xre) and a temperature-sensitive allele from the Bacillus subtilis prophage, PBSX. Gene 96:83-88. [DOI] [PubMed] [Google Scholar]
- 45.Yamada, A., N. Tsukagoshi, S. Udaka, T. Sasaki, S. Makino, S. Nakamura, C. Little, M. Tomita, and H. Ikezawa. 1988. Nucleotide sequence and expression in Escherichia coli of the gene coding for sphingomyelinase of Bacillus cereus. Eur. J. Biochem. 175:213-220. [DOI] [PubMed] [Google Scholar]