Abstract
Signaling via the pre-T cell receptor (TCR) is required for the proliferative expansion and maturation of CD4−CD8− double-negative (DN) thymocytes into CD4+CD8+ double-positive (DP) cells and for TCR-β allelic exclusion. The adaptor protein SH2 domain–containing leukocyte protein (SLP)-76 has been shown to play a crucial role in thymic development, because thymocytes of SLP-76−/− mice are arrested at the CD25+CD44− DN stage. Here we show that SLP-76−/− DN thymocytes express the pre-TCR on their surfaces and that introduction of a TCR-α/β transgene into the SLP-76−/− background fails to cause expansion of DN thymocytes or developmental progression to the DP stage. Moreover, analysis of TCR-β rearrangement in SLP-76−/− TCR-transgenic mice or in single CD25+CD44− DN cells from SLP-76−/− mice indicates an essential role of SLP-76 in TCR-β allelic exclusion.
Keywords: SLP-76, allelic exclusion, thymocyte development, pre-TCR, TCR signaling
The pre-TCR controls the survival of CD4−CD8− double-negative (DN)1 thymocytes and their intense proliferation and maturation into CD4+CD8+ double-positive (DP) thymocytes by mostly unknown mechanisms 1. It is clear that this control requires the pre–TCR-α chain (pTα), the TCR-β chain, signal-transducing molecules of the CD3 complex, Src kinases, and Zap-70 and Syk kinases 2 3 4 5 6. Some experiments suggest that exit from the endoplasmic reticulum is essential for pre-TCR function 7, but other data have been interpreted to indicate that binding to a ligand may not be an essential event 8. In fact, surface expression of the pre-TCR on the CD25+ CD44− population of thymocytes that require the pre-TCR for further maturation has not been well documented. It is also not clear whether the function of the pre-TCR includes mainly rescue from cell death, allowing an already imprinted program of expansion and maturation to take place, or whether the pre-TCR directly controls cell cycle progression as well as expression of genes encoding various cell surface molecules. Experiments addressing this question have given conflicting results, as in bcl-2–transgenic SCID mice, antiapoptotic bcl-2–dependent signals were insufficient to permit developmental changes, whereas overexpression of bcl-2 in DN thymocytes of recombination activating gene (RAG)−/− mice promoted differentiation to the CD4+CD8+ DP stage 9 10.
Another event associated with pre-TCR signaling is the allelic exclusion of the TCR-β locus, i.e., the feedback inhibition of the rearrangement on one allele by the TCR-β protein encoded by the other allele. Effective allelic exclusion as it is observed in α/β T cells requires not only this feedback inhibition but also asynchronous Vβ rearrangement on the two alleles 11. Direct analysis of TCR-β rearrangement in single cells at the CD25+44− stage of development, i.e., before selection by the pre-TCR, has shown that the pre-TCR has an essential function in this process 12. Although these data were obtained by single-cell PCR analysis in pTα−/− mice, experiments in TCR-β–transgenic mice have shown that TCR-β transgenes can inhibit Vβ rearrangement in the absence of the pTα protein, most likely because of their high and early expression 13. Whatever the mechanism of this suppression, it also appears to depend on signals transduced via CD3, because it is not operative in CD3∈-deficient mice 14.
Experiments in mice expressing active p56lck indicate that Src kinases are also involved in TCR-β allelic exclusion, as Vβ rearrangement is inhibited in such mice 15. Recent data, however, suggest that TCR-β allelic exclusion may require signaling pathways that are distinct from signals controlling proliferation and maturation, indicating that the pre-TCR has functions in addition to those responsible for rescue from programmed cell death. Active Ras, known to activate the mitogen-activated protein kinase pathway, was shown to drive proliferation and maturation in immature thymocytes but, in contrast to active p56lck, failed to suppress TCR-β rearrangement 16. This means that some bifurcation of signal transduction pathways downstream of p56lck is responsible for proliferation and maturation on the one hand and allelic exclusion on the other.
Recently, it has been shown that the adaptor protein SH2 domain–containing leukocyte protein (SLP)-76 is involved in several distinct signaling pathways and represents an essential component in the signaling cascade that is involved in the proliferation and maturation of α/β T cells 17 18. Although SLP-76 lacks intrinsic enzymatic activity, it facilitates through its tyrosine phosphorylation the propagation of signals emanating from the TCR 19. Biochemical characterization reveals three domains in SLP-76 that direct intramolecular interactions with Zap-70, Vav, Grb2, SLP-76–associated phosphoprotein, 130 kD (SLAP-130), and other proteins 20. Overexpression of SLP-76 augments TCR-mediated IL-2 transcription 21, whereas extinction of SLP-76 expression in a mutant variant of the Jurkat T cell line leads to attenuated calcium mobilization and mitogen-activated protein kinase activation 22. Furthermore, mice deficient in SLP-76 demonstrate a total block in early T cell development, implicating the SLP-76 linker protein in the pre-TCR signaling cascade 17 18. Here we have analyzed the role of SLP-76 in the regulation of expression of the TCR-β locus as well as in TCR-β allelic exclusion. Our results indicate that SLP-76 has an essential role in all known phenotypic changes that are mediated by the pre-TCR.
Materials and Methods
Cells, Antibodies, and Flow Cytometric Analysis.
The SCB29 cell line, which expresses the pre-TCR on the cell surface, was described previously 23. Single-cell suspensions from thymi were prepared by compression between ground glass slides followed by centrifugation on a density gradient of Lympholyte-M (Cedarlane Labs., Inc.). Streptavidin–Cy-Chrome, streptavidin–PE, and mAbs to the following mouse antigens were purchased from PharMingen: CD3∈ (clone 145-2C11), CD4 (L3T4), CD8α (53-6.7), CD25 (7D4), CD44 (IM7), Thy1.2 (53-2.1), Mac-1 (M1/70), Gr-1 (RB6-8C5), and TCR-β (H57-597). Anti-CD25 mAb (3C7) conjugated to PE and anti-CD44 mAb conjugated to FITC were purchased from Sigma Chemical Co. Anti-pTα mAb 2F5 was raised against the extracellular (Ig-like) domain as described previously 24. In the case of pre-TCR surface detection, thymocytes were enriched for the CD4−CD8− subset by depletion of CD4/CD8-positive cells using Dynabeads (Dynal). pTα (2F5) and TCR-β (H-57) biotinylated antibodies were revealed with streptavidin–PBXL-3 (Martek Biosciences). NK and NK-T cells were gated out with DX-5, NK1.1, or 2B4 antibodies (PharMingen). Cytoplasmic staining for TCR-β was performed as previously described 25. Cells were surface stained and analyzed on a FACSCalibur™ flow cytometer (Becton Dickinson) as previously described 26. Intracellular staining for TCR-β was performed on CD25+ sorted cells as previously described 12. Data on 5 × 105–2.0 × 106 viable, nonerythroid cells (as determined by forward versus side scatter) were collected for each sample. FACS™ analysis was performed on cells from groups of at least three mice aged 2–4 wk.
Generation of SLP-76−/− TCR-α/β–transgenic Mice.
The generation of TCR-α−/−, pTα−/−, RAG-2−/−, and SLP-76−/− mice was previously described 2 4 17 27. Mice transgenic for the TCRVα13 and TCRVβ8.2 chains of the OVA-specific mouse T cell hybridoma DO11.10 were a gift of Dr. Dennis Loh (Hoffmann-LaRoche, Inc., Nutley, NJ; reference 28). Screening for expression of the transgenic TCR was done by PCR analysis of tail DNA, using primer sequences provided by Dr. K. Murphy (Washington University, St. Louis, MO), and by FACS™ analysis, using the antiidiotypic mAb KJ126, provided by Dr. K. Murphy. Screening for disruption of the SLP-76 gene was done as previously described 17.
PCR Analysis of TCR-β Gene Rearrangements in Thymocytes.
Genomic DNA was isolated according to published procedures 29. Vβ to DJβ rearrangements were assessed as described in detail in reference 16. In brief, upstream primers were located on the Vβ segments 5, 8, and 10, and the downstream primer was located 3′ of Jβ2.7. After amplification of the rearrangements by PCR, the products were separated on agarose gels, transferred to nylon membranes, and hybridized with a Jβ2.7-specific, 32P-labeled oligonucleotide probe. Hybridizing bands were scanned using a PhosphorImager (Molecular Dynamics). Amplification of a fragment of the nonrearranging Cμ gene served as a loading control.
Single-Cell PCR.
TCR-β+ icCD25+ small single cells from SLP-76−/− mice were sorted using a FACSVantage™ equipped with an automatic cell deposition unit (Becton Dickinson). DNA from single cells was prepared as previously described 12. TCR-β gene rearrangements were amplified by a seminested two-step PCR protocol 12 30. In the first step, both alleles were amplified simultaneously by addition to each tube of 35 μl of a mixture containing dNTPs, buffer, and Taq polymerase at 0.5 U/sample (Perkin-Elmer Corp.). 18 5′ primers (3 pmol of each) homologous to 16 Vβ gene families and Dβ1 and Dβ2 genes, and 2 3′ primers (3 pmol of each) that primed downstream of the Jβ1 and Jβ2 cluster sequences, respectively, were used 12. In addition to the previously described primers, 5′ primers specific for Vβ3 (5′-ACGattctctgctgagtgtcctcc-3′), Vβ9 (5′-gaacagg-gaagctgacacttttgag-3′), and Vβ17 (5′-gtcctgaaaaagggcacactgcct-3′) were used. The first round of amplification was performed in a final volume of 60 μl for five cycles, in which the annealing temperature decreased from 68 to 60°C, followed by 25 cycles of amplification (30 s at 94°C, 1 min at 58°C, 1 min at 72°C), and finally 5 min at 72°C. For the second round of amplification, 1 μl of the first PCR product was transferred into separate tubes, each containing a single 5′ primer in combination with the nested Jβ2 or Jβ1 3′ primer (10 pmol of each), dNTP, reaction buffer, and 1 U of Taq polymerase in a final volume of 20 μl. Amplification was then carried out for 35 cycles, following the procedure of the first PCR. Vβ and Jβ were identified by migration of the total PCR product on a 1.5% ethidium bromide–stained agarose gel, and positives were purified using Geneclean III (Bio 101). Direct sequencing of the PCR products was performed using the Ready Reaction DyeDeoxy Terminator Cycle sequencing kit (Applied Biosystems, Inc.) and sequenced by automated sequencing (Applied Biosystems, Inc.).
Results
The Pre-TCR Is Expressed on the Surfaces of SLP-76−/− Thymocytes.
Surface expression of the pre-TCR is required for TCR-β allelic exclusion (for review see reference 31). In the analysis of allelic exclusion in SLP-76−/− mice, it was therefore important to ascertain expression of the pre-TCR by SLP-76–deficient thymocytes. In previous experiments, we have detected low levels of expression of CD3 and TCR-β on SLP-76−/−CD4−CD8− DN thymocytes, suggesting that the pre-TCR is expressed on the cell surfaces of these immature thymocytes 17. To extend these observations, we repeated these experiments with a fluorochrome reagent, streptavidin–PBXL-3 (Martek Biosciences), that gives much higher signal intensity than conventional reagents (i.e., streptavidin–FITC or –APC) when used in combination with biotinylated antibodies. This can be clearly seen by staining the SCB29 cell line, which expresses the pre-TCR on the cell surface 23, with biotinylated mAb to TCR-β (H57) or pTα (2F5), followed by incubation with either streptavidin–FITC or –PBXL-3 (Fig. 1). Incubation with irrelevant mAbs of the same Ig class produces some slight background that, however, is clearly distinct from the staining obtained with antibodies specific for cell surface–expressed proteins (Fig. 1).
Figure 1.
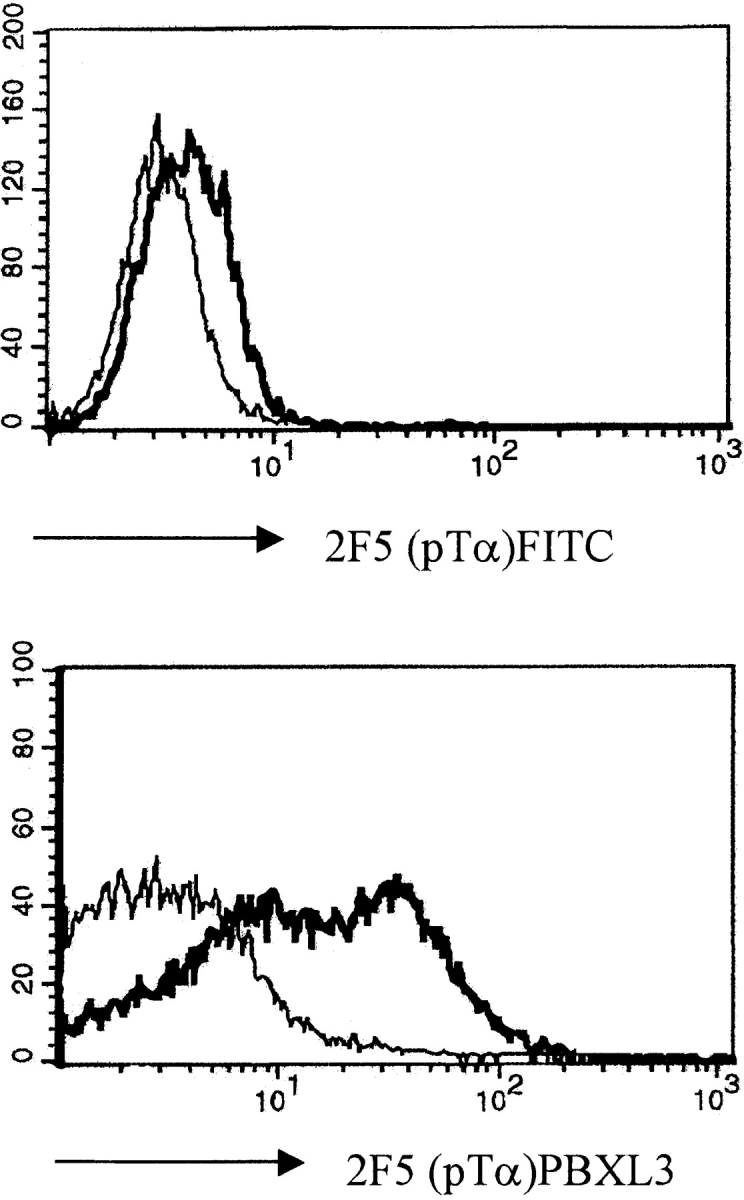
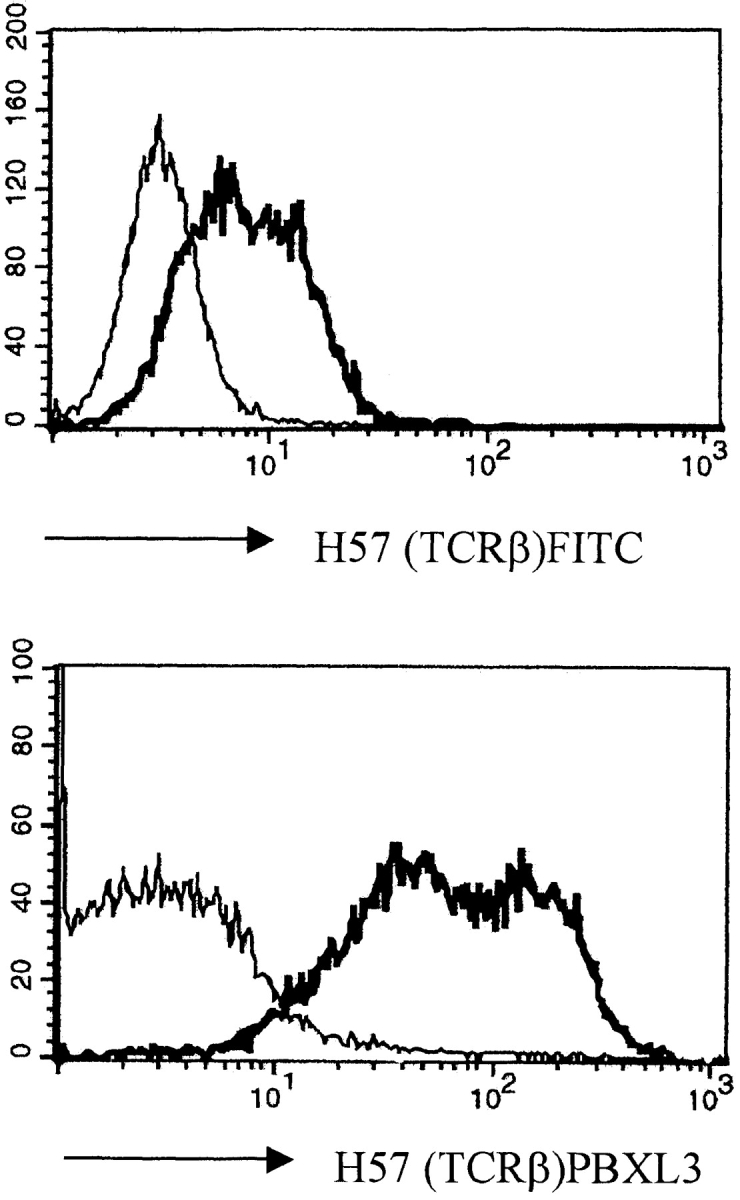
Pre-TCR expression on the surface of the SCB29 cell line and comparison of the labeling intensity between streptavidin–PBXL-3 and streptavidin–FITC. SCB29 cells were initially incubated with biotinylated 2F5 (pTα) and H57 (pan-TCR-α/β) antibodies (thick line) or isotype-matched controls (thin line), and surface expression was revealed by streptavidin conjugated to the secondary reagents. Similar results were obtained in two other experiments.
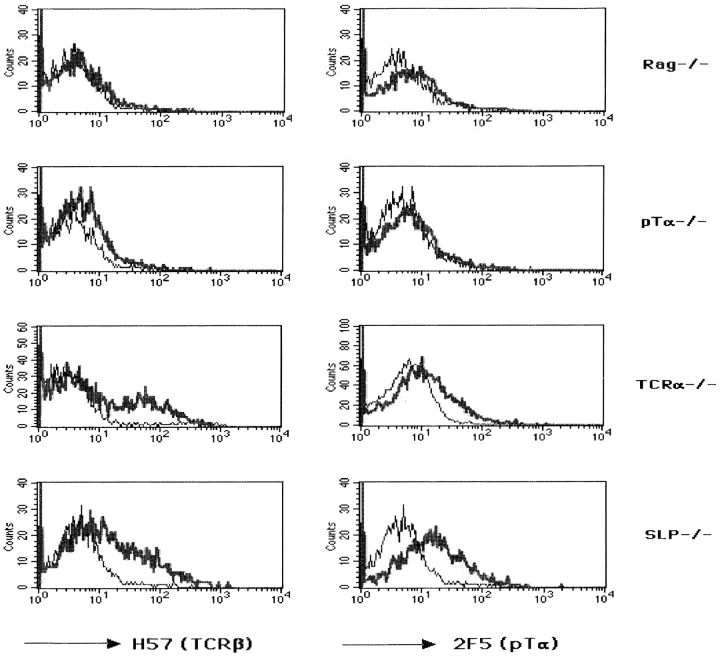
In subsequent experiments, CD4−CD8− DN thymocytes from wild-type (WT), TCR-α−/−, SLP-76−/−, pTα−/−, and RAG-2−/− mice were examined for TCR-β and pTα expression. Thymocyte development in SLP-76−/− mice is blocked at the stage of CD25+CD44− DN cells. CD25− CD44− cells, which represent a more mature stage of DN thymocytes, are absent in SLP-76−/− mice 17. We therefore focused our analysis on CD25+CD44− DN cells. DN thymocytes were stained with directly labeled antibodies against CD44 and CD25, as well as biotinylated antibodies against either TCR-β or pTα followed by streptavidin–PBXL-3, and gated populations of CD25+CD44− cells were analyzed for TCR-β and pTα expression. The histograms in Fig. 2 show that, as expected, mAb to TCR-β did not stain thymocytes from RAG-2−/− mice and mAb to pTα did not stain thymocytes from pTα−/− mice when compared with control antibodies. This further confirms the specificity of the staining. There was negligible, if any, staining of RAG-2−/− thymocytes with anti-pTα mAb or of pTα−/− thymocytes with anti–TCR-β mAb. CD25+CD44− DN thymocytes from SLP-76−/− mice stained with both antibodies to the same or slightly higher level as CD25+ CD44− DN cells from TCR-α−/− mice (Fig. 2). DN thymocytes from TCR-α−/− mice were used as controls to eliminate the possibility of staining TCR-β proteins that are transported to the cell surface as α/β TCRs. The surface expression of TCR-β and pTα chains on CD25+CD44− as well as CD25−CD44− thymocytes from TCR-α−/− mice and WT controls was identical (data not shown).
Figure 2.
Expression of the pre-TCR complex on the surfaces of CD25+CD44− CD4− CD8− DN thymocytes from RAG-2−/−, pTα−/−, TCR-α−/−, and SLP-76−/− mice. CD4+ and CD8+ thymocytes and NK cells were depleted with Dynabeads, and DN cells were stained with CD25, CD44, 2.4G2 (Fc-block), H57 (TCR-β), or 2F5 (pTα) antibodies. Analysis of gated CD25+ CD44− cells is shown. Results are representative of three mice in each group.
We next examined intracellular TCR-β protein expression in SLP-76−/− thymocytes. Fig. 3 shows that SLP-76–deficient CD25+ thymocytes expressed lower levels of intracellular TCR-β protein than WT CD25+ thymocytes (Fig. 3). Decreased levels of expression of intracellular TCR-β protein were also observed in CD25+ thymocytes from pTα−/− mice, which are also blocked at the CD25+CD44− DN stage 2. The CD25−TCR-β+ cells in SLP-76−/− mice may represent γ/δ cells that express cytoplasmic TCR-β chain, as has been shown recently in pTα−/− mice 32. These results suggest that SLP-76 is not required for assembly and surface expression of TCR-β chains. However, it is required for the generation of CD25+ DN cells that express high levels of TCR-β protein intracellularly.
Figure 3.
Intracellular TCR-β expression in CD4−CD8− DN cells from WT (B6), pTα−/−, and SLP-76−/− mice. DN cells were surface stained for CD25 and subsequently cytoplasmically labeled with TCR-β antibodies. Results are representative of three mice in each group.
Introduction of TCR-α/β Transgenes Does Not Overcome the Block in Thymic Development in SLP-76−/− Mice.
Introduction of a TCR-β transgene into SCID or RAG-deficient mice drives the development of DN thymocytes into DP cells 33 34. To examine the effect of introduction of TCR transgenes on thymic development in SLP-76−/− mice, they were bred with mice transgenic for the TCRVα13 and TCRVβ8.2 chains of the OVA-specific mouse T cell hybridoma DO11.10. The transgenes are concordantly expressed early in thymocyte development at the DN stage 28, and their introduction into a RAG-deficient background drives T cell development to the DN and single-positive stage 33 34. SLP-76−/− TCR-α/β–transgenic (SLP-76−/−-tg) F2 mice were identified by PCR analysis of tail DNA.
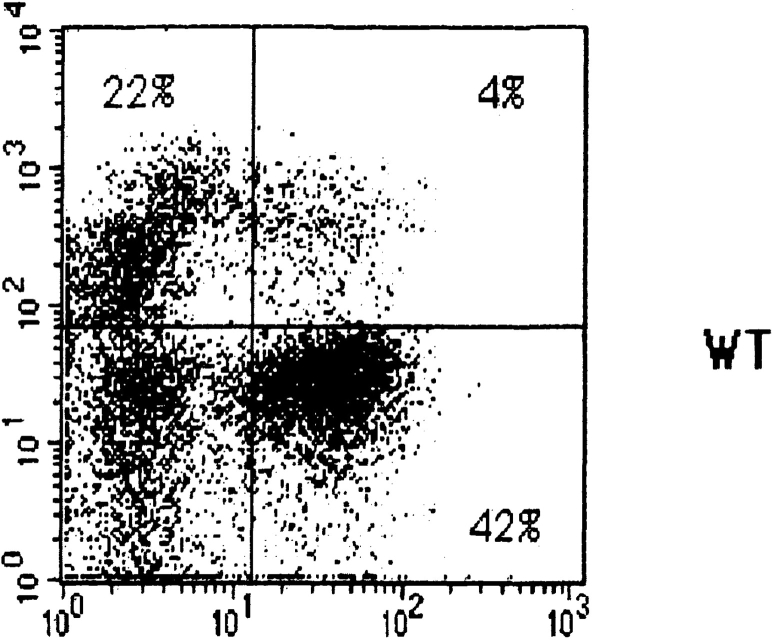
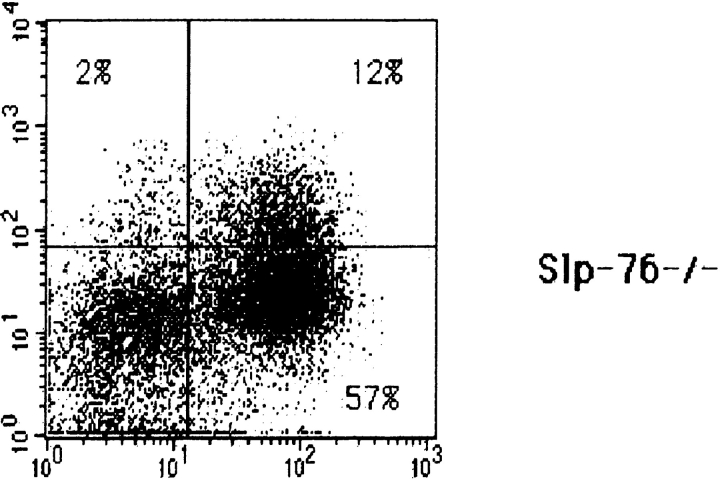
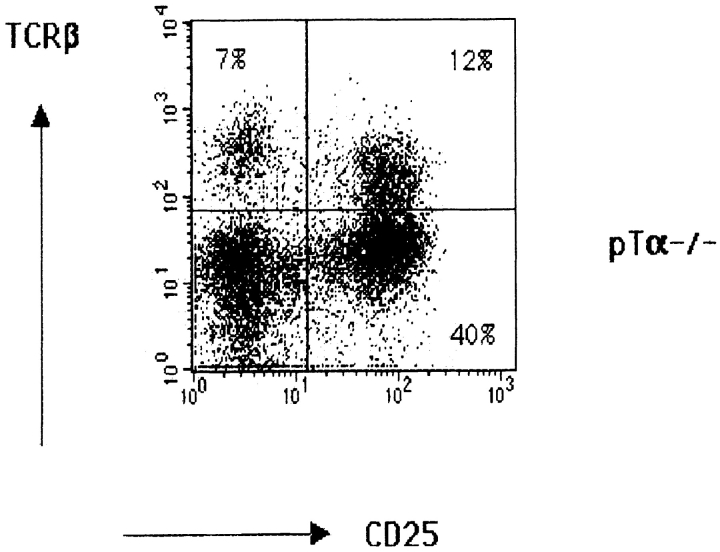
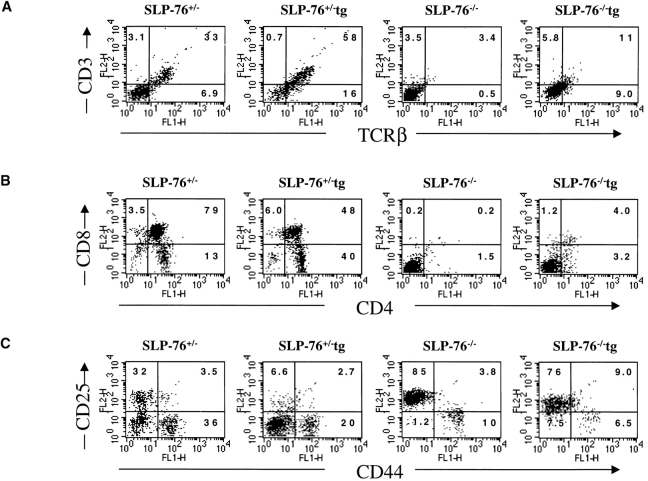
The effect of introduction of the DO11.10 TCR-α/β transgenes on thymic development in SLP-76−/− mice was assessed by examining the thymi of SLP-76−/−-tg mice for cellularity, expression of CD3, TCR-β, CD4, and CD8, and distribution of CD25+ and CD44+ DN subsets. As previously described, thymic cellularity in SLP-76−/− mice was severely reduced to ∼1% of that of heterozygous littermates, and there were no detectable DP nor single-positive thymocytes 17. Introduction of the transgenic TCR into the SLP-76−/− background did not increase thymic cellularity (1.30 ± 0.18 × 106 cells in SLP-76−/−-tg mice versus 1.62 ± 0.75 × 106 cells in SLP-76−/− mice; n = 4 in each group), but resulted in an increase in CD3+TCR-β+ thymocytes, consistent with surface expression of the transgene (Fig. 4 A). The transition from DN to DP cells remained severely impaired, as we could detect no or very few DP thymocytes in SLP-76−/−-tg mice (Fig. 4 B). Analysis of the B220−Mac-1−Gr-1−CD3−CD4−CD8− DN compartment for the expression of CD25 and CD44 revealed that introduction of the transgenic TCR failed to overcome the block in thymic development at the CD25+CD44− stage present in SLP-76−/− mice (Fig. 4 C). These results suggest that the failure of DN cells from SLP-76−/− mice to progress to the DP stage is due to deficient SLP-76–mediated signaling and cannot be overcome by surface expression of the transgenic TCR.
Figure 4.
Flow cytometric analysis of thymocytes from TCR-transgenic SLP-76−/− and SLP-76+/− littermates. Surface expression on thymocytes of (A) CD3 (anti-CD3∈–PE) versus TCR-β (anti–TCR-β–FITC), (B) CD4 (anti-CD4–FITC) versus CD8 (anti-CD8–PE), and (C) CD44 and CD25 on DN thymocytes from SLP-76−/− and SLP-76+/− littermates. Cells were triple stained with anti-CD44–FITC, anti-CD25–PE, and a cocktail of biotin-conjugated mAbs to CD3, CD4, CD8, B220, Mac-1, and Gr-1, followed by streptavidin–Cy-Chrome. Analysis was performed on gated Cy-Chrome–negative cells. The percentage of cells found in each quadrant is indicated. In all FACS™ analyses, results from SLP-76+/− and WT mice were similar. Therefore, only data of SLP-76+/− mice is shown. Results are representative of four mice examined in each group.
Introduction of a TCR-α/β Transgene Does Not Inhibit Endogenous Rearrangement at the TCR-β Locus in SLP-76−/− Mice.
Having established that the pre-TCR is expressed on DN thymocytes from SLP-76−/− mice, we proceeded to address the issue of whether TCR-β allelic exclusion takes place in the absence of SLP-76. There are two ways in which feedback inhibition by the pre-TCR resulting in TCR-β allelic exclusion has been analyzed: one is the inhibition of endogenous Vβ rearrangements by TCR-β transgenes, and the other is the analysis of both TCR-β alleles in single cells. We initially examined inhibition of Vβ rearrangements by TCR transgenes.
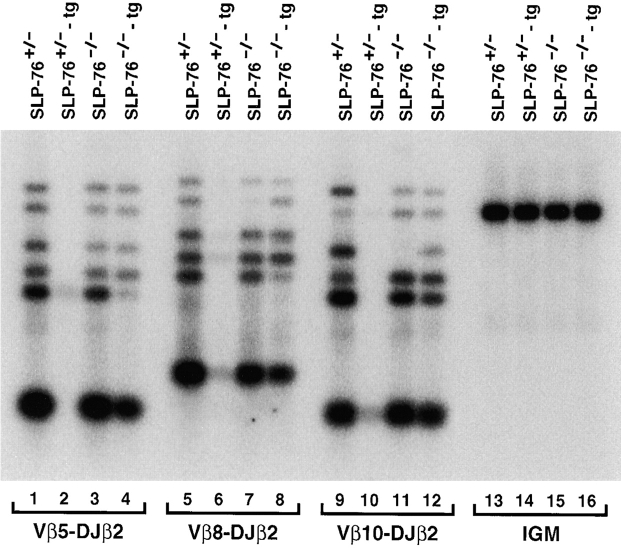
In normal thymocytes, introduction of functionally rearranged TCR-β transgenes leads to inhibition of rearrangements in the endogenous TCR-β locus at the V to DJ step 35. Furthermore, introduction of a TCR-β transgene in pTα−/− and p56lck−/− also leads to inhibition of endogenous TCR-β rearrangements 13 36. To examine if introduction of the transgenic TCR into SLP-76–deficient thymocytes could also inhibit endogenous Vβ to DJβ rearrangements, we used a semiquantitative PCR assay as previously described 16 37. In these experiments, Vβ to DJβ2 rearrangements involving representative Vβ segments (Vβ5, Vβ8, and Vβ10) were amplified from genomic DNA samples isolated from DN thymocytes of TCR-transgenic or nontransgenic SLP-76+/− or SLP-76−/− mice. The identity of the resulting DNA fragments corresponding to rearrangements between a particular Vβ and one of the Jβ segments was confirmed by Southern blot analysis with a Jβ2.7-specific probe. In agreement with our previously published observations 17, Vβ to DJβ rearrangements were readily detectable in both SLP-76+/− and SLP-76−/− thymocytes (Fig. 5, lanes 1, 3, 5, 7, 9, and 11). As expected, introduction of the TCR-α/β transgene into SLP-76+/− thymocytes almost completely blocked Vβ to DJβ rearrangements of the endogenous TCR-β genes (Fig. 5, lanes 1, 2, 5, 6, 9, and 10). In contrast, rearrangements of the endogenous TCR-β genes occurred at comparable levels in both TCR-transgenic and nontransgenic SLP-76−/− thymocytes (Fig. 5, lanes 3, 4, 7, 8, 11, and 12). These results suggest that SLP-76 is essential for transgene-mediated inhibition of TCR-β locus rearrangement.
Figure 5.
Thymocytes from TCR-α/β–transgenic SLP-76−/− mice fail to suppress V to DJ rearrangements in the endogenous TCR-β locus. 20 ng of genomic DNA isolated from DN thymocytes of SLP-76+/−, SLP-76+/−-tg, SLP-76−/−, and SLP-76−/−-tg was amplified with primers that specifically detect rearrangements among Vβ5, Vβ8, Vβ10, and DJβ2. Equivalent loading was confirmed by amplification of a fragment of the Cμ gene (IgM). The PCR products were separated on a 1.5% agarose gel, transferred to a nylon membrane, and hybridized with radiolabeled oligonucleotide probes specific for Jβ2.7 and Cμ. For each Vβ, there are six bands that correspond to rearrangements to the DJβ2 segments DJβ2.1 through DJβ2.7 (Jβ2.6 is a pseudogene and is not rearranged). Similar results were obtained in two other experiments.
The SLP-76 Adaptor Protein Is Required for TCR-β Allelic Exclusion.
Introduction of a TCR-β transgene into pTα−/− mice 13 inhibits rearrangements of endogenous Vβ segments, although single-cell PCR analysis of Vβ rearrangements reveals a failure of TCR-β allelic exclusion in pTα−/− mice 12. This suggests that expression of the transgene provides signals for inhibition of TCR-β rearrangement that differ quantitatively and/or qualitatively from physiologic signals delivered via the pre-TCR and may not reflect authentic allelic exclusion. To assess the status of TCR-β allelic exclusion in unmanipulated SLP-76−/− mice, we examined Vβ rearrangements in single CD25+CD44− thymocytes from these mice.
In WT mice, only ∼40% of CD25+TCR-β+ thymocytes rearrange Vβ on both alleles due to the feedback inhibition by the first productively rearranged allele that prevents further rearrangement on the other allele 12. In pTα−/− mice, this feedback inhibition fails, and hence a greater population of cells (60%) exhibits two completely rearranged VDJβ alleles 12. When single CD25+CD44− DN cells from SLP-76−/− mice that expressed cytoplasmic TCR-β chains were sorted and analyzed, a similar proportion of the cells (60%) was found to contain rearranged VDJβ on both alleles (Table ).
Table 1.
Number of CD25+44− Thymocytes from SLP-76−/− Mice According to the Type of Rearrangements on Both TCR-β Alleles
| Type of rearrangement | |||
|---|---|---|---|
| V(D)J/DJ | V(D)J/V(D)J | V(D)J+/V(D)J+ | V(D)J+/V(D)J− |
| 18 (40%) | 27 (60%) | 8 (31%) | 18 (69%) |
T cells with two productive Vβ rearrangements are a rare event in WT mice; <5% of normal cells with two Vβ rearrangements contain two productive alleles 12. When the alleles from SLP-76−/− thymocytes with two Vβ rearrangements were sequenced, it was found that 31% of the cells with two Vβ rearrangements had two productive alleles (Table ). The Vβ usage and the sequences of the two in-frame alleles are shown in Table and Table . They provide no indication for the selection of certain Vβ alleles in allelically included cells, even though it is difficult to entirely rule out this possibility due to the relatively small sample size. Taken together, the data obtained by the single-cell PCR analysis indicates that allelic exclusion at the TCR-β locus requires the SLP-76 adaptor protein.
Table 2.
Junctional Sequences of SLP-76−/−CD25+44− Thymocytes Bearing Two Productive TCR-β Alleles
| 3′ Vβ | N region | 5′ Jβ | Vβ/Jβ |
|---|---|---|---|
| 97 | |||
| TGT GCC AGC AG……A A…CT GGA AAT ACG CTC TAT TTT GGA | V10J1.3 | ||
| TGT GCC AGC AGC C……CA CCG GGA CAG TC…A AAC ACA GAA GTC TTC TTT GGT | V1JI.1 | ||
| 118 | |||
| TGT GCC AGC AGC CA……A G…AC CAA GAC ACC CAG TAC TTT GGG | V1J2.5 | ||
| TGT GCC AGC AGT AT……G GCT GGG GGG GC…G CAA GAC ACC CAG TAC TTT GGG | V6J2.5 | ||
| 120 | |||
| TGT GCC AGC TCT C……CC CAG ACT GGG GGG GC…A GAC ACC CAG TAC TTT GGG | V5J2.5 | ||
| TGT GCC AGC AGC T……T…C TCC TAT GAA CAG TAC TTC GGT | V10J2.6 | ||
| 131 | |||
| TGT GCC AGC TC……C CCT ACT GGG GGA GGG…AGT CAA AAC ACC TTG TAC TTT GGT | V5J2.4 | ||
| TGT GCC AGC GGT GAT G……CA GGG GAA TGG AGC…GAA AGA TTA TTT TTC | V8J1.4 | ||
| 143 | |||
| TGT GCC AGC GG……C TAT ACA GGG GTG TC…C ACC GGG CAG CTC TAC TTT GGT | V8J2.2 | ||
| TGT GCA AGC AG……A AAG GGG GG…T GCA GAA ACG CTG TAT TTT GGC | V11J2.3 | ||
| 114 | |||
| TGT GCC AGC AGC CA……A GAT CCG CGG GAC AGG GGG…TAT GAA CAG TAC TTC GGT | V1J2.6 | ||
| TGT GCC AGC GG…C GGA CTG GGG GGG CGG TT…T GAA CAG TAC TTC GGT | V8J2.6 | ||
| 11 | |||
| TGC ACC TGC AGT G……TC CGG GAC AGG GG…C AAC GAA AGA TTA TTT TTC | V2J1.4 | ||
| TGT GCC AGC AGT GAT G……AG GGA GAA GGG…GAC ACC CAG TAC TTT GGG | V8J2.5 | ||
| 39 | |||
| TGT GCC AGC AGC CA……A AAA AAG GCC GGG ACT GGG GGG GCG A…AA CAG TAC TTC GGT | V1J2.6 | ||
| TGT GCC AGC AGT G……GT CCG AGG GGG AG…A GAA GTC TTC TTT GGT | V8J1.1 | ||
Table 3.
Characterization of TCR-β Rearrangements on Both Alleles of CD25+CD44− Thymocytes Obtained from SLP-76−/− Mice
| Code | Rearrangement | Code | Rearrangement |
|---|---|---|---|
| 10 | V15J2/D1J1 | 34 | V11J2/V4J2 |
| 4 | V16J1/D2J2 | 39 | V8J1/V1J2 |
| 15 | V11J1/D2J2 | 40 | V4J2/V10J |
| 29 | V5J1/D2J2 | 49 | V10J2/V15J1 |
| 41 | V4J2/D1JI | 54 | V12J2/V4J2 |
| 43 | V1J1/D2J2 | 62 | V11J2/V1J1 |
| 45 | V8J2/D1J1 | 64 | V12J2/V10J2 |
| 47 | V14J1/D1J1 | 65 | V8J1/V12J1 |
| 52 | V1J2/D1J2 | 69 | V5J2/V17J2 |
| 58 | V3J1/D1J2 | 74 | V1J2/V5J2 |
| 68 | V14J1/D1J2 | 76 | V1J2/V5J2 |
| 73 | V8J2/D1J2 | 93 | V15J1/V14J2 |
| 79 | V8J2/D1J2 | 97 | V1J1/V10J1 |
| 87 | V4J1/D2J2 | 104 | V8J1/V5J1 |
| 100 | V6J1/D2J2 | 114 | V1J2/V8J2 |
| 103 | V8J2/D2J2 | 118 | V1J2/V6J2 |
| 119 | V5J2/D1J1 | 120 | V5J2/V10J2 |
| 134 | V11J2/D1J1 | 131 | V8J1/V5J2 |
| 94 | V5J2/V15J2 | 138 | V10J2/V15J1 |
| 7 | V3J1/V17J2 | 141 | V7J2/V1J1 |
| 11 | V8J2/V2J1 | 143 | V8J2/V11J2 |
| 19 | V8J2/V4J2 | 2 | V2J2/V9J2 |
| 32 | V7J1/V12J2 |
Discussion
The results of this study indicate that the adaptor protein SLP-76 is not only essential for thymocyte expansion and maturation from the DN to the DP stage but is also essential for TCR-β allelic exclusion. Despite expression of surface pre-TCR, thymocytes from SLP-76−/− mice, which are blocked at the CD25+CD44− DN stage, failed to exhibit TCR-β allelic exclusion, as evidenced by single-cell PCR. Neither the defect in the feedback inhibition of endogenous TCR-β locus rearrangements nor the block in thymic development could be overcome by introduction of a functional TCR-α/β transgene.
We have ascertained that CD25+CD44− DN thymocytes from SLP-76−/− as well as WT mice express the pTα and TCR-β chains of the pre-TCR complex (Fig. 2). As the 2F5 antibody we used is directed against the extracellular Ig-like domain of pTα, it is also clear from our studies that it is the form of pTα that expresses the extracellular Ig-like domain that reaches the cell surface. DN thymocytes from SLP-76−/− mice expressed equal or slightly higher amounts of TCR-β chains on their surfaces compared with TCR-α−/− (Fig. 2) or WT mice (data not shown). The fact that DN thymocytes from both SLP-76−/− and pTα−/− mice express lesser amounts of intracellular TCR-β chains than WT mice raises the possibility that signals through the pre-TCR mediated by SLP-76 may be important in upregulating TCR-β chain expression. The presence of a very small population of CD25− TCR-β+ DN thymocytes in SLP-76−/− thymocytes (∼2%) is consistent with the presence of a very small population of CD25−CD44− triple-negative cells in these mice (1.2%; Fig. 4 C). The equal or slightly higher surface expression of TCR-β and pTα in SLP-76−/−CD25+ CD44− DN thymocytes may then be accounted for by the developmental arrest that results in the accumulation of small, noncycling CD25+ CD44− DN cells, allowing these cells to bring a relatively higher amount of the pre-TCR to the cell surface. Alternatively, in the absence of SLP-76, internalization and subsequent degradation of the pre-TCR, which is dependent on activation of serine/threonine and tyrosine kinases 38, may be retarded.
Introduction of TCR-β transgenes into rearrangement-deficient mice drives proliferative expansion and differentiation of DN to DP cells 33 34. In contrast, introduction of functional TCR-β transgenes into SLP-76−/− mice failed to increase thymic cellularity and failed to overcome the block in the development of CD25−CD44− DN cells and in the transition from DN to DP cells (Fig. 4). We had previously shown that treatment of SLP-76−/− mice with anti-CD3∈ mAb induced the appearance of only a few DP thymocytes 17, suggesting that an SLP-76–independent pathway for DN to DP transition may exist. Such a pathway could involve linker for activation of T cells (LAT)-Grb2-Sos–mediated activation of Ras, as a transgene encoding an active form of Ras (RasV12) has been shown to drive the development of DP cells in the RAG−/− background 39. It appears from our data that expression of a transgenic TCR may not be sufficient to activate an SLP-76–independent pathway of thymocyte maturation.
Introduction of TCR-α/β transgenes into the SLP-76 background failed to suppress endogenous TCR-β rearrangement (Fig. 5). It has been suggested that inhibition of endogenous TCR-β rearrangement by TCR-β transgenes in pTα−/− mice might be artifactual and might reflect activation of downstream pathways not ordinarily engaged by the endogenous pre-TCR 12. Indeed, whereas introduction of a TCR-β transgene into the pTα−/− background shuts off endogenous TCR-β rearrangement, single-cell analysis in pTα−/− mice reveals failure of TCR-β allelic exclusion 12 13. As introduction of TCR-β transgenes into pTα−/− mice also drives the maturation of DN cells into DP cells and their proliferation, premature transgene-generated signals might accelerate development of normal cells through stages at which the endogenous TCR-β genes are assembled, leading to a block in rearrangement 40. The failure of TCR-α/β transgenes to suppress endogenous TCR-β rearrangement and induce maturation and expansion of DN cells in SLP-76−/− mice suggests that transgene-generated signals that lead to maturation and expansion of DN cells and to feedback inhibition of endogenous TCR-β rearrangement are strictly dependent on SLP-76.
Normally, 60% of surviving thymocytes carry only a single Vβ to DJβ rearrangement, whereas the remaining 40% carry one nonproductive and one productive Vβ to DJβ rearrangement 35. The presence of >40% of cells with Vβ to DJβ rearrangement on both alleles indicates a violation of TCR-β allelic exclusion. Single-cell PCR analysis revealed that 60% of CD25+CD44−TCR-β+ DN thymocytes from SLP-76−/− mice have undergone Vβ to DJβ rearrangement on both TCR-β alleles (Table ). This indicates that TCR-β allelic exclusion is defective in the absence of SLP-76 and suggests that SLP-76 is essential for the transduction of the physiologic pre-TCR signal that inhibits V to DJ rearrangement on the second TCR-β allele.
Due to feedback inhibition of TCR-β locus rearrangement by a productively rearranged allele 41, <3% of normal T cells carry two productive Vβ gene rearrangements 12. In the absence of feedback inhibition, it is expected that in 20% of cells that rearrange two alleles, both alleles are rearranged productively. When the alleles from SLP-76−/− thymocytes with two Vβ rearrangements were sequenced, it was found that in 31% of the cells, both rearrangements were productive, i.e., in frame (Table ). This is almost exactly the same percentage that was found in pTα−/− mice 12. The discrepancy between the observed and the predicted fraction of cells with two productive Vβ rearrangements may be due to selection against cells that make a productive rearrangement on the second allele only, due to lower TCR-β staining intensity, when cells are sorted for expression of cytoplasmic TCR-β chains. There was no indication for the selection of specific Vβ alleles in allelically included cells in either SLP-76−/− mice (Table and Table ) or pTα−/− mice 12. The presence of a significant proportion of SLP-76−/− thymocytes with two productively rearranged TCR-β alleles documents an essential role of SLP-76 in allelic exclusion at the TCR-β locus. The fact that similar percentages of cells with two in-frame alleles were found in SLP-76−/− and pTα−/− mice suggests that the pre-TCR is the most important receptor that mediates feedback inhibition of Vβ rearrangement under physiological conditions.
Recent data suggests that activation of Ras results in differentiation and expansion of DN thymocytes but not in TCR-β allelic exclusion 16. Thus, suppression of TCR-β gene rearrangements appears to require the activity of an additional and/or complementary pathway. As SLP-76−/− thymocytes are arrested at the DN stage and also fail to suppress endogenous Vβ to DJβ rearrangements, even after introduction of TCR-α/β transgenes, SLP-76 is likely to be critical for TCR-mediated activation of both Ras-dependent and Ras-independent pathways.
Acknowledgments
We thank Dr. O. Azogui for helpful discussions and Ms. O. Pivniouk and Ms. S. Freeman for excellent technical assistance.
This work was supported by National Institutes of Health grant AI-35714, grants from Baxter Healthcare, Caremark Corporation, and Olsten Corporation (to R.S. Geha), and a grant from the Howard Hughes Medical Institute (to F.W. Alt). I. Aifantis is the recipient of a grant from the Fondation Pour La Recherche Médicale (FRM). V. Pivniouk is a recipient of the Charles A. King Trust Fellowship Award. H. von Boehmer is supported by the Institut Universitaire de France, the Juvenile Diabetes Foundation, and the Körber Foundation (Germany). Supported in part by the Institut National de la Santé et de la Recherche Médicale, Paris, and by the Faculté Necker Enfants Malades, Descartes Université, Paris.
Footnotes
1used in this paper: DN, double-negative; DP, double-positive; RAG, recombination activating gene; SLP, SH2 domain–containing leukocyte protein; WT, wild-type
I. Aifantis, V. Pivniouk, and F. Gärtner contributed equally to this work.
References
- von Boehmer H., Fehling H.J. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova A., Saint-Ruf C., von Boehmer H. Crucial role of the pre-T-cell receptor a gene in development of αβ but not γδ T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- Malissen M., Gillet A., Rocha B., Trucy J., Vivier E., Boyer C., Kontgen F., Brun N., Mazza G., Spanopoulo E. T cell development in mice lacking the CD3 ζ/η gene. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:4347–4355. doi: 10.1002/j.1460-2075.1993.tb06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Clarke A.R., Rudnicki M.A., Iacomini J., Itohara S., Lafaille J.J., Wang L., Ichikawa Y., Jaenisch R., Hooper M.L. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- Molina T.J., Kishihara K., Siderovski D.P., van Ewijk W., Narendran A., Timms E., Wakeham A., Paige C.J., Hartmann K.-U., Veilette A. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- van Oers N.S.C., Lowin-Kropf B., Finlay D., Connolly K., Weiss A. αβ T cell development is abolished in mice lacking both Lck and Fyn protein tyrosine kinases. Immunity. 1996;5:429–436. doi: 10.1016/s1074-7613(00)80499-9. [DOI] [PubMed] [Google Scholar]
- O'Shea C.C., Thornell A.P., Rosewell I.R., Hayes B., Owen M.J. Exit of the pre-TCR from the ER/cis-Golgi is necessary for signaling differentiation, proliferation, and allelic exclusion in immature thymocytes. Immunity. 1997;7:591–599. doi: 10.1016/s1074-7613(00)80380-5. [DOI] [PubMed] [Google Scholar]
- Irving B.A., Alt F.W., Killeen N. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 1998;280:905–908. doi: 10.1126/science.280.5365.905. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Corcoran L.M., Cory S. Bcl-2 expression promotes B- but not T-lymphoid development in scid mice. Nature. 1994;368:457–460. doi: 10.1038/368457a0. [DOI] [PubMed] [Google Scholar]
- Linette G.P., Grusby M.J., Hedrick S.M., Hansen T.H., Glimcher L.H., Korsmeyer S.J. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity. 1994;1:197–205. doi: 10.1016/1074-7613(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Alt F.W., Yancopoulos G.D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO (Eur. Mol. Biol. Organ.) J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aifantis I., Buer J., von Boehmer H., Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 1997;7:601–607. doi: 10.1016/s1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- Krotkova A., von Boehmer H., Fehling H.J. Allelic exclusion in pTα-deficient miceno evidence for cell surface expression of two T cell receptor (TCR)-β chains, but less efficient inhibition of endogeneous Vβ→(D)Jβ rearrangements in the presence of a functional TCR-β transgene. J. Exp. Med. 1997;186:767–775. doi: 10.1084/jem.186.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardouin L., Ismaili J., Malissen B., Malissen M. The CD3-γδ∈ and CD3-ζ/η modules are each essential for allelic exclusion at the T cell receptor β locus are both dispensable for the initiation of V to (D)J recombination at the T cell receptor-β, -γ, and -δ loci. J. Exp. Med. 1998;187:105–116. doi: 10.1084/jem.187.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.J., Levin S.D., Perlmutter R.M. Protein tyrosine kinase p56lck controls allelic exclusion of T-cell receptor beta-chain genes. Nature. 1993;365:552–554. doi: 10.1038/365552a0. [DOI] [PubMed] [Google Scholar]
- Gartner F., Alt F.W., Monroe R., Chu M., Sleckman B.P., Davidson L., Swat W. Immature thymocytes employ distinct signaling pathways for allelic exclusion versus differentiation and expansion. Immunity. 1999;10:537–546. doi: 10.1016/s1074-7613(00)80053-9. [DOI] [PubMed] [Google Scholar]
- Pivniouk V., Tsitsikov E., Swinton P., Rathbun G., Alt F.W., Geha R.S. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- Clements J.L., Yang B., Ross-Barta S.E., Eliason S.L., Hrstka R.F., Williamson R.A., Koretzky G.A. Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science. 1998;281:416–419. doi: 10.1126/science.281.5375.416. [DOI] [PubMed] [Google Scholar]
- Clements J.L., Koretzky G.A. Recent developments in lymphocyte activationlinking kinases to downstream signaling events. J. Clin. Invest. 1999;103:925–929. doi: 10.1172/JCI6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretzky G. The role of Grb2-associated proteins in T-cell activation. Immunol. Today. 1997;18:401–406. doi: 10.1016/s0167-5699(97)01088-8. [DOI] [PubMed] [Google Scholar]
- Motto D.G., Ross S.E., Wu J., Hendricks-Taylor L.R., Koretzky G.A. Implication of the GRB2-associated phosphoprotein SLP-76 in T cell receptor–mediated interleukin 2 production. J. Exp. Med. 1996;183:1937–1943. doi: 10.1084/jem.183.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonski D., Kuhne M.R., Kadlecek T., Weiss A. Uncoupling of nonreceptor tyrosine kinases from PLC-γ1 in an SLP-76-deficient T cell. Science. 1998;281:413–416. doi: 10.1126/science.281.5375.413. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Ungewiss K., Azogui O., Palacios R., Owen M.J., Hayday A.C., von Boehmer H. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- Aifantis I., Azogui O., Feinberg J., Saint-Ruf C., Buer J., von Boehmer H. On the role of the pre-T cell receptor in αβ versus γδ T lineage commitment. Immunity. 1998;9:649–655. doi: 10.1016/s1074-7613(00)80662-7. [DOI] [PubMed] [Google Scholar]
- Buer J., Aifantis I., DiSanto J.P., Fehling H.J., von Boehmer H. T-cell development in the absence of the pre-T-cell receptor. Immunol. Lett. 1997;57:5–8. doi: 10.1016/s0165-2478(97)00078-3. [DOI] [PubMed] [Google Scholar]
- Hollander G., Castigli E., Kulback R., Su M., Burakoff S., Gutierrez-Ramos J.-C., Geha R. Induction of alloantigen-specific tolerance by B cells from CD40 deficient mice. Proc. Natl. Acad. Sci. USA. 1996;93:4994–4998. doi: 10.1073/pnas.93.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M. RAG-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Murphy K.M., Heimberger A.B., Loh D.Y. Induction by antigen of intrathymic apoptosis of CD4+CD8+ TCRlo thymocytes in vivo. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- Laird P.W., Zijderveld A., Linders K., Rudnicki M.A., Jaenisch R., Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffert D., Ehlich A., Muller W., Rajewsky K. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity. 1996;4:133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- von Boehmer H., Aifantis I., Feinberg J., Lechner O., Saint-Ruf C., Walter U., Buer J., Azogui O. Pleiotropic changes controlled by the pre-T cell receptor. Curr. Opin. Immunol. 1999;11:135–142. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- Aifantis I., Feinberg J., Fehling H.J., Di Santo J.P., von Boehmer H. Early T cell receptor β gene expression is regulated by the pre-T cell receptor–CD3 complex. J. Exp. Med. 1999;190:141–144. doi: 10.1084/jem.190.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. Developmental biology of T cells in T cell receptor transgenic mice. Annu. Rev. Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Koyasu S., Nakayama K., Murphy K.M., Loh D.Y., Reinherz E.L., Alt F.W. Restoration of T cell development in RAG2-deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- Malissen M., Trucy J., Jouvin-Marche E., Cazenave P.A., Scollay R., Malissen B. Regulation of TCR alpha and beta gene allelic exclusion during T-cell development. Immunol. Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- Xu Y., Davidson L., Alt F.W., Baltimore D. Function of the pre-T-cell receptor alpha chain in T-cell development and allelic exclusion at the T-cell receptor beta locus. Proc. Natl. Acad. Sci. USA. 1996;93:2169–2173. doi: 10.1073/pnas.93.5.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meerwijk J.P., Bluthmann H., Steinmetz M. T-cell specific rearrangement of T-cell receptor β transgenes in mice. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:1057–1062. doi: 10.1002/j.1460-2075.1990.tb08210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luton F., Legendre V., Gorvel J.P., Schmitt-Verhulst A.M., Boyer C. Tyrosine and serine protein kinase activities associated with ligand-induced internalized TCR/CD3 complexes. J. Immunol. 1997;158:3140–3147. [PubMed] [Google Scholar]
- Swat W., Shinkai Y., Cheng H.-W., Davidson L., Alt F.W. Activated Ras signals differentiation and expansion of CD4+8+ thymocytes. Proc. Natl. Acad. Sci. USA. 1996;93:4683–4687. doi: 10.1073/pnas.93.10.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu Y., Ryser S., Dembic Z., Borgulya P., Krimpenfort P., Berns A., von Boehmer H., Steinmetz M. In transgenic mice the introduced functional T cell receptor beta gene prevents expression of endogenous beta genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Khor B., Monroe R., Alt F.W. Assembly of productive T cell receptor δ variable region genes exhibits allelic inclusion. J. Exp. Med. 1998;188:1465–1471. doi: 10.1084/jem.188.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden B., Clark S.P., Kabelitz D., Mak T.W. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- Clark S.P., Yoshikai Y., Taylor S., Siu G., Hood L., Mak T.W. Identification of a diversity segment of human T-cell receptor beta-chain, and comparison with the analogous murine element. Nature. 1984;311:387–389. doi: 10.1038/311387a0. [DOI] [PubMed] [Google Scholar]
- Malissen M., Minard K., Mjolsness S., Kronenberg M., Goverman J., Hunkapiller T., Prystowsky M.B., Yoshikai Y., Fitch F., Mak T.W. Mouse T cell antigen receptorstructure and organization of constant and joining gene segments encoding the beta polypeptide. Cell. 1984;37:1101–1110. doi: 10.1016/0092-8674(84)90444-6. [DOI] [PubMed] [Google Scholar]