Abstract
TECK (thymus-expressed chemokine), a recently described CC chemokine expressed in thymus and small intestine, was found to mediate chemotaxis of human G protein–coupled receptor GPR-9-6/L1.2 transfectants. This activity was blocked by anti–GPR-9-6 monoclonal antibody (mAb) 3C3. GPR-9-6 is expressed on a subset of memory α4β7high intestinal trafficking CD4 and CD8 lymphocytes. In addition, all intestinal lamina propria and intraepithelial lymphocytes express GPR-9-6. In contrast, GPR-9-6 is not displayed on cutaneous lymphocyte antigen–positive (CLA+) memory CD4 and CD8 lymphocytes, which traffic to skin inflammatory sites, or on other systemic α4β7−CLA− memory CD4/CD8 lymphocytes. The majority of thymocytes also express GPR-9-6, but natural killer cells, monocytes, eosinophils, basophils, and neutrophils are GPR-9-6 negative. Transcripts of GPR-9-6 and TECK are present in both small intestine and thymus. Importantly, the expression profile of GPR-9-6 correlates with migration to TECK of blood T lymphocytes and thymocytes. As migration of these cells is blocked by anti–GPR-9-6 mAb 3C3, we conclude that GPR-9-6 is the principal chemokine receptor for TECK. In agreement with the nomenclature rules for chemokine receptors, we propose the designation CCR-9 for GPR-9-6. The selective expression of TECK and GPR-9-6 in thymus and small intestine implies a dual role for GPR-9-6/CCR-9, both in T cell development and the mucosal immune response.
Keywords: chemokine, α4β7, cutaneous lymphocyte antigen, memory, intestinal
Chemokines are a large and growing family of nearly 40 6–14-kD (nonglycosylated) heparin binding proteins that mediate a wide range of biological functions 1. The chemokines are divided into four families based on the position of four cysteine residues that form two disulfide bonds. Chemokine receptors are currently divided into four families based on the type of chemokine they bind, although no clear structural differences have been identified that distinguish the receptor subfamilies 2. There are also a number of “orphan” receptors with significant sequence similarity to chemokine receptors whose agonists and functions have yet to be defined. Chemokines play a vital role in leukocyte adhesion and extravasation. In various in vitro assays, chemokines support the chemotaxis or transendothelial migration of leukocytes 1, while in vivo injection 3 or overexpression of chemokines 4 results in leukocyte accumulation at the site of chemokine expression. Antagonists of chemokines prevent leukocyte trafficking 5 and have beneficial effects in several acute and chronic inflammatory models 6 7. Chemokines also modulate angiogenesis 8, hematopoiesis 1, and T lymphocyte activation 9 10, and several act as coreceptors with CD4 for entry of M-tropic and T-tropic HIV-1 11 12.
Subsets of CD4 lymphocytes can be defined based on their expression of adhesion molecules CD62L, CD49d, cutaneous lymphocyte antigen (CLA), and α4β7, which mediate lymphocyte trafficking to different physiological sites 13. For example, CLA+ memory CD4 lymphocytes are thought to traffic to the skin 14, whereas the mutually exclusive subset of α4β7+ memory CD4 lymphocytes traffics to intestinal sites 15. Leukocyte adhesion to endothelium is thought to involve several overlapping steps of rolling, activation, and arrest, in which exposure of rolling leukocytes to factors expressed at the site of adhesion activates leukocytes and upregulates integrin-mediated adhesion. As a result of this interaction, leukocytes arrest on the endothelium 5 16. Leukocyte activation and upregulation of integrin molecules occur via a pertussis toxin–sensitive mechanism that is thought to involve chemokine receptors 8 16 17. A growing body of evidence indicates that leukocyte subsets will also be defined by the expression of chemokine receptors required to traffic to various physiological sites 18.
To date, several chemokine receptors have been shown to subdivide memory CD4 lymphocytes. CXC chemokine receptor (CXCR)3, CC chemokine receptor (CCR)2, and CCR5 19 20 21 are all expressed on subsets of memory CD4 lymphocytes, whereas certain chemokines act preferentially on naive T lymphocytes 8 22. Further, several chemokines that interact with these receptors are expressed in inflammatory sites and LNs 23 24. In addition, in vitro–derived Th1 lymphocyte lines selectively express CXCR3 and CCR5, whereas Th2 lymphocyte lines selectively express CCR3, CCR4, and CCR8 25 26 27 28 29. Interestingly, in some cases the chemokines that bind to these respective chemokine receptors, such as macrophage-derived chemokine (MDC) for CCR4 and IFN-γ–inducible 10-kD protein (IP-10) for CXCR3, are induced by cytokines associated with a Th1 or Th2 environment 28 30.
Based on the assumption that the expression pattern of chemokine receptors on lymphocytes will prove informative of their role in leukocyte trafficking, we have prepared mAbs to orphan chemokine receptors to determine which subsets of leukocytes express these receptors. In this way, we hope to better understand the role these orphan chemokine receptors play in leukocyte trafficking. One such orphan chemokine receptor, human G protein–coupled receptor GPR-9-6, is described here. Based on the selective expression of GPR-9-6 on intestinal homing T lymphocytes and resident intestinal intraepithelial lymphocytes (IELs) and lamina propria lymphocytes (LPLs), we propose that the receptor will play an important role in T lymphocyte trafficking to mucosal sites and in T lymphocyte development.
Materials and Methods
Purification of Cell Populations.
Human peripheral blood was collected in 10% (vol/vol) 0.1 M EDTA, layered onto 1-Step Polymorphs gradient (Nycomed Pharma) and centrifuged at 400 g for 30 min at room temperature. Neutrophil and mononuclear cell layers were collected, resuspended in Dulbecco's PBS (DPBS) without calcium and magnesium (GIBCO BRL), and centrifuged for 15 min at ∼750 g. Red blood cells were lysed in the neutrophil fraction by resuspending the pellet in E-Lyse (Cardinal Associates) for 5 min on ice. Both cell fractions were washed two times with ice-cold DPBS. The mononuclear cells were allowed to adhere to protein-coated plastic for 2–3 h, and nonadherent cells were gently washed off the plate. After an additional 12 h, the nonadherent dendritic cells were washed off the plate and depleted of B lymphocytes and T lymphocytes with anti-CD19 and anti-CD2 Dynabeads (5 beads per cell; Dynal). The remaining cells were cultured in 50 ng/ml GM-CSF, 40 ng/ml IL-4 DMEM, and 10% FCS plus additives for 7 d to generate immature DCs, and in some cases 24 h additional culture in 10 ng/ml LPS was used to mature the dendritic cells. CD4, CD8, CD14, CD56, and CD19 populations were purified from mononuclear cells with the relevant microbeads (Miltenyi Biotec) using 20 μl of beads for 107 mononuclear cells in PBS, 1% BSA, 5 mM EDTA at 5 × 107 cells/ml for 30 min at 4°C. They were then pelleted, resuspended in PBS with 1% BSA and 5 mM EDTA at 5 × 107 cells/ml, and passed over a VS column (Miltenyi Biotec) in a magnetic field to remove nontagged cells. Cells were removed by forcing 20 ml of PBS with 1% BSA and 5 mM EDTA over the VS column, outside the magnetic field.
Human small intestine LPLs and IELs were isolated from patients undergoing gastric bypass surgery for morbid obesity as described previously 31. IEL preparations were on average 80% CD3+, 70% CD8+, 5% CD4+, whereas LPL preparations were 85% CD3+, 40% CD8+, 45% CD4+, consistent with previous analysis 31. All human subject protocols were approved by the Human Resources Committee at the Robert Wood Johnson Medical School, Brigham and Women's Hospital, or Massachusetts General Hospital.
Antibodies and Reagents.
Anti-CD4, -CD8, -CD14, -CD19, -CD49d, -CD56, -CD62L, -CLA, -CD45RA, and -CD45RO dye-linked mAbs for immunofluorescence studies were all obtained from PharMingen, while anti-αE–FITC was obtained from Coulter Pharmaceuticals. Anti-CCR4 mAb (2B10) was generated in the laboratory to CCR4/L1.2 transfectants. OKT3, an anti–human CD3 mAb, was obtained from American Type Culture Collection, and anti–human CD28 mAb was purchased from Becton Dickinson. IP-10, eotaxin-1, B cell–attracting chemokine 1 (BCL-1), IL-8, IFN-inducible T cell α chemoattractant (I-TAC), regulated on activation, normal T cell expressed and secreted protein (RANTES), fractalkine, leukotactin, liver-expressed chemokine (LEC), eotaxin-3, CKα2, macrophage inflammatory protein (MIP)-1α, monocyte chemoattractant protein (MCP)-1, MIP-4, I-309, and MDC were synthesized at LeukoSite, Inc., by f-moc chemistry (433A automated peptide synthesizer; Perkin-Elmer Biosystems). These chemokines were purified and folded as described previously 32. For biotinylated thymus-expressed cytokine (TECK), the chemokine domain of TECK was synthesized (TECK 24–99). Lys 99 was derivitized during the synthesis with an aminocapropyl-biotin moiety. A COOH-terminal glycine was added for synthesis convenience. The TECK chemokine domain had the same potency as full-length TECK for chemotaxis of GPR-9-6/L1.2 transfectants (data not shown). All other recombinant chemokines were obtained from either PeproTech or R&D Systems. The human endothelial cell line ECV304 was purchased from American Type Culture Collection. All cytokines were obtained from R&D Systems.
Generation of Anti–GPR-9-6 mAbs.
A peptide consisting of the NH2 terminus of GPR-9-6 was generated by American Peptide Company having the sequence NH2-MADDYGSESTSSMEDYVNFNFTDFYC. BALB/c mice were immunized intraperitoneally with 10 μg each of GPR-9-6 peptide/KLH conjugate prepared in complete Freund's adjuvant (CFA) at day 1, IFA at day 20, and PBS at day 40. At day 60, the mice were boosted with 10 μg of GPR-9-6 peptide/KLH in PBS, and after 4 d the spleens were removed and fused to SP2/0 myeloma cells (American Type Culture Collection). Fusions were screened by ELISA with plates coated with GPR-9-6 peptide. Hybridomas producing anti–GPR-9-6 mAbs were checked for reactivity with GPR-9-6 transfectants and subcloned for further characterization.
Preparation of Chronically Activated Th1 and Th2 Lymphocytes.
As described previously 33, 6-well Falcon plates were coated overnight with 10 μg/ml anti-CD28 and 2 μg/ml OKT3, then washed twice with PBS. Umbilical cord blood CD4 lymphocytes (Poietic Systems) were cultured at 105–106 cells/ml in DMEM with 10% FCS and IL-2 (4 ng/ml). IL-12 (5 ng/ml) and anti–IL-4 (1 μg/ml) were used to direct to Th1, while IL-4 (5 ng/ml) and anti–IFN-γ (1 μg/ml) were used to direct to Th2. After 4–5 d, the activated Th1 and Th2 lymphocytes were washed once in DMEM and cultured for 4–7 d in DMEM with 10% FCS and IL-2 (1 ng/ml). After this, the activated Th1 and Th2 lymphocytes were restimulated for 5 d with anti-CD28/OKT3 and cytokines as described above, but with the addition of anti-CD95L (1 μg/ml) to prevent apoptosis. After 4–5 d, the Th1 and Th2 lymphocytes were washed and then cultured again with IL-2 for 4 d. Activated Th1 and Th2 lymphocytes were maintained in this way for a maximum of three cycles. Direction of the Th1 and Th2 lines was confirmed by intracellular cytokine staining and ELISAs for IL-4 and IFN-γ.
ECV304 Transmigration and Chemotaxis Assays.
3-μM pore diameter Transwell inserts were generally used, with the exception of the bulk migration experiments for the phenotyping of migrated cells, in which 5-μM inserts were used. The cells under study were washed once in RPMI and resuspended at 4 × 106 cells/ml for Th1/Th2 lymphocytes, cell lines, and transfectants, and at 107 cells/ml for resting CD4 lymphocytes, in RPMI with 0.5% heat-shock antigen (HSA) and 10 mM Hepes. An aliquot of 200 μl of cell suspension (input of 8 × 105 and 2 × 106 cells, respectively) was added to each insert. Chemokine in 500–600 μl of RPMI with 0.5% HSA and 10 mM Hepes was added to the lower well. After 2–4 h, the inserts were removed and the number of cells which had migrated through the insert to the lower well was counted for 30–60 s on a Becton Dickinson FACScan™ with the gates set to acquire the cells of interest. Using this technique, 100% migration would be 25,000 cells for Th1/Th2 cells and 75,000 cells for resting CD4 lymphocytes, where this number represents the cells in the lower well counted on the FACScan™ over 30–60 s. In all cases, the data points were the result of duplicate wells, with the mean value shown and the error bars representing the sample standard deviation.
To determine the migration of lymphocyte subsets, the number of cells in each subtype was determined by multi-color FACScan™ analysis for the starting population of each chemotactic assay 34. The number of cells belonging to the same subtype was determined in the migrated population, and the percentage of migration was determined from these two numbers. Eight wells were used for each treatment and pooled for FACScan™ analysis.
As it has been demonstrated previously that most chemokine receptors couple to the Gi class of G proteins, we wanted to determine if GPR-9-6 also couples to the Gi isoform. Pertussis toxin was used to block Gi-mediated cellular responses. 2 × 106 cells/ml in prewarmed RPMI plus 10% FCS were incubated with pertussis toxin (GIBCO BRL) at a final concentration of 100 ng/ml for 2 h at 37°C, washed twice with medium, and resuspended in chemotaxis medium. Migration was assayed as described above.
Immunofluorescence Staining.
Cells were resuspended at 107 cells/ml in FACS® buffer (PBS with 5% FCS, 0.1% azide, and 10% human serum to block Fc receptors), and primary mAb was added. After 20 min, the cells were washed twice in FACS® buffer, and F(ab)2 anti–mouse IgG–PE (absorbed against human Ig) was added. After 20 min, the cells were washed twice in FACS® buffer and then blocked in 10% mouse serum before the second mAb directly linked to a fluorochrome was added. After 20 min, the cells were washed three times in FACS® buffer and analyzed. Throughout the staining, the cells were kept on ice. We used an IgG2b isotype control followed by F(ab)2 anti–mouse IgG–PE as a negative control, and isotype control staining always fell below 10 FL1 or FL2 units.
Binding Assay with Biotinylated TECK.
4 × 105 cells in 25 μl of PBS with 10% rabbit serum, 1% FCS, and 0.1% sodium azide (assay buffer) were preincubated with either assay buffer (no treatment), 1,000 nM TECK/MCP-1, or 50 μg/ml 3C3/IgG2b for 30 min. 25 μl of biotinylated TECK at a final concentration ranging up to 100 nM was then added, and after 1 h, the cells were washed twice with 200 μl of wash buffer (PBS with 1% FCS and 0.1% azide) and resuspended in 25 μl of Wallac Eμ-labeled streptavidin diluted 1:400 in assay buffer. After 30 min at room temperature, the cells were washed in wash buffer and transferred to a 96-well solid white nonspecific binding plate (3600; Corning) and, after spinning to remove supernatant, 200 μl of Wallac Enhancement Solution was added. The fluorescence was counted on a multilabel counter (model 1420; Wallac) at 340/613 nm and reported in relative fluorescence units (RFUs).
Calcium Mobilization with Molt-4 Cells.
107 cells/ml in DPBS were labeled for 30 min with Fura-2 AM (Molecular Probes) at 2 μM, washed three times in DPBS, and resuspended at 106 cells/ml in DPBS containing 1 mM CaCl2, 0.5 mM MgCl2, 10 mM Hepes (pH 7.2), and 5.5 mM glucose. Chemokine was then added as indicated in the figure legend, and the calcium flux was measured on a fluorimeter using 10% NP-40 and 10 mM EGTA to establish the maximum and minimum Ca2+ mobilized.
Recombinant DNA Methods.
Plasmid DNA was isolated using Genomic tips as recommended by the manufacturer (Qiagen). DNA ligations, restriction endonuclease digestions, and gel electrophoresis were performed as described previously 35. DNA purification through agarose gel extraction was performed using the QIAEXII Gel Extraction kit as recommended by the manufacturer (Qiagen). Plasmid DNA was introduced into Escherichia coli by chemical transformation (GIBCO BRL). Enzymes were purchased from New England Biolabs, GIBCO BRL, or Boehringer Mannheim. RNA was isolated from frozen tissues or cells using either the standard guanidinium isothiocyanate method 35 or the RNeasy kit as recommended (Qiagen). DNA sequencing was performed by MacroMolecular Resources (Colorado State University, Fort Collins, CO) using an ABI DyeRhodamine Terminator cycle sequencing kit and an ABI Prism DNA Sequencer (model 377; Perkin-Elmer Applied Biosystems) according to the manufacturer's specifications. Sequences were analyzed using SeqMan (DNASTAR). Primers were synthesized by MacroMolecular Resources or by Operon Technologies, Inc.
PCR.
Primers were designed for use in the PCR to amplify the coding region of GPR-9-6 based on the nucleotide sequence deposited in EMBL/GenBank/DDBJ (accession no. U45982). BamHI and XbaI sites were incorporated into primer pair BAZ201 5′-TCGAAGGGATC CCTAACATGGCTGATGACTATGGC-3′ and BAZ202 5′-AAGAAGTCTAGAACCCCTCAGAGGGAGAGTGCTCC-3′ for directional cloning (bold, coding sequence; italic, enzyme site). 5 μg of total human genomic DNA (Clontech) was used as the template in the Pfu PCR cycles, with 60 mM Tris-HCl (pH 9.5), 1.5 mM MgCl2, 100 pmol primers, 200 μM dNTP, and 5 U PfuI polymerase (Invitrogen) in 100 μl volume. The cycle parameters were an initial melt at 95°C for 2 min, then 35 cycles: 95°C, 30 s; 55°C, 30 s; 72°C, 2 min 15 s, followed by a final extension of 72°C, 7 min in a DNA thermal cycler (Perkin-Elmer Corp.).
Primers were designed to amplify the complete coding region of TECK (sequence data available from EMBL/GenBank/DDBJ under accession no. U86358) based on the published nucleotide sequence. HindIII and XbaI sites were incorporated into the primer pairs for directional cloning. BAZ203 5′-TCGAAGAAGCTT ATGAACCTGTGGCTCCTG-3′ and BAZ204 5′-AAGAAGTCTAGA TCACAGTCCTGAATTAGC-3′ were used to amplify TECK (bold, coding sequence; italic, enzyme site). 5 μg of human thymus RNA was reverse transcribed with oligo(dT) in 20 μl volume. The cDNA was mixed with 200 μM dNTP, 100 pmol primers, 60 mM Tris-HCl (pH 9.5), 1.5 mM MgCl2, and 10 U AmpliTaq in 50 μl volume. The cycle parameters were an initial melt at 95°C for 2 min, then 35 cycles: 95°C, 30 s; 55°C, 30 s; 72°C, 1 min, followed by a final extension of 72°C, 7 min. The human thymus was obtained from Boston Children's Hospital (Boston, MA).
Reverse transcription (RT)-PCR amplifying TECK (BAZ203, BAZ204) and GPR-9-6 (BAZ201, BAZ202) was performed using equal amounts (0.5 ng) of cDNA template from thymus, small intestine, colon, and brain, as well as 5 μg genomic DNA (Clontech). The RNA used to prepare the cDNA template was triple oligo(dT) column purified. The same conditions and PCR profile were used as the AmpliTaq PCR cycle described above, except that 30 cycles were performed. Amplification with glyceraldehyde 3-phosphate dehydrogenase (G3PDH) primers (Clontech) was used to demonstrate equivalence of template. Amplification with G3PDH intron B specific primers (BAZ205 5′-TCCCCTGCCAGCCTAGCGTTGACC-3′ and BAZ206 5′-CCCCACTATGCCACCCCAGGAATG-3′) was used to confirm the absence of contaminating genomic DNA in the Clontech cDNAs.
After agarose gel electrophoresis, the PCR products were visualized in the presence of ethidium bromide with an ultraviolet light source. DNA fragments of predicted size (∼450 bp for TECK and ∼1 kb for GPR-9-6) were isolated and cloned into pBluescript II KS+ (Stratagene) and pcDNA3 (Invitrogen) for sequence analysis and further manipulation.
Expression Vector Construction and Generation of a GPR-9-6–expressing Stable Cell Line.
The coding region of GPR-9-6 was amplified by PCR and directionally cloned into the BamHI/XbaI sites of pcDNA3. Transfectants were generated in the murine pre-B lymphoma cell line L1.2 as described previously 36, and maintained in RPMI 1640 supplemented with 10% FCS, 2 mM l-glutamine, 50 U/ml penicillin/streptomycin, 0.55 mM β-mercaptoethanol, 10 mM Hepes, 1 mM sodium pyruvate, and 1 mg/ml G418 (Geneticin) for selection. The transfectants were then stained by mAbs with reactivity against the GPR-9-6 peptide (see below) and analyzed by FACScan™ to confirm surface expression of GPR-9-6, then cloned by limiting dilution. Transfected cells were treated with 5 mM n-butyric acid for 24 h before experimentation.
Northern Blot Analysis.
Northern blots were either purchased from Clontech or prepared as follows: total RNA was separated by electrophoresis on 1.2% formaldehyde agarose gels and transferred to a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech) by the capillary method as described previously 34 and cross-linked using a Stratalinker (Stratagene). Full-length gel-purified TECK, GPR-9-6, and β-actin (Clontech) were radiolabeled using High Prime reagents (Boehringer Mannheim) according to the manufacturer's specifications. Hybridizations were performed in ExpressHyb Solution (Clontech) using the manufacturer's suggested protocol. Length of autoradiography exposure is described in appropriate figure legends.
Results
An mAb Raised against GPR-9-6 Selectively Reacts with GPR-9-6 Transfectants.
Due to its close phylogenetic association with other known leukocyte chemokine receptors such as CCR6 and CCR7, we cloned GPR-9-6 by PCR, using primers designed from the deposited EMBL/GenBank/DDBJ sequence. GPR-9-6/L1.2 transfectants were prepared and stained with mAbs raised against the first 30 amino acids of the NH2 terminus of GPR-9-6 coupled to KLH. One mAb, designated 3C3, reacted with GPR-9-6/L1.2 transfectants but not with parental L1.2 cells. 3C3 was found to have an IgG2b isotype. In cross-reactivity studies, 3C3 did not cross-react with CCR1–7 or CXCR1–4 transfectants. We show the data for CCR6 here, as it is one of the more closely related chemokine receptors to GPR-9-6 (Fig. 1). Also, the NH2-terminal peptide of GPR-9-6 blocked the binding of mAb 3C3 to GPR-9-6 transfectants (data not shown), further validating the specificity of this mAb.
Figure 1.

The mAb 3C3 binds to GPR-9-6 transfectants. (A) GPR-9-6/L1.2 transfectants were stained with mAb 3C3 (black profile), an IgG2b control mAb (white), and an anti-CCR6 mAb (gray). (B) CCR6/L1.2 transfectants were stained with the same anti-CCR6 (gray), 3C3 (black), and IgG2b (white) mAbs (n = 2).
GPR-9-6 Is Expressed on Molt T Cell Lines, Subsets of CD4, CD8, and B Lymphocytes in Peripheral Blood, and Thymocytes.
mAb 3C3 was used to identify surface expression of GPR-9-6 on various cell lines and primary cells. Out of a panel of cell lines, only Molt-4 and Molt-13 T cell lines expressed GPR-9-6 (Table ). In addition, GPR-9-6 was expressed on a small subset of CD4 lymphocytes (2–4%) as well as on a small subset of CD8 lymphocytes (Fig. 2). A subset of B lymphocytes also expressed GPR-9-6. Monocytes, basophils, eosinophils, neutrophils, immature and mature dendritic cells, and NK cells did not express GPR-9-6 (Fig. 2, and data not shown). GPR-9-6 was expressed on >90% of thymocytes that expressed all levels of TCR, although a small subset of TCRhighGPR-9-6− thymocytes was evident. In three-color experiments, GPR-9-6 was found on the majority of CD4+, CD8+, and CD4+CD8+ thymocytes and on ∼50% of immature CD4−CD8− thymocytes (data not shown).
Table 1.
Distribution of GPR-9-6 on Cell Lines
| Cell line | GPR-9-6 | CXCR4 | Cell | Chemotaxis |
|---|---|---|---|---|
| Molt-4 | + | + | T | + |
| Molt-13 | + | + | T | + |
| CEM | − | + | T | − |
| Peer | − | + | T | − |
| Hut-78 | − | + | T | ND |
| PM1 | − | + | T | ND |
| SKW3 | − | + | T | ND |
| Jurkat | − | + | T | ND |
| Ramos | − | + | B | ND |
| Raji | − | + | B | ND |
| JY | − | + | B | − |
| THP1 | − | − | M | ND |
| U937 | − | + | M | − |
| HL60 | − | +/− | N | ND |
| KG1 | − | + | ML | ND |
| KU812 | − | − | ML | ND |
| EOL | − | − | Eos | ND |
Several cell lines representing T lymphocytes (T), B lymphocytes (B), neutrophils (N), eosinophils (Eos), monocytes (M), and myelogenous leukemia cells (ML) were stained with mAb 3C3, as well as with an anti-CXCR4 control mAb since most cell lines express CXCR4. The cell lines that expressed GPR-9-6, as well as several negative lines, were examined for their chemotaxis to 150 nM TECK.
Figure 2.
GPR-9-6 is expressed on subsets of CD4 and CD8 lymphocytes as well as on most thymocytes. The mAb 3C3 was used in two-color studies on mononuclear cells along with a F(ab)2 anti–mouse IgG–PE second stage and anti-CD4–FITC, anti-CD8–FITC, anti-CD19–FITC, anti-CCR3–FITC, and anti-CD56–Cychrome mAbs. For thymocytes, two-color studies were performed with 3C3 and anti-TCR–Cychrome. The percentage of cells falling within each quadrant is shown at the upper right. Staining with an isotype control IgG2b mAb followed by F(ab)2 anti–mouse IgG–PE was performed and gave FL2 values <10 U (n = 6).
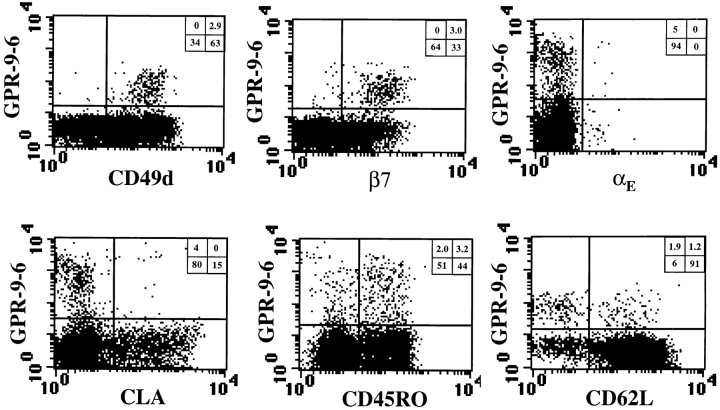
The CD4 and CD8 Lymphocyte Subsets That Express GPR-9-6 Also Express High Levels of Mucosal Lymphoid Homing Receptor α4β7 but Not Skin Homing Receptor CLA.
To determine which subset of CD4 and CD8 lymphocytes expressed GPR-9-6, three-color flow cytometry was performed (Fig. 3). The CD4 lymphocytes that expressed GPR-9-6 were mainly of memory phenotype, and those cells that expressed the highest levels of GPR-9-6 were all of memory phenotype. Interestingly, almost all of the GPR-9-6+ CD4 lymphocytes expressed high levels of the α4β7 integrin, which is expressed on gut-trafficking T lymphocytes, and lacked expression of CLA, which mediates T lymphocyte trafficking to inflamed skin. The subset of memory CD4 lymphocytes defined by expression of αE and CD62L was also subdivided into GPR-9-6 positive and negative subpopulations.
Figure 3.
GPR-9-6 is expressed on β7highCLA− memory CD4 lymphocytes. Mononuclear cells were stained in three-color experiments using anti-CD4–Tricolor to gate on CD4 lymphocytes. The cells were also stained with anti–GPR-9-6 mAb 3C3 followed by F(ab)2 anti–mouse IgG–PE or –FITC to study GPR-9-6 expression on subsets defined with dye-linked mAbs: anti-αE–FITC (HML1), anti-β7–PE (Fib504), anti-CD49d–PE (HP2/1), anti-CLA–FITC (HECA 452), anti-CD45RO–FITC (UCLH1), and anti-CD62L–PE (Dreg 56). Staining with an isotype control IgG2b mAb followed by F(ab)2 anti–mouse IgG–PE/FITC was performed and gave FL1/FL2 values <10 U. The percentage of cells falling within each quadrant is shown in the upper right (n = 5).
Upon analysis over multiple donors, a subset of CD4 lymphocytes that expressed low levels of GPR-9-6 was evident. This subset was more apparent when we used four-color analysis and gated on memory and naive CD4 lymphocytes, as defined by CD45RO expression (Fig. 4). Expression of GPR-9-6 on memory CD4 lymphocytes was more than threefold higher than that on naive CD4 lymphocytes, as shown in Fig. 4 where the ratio of the mean fluorescent intensities of GPR-9-6 on naive and memory CD4 lymphocytes approaches 3.5 when an anti–mouse IgG–PE second stage was used. GPR-9-6low CD4 lymphocytes expressed CD62L, a homing receptor involved in lymphocyte trafficking to organized lymphoid tissues via high endothelial venules (HEVs), and low levels of β7. In contrast, the majority of the GPR-9-6high memory CD4 lymphocyte subset did not express CD62L, but expressed high levels of β7.
Figure 4.

Characterization of GPR-9-6 expression on memory and naive CD4 lymphocytes: Mononuclear cells were stained in four-color experiments using anti-CD4–TC and anti-CD45RO–allophycocyanin to gate on memory and naive CD4 lymphocytes. The cells were also stained with anti–GPR-9-6 mAb 3C3 followed by F(ab)2 anti–mouse IgG–PE or –FITC to study GPR-9-6 expression on subsets defined with dye-linked mAbs: anti-αE–FITC (HML1), anti-β7–PE (Fib504), anti-CD49d–PE (HP2/1), anti-CLA–FITC (HECA 452), and anti-CD62L–PE (Dreg 56). Staining with an isotype control IgG2b mAb followed by F(ab)2 anti–mouse IgG–PE/FITC was performed and gave FL1/FL2 values <10 U. The percentage of cells falling within each quadrant is shown in the upper right. To directly compare the levels of GPR-9-6 on naive and memory subsets defined by expression of GPR-9-6, we show the ratio of the mean fluorescence intensity for memory/naive subset for the relevant quadrants (Q) (n = 3).
Like GPR-9-6+ CD4 lymphocytes, GPR-9-6+ CD8 lymphocytes also expressed high levels of β7 and were CLA− (Fig. 5). CD45RO and CD45RA are not as extensively used as memory markers for CD8 lymphocytes. However, GPR-9-6 was expressed on CD45RAlow CD8 lymphocytes. Also, as observed for CD4 lymphocytes, a subset of GPR-9-6low cells was evident that was CD45RAhighCD62L+, whereas GPR-9-6high CD8 lymphocytes were CD62L−. Finally, the majority of αE+ CD8 lymphocytes expressed GPR-9-6, whereas GPR-9-6low CD8 lymphocytes were mainly αE−. Further, αE+ CD8 lymphocytes that did not express GPR-9-6 expressed lower levels of αE than αE+GPR-9-6+ CD8 lymphocytes.
Figure 5.
GPR-9-6 is expressed on β7high CLA− memory CD8 lymphocytes and predominantly αE+ CD8 lymphocytes. Mononuclear cells were stained in three-color experiments using anti-CD8–TC to gate CD8 lymphocytes. The cells were also stained with anti–GPR-9-6 mAb 3C3 followed by F(ab)2 anti–mouse IgG–PE or –FITC to study GPR-9-6 expression on subsets defined with dye-linked mAbs: anti-αE–FITC (HML1), anti-β7–PE (Fib504), anti-CD49d–PE (HP2/1), anti-CLA–FITC (HECA 452), anti-CD45RA–FITC, and anti-CD62L–PE (Dreg 56). Staining with an isotype control IgG2b mAb followed by F(ab)2 anti–mouse IgG–PE/FITC was performed and gave values <10 U. The percentage of cells falling within each quadrant is shown in the upper right (n = 4).
GPR-9-6 Is Expressed on Intestinal IELs and LPLs.
The selective expression of GPR-9-6 on a subset of intestinal trafficking CD4 and CD8 blood lymphocytes suggests that the receptor may be involved in lymphocyte homing to the intestine. Therefore, we examined the expression of GPR-9-6 on T lymphocytes isolated from the small intestine. Flow cytometric analysis revealed that GPR-9-6 was expressed on all intestinal IELs and LPLs (Fig. 6). Compared with GPR-9-6 expression on PBL lymphocytes, the chemokine receptor was found to be significantly enriched on T lymphocytes in intestinal tissue.
Figure 6.
GPR-9-6 is expressed on intestinal IELs and LPLs. Freshly isolated LPLs and IELs were stained with anti–GPR-9-6 mAb 3C3 (gray profile) or with a mouse IgG2b control mAb (white). PBLs were also stained as an internal control from a separate donor. Similar results were obtained from four different mucosal cell populations.
Activation of Umbilical CD4 Lymphocytes to Th1 or Th2 Effector T Lymphocytes Did Not Induce Expression of GPR-9-6, Whereas TCR Cross-linking Resulted in Downregulation of GPR-9-6 on T Lymphocytes.
As GPR-9-6 was expressed on a subset of memory T lymphocytes, we attempted to induce expression of GPR-9-6 on umbilical CD4 lymphocytes by activation in the presence of directing cytokines to generate Th1 and Th2 lymphocytes. Although the Th1 and Th2 lymphocyte lines showed the appropriate differential production of IL-4, IL-13, and IFN-γ upon activation (data not shown), chronic activation of these cells in the presence of IL-12 or IL-4 to generate Th1 or Th2 lymphocytes failed to induce the expression of GPR-9-6 (Fig. 7 B). Further, chronic activation of umbilical CD4 lymphocytes in the presence of IL-1–13, IL-15, IL-17, IL-18, and TGF-β did not induce expression of GPR-9-6 (data not shown). As expected, CXCR3 was upregulated selectively on Th1 lymphocytes (Fig. 7 A). Interestingly, activation of T lymphocytes through cross-linking of TCR resulted in transient downregulation of GPR-9-6 expression (Fig. 6 C), with reexpression of GPR-9-6 observed upon removal of the TCR stimulation and culture in IL-2. This effect was not specific to the chemokine receptor GPR-9-6, as the other chemokine receptors expressed by T lymphocytes that we examined (CCR5 and CCR6) were also transiently downregulated upon TCR cross-linking (Fig. 7 D). Expression by T lymphocytes of other CD molecules such as CD29, CD44, and CD49d was unaffected by T lymphocyte activation (data not shown).
Figure 7.
Modulation of GPR-9-6 on lymphocytes by cytokines and upon T lymphocyte activation. (A and B) Chronically activated Th1 (dark line) and Th2 lymphocytes (thin line) at second stage activation were stained with anti-CXCR3 and anti–GPR-9-6 mAb 3C3. Staining of Th1 lymphocytes with an isotype control IgG2b mAb served as a negative control (dashed line). (C and D) Mononuclear cells were activated with plate-bound anti-TCR mAb OKT3 for 4 d followed by expansion with IL-2 at 5 ng/ml. Aliquots of cells were stained at the indicated times with anti–GPR-9-6, anti-CCR5, and anti-CCR6 to determine the effect of T lymphocyte activation upon expression of GPR-9-6 (C) and other chemokine receptors (D), using an anti-CD3 mAb to directly label the T lymphocytes (n = 2).
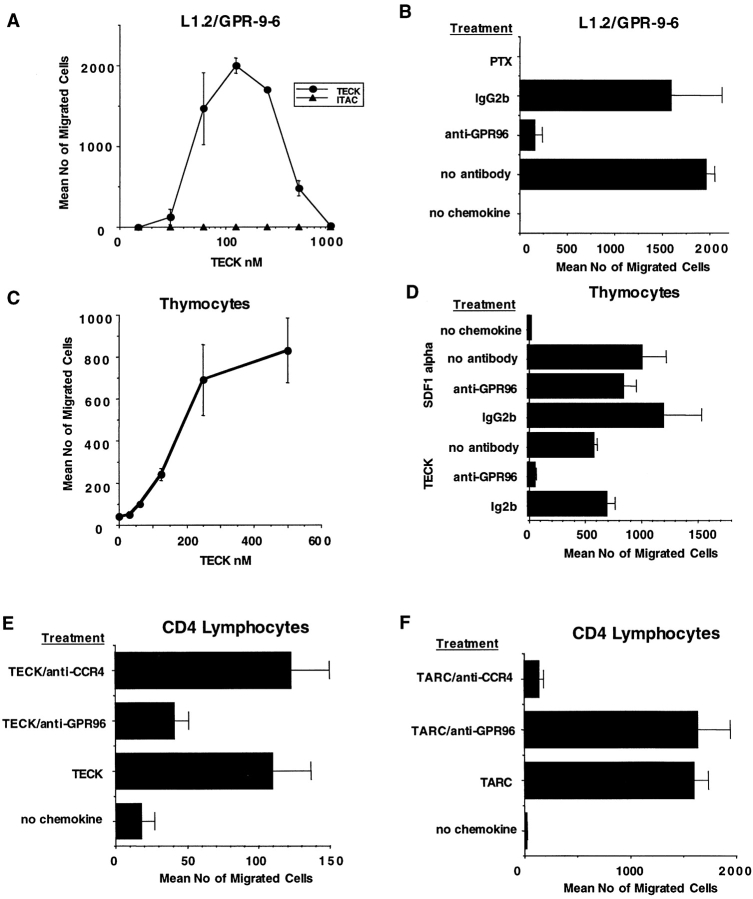
The GPR-9-6 Chemokine Receptor Specifically Interacts with TECK and Mediates Thymocyte and CD4 Lymphocyte Chemotaxis.
From a panel of chemokines including MCP-1–4, MIP-1αβ, MIP-3αβ, eotaxin-1–3, RANTES, I-309, thymus and activation-regulated chemokine (TARC), MDC, liver-expressed chemokine (LEC), CKα2, secondary lymphoid-tissue chemokine (SLC), human CC chemokine 1, fractalkine, lymphotactin, monokine induced by IFN-γ (MIG), IP-10, I-TAC, B cell–attracting chemokine 1 (BCL-1), IL-8, gro-αβψ, leukotactin, stromal cell–derived factor (SDF)-1α/β, MIP-3, and MIP-4, only TECK induced chemotaxis of GPR-9-6/L1.2 transfectants (Fig. 8 A, and data not shown). TECK-mediated chemotaxis of L1.2/GPR-9-6 transfectants was inhibited by anti–GPR-9-6 mAb 3C3 (Fig. 8 B), while pretreatment of the transfectants with pertussis toxin also inhibited migration to TECK (Fig. 8 B). The observation that pertussis toxin abolished TECK-mediated transfectant chemotaxis indicates that chemokine receptor GPR-9-6 couples to the Gi class of G proteins. TECK did not act on any of the other transfectants tested (CCR1,2,4–7 and CXCR1–4), with the exception of CCR3/L1.2 transfectants, for which high concentrations of TECK were weakly chemotactic (data not shown). Also in examining our panel of cell lines, we found that only GPR-9-6+ Molt-4 and Molt-13 cell lines migrated to TECK (Table ). Migration of Molt-4 and Molt-13 cells to TECK was blocked by anti–GPR-9-6 mAb 3C3 (data not shown).
Figure 8.
GPR-9-6 is a chemokine receptor for TECK. (A) GPR-9-6 transfectants were examined for chemotactic responses to various concentrations of TECK and I-TAC ranging from 10 to 1,000 nM. (B) The effect of anti–GPR-9-6 mAb 3C3 and pertussis toxin (PTX) on the migration was examined. All mAb treatments were for 10 min at 4°C before the migration assay. (C) Thymocytes were examined for their chemotaxis to various concentrations of TECK. (D) The effect of anti–GPR-9-6 mAb 3C3 and an IgG2b control on TECK- and SDF-1α–induced thymocyte migration was also examined. The effect of anti–GPR-9-6 mAb and anti-CCR4 mAb 2B10 on the chemotaxis of CD4 lymphocytes to TECK (E) and TARC (F) was also tested (n = 2).
Primary thymocytes were also chemotaxic for TECK in a dose-dependent manner (Fig. 8 C), and this migration was blocked by anti–GPR-9-6 mAb 3C3, but not by an IgG2b control (Fig. 8 D). In contrast, anti–GPR-9-6 did not block thymocyte migration to SDF-1α, which binds to CXCR4 on thymocytes (Fig. 8 D). We also found that a small subset of CD4 lymphocytes (<0.5% of input) migrated to TECK and that this chemotaxis was blocked by anti–GPR-9-6 mAb 3C3, but not by an anti-CCR4 mAb, 2B10. In contrast, 3C3 mAb had no effect on TARC-induced CD4 lymphocyte chemotaxis (Fig. 8E and Fig. F). In our experiments, TECK was not chemotactic for other total cell populations in peripheral blood, including B lymphocytes, CD8 lymphocytes, neutrophils, monocytes, and eosinophils (data not shown).
The α4β7highCLA−CD45RA− CD4 and αE+α4β7+ CD45RAlow/− CD8 Lymphocytes Migrate to TECK.
In our preliminary chemotaxis studies, we were able to demonstrate that TECK induced significant but very low levels of migration of peripheral blood CD4 lymphocytes. This reflects the low percentage of CD4 lymphocytes that express GPR-9-6 and the background migration of the total population of CD4 lymphocytes. As GPR-9-6 was expressed at high levels mainly on small subsets of memory β7high CD4 and CD8 lymphocytes, we examined the phenotype of the memory CD4 and CD8 lymphocytes that migrate to TECK using multi-color FACScan™ analysis with anti-CD4, anti-CD8, anti-CD45RA, anti-CLA, anti-α4β7 mAb Act1, and anti-αE (Fig. 9). Act1 is an mAb that recognizes a combinatorial epitope on α4β7 and therefore can be used to specifically examine expression of this integrin on leukocytes. The gut-homing α4β7+CLA− memory CD4 lymphocytes were enriched in the migrated population, whereas skin-homing α4β7−CLA+ memory CD4 lymphocytes and α4β7−CLA− memory CD4 lymphocytes were not (Fig. 10 A). Therefore, the memory CD4 lymphocytes that migrate to TECK are contained in the gut-homing α4β7+ CLA− memory CD4 lymphocyte subpopulation. For the CD8 lymphocytes, the anti-CD45RA mAb was used to define the memory CD8 lymphocytes as CD45RAlow/−. As GPR-9-6 was expressed at high levels on CD8 lymphocytes that express high levels of αE or β7, we analyzed the expression of αE and α4β7 on the migrated CD8 lymphocytes. Higher random migration of CD45RAlow/− CD8 lymphocytes was evident, but the cells responsive to TECK were greatly enriched in the αE+α4β7+CD45RAlow/− CD8 lymphocyte subset (Fig. 9 B).
Figure 9.

TECK preferentially attracts gut-homing memory CD4 lymphocytes and αE+α4β7+CD45RAlow/− CD8 lymphocytes. Mononuclear cells from two donors were allowed to migrate to 1 μM TECK (black bars) and also to medium (white). (A) The starting and migrated populations were analyzed by multiparameter flow cytometry to determine the percentages of skin-homing CLA+α4β7−, gut-homing memory CLA−α4β7+, and other systemic CLA−α4β7− memory CD4 lymphocytes in the starting and migrated populations. (B) The starting and migrated populations were analyzed to determine the percentage of αE+α4β7+, αE−α4β7+, and αE−α4β7−CD45RAlow/− CD8 lymphocytes in the starting and migrated populations. With knowledge of the total input and the number of migrated cells, these values are expressed as percentage of input (n = 3).
Figure 10.
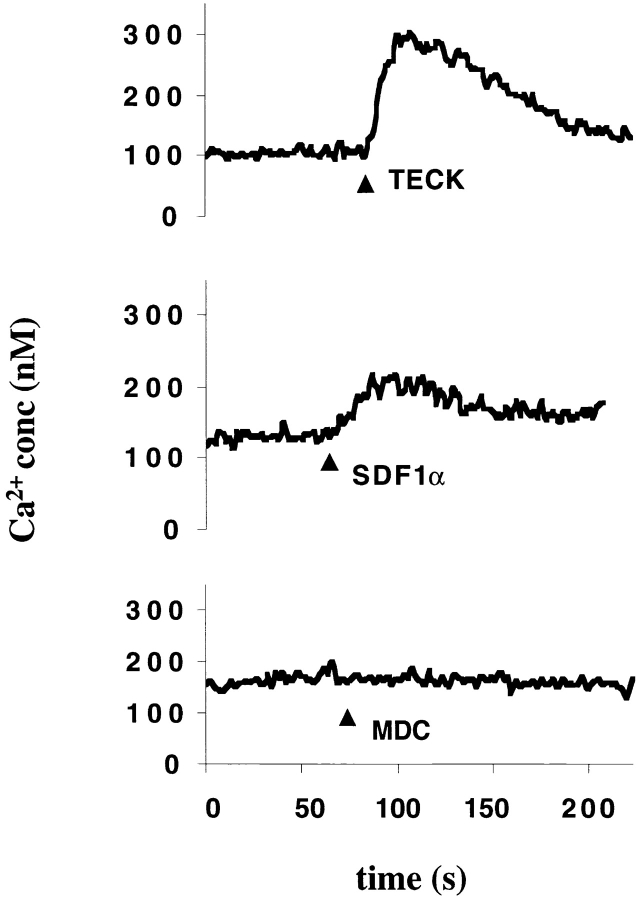
TECK induces Ca2+ flux in GPR-9-6+ Molt-4 cells. The GPR-9-6–expressing cell line Molt-4 was loaded with Ca2+ dye Fura-2 and then tested for the ability to mobilize Ca2+ in response to 150 nM TECK, 100 nM SDF-1α, and 100 nM MDC (n = 2).
TECK Induces Ca2+ Flux in GPR-9-6+ Cell Line Molt-4.
In addition to its ability to induce cell migration, TECK also induced a more robust Ca2+ flux in Molt-4 cells than was observed with SDF-1α, whose receptor (CXCR4) was also expressed by Molt-4 cells (Fig. 10). MDC did not induce Ca2+ flux in Molt-4 cells, which do not express CCR4, the receptor for MDC.
TECK Binds to GPR-9-6+ Molt-4 Cells and Blocks the Binding of Anti–GPR-9-6 mAb 3C3 to GPR-9-6+ Cells.
As TECK was chemotactic for GPR-9-6–expressing cells, we evaluated whether TECK binds to GPR-9-6. Although tyrosine-iodinated TECK did not bind to GPR-9-6+ Molt-4 cells, biotinylated TECK bound in a dose-dependent and saturable manner. In contrast, biotinylated TECK did not bind to GPR-9-6− CEM cells (Fig. 11 A). Binding of biotinylated TECK to Molt-4 cells was inhibited by either 500 nM TECK or by the anti–GPR-9-6 mAb 3C3, but not by either 500 nM MCP-1 or an IgG2b control mAb (Fig. 11 B).
Figure 11.
Biotinylated TECK binds to Molt-4 cells. (A) Molt-4 and CEM cells were tested for their ability to bind to biotinylated TECK used at concentrations of 100, 50, 25, 12.5, 6.25, and 3.125 nM. (B) We also examined the effect of preincubating the cells for 10 min with 500 nM TECK, 500 nM MCP-1, and 50 ug/ml anti–GPR-9-6 mAb 3C3 or an IgG2b mAb on the binding of Molt-4 and CEM cells with biotinylated TECK (n = 2).
Preincubation of GPR-9-6 transfectants, CD4 cells, and B lymphocytes with TECK inhibited the staining with anti–GPR-9-6 mAb 3C3, whereas MDC had no effect (Fig. 12). Although these experiments were performed at 4°C and with azide, in the absence of a nonblocking anti–GPR-9-6 mAb, we cannot be certain of whether these results reflect either blocking or downmodulation of the chemokine receptor. However, these data do demonstrate that the staining pattern we observe with mAb 3C3 is not due to nonspecific binding of mAb 3C3 to lymphocytes.
Figure 12.
TECK blocks the binding of mAb 3C3 to GPR-9-6 on transfectants and lymphocytes. (A) L1.2/GPR-9-6 transfectants were preincubated with various concentrations of TECK or MDC on ice in PBS with 5% FCS, 10% human serum, and 0.2% azide for 10 min before the addition of mAb 3C3. (B) Mononuclear cells were treated in a similar manner but with either 500 nM MDC or 500 nM TECK. Cells were then stained as described in Materials and Methods (n = 2).
Tissue Distribution of TECK and GPR-9-6 Transcripts.
Due to the expression of GPR-9-6 on mucosal homing lymphocytes, we examined the distribution of TECK and GPR-9-6 transcripts in lymphoid and mucosal tissues (Fig. 13). TECK was selectively expressed in thymus and small intestine (Fig. 13 A). GPR-9-6 was expressed at high levels in thymus and weakly in spleen and PBLs (Fig. 13 B). Although we could not detect GPR-9-6 transcripts by Northern blot analysis in small intestine, we were able to detect GPR-9-6 message in small intestine and thymus using the more sensitive technique of RT-PCR (Fig. 13 C). Message for either gene was not detected in brain or colon. Also as predicted from our expression data on cell lines with mAb 3C3, GPR-9-6 was expressed by Molt-13 cells (and Molt-4 cells, data not shown) but not in GPR-9-6− JY, KU812, and EOL cells. In additional Northern blot analysis, TECK and GPR-9-6 were not detected in Th1, Th2, or T regulatory type 1 lymphocytes, LAK (lymphokine-activated NK) cells, monocytes, CD34-derived dendritic cells, monocyte-derived dendritic cells, astrocytes, high umbilical vein endothelial cells, or pulmonary vein endothelial cells (data not shown).
Figure 13.
Tissue distribution of TECK and GPR-9-6. (A) TECK hybridizations: 32P-labeled TECK DNA was used to probe multitissue Northern blot filters (2 μg poly-A RNA per lane, 24-h exposure; Clontech). (B) GPR-9-6 hybridizations: 32P-labeled GPR-9-6 DNA was used to probe the same filters as in A as well as a multi-cell line Northern blot filter (72-h exposure). (C) Equivalent amounts (0.5 ng) of primary cDNA (Clontech) from colon, small intestine, brain, and thymus, as well as 500 ng genomic DNA were amplified in PCR (30 cycles) using GPR-9-6 and TECK primers. G3PDH primers and G3PDH intron B primers were used as controls.
Discussion
Selective expression of adhesion molecules on T lymphocyte subsets mediates trafficking to distinct physiologic locations, such as peripheral LNs 37, intestinal sites 15 38, and different tissue inflammatory sites 13 39 40 41. It is thought that specific chemokine receptors expressed on these lymphocyte subsets interact with chemokines expressed in these areas to mediate leukocyte activation, arrest, and transendothelial migration. Therefore, it is possible that lymphocyte subsets defined by their expression of certain adhesion molecules may also express known, orphan, or as yet undiscovered chemokine receptors that will be important in trafficking of the lymphocytes into these sites. Our work describes one such chemokine receptor that is selectively expressed at high levels on thymocytes, gut-homing memory CD4 and CD8 lymphocytes, and intestinal mucosal lymphocytes.
GPR-9-6 was originally chosen as a potentially interesting orphan chemokine receptor due to its strong phylogenetic linkage with other known chemokine receptors including CCR6 and CCR7. To evaluate its function, we generated GPR-9-6 transfectants and performed studies to determine which chemokines induced chemotaxis of GPR-9-6+ cells. Out of all the chemokines tested, only TECK 42 acted as a chemoattractant for GPR-9-6/L1.2 transfectants. In contrast, TECK did not induce chemotaxis of L1.2 cells transfected with CCR1,2,4–7 or 9 or CXCR1–4 or 5. Although TECK also induced chemotaxis of CCR3/L1.2 transfectants, this was a relatively weak effect, inducing only 20% of the chemotactic mobility observed with eotaxin-1. Thus, TECK appears to have a restricted effect. In addition, in examining tumor cell lines for chemotaxis to TECK, GPR-9-6 expression directly correlated with chemotaxis. TECK-mediated chemotaxis of GPR-9-6+ transfectants, tumor cell lines, thymocytes, and CD4 lymphocytes was inhibited by anti–GPR-9-6 mAb 3C3. Thus, GPR-9-6 appears to represent the main chemokine receptor through which TECK acts on these cell populations, and to be a selective receptor for TECK. Like other chemokine receptors, the chemotaxis of GPR-9-6/L1.2 transfectants to TECK appears to be mediated exclusively through Gi signaling, as treatment with pertussis toxin abolishes transfectant migration to TECK.
In dose–response curves, 150 nM TECK resulted in optimal chemotaxis of GPR-9-6–expressing transfectants. This falls into the range of 1 nM to 1 μM at which other leukocyte chemokines are active. However, as a recombinant TECK produced by E. coli (PeproTech) was used for these studies, it remains possible that proper posttranslational modifications of the chemokine may not have occurred and that the recombinant TECK has a somewhat different activity than naturally produced TECK. In addition, further cleavage of TECK by factors outside the cell in vivo could generate more active fragments, as is the case for CKβ8 43. Thus, the actual dose–response curve in vivo may differ from that determined using the reagents currently available.
In published reports and in our own analysis, TECK is prominently expressed in the thymus. Therefore, we evaluated whether GPR-9-6 is expressed on thymocytes. Indeed, GPR-9-6 mRNA was detected prominently in thymic tissue based on Northern blot analysis. In addition, based on two- and three-color flow cytometry, GPR-9-6 polypeptide was expressed on the cell surface of the majority of thymocytes bearing all levels of TCR. Thus, GPR-9-6 is apparently expressed at all stages of T lymphocyte development. However, there was a small subpopulation of TCRhigh thymocytes that lacked GPR-9-6 expression. Further, most naive peripheral blood T lymphocytes derived from adults were either GPR-9-6− or expressed low levels of GPR-9-6 based on FACS® analysis. Thus, it appears likely that GPR-9-6 is downregulated near the time of thymocyte exit to the periphery. The expression of both GPR-9-6 and its ligand in the thymus demonstrates that TECK and GPR-9-6 probably interact in vivo as well as in vitro. This chemokine receptor–ligand pair is likely to play a role in thymocyte development, perhaps through effects on thymocyte localization or activation.
TECK was originally cloned from fetal intestine 42, and our Northern blot analysis revealed abundant TECK mRNA expressed in small intestine tissue derived from adults. Thus, it seemed possible that TECK might also function to recruit circulating lymphocytes to the intestine. To evaluate this possibility, we determined the expression of GPR-9-6 on PBLs. Although most PBLs lacked cell surface GPR-9-6, there was a small subset of peripheral T lymphocytes that was GPR-9-6+. In three-color experiments, GPR-9-6 was found predominantly on memory CD4 and CD8 lymphocytes that coexpressed high levels of the gut-homing receptor, α4β7. However, in the case of CD8 lymphocytes, we observed a greater correlation of GPR-9-6 and αE expression. GPR-9-6+ T lymphocytes also lack expression of CLA, which is consistent with a possible role of TECK in gut T lymphocyte localization. Furthermore, TECK induced chemotaxis of a small subset of CD4 lymphocytes that were enriched in the β7+ memory CD4/CD8 subset that expresses GPR-9-6. In addition, although we did not detect chemotaxis of total CD8 lymphocytes to TECK, we were able to identify a subset of αE+α4β7+CD45RAlow/− CD8 lymphocytes that was enriched in the population of cells that migrated in response to TECK, suggesting that TECK induced migration of this CD8 lymphocyte subset. Thus, the GPR-9-6 expressed on memory CD4 and CD8 lymphocytes is functional to induce chemotaxis. In contrast to GPR-9-6 expression, CCR4 is expressed on CLA+ or CLA− α4β7− circulating memory CD4 lymphocytes 13 38 44, but not on α4β7high cells. Thus, CCR4 and GPR-9-6 may define nonoverlapping subsets of memory CD4 lymphocytes that traffic to different sites.
GPR-9-6 was also expressed on a substantial proportion of B lymphocytes in peripheral blood, both with 3C3 and also with a second anti–GPR-9-6 mAb (data not shown), and staining of B lymphocytes with 3C3 was blocked by TECK. However, B lymphocytes were not chemotaxic for TECK. This may reflect either the reduced motility of these cells or the failure of GPR-9-6 to mediate chemotaxis of B lymphocytes, as observed for CCR1 on neutrophils. Although the evidence suggests that a subset of B lymphocytes that express GPR-9-6 does exist, the function of GPR-9-6 on these cells is not clear.
If GPR-9-6 plays a role in the localization of T lymphocytes to the intestine, its expression might be enhanced on T lymphocytes in this tissue site. Indeed, while we did not detect GPR-9-6 transcripts in small intestine using relatively less sensitive Northern blot analysis, GPR-9-6 transcripts were detected in small intestinal tissue via RT-PCR. Further, flow cytometry revealed GPR-9-6 expression on almost all intestinal IELs and lamina propria T lymphocytes isolated from normal individuals undergoing gastric bypass surgery. Thus, expression of GPR-9-6 was greatly enriched on CD4 and CD8 lymphocytes present in intestinal tissue compared with its expression on peripheral T lymphocytes.
The expression of TECK in small intestine and of GPR-9-6 on a subset of intestinal trafficking T lymphocytes indicates that TECK may play a role in the trafficking of GPR-9-6+ CD4 and CD8 lymphocytes to intestinal sites. It is possible that expression of TECK by postcapillary venules in the lamina propria or by Peyer's patch HEVs may activate GPR-9-6+ T lymphocytes and lead to leukocyte arrest via α4β7–mucosal addressin cell adhesion molecule 1 (MAdCAM-1) interactions, and thus in the selective extravasation of memory intestinal T lymphocytes within the intestine. Additionally, as the majority of resident intestinal IELs and LPLs express GPR-9-6, TECK may play a role in their localization within intestinal tissue or effector action at these sites. For example, it is possible that selective expression of TECK by epithelial or other cells in the small intestine could direct GPR-9-6+ T lymphocyte migration after the cells have crossed the endothelium and entered the intestinal tissue. Finally, although the majority of CD4 lymphocytes that express GPR-9-6 are memory cells, a small subset of naive phenotype CD4 lymphocytes also expresses GPR-9-6, albeit at lower levels. These may represent T lymphocytes that have recently emigrated from the thymus. Alternatively, a subpopulation of naive T lymphocytes may traffic to sites of TECK expression in the small intestine. It is of interest that while most GPR-9-6high memory CD4 lymphocytes do not express CD62L, GPR-9-6low naive CD4 lymphocytes do express CD62L. High levels of expression of α4β7 (such as those observed on memory CD4 lymphocytes) are thought to be sufficient to mediate lymphocyte tethering and rolling via MAdCAM-1 on lamina propria as well as HEVs in Peyer's patches. However, low levels of expression of this integrin (such as those observed on naive CD4 lymphocytes) are not thought to be sufficient 16 17. It is possible that the coexpression of CD62L and low levels of α4β7 on GPR-9-6low naive CD4 lymphocyte could allow initiation of rolling on Peyer's patch HEVs, with resulting arrest after lymphocyte activation and LFA-1 and α4β7 avidity upregulation.
We postulate that factors present in the mucosal environment lead to the induction of GPR-9-6 on T lymphocytes as well as TECK expression. Cytokines present in Th1/Th2 environments, at least in vitro, induce expression of certain chemokine receptors, such as CCR4 on Th2 and CXCR3 on Th1 lymphocytes, as well as the production of the chemokines that bind these receptors 25 26 27 28 29 30. However, these conditions did not upregulate GPR-9-6 expression on T lymphocytes. Also, our attempts to induce expression of GPR-9-6 on activated umbilical CD4 lymphocytes with cytokines IL-1–13, IL-15, IL-17, IL-18, and TGF-β, previously shown to induce αE on T lymphocytes 45, failed to identify a cytokine that upregulates GPR-9-6 expression. Therefore, there must be an as yet undefined mechanism by which GPR-9-6 expression is controlled on T lymphocytes.
In addition to studying the effect of cytokines on chemokine receptor expression, we wanted to determine if GPR-9-6 was modulated after T lymphocyte stimulation via antigen receptor cross-linking. Upon activation via TCR cross-linking, expression of GPR-9-6 is downregulated, as are other chemokine receptors such as CCR5, CCR6, and CXCR4 46. When the TCR is disengaged, GPR-9-6 is reexpressed by the T lymphocyte. As TCR cross-linking mimics antigen presentation, we conclude that upon TCR cross-linking via appropriate MHC class II–peptide complexes expressed by APCs, T lymphocytes will downregulate chemokine receptors such as GPR-9-6. By this method, T lymphocytes will be held in association with either dendritic cells or B lymphocytes until the relevant signals involved in antigen presentation or T–B cognate interactions have occurred.
In summary, we have shown that orphan chemokine receptor GPR-9-6 is expressed on the majority of thymocytes, intestinal IELs, LPLs, and on discrete subsets of T lymphocytes that traffic to intestinal sites. The CC chemokine TECK interacts with GPR-9-6 and mediates the chemotaxis of cells bearing this receptor. Based on the demonstration that GPR-9-6 induces chemotaxis of cell populations in a dose-dependent fashion in response to a chemokine, we propose to name this orphan chemokine receptor CCR-9. These findings, together with the selective expression of TECK and GPR-9-6 in thymus and small intestine, imply a dual role for GPR-9-6, both in T cell development and in the intestinal immune response.
While this manuscript was in submission, Zaballos et al. 47 reported that TECK acts as a chemoattractant for HEK293/human GPR-9-6 transfectants as well as for the Molt-4 cell line.
Acknowledgments
We thank Dr. Walter Newman for his advice and critical analysis of this manuscript. We thank Dr. Steve Roth of Boston Children's Hospital for assistance in obtaining thymus samples. We would like to acknowledge Suzanne Densmore, Sam Massoni, and Erin Scanlon for their contributions in chemokine synthesis, and Vilmos Csizmadia, Melissa Goodwille, and April Miao for their technical advice. We also acknowledge Dr. Yieh-Ping Wan for his efforts in generating radiolabeled TECK.
This work was supported in part by a grant from the Swedish Foundation for International Cooperation in Research and Higher Education (STINT) to W.W. Agace, and by grants from the National Institutes of Health to C.M. Parker (DK52978), E.C. Ebert (DK42166), and E.C. Butcher (AI37832, GM37734).
Footnotes
Abbreviations used in this paper: CLA, cutaneous lymphocyte–associated antigen; G3PDH, glyceraldehyde 3-phosphate dehydrogenase; GPR, human G protein–coupled receptor; HEV, high endothelial venule; IEL, intraepithelial lymphocyte; IP-10, IFN-γ–inducible 10-kD protein; I-TAC, IFN-inducible T cell α chemoattractant; LPL, lamina propria lymphocyte; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIG, monokine induced by IFN-γ; MIP, macrophage inflammatory protein; RANTES, regulated upon activation, normal T cell expressed and secreted protein; SDF, stromal cell–derived factor; TARC, thymus and activation-regulated chemokine; TECK, thymus-expressed chemokine.
B.A. Zabel and W.W. Agace contributed equally to this work.
References
- Taub D.D., Oppenheim J.J. Chemokines, inflammation and the immune system. Ther. Immunol. 1994;1:229–246. [PubMed] [Google Scholar]
- Mackay C.R. Chemokine receptors and T cell chemotaxis. J. Exp. Med. 1996;184:799–802. doi: 10.1084/jem.184.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub D.D., Anver M., Oppenheim J.J., Longo D.L., Murphy W.J. T lymphocyte recruitment by interleukin-8 (IL-8) J. Clin. Invest. 1996;97:1931–1941. doi: 10.1172/JCI118625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes M.E., Durham S.K., Swerdel M.R., Lewin A.C., Barton D.S., Megil J.R., Bravo R., Lira S.A. Controlled recruitment of monocytes and macrophages to specific organs through transgenic expression of monocyte chemoattractant protein-1. J. Immunol. 1995;155:5769–5776. [PubMed] [Google Scholar]
- Bargatze R.F., Butcher E.C. Rapid G protein–regulated activation event involved in lymphocyte binding to high endothelial venules. J. Exp. Med. 1993;178:367–372. doi: 10.1084/jem.178.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido N., Mukaida N., Harada A., Nakanishi I., Watanabe Y., Matsushima K. Prevention of lung reperfusion injury in rabbits by a monoclonal antibody against interleukin-8. Nature. 1993;365:654–657. doi: 10.1038/365654a0. [DOI] [PubMed] [Google Scholar]
- Karpus W.J., Lukacs N.W., McRae B.L., Strieter R.M., Kunkel S.L., Miller S.D. An important role for the chemokine macrophage inflammatory protein-1 alpha in the pathogenesis of the T cell-mediated autoimmune disease, experimental autoimmune encephalomyelitis. J. Immunol. 1995;155:5003–5010. [PubMed] [Google Scholar]
- Gupta S.K., Hassel T., Singh J.P. A potent inhibitor of endothelial cell proliferation is generated by proteolytic cleavage of the chemokine platelet factor 4. Proc. Natl. Acad. Sci. USA. 1995;92:7799–7803. doi: 10.1073/pnas.92.17.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Kim Y.J., Pollok K., Hurtado J., Lee J.L., Broxmeyer H.E., Kwon B.S. Macrophage inflammatory protein-1 alpha rapidly modulates its receptors and inhibits the anti-CD3 mAb-mediated proliferation of T lymphocytes. J. Immunol. 1993;151:4333–4341. [PubMed] [Google Scholar]
- Taub D.D., Turcovski-Corrales S.M., Key M.L., Longo D.L., Murphy W.J. Chemokines and T lymphocyte activation. I. Beta chemokines costimulate human T lymphocyte activation in vitro . J. Immunol. 1996;156:2095–2103. [PubMed] [Google Scholar]
- Choe H., Farzan M., Sun Y., Sullivan N., Rollins B., Ponath P.D., Wu L., Mackay C.R., LaRosa G., Newman W. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Feng Y., Broder C.C., Kennedyu P.E., Berger E.A. HIV-1 entry cofactorfunctional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Butcher E.C., Picker L.J. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- Berg E.L., Yoshino T., Rott L.S., Robinson M.K., Warnock R.A., Kishimoto T.K., Picker L.S., Butcher E.C. The cutaneous lymphocyte antigen is a skin lymphocyte homing receptor for the vascular lectin endothelial cell leukocyte adhesion molecule-1. Nature. 1991;174:1461–1466. doi: 10.1084/jem.174.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman A., Andrew D.P., Jablonski-Westrich D., Holzmann B., Butcher E.C. Role of α4 integrins in lymphocyte homing to mucosal tissues in vivo . J. Immunol. 1994;152:3282–3292. [PubMed] [Google Scholar]
- Bargatze R.F., Jutila M.A., Butcher E.C. Distinct roles of L-selectin and integrins α4β7 and LFA-1 in lymphocyte homing to Peyer's patch-HEV in situthe multistep model confirmed and refined. Immunity. 1995;3:99–108. doi: 10.1016/1074-7613(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Campbell J.J., Hedrick J., Zlotnik A., Siani M.A., Thompson D.A., Butcher E.C. Chemokines and the arrest of lymphocytes rolling under flow conditions. Science. 1998;279:381–383. doi: 10.1126/science.279.5349.381. [DOI] [PubMed] [Google Scholar]
- Chang K.H., Broxmeyer H.E. Chemokinessignal lamps for trafficking of T and B cells for development and effector function. J. Leukoc. Biol. 1999;65:6–15. doi: 10.1002/jlb.65.1.6. [DOI] [PubMed] [Google Scholar]
- Qin S., LaRosa G., Campbell J.J., Smith-Heath H., Kassam N., Shi X., Zeng L., Butcher E.C., Mackay C.R. Expression of monocyte chemoattractant protein-1 and IL-8 receptors on subsets of T cellscorrelation with transendothelial chemotactic potential. Eur. J. Immunol. 1996;26:640–647. doi: 10.1002/eji.1830260320. [DOI] [PubMed] [Google Scholar]
- Qin S., Rottman J.B., Myers P., Kassam N., Weinblatt M., Loetscher M., Koch A.E., Moser B., Mackay C.R. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Invest. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F., Rabin R.L., Smith C.R., Sharma G., Nutman T.B., Farber J.M. CC-chemokine receptor 6 is expressed on diverse memory subsets of T cells and determines responsiveness to macrophage inflammatory protein 3α. J. Immunol. 1999;162:186–194. [PubMed] [Google Scholar]
- Gunn M.D., Tangemann K., Tam C., Cyster J.G., Rosen S.D., Williams L.T. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc. Natl. Acad. Sci. USA. 1998;95:258–263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo J.A., Lloyd C.M., Kremer L., Finger E., Martinez-A C., Siegelman M.H., Cybulsky M., Gutierrez-Ramos J.C. Eosinophil recruitment to the lung in a murine model of allergic inflammation. The role of T cells, chemokines, and adhesion receptors. J. Clin. Invest. 1996;98:2332–2345. doi: 10.1172/JCI119045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedla N., Wang H.W., McNeil H.P., Di Girolamo N., Hampartzoumian T., Wakefield D., Lloyd A. Regulation of T lymphocyte trafficking into lymph nodes during an immune response by the chemokines macrophage inflammatory protein (MIP)-1α and MIP-1β. J. Immunol. 1998;161:5663–5672. [PubMed] [Google Scholar]
- Bonecchi R., Bianchi G., Bordignon P.P., D'Ambrosio D., Lang R., Borsatti A., Sozzani S., Allavena P., Gray P.W., Mantovani A., Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Mackay C., Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Mackay C.R., Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- Andrew D.P., Chang M.S., McNinch J., Walthen S.T., Rihanek M., Tseng J., Spellberg J.P., Elias C.G., III. STCP-1 (MDC) CC chemokine acts specifically on chronically activated Th2 lymphocytes and is produced by monocytes on stimulation with Th2 cytokines IL-4 and IL-13. J. Immunol. 1998;161:5027–5038. [PubMed] [Google Scholar]
- Zingoni A., Soto H., Hedrick J.A., Stoppacciaro A., Storlazzi C.T., Sinigaglia F., D'Ambrosio D., O'Garra A., Robinson D., Rocchi M. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J. Immunol. 1998;161:547–555. [PubMed] [Google Scholar]
- Luster A.D., Unkeless J.C., Ravetch J.V. Interferon gamma transcriptionally regulates an early response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- Ebert E.C., Roberts A.I., Brolin R.E., Raska K. Examination of the low proliferative capacity of human jejunal intra-epithelial lymphocytes. Clin. Exp. Immunol. 1986;65:148–157. [PMC free article] [PubMed] [Google Scholar]
- Clark-Lewis I., Moser B., Walz A., Baggiolini M., Scott G.J., Aebersold R. Chemical synthesis, purification, and characterization of two inflammatory proteins, neutrophil activating peptide 1 (interleukin-8) and neutrophil activating peptide. Biochemistry. 1991;30:3128–3135. doi: 10.1021/bi00226a021. [DOI] [PubMed] [Google Scholar]
- Murphy E., Shibuya K., Hosken N., Openshaw P., Maino V., Davis K., Murphy K., O'Garra A. Reversibility of T helper 1 and 2 populations is lost after long term stimulation. J. Exp. Med. 1997;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.J., Bowman E.A., Murphy K., Youngman K.R., Siani M.A., Thompson D.A., Wu L., Zlotnik A., Butcher E.C. 6-C-kine (SLC), a lymphocytes adhesion-triggering chemokine expressed by high endothelium, is an agonist for the MIP-3β receptor CCR7. J. Cell Biol. 1998;141:1053–1059. doi: 10.1083/jcb.141.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular CloningA Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; Plainview, NY: 1989. [Google Scholar]
- Ponath P.D., Qin S., Post T.W., Wang J., Wu L., Gerard N.P., Newman W., Gerard C., Mackay C. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J. Exp. Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallitin W.M., Weissman I.L., Butcher E.C. A cell surface molecule involved in organ specific homing of lymphocytes. Nature. 1983;304:30–34. doi: 10.1038/304030a0. [DOI] [PubMed] [Google Scholar]
- Andrew D.P., Rott L.S., Kilshaw P.J., Butcher E.C. Distribution of α4β7 and αEB7 integrins on thymocytes, intestinal epithelial lymphocytes and peripheral lymphocytes. Eur. J. Immunol. 1996;26:897–905. doi: 10.1002/eji.1830260427. [DOI] [PubMed] [Google Scholar]
- Frenette P.S., Mayadas T.N., Reyburn H., Hynes R.O., Wagner D.D. Susceptibility to infection and altered hematopoiesis in mice deficient in both P and E selectins. Cell. 1996;84:563–574. doi: 10.1016/s0092-8674(00)81032-6. [DOI] [PubMed] [Google Scholar]
- Tietz W., Allemand Y., Borges E., von Laer D., Hallmann R., Vestweber D., Hamann A. CD4 T cells migrate into inflamed skin only if they express ligands for E- and P-selectin. J. Immunol. 1998;161:963–970. [PubMed] [Google Scholar]
- Picker L.J., Terstappen L.W., Rott L.S., Streeter P.R., Stein H., Butcher E.C. Differential expression of homing associated adhesion molecules by T cell subsets in man. J. Immunol. 1990;145:3247–3255. [PubMed] [Google Scholar]
- Vicari A.P., Figuero D.J., Hedrick J.A., Foster J.S., Singh K.P., Menan S., Copeland N.G., Gilbert D.J., Jenkins N.A., Bacon K.B., Zlotnik A. TECKa novel CC chemokine specifically expressed by thymic dendritic cells and potentially involved in T cell development. Immunity. 1997;7:291–301. doi: 10.1016/s1074-7613(00)80531-2. [DOI] [PubMed] [Google Scholar]
- Macphee C.H., Appelbaum E.R., Johanson K., Moores K.E., Imburgia C.S., Fornwald J., Berkhout T., Brawner M., Groot P.H.E., O'Donnell K. Identification of a truncated form of the CC chemokine CKB-8 demonstrating greatly enhanced biological activity. J. Immunol. 1998;161:6273–6279. [PubMed] [Google Scholar]
- Campbell J.J., Haraldsen G., Pan J., Rottman J., Qin S., Ponath P., Andrew D.P., Warnke R., Ruffing N., Kassam N., Butcher E.C. The chemokine receptor CCR4 in vascular recognition by cutaneous but not intestinal memory T cells. Nature. 1999;400:776–780. doi: 10.1038/23495. [DOI] [PubMed] [Google Scholar]
- Kilshaw P.J., Murant S.J. Expression and regulation of β7(βp) integrins on mouse lymphocytesrelevance to the mucosal immune system. Eur. J. Immunol. 1991;21:2591–2597. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- Bermejo M., Martin-Serrano J., Oberlin E., Pedraza M.-A., Serrano A., Santiago B., Caruz A., Loetscher P., Baggiolioni M., Arenzana-Seisdedos F., Alcami J. Activation of blood T lymphocytes down-regulates CXCR4 expression and interferes with propagation of X4 HIV strains. J. Immunol. 1998;28:3192–3204. doi: 10.1002/(SICI)1521-4141(199810)28:10<3192::AID-IMMU3192>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Zaballos A., Gutierrez J., Varona R., Ardavin C., Marquez G. Identification of the orphan chemokine receptor GPR-9-6 as CCR9, the receptor for the chemokine TECK. J. Immunol. 1999;162:5671–5675. [PubMed] [Google Scholar]