Abstract
Mycobacterium tuberculosis, the causative agent of tuberculosis, possesses a class Ib ribonucleotide reductase (RNR), encoded by the nrdE and nrdF2 genes, in addition to a putative class II RNR, encoded by nrdZ. In this study we probed the relative contributions of these RNRs to the growth and persistence of M. tuberculosis. We found that targeted knockout of the nrdF2 gene could be achieved only in the presence of a complementing allele, confirming that this gene is essential under normal, in vitro growth conditions. This observation also implied that the alternate class Ib small subunit encoded by the nrdF1 gene is unable to substitute for nrdF2 and that the class II RNR, NrdZ, cannot substitute for the class Ib enzyme, NrdEF2. Conversely, a ΔnrdZ null mutant of M. tuberculosis was readily obtained by allelic exchange mutagenesis. Quantification of levels of nrdE, nrdF2, nrdF1, and nrdZ gene expression by real-time, quantitative reverse transcription-PCR with molecular beacons by using mRNA from aerobic and O2-limited cultures showed that nrdZ was significantly induced under microaerophilic conditions, in contrast to the other genes, whose expression was reduced by O2 restriction. However, survival of the ΔnrdZ mutant strain was not impaired under hypoxic conditions in vitro. Moreover, the lungs of B6D2/F1 mice infected with the ΔnrdZ mutant had bacterial loads comparable to those of lungs infected with the parental wild-type strain, which argues against the hypothesis that nrdZ plays a significant role in the virulence of M. tuberculosis in this mouse model.
Mycobacterium tuberculosis is a formidable human pathogen that is estimated to infect one-third of the world's population (2). The success of this pathogen is attributable to its remarkable ability to persist for prolonged periods in a clinically latent state from which it is able to reactivate and cause disease. During the course of infection in humans, the tubercle bacillus is likely to encounter environments where there is limited O2 availability, most notably the fibrous granulomas (9). This notion has underpinned efforts to identify the metabolic changes that occur in M. tuberculosis in response to hypoxia as a means of modeling the changes that may be associated with clinical latency (13, 37, 44). The value of this approach was underscored by a recent report (38) suggesting that during stationary infection in mice, the bacilli may be in a physiological state that approximates the nonreplicating persistence achieved in the widely used model of adaptation of M. tuberculosis to hypoxia in vitro developed by Wayne and coworkers (44, 45).
An important class of enzymes that includes both O2-dependent and O2-independent forms is the ribonucleotide reductases (RNRs), which catalyze the reduction of ribonucleotides to deoxyribonucleotides. These enzymes perform an essential role in the cycling of nucleotides in the cell and during replication of the chromosome in all organisms and provide attractive targets for antiproliferative drugs (40) and subunit vaccines (12). Although all RNRs contain an active free radical that is critical for catalytic activity, the essential metal cofactors have not been evolutionarily conserved. The oxygen-dependent, class I RNR enzymes are subdivided into classes Ia and Ib based on allosteric regulation and utilization of different electron donors (19). The class I RNRs consist of two homodimers in a α2β2 subunit structure, and the nrdAB and nrdEF genes encode the large α-chain (NrdA or NrdE) and small β-chain (NrdB or NrdF) subunits of class Ia and class Ib RNRs, respectively. In contrast, nrdJ-encoded class II RNRs are O2-independent α or α2 forms and use adenosylcobalamin as a radical generator, whereas the nrdDG-encoded class III RNRs contain an O2-sensitive glycyl radical generated by using S-adenosylmethionine. Despite these differences, common catalytic and allosteric mechanisms, as well as retention of critical residues in the protein sequence, suggest that the tertiary structures of all RNRs are similar and that all RNRs had a common evolutionary origin (reviewed in reference 34).
It has been proposed that the division of the classes based on O2 sensitivity and the presence in many organisms of more than one RNR may allow adaptation to different O2 levels in the environment (32, 34). Consistent with this notion is the documented responsiveness of expression of certain bacterial RNR-encoding genes to O2 availability (14, 24, 43); the regulation of this is poorly understood, but in the case of Pseudomonas stutzeri nrdD, expression appears to be controlled by an FNR-type regulator (43). However, the simultaneous presence of more than one class of active RNR in some organisms suggests that this may be a simplistic approach; for example, Streptomyces clavuligerus and Pseudomonas aeruginosa use both their class I and class II RNRs during aerobic growth (4, 21). Further complexity can also be provided by the presence of more than one enzyme belonging to the same class or subclass, such as the class Ia and class Ib RNRs of Escherichia coli (27), or by the presence of more than one large or small RNR subunit with nonoverlapping functions (17).
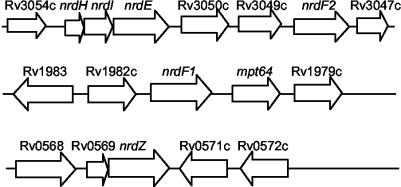
M. tuberculosis possesses a class Ib RNR encoded by nrdE (Rv3051c) and nrdF2 (Rv3048c) (5, 46), as well as a putative alternate small subunit encoded by nrdF1 (Rv1981c), which contains key catalytic residues but cannot associate with NrdE to form a functional RNR (46). In addition to these class Ib RNR-encoding genes, M. tuberculosis also contains a gene, nrdZ (Rv0570), which encodes a putative class II RNR (7) (Fig. 1). Significantly, transcription of this gene was shown recently to be dependent on DosR/DevR, the primary regulator of the hypoxic response in M. tuberculosis (31), suggesting that this organism might be capable of modulating deoxynucleoside triphosphate (dNTP) biosynthesis in response to changes in O2 tension. To investigate this possibility, we analyzed the contributions of the class Ib and class II RNRs to growth of M. tuberculosis by targeted knockout of the nrdF2 and nrdZ genes, nrd gene expression analysis by real-time, quantitative reverse transcription (RT)-PCR, and phenotypic characterization of a nrdZ mutant strain in a murine infection model.
FIG. 1.
Genomic organization of RNR-encoding genes in M. tuberculosis H37Rv. The gene notation is that of TubercuList (http://genolist.pasteur.fr/TubercuList/).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are shown in Table 1. The vectors pNRDF2KO and pΔNRDZ were used for allelic replacement experiments, and pNRDF2 was used for genetic complementation. pNRDF2KO comprised a 5,263-bp EcoRI-NotI fragment derived from an EcoR1 library of M. tuberculosis Erdman (46) that was subcloned into p2NIL (30). The nrdF2 allele was inactivated by insertion of the hygromycin B resistance cassette (hyg) from pIJ963 (1) into the unique BglII site 334 bp downstream from the start codon of the nrdF2 gene. The markers sacB and lacZ were cloned into this construct as a PacI cassette from pGOAL17 (30). A PCR-generated nrdZ amplicon was used to probe a gridded plasmid library of the M. tuberculosis genome (kindly provided by M. Everett, GlaxoSmithKline, Stevenage, United Kingdom). The pDWZ04 clone was found to contain the entire 2,076-bp nrdZ gene plus a 3,286-bp sequence downstream of the stop codon and 1,260 bp upstream of the start codon. A 1,568-bp PstI-SalI fragment and a 2,205-bp SalI-KpnI fragment were excised from pDWZ04 and cloned into the corresponding sites of p2NIL to obtain a vector containing a ΔnrdZ allele lacking the internal 871-bp SalI fragment that encodes three of the five cysteine residues shown to be essential for RNR activity in Lactobacillus leichmannii (3). The hyg-lacZ-sacB cassette from pGOAL19 (30) was cloned into the PacI site to obtain pΔNRDZ. The complementing plasmid, pNRDF2, was constructed by cloning a 1,876-bp SalI-HindIII fragment containing nrdF2 plus a flanking sequence consisting of 586 bp 5′ of nrdF2 and 331 bp 3′ of nrdF2 between the XhoI and HindIII sites of pGINT, which is a gentamicin-resistant (Gmr) derivative of the integrative vector, pHINT (29).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Promega, Madison, Wis. |
| H37Rv | Virulent laboratory isolate (ATCC 25618) | Laboratory collection |
| ΔnrdZ | Mutant of H37Rv carrying a deletion in the nrdZ gene (ΔnrdZ) | This study |
| Plasmids | ||
| p2NIL | Cloning vector; Kmr | 30 |
| pGOAL17 | Plasmid carrying lacZ and sacB genes as a PacI cassette; Apr | 30 |
| pGOAL19 | Plasmid carrying hyg, lacZ, and sacB genes as a PacI cassette; Apr | 30 |
| pHINT | E. coli-Mycobacterium integrating shuttle vector; Hygr Apr | 29 |
| pGINT | Derivative of pHINT carrying gentamicin resistance cassette; Gmr Apr | S. Durbach |
| pNRDF2KO | Knockout vector carrying nrdF2::hyg allele; Hygr Kmr | This study |
| pNRDF2 | Integrative complementing vector carrying nrdF2 cloned in pGINT; Gmr Apr | This study |
| pDWZ04 | Subclone from gridded library of M. tuberculosis cloned in pBluescript carrying nrdZ gene and flanking sequences; Apr | This study |
| pΔNRDZ | Knockout vector carrying ΔnrdZ allele; Hygr Kmr | This study |
Bacterial culture conditions.
M. tuberculosis strains were grown in Middlebrook 7H9 medium supplemented with 0.2% glycerol and 0.05% Tween 80 (18) in roller bottles or as stirred cultures. The antibiotic supplements used were kanamycin (25 μg/ml), hygromycin B (50 μg/ml), and gentamicin (10 μg/ml). E. coli DH5α was used for all cloning procedures and was grown in Luria broth or Luria agar with 100 μg of ampicillin per ml when necessary. The electroporation conditions used and the method used for selection of merodiploid and allelic replacement strains were the conditions and method described by Gordhan and Parish (15). Wayne model conditions were produced in round-bottom Pyrex tubes (capacity, 30 ml) which were filled with 20 ml of Dubos albumin broth (Difco) with 0.05% Tween 80 as described by Wayne and Sohaskey (44) to generate a headspace-to-volume ratio of 0.5. The Wayne model for adaptation of M. tuberculosis to hypoxia induces a sequential shiftdown of the organism through two stages of nonreplicating persistence (NRPI and NRPII) in response to a self-generated, temporal O2 gradient (45). In this model, the sealed culture replicates in a logarithmic fashion (log), but as O2 becomes limiting (concentration, <1%), replication ceases, and although the optical density continues to increase, the CFU counts do not (NRPI). The culture enters NRPII at O2 concentrations less than 0.06%, when no further increase in optical density is observed. The 21 tubes used were inoculated with a 1/100 dilution of a log-phase culture in fresh Dubos medium and sealed with Parafilm internally and externally. One tube contained 0.5 μg of methylene blue per ml as an indicator. These cultures were stirred slowly with 5-mm magnetic stir bars (Fisher Scientific, Pittsburgh, Pa.). Tubes were harvested in duplicate or triplicate and chilled on ice prior to measurement of the optical density at 600 nm and harvesting of cells for RNA isolation. MICs of hydroxyurea (HU) for parental and ΔnrdZ strains were determined by using BACTEC 460-TB methodology (33) and BACTEC 12B vials (Becton-Dickinson, Towson, Md.) containing cyanocobalamin (Sigma-Aldrich, St. Louis, Mo.) at a concentration of 1 μg/ml and concentrations of HU (Sigma-Aldrich) ranging from 1 to 50 mM.
Isolation of RNA.
RNA was isolated from 20-ml aliquots as described by Manganelli et al. (23) and was resuspended in 100 μl of diethyl pyrocarbonate-treated water. RNA was diluted approximately 10-fold prior to RT.
Quantification of RNA levels.
Primers and molecular beacons were designed by using the Primer3 software (35), the virtual PCR amplification software AMPLIFY (11), and the DNA M-fold program (36). All primers (Table 2) were tested for the ability to amplify 102 to 105 genome equivalents in a concentration-dependent manner. Molecular beacons (Eurogentec, Seraing, Belgium) were synthesized with a 5′ B-FAM group and a 3′-[4-(4-dimethylaminophenylazo)benzoic acid]succinimidyl ester group. RT primers (2.5 pmol each) were annealed to 4 μg of RNA by incubation at 94°C for 90 s, followed by 65°C for 3 min and 57°C for 3 min. RNA was reverse transcribed at 60°C for 30 min by using a Carboxydothermus hydrogenoformans two-step RT kit (Roche Molecular Biochemicals, Mannheim, Germany), and the enzyme was inactivated by incubation at 95°C for 5 min. One-tenth of the cDNA was used for real-time PCR analysis (duplicate 20-μl reaction mixtures) with each molecular beacon and amplification primer pair by using the Roche LightCycler system and FastStart DNA polymerase (both obtained from Roche Molecular Biochemicals) according to the manufacturer's instructions, and the amount was quantified by using genomic standards containing from 105 to 102 genome equivalents. HspX was quantified by using a LightCycler FastStart DNA Master SYBR Green 1 kit (Roche Molecular Biochemicals). The cycle conditions were as follows: 95°C for 10 min, followed by 15 cycles of 95°C for 0 s, 65°C for 10 s, and 72°C for 10 s and then 25 cycles of 95°C for 0 s, 57°C for 10 s, and 72°C for 10 s.
TABLE 2.
Oligonucleotides used in this study
| Application | Primer | Sequence (5′-3′)a |
|---|---|---|
| RT | nrdE-RT | cggcgtagacacttgcagga |
| nrdF2-RT | gcctcatagccgaggttcatc | |
| nrdF1-RT | acgatcgaatgcaggctggt | |
| nrdZ-RT | tcggtcacaccaaccgatag | |
| hspX-RT | aaccgccaccgacacagt | |
| sigA-RT | ctgacatgggggcccgctacgttg | |
| Amplification primer pairsb | nrdE-F1 | gatcaaggcacgggagttctt |
| nrdE-R1 | ttggattagcgcgattgacg | |
| nrdF2-F1 | gtctggcgttggttgacgac | |
| nrdF2-R1 | cgtcgtagaggtcctgggtgt | |
| nrdF1-F1 | gaccaccgcgaatacacctg | |
| nrdF1-R1 | cgcatgtagggcaaaacgtc | |
| nrdZ-F1 | cggaccggtgtcgtttctac | |
| nrdZ-R1 | cttggcggtgacgaaatcac | |
| hspX-F1 | ccgagcgcaccgagcagaag | |
| hspX-R1 | ggtggccttaatgtcgtcctcgtc | |
| sigA-F1 | tgcagtcggtgctggacac | |
| sigA-R1 | cgcgcaggacctgtgagcgg | |
| Molecular beacons | nrdE-MB | CCTCGCgagtccggctacccctatatcatgGCGAGG |
| nrdF2-MB | GCAGCGccgagctcaaggactacacctaCGCTGC | |
| nrdF1-MB | GCTCCCatcgactatgcgcacgacttgtacGGGAGC | |
| nrdZ-MB | GGACCCctgtatggctgtgcttgatgtgtcGGGTCC | |
| sigA-MB | CCTCGCgtcgaagttgcgccatccgaGCGAGG |
Lowercase letters indicate bases complementary to the M. tuberculosis sequence, whereas uppercase letters indicate bases added to form the stem of the molecular beacon.
F1, forward primer; R1, reverse primer.
Infection of mice, bacterial load, and survival.
Ten-week-old female B6D2/F1 mice (Charles River Laboratories, Wilmington, Mass.) were infected via the respiratory route as described previously (28). Mice were inoculated by exposure to an aerosolized suspension of the parental or ΔnrdZ strains of M. tuberculosis by using a nose-only exposure system (In-Tox Products, Albuquerque, N.M.) (42). This procedure resulted in implantation of approximately 100 organisms into the lungs of each mouse, which was confirmed by plating lung homogenates on Middlebrook 7H11 plates 3 h postinfection. At different times four mice from each group were sacrificed, and organs were harvested. Lungs, livers, and spleens were homogenized and plated to determine bacterial loads. A portion of the upper right lung was used for histological analysis. Five mice from each group were monitored for survival.
Statistical analysis.
Culture tubes from the Wayne model analysis were harvested in triplicate unless otherwise indicated. RT reactions were performed twice with each RNA preparation, and PCRs were performed in duplicate. One-way analysis of variance was used to determine the significance of differences between data sets.
RESULTS
Targeted knockout of the nrdF2 and nrdZ genes.
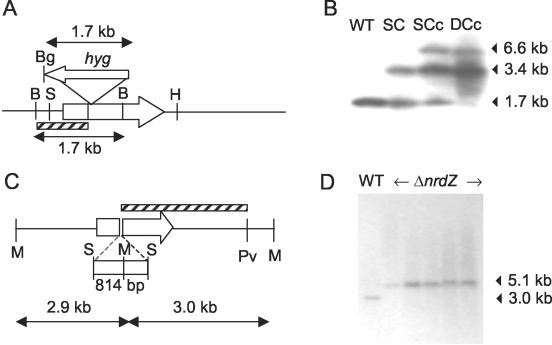
To investigate the physiological role of the NrdEF2 class Ib enzyme, allelic exchange mutagenesis of nrdF2 was attempted by using an nrdF2::hyg allele, which was constructed by insertion of a hygromycin resistance marker 333 bp downstream of the start codon of the gene (Fig. 2A). As expected, delivery of the mutant allele into the chromosome of M. tuberculosis H37Rv on the suicide plasmid pNRDF2KO yielded products of site-specific, single-crossover events. However, counterselection against sacB-containing single crossovers by plating on sucrose yielded no double crossovers. All sucrose-resistant clones were found to be spontaneous sacB mutants based on the following evidence: (i) all 27 clones were positive in a PCR amplification test for the Kmr cassette on the delivery vector (data not shown); and (ii) more than 200 clones produced blue colonies when they were plated on media containing the indicator X-Gal (5-bromo-4-chloro-3- indolyl-β-d-galactopyranoside), confirming the presence of a lacZ gene from the delivery vector (30). These results suggested that nrdF2 might encode an essential function. To test this hypothesis, a single functional copy of nrdF2 under the control of its own promoter was inserted at the attB locus. In the presence of the complementing gene, double crossovers were readily obtained at the chromosomal locus, as shown by the loss of the wild-type allele and the retention of the inactivated and complementing alleles in all 24 sucrose-resistant mutants (Fig. 2B). This result confirmed that nrdF2 is essential for growth of M. tuberculosis in vitro and was consistent with previous biochemical evidence suggesting that NrdF1 cannot substitute for NrdF2 to form a functional class Ib RNR in association with the large NrdE subunit (46).
FIG. 2.
Targeted knockout of RNR-encoding genes in M. tuberculosis. (A) Schematic representation of the inactivated nrdF2 allele showing the site of insertion of the hygromycin cassette, the position of the 1,126-bp BamHI-BglII probe (striped box) used for Southern blot analysis, and the extent of homologous DNA used in the knockout construct. (B) Southern blot analysis of nrdF2 recombinant strains. Chromosomal DNA from wild-type M. tuberculosis strain H37Rv (WT), a single-crossover recombinant carrying tandem copies of the inserted and wild-type alleles (SC), a single-crossover recombinant carrying a functional nrdF2 gene integrated at the attB locus (SCc), and a double-crossover recombinant with a crossover at the chromosomal locus of the nrdF2 allele arising from an SCc background (DCc) were digested with BamHI and probed with the BamHI-BglII probe containing the 5′ portion of the nrdF2 gene. (C) Schematic representation of the inactivated ΔnrdZ allele showing the 814-bp SalI deletion and the positions of the 2,011-bp SalI-PvuII probe (striped box) and the MluI sites used for Southern blot analysis. (D) Southern blot analysis of five ΔnrdZ allele replacement strains. Chromosomal DNA from wild-type M. tuberculosis H37Rv (WT) and five ΔnrdZ isolates were digested with MluI and probed with the SalI-PvuII probe shown in panel C. Abbreviations: Bg, BglII; H, HindIII; S, SalI; M, MluI; Pv, PvuII.
An additional implication of the finding that nrdF2 is essential is that NrdZ does not provide sufficient RNR activity under standard in vitro culture conditions to allow M. tuberculosis to grow in the absence of NrdEF2. The nrdZ gene was shown to be dispensable for growth of M. tuberculosis under these conditions by the successful recovery of allelic exchange mutants carrying an unmarked deletion allele, ΔnrdZ (Fig. 2C and D), in which an internal gene segment spanning the region encoding three of the five cysteine residues identified as important for catalysis in Lactobacillus leichmannii class II RNR (3) was deleted. The growth rates of the ΔnrdZ mutant and its parental wild type were indistinguishable in all phases under standard culture conditions (data not shown).
Comparative sensitivities of ΔnrdZ and wild-type strains to HU.
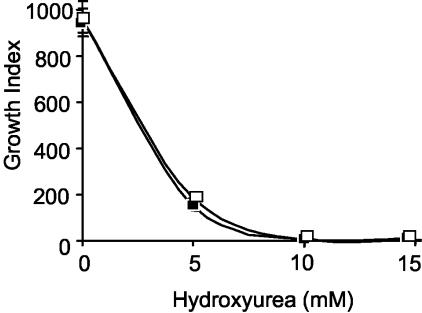
Class II RNRs require adenosylcobalamin as a cofactor. Several mycobacterial species, including Mycobacterium smegmatis, have been reported to synthesize cobalamin (22). Although bioinformatic analysis of the M. tuberculosis genome sequence has revealed the presence of homologues of most of the genes required to make this complex cofactor, it is not known if M. tuberculosis can actually synthesize cobalamin. To further investigate the contribution, if any, of nrdZ to growth and survival of M. tuberculosis under aerobic conditions, we compared the growth kinetics of the M. tuberculosis ΔnrdZ mutant to that of its parental wild type under conditions in which the class Ib activity was inhibited by the potent class I RNR inhibitor HU and cyanocobalamin was included in the medium in an attempt to ensure that there was sufficient adenosylcobalamin cofactor. The growth kinetics in the presence of HU (1 to 50 mM) were assessed radiometrically by using the BACTEC system. This analysis confirmed that HU inhibits the growth of M. tuberculosis H37Rv, with an MIC of 5 mM (Fig. 3), which is consistent with the susceptibility of the M. tuberculosis NrdEF2 enzyme to inhibition by this drug (46). However, no differential susceptibility to HU was observed for the two strains.
FIG. 3.
Inhibition of M. tuberculosis strains H37Rv and ΔnrdZ by HU, as determined by BACTEC susceptibility testing. Cultures were inoculated into Middlebrook 7H12 test medium supplemented with cyanocobalamin containing different concentrations of HU. Growth (which was directly proportional to the growth index) was monitored for 10 days. Symbols: ▪, H37Rv; □, ΔnrdZ.
Quantification of nrd RNA levels.
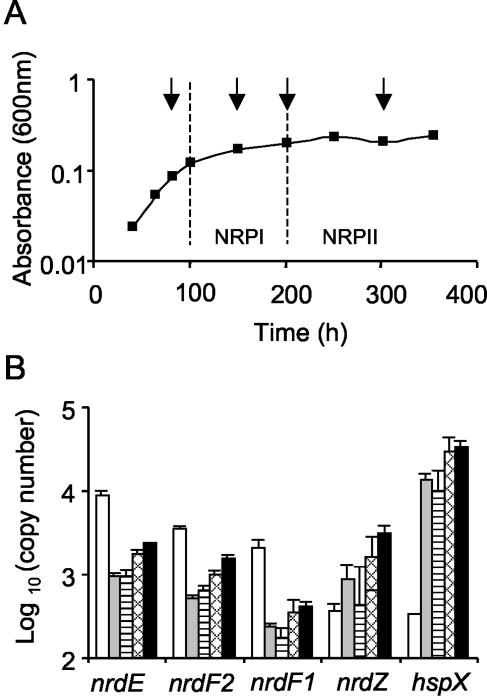
Since the O2 requirements of the class I and II RNRs are different, we examined the responses of expression of the various nrd genes to changes in the O2 tension of the culture media. Baseline levels of nrd expression were obtained from cultures of M. tuberculosis H37Rv grown with agitation (shaking) to the mid-log phase. RNA from such a mid-log-phase culture was subjected to real-time RT-PCR analysis with molecular beacons for all four nrd genes (Table 2). The hspX gene was included as a positive control for induction of message under low-O2-tension conditions (8, 10, 37). nrdF2 and nrdF1 were expressed at similar levels, but the levels were almost 2.5-fold lower than the levels of nrdE expression, whereas the level of nrdZ expression was 10-fold lower than the levels of expression of nrdF2 and nrdF1. RNA was then isolated at various stages from cultures grown under the conditions of the Wayne model (44, 45) (Fig. 4A). The mRNA levels were normalized by comparison with the levels of transcription of sigA, a housekeeping gene that has been shown to be constitutively transcribed under a number of environmental stress conditions, including O2 deprivation (16), and the significance was analyzed by using one-way analysis of variance (Fig. 4B). Interestingly, the levels of nrdE, nrdF1, and nrdF2 in the log phase and NRPI were markedly lower than the levels obtained with well-aerated cultures (ninefold, ninefold, and sevenfold lower, respectively; P < 0.001). In contrast, nrdZ transcription was induced such that the nrdE and nrdZ levels were approximately equal. Interestingly, despite an initial drop, transcription of nrdF2 and nrdE increased significantly, albeit slightly, as the O2 in the culture became progressively depleted through NRPI/II and NRPII (NRPI versus NRPII, P < 0.001), and the ratios of nrdE, nrdF2, and nrdZ remained constant. Although there were net reductions in the levels of nrdE, nrdF1, and nrdF2 in NRPII compared with the unstressed levels, nrdZ was upregulated eightfold compared with the unstressed levels. As expected, transcription of hspX responded positively to O2 depletion.
FIG. 4.
Quantitative analysis of nrd gene expression in M. tuberculosis cultured under the Wayne model conditions (44, 45). (A) Growth of M. tuberculosis H37Rv in slowly stirred sealed tubes, showing a gradual cessation of growth as available oxygen becomes limiting. Three independent tubes were harvested at most of the times indicated by arrows; the exception was the 202-h time point, when two tubes were harvested. (B) Relative abundance of message as determined by quantitative RT-PCR by using molecular beacons in aerated (stirred) Dubos medium (open bars) and under Wayne model conditions. Copies of mRNA were normalized to 1,000 copies of sigA mRNA and the values shown are the means and standard errors. Log-phase samples were taken at 83 h (grey bars), NRPI samples were taken at 151 h (striped bars), NRPI/II samples were taken at 202 h (cross-hatched bars), and NRPII samples were taken at 302 h (solid bars).
Survival of the ΔnrdZ mutant under microaerophilic conditions.
The induction of nrdZ under low-oxygen conditions led us to examine whether the ΔnrdZ mutant was impaired for survival under the conditions described for the stirred or settled Wayne model. However, the mutant displayed no impairment of survival compared with the parental wild-type strain in either model, as judged by plating efficiencies of aliquots taken 1 week after entry into NRPII in the stirred Wayne model or 6 months after entry into NRPII in the settled model (data not shown).
Survival of the ΔnrdZ mutant in mice.
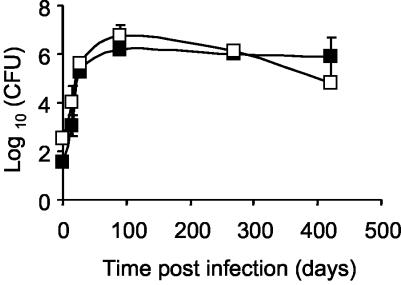
We next investigated whether loss of nrdZ had implications for survival of M. tuberculosis in mice infected by the aerosol route. B6D2/F1 mice, which are known to be resistant to infection with M. tuberculosis (26), were used in order to amplify a possible survival defect in ΔnrdZ. Approximately 100-CFU portions of the parental or ΔnrdZ strain of M. tuberculosis were implanted into the lungs of mice (Fig. 5). The numbers of CFU in the lungs of mice in both groups increased progressively until day 90 postinfection, and the levels reached about 6 log10 CFU. High numbers of viable mycobacteria persisted in the lungs for up to 14 months. No significant difference in the bacillary loads in the lungs was seen when we compared mice infected with the ΔnrdZ strain and mice infected with the wild-type M. tuberculosis H37Rv strain up to 14 months postinfection. Similarly, in both groups there were no significant differences in the numbers of CFU in the spleens and livers of the mice (data not shown), and histological analysis of lung samples obtained after 90 days and 9 months revealed comparable pathologies (data not shown).
FIG. 5.
Growth of the M. tuberculosis H37Rv and ΔnrdZ strains in B6D2/F1 mice. Mice were infected with M. tuberculosis by using an aerosol, and bacillary loads in the lungs were determined over a 14-month infection period, as described in Materials and Methods. Symbols: ▪, mice infected with the parental strain; □, mice infected with the ΔnrdZ strain. Each point represents the mean for four mice, and the error bars indicate the standard deviations.
DISCUSSION
In this study we used a combination of gene knockout and expression analysis to functionally characterize RNR-encoding genes in M. tuberculosis. The inability to inactivate nrdF2 in the absence of a complementing copy of this gene confirmed that the class Ib enzyme NrdEF2 is essential for growth of M. tuberculosis and thus represents a valid target for novel antitubercular drug development. The finding that nrdF2 is essential in M. tuberculosis is consistent with previous biochemical data for recombinant proteins, which suggested that NrdF1 could not substitute for NrdF2 to form a functional class Ib enzyme in association with NrdE (46). It has been shown that in Saccharomyces cerevisiae a second small subunit of the class Ib RNR, RNR4, although apparently without catalytic activity, is nonetheless essential for viability (17). Huang and Elledge postulated that this subunit has a structural or nonenzymatic regulatory role and suggested that the holoenzyme has a α2ββ′ form. However, an insertion in nrdF1 has been identified in a transposon library of M. tuberculosis (25), suggesting that the situation in M. tuberculosis does not parallel that in S. cerevisiae. The nrdF1 gene is expressed in M. tuberculosis at a level comparable to that of nrdF2, suggesting that it may serve a cellular function, albeit not a function directly associated with ribonucleotide reduction. Interestingly, although the genes encoding the two subunits of the class Ib RNR are not arranged in an operon, they appear to be coordinately regulated in response to increasing oxygen depletion, in contrast to nrdF1, which did not show the same response. This observation is analogous to the coordinate regulation of the nonoperon class Ib RNR-encoding genes that has been found to occur in Corynebacterium ammoniagenes (41).
The putative class II RNR found in M. tuberculosis (NrdZ) is more closely related to the cobalamin-dependent class II enzymes found in archaeal organisms, such as Archaeoglobus fulgidus (52% identity over 562 amino acids), than to eubacterial class II enzymes. The nrdZ gene also has a notable distribution within the genus Mycobacterium; genome sequencing has revealed identical genes in M. tuberculosis and M. bovis but no homologs in M. leprae, M. smegmatis, or M. avium. Comparison of the amino acid sequence of M. tuberculosis NrdZ with the amino acid sequences of functionally characterized archaeal and eubacterial class II enzymes revealed conservation of residues essential for catalysis, but in spite of its predicted activity as a functional RNR enzyme, NrdZ was unable to compensate for the loss of NrdEF2 in M. tuberculosis under normal in vitro growth conditions. This finding contrasts with the results obtained for S. clavuligerus (4), in which the class II enzyme NrdJ supplies most of the RNR activity during vegetative growth. Similarly, the class II RNR provides most of the activity in Deinococcus radiodurans, which also possesses a class Ib RNR (20). The expression level of the M. tuberculosis nrdZ gene under aerobic, in vitro growth conditions was 10-fold lower than that of the nrdF2 gene, suggesting that the failure of NrdZ to compensate for a loss of NrdEF2 might be due to inadequate production of a functional RNR enzyme. Alternatively, the M. tuberculosis nrdZ promoter might be responsive to signals other than changes in the dNTP pool, by analogy with the situation in E. coli, in which the nrdAB and nrdEF operons are regulated differently and in response to different environmental signals (27).
The differences in the expression levels of the nrd and hspX genes observed in stirred, continuously aerated cultures and in the log phase in the Wayne model indicate that M. tuberculosis modulates gene expression in response to even small changes in oxygen availability. Interestingly, after the initial precipitous drop in the nrdE and nrdF2 levels in the log phase of the Wayne model, slight but significant upregulation of both nrdE and nrdF2 was observed during passage from NRPI to NRPII, suggesting that the enzyme may still contribute activity even under extreme O2 limitation conditions. In contrast, nrdZ was highly and progressively upregulated in all stages of the Wayne model, with the highest level of nrdZ expression occurring in NRPII as the culture approached anaerobiosis. The levels of nrdZ remained elevated in NRPII and returned to normal after exit from this stage (data not shown). The nrdZ gene is located 29 bp downstream of a gene encoding a putative transcriptional regulator, Rv0569, and both genes are highly induced at an O2 tension of 0.2% (31, 37), suggesting that the two genes may form a hypoxia-inducible operon.
In the Wayne model, DNA synthesis ceases in M. tuberculosis as the O2 tension drops below 1% (45). Although DNA synthesis in the obligate anaerobe Bacteroides fragilis ceases in an analogous way after exposure to O2, this organism possesses a class I RNR that is involved in survival during exposure to O2 and that plays a role in maintaining dNTP pools for DNA repair and recovery following reintroduction into anaerobic growth conditions (39). We hypothesized that the reverse may be true in the obligate aerobe M. tuberculosis, with the O2-independent RNR playing an analogous dNTP maintenance role under hypoxic conditions generated in vitro in the Wayne model or encountered in an in vivo infection. During nonreplicating persistence of M. tuberculosis, DNA replication is predicted to occur intermittently (if at all), which should reduce the demand for dNTPs. However, there is evidence which suggests that repair synthesis occurs during persistent infections (5), implying that under these conditions, dNTP pools must be maintained in M. tuberculosis, possibly even at elevated levels (6), to serve the repair function. The M. tuberculosis ΔnrdZ gene exhibited no survival phenotype in the Wayne model, suggesting that anaerobiosis per se, in the absence of any external stress such as that imposed by the immune response of the host, may not damage DNA to a sufficient extent to result in a survival phenotype. We therefore challenged a relatively M. tuberculosis-resistant strain of mice with the ΔnrdZ mutant and assessed its ability to proliferate and survive under immune surveillance conditions. However, the mutant displayed little or no in vivo growth phenotype during the acute and chronic stages of infection, suggesting that NrdZ does not contribute significantly to the pathogenesis of mouse tuberculosis infection under the conditions employed in this study.
There are several possible explanations for the lack of a phenotype in the ΔnrdZ mutant. On the one hand, nrdZ might not encode a functional class II RNR. Alternatively, NrdZ may be functional, but its activity could be compromised by insufficient levels of cobalamin in M. tuberculosis under the growth conditions employed in this study. We have not excluded the possibility that in addition to the possible inability of M. tuberculosis to synthesize adenosylcobalamin, this bacterium cannot transport and/or convert cyanocobalamin to adenosylcobalamin. Alternatively, the NrdEF2 enzyme alone may provide sufficient activity for maintaining dNTP pools even under severely O2-limiting conditions (19, 24), thus obscuring any possible contribution made by NrdZ. Work is currently under way to investigate these various possibilities.
Acknowledgments
This work was supported by NIH grant R01-AI43420 (to H.R.) and by grants from the Medical Research Council of South Africa, the National Research Foundation, and the National Health Laboratory Service. V.M. was also supported by an International Research Scholar grant from the Howard Hughes Medical Institute, and S.D. was supported by a traineeship from the Columbia University-Southern African Fogarty AIDS International Research and Training Programme (grant 5 D43 TWOO231 funded by the Fogarty International Center of the National Institutes of Health).
We thank Martin Everett for providing the gridded M. tuberculosis plasmid library and for recovering clones, Sue Andersen and Steven Durbach for technical assistance, and Minty van der Meulen for assistance with the BACTEC assays.
Editor: J. N. Weiser
REFERENCES
- 1.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, B. R., and P. M. Small. 1998. The evolving relation between humans and Mycobacterium tuberculosis. N. Engl. J. Med. 338:677-678. [DOI] [PubMed] [Google Scholar]
- 3.Booker, S., S. Licht, J. Broderick, and J. Stubbe. 1994. Coenzyme B12-dependent ribonucleotide reductase: evidence for the participation of five cysteine residues in ribonucleotide reduction. Biochemistry 33:12676-12685. [DOI] [PubMed] [Google Scholar]
- 4.Borovok, I., R. Kreisberg-Zakarin, M. Yanko, R. Schreiber, M. Myslovati, F. Aslund, A. Holmgren, G. Cohen, and Y. Aharonowitz. 2002. Streptomyces spp. contain class Ia and class II ribonucleotide reductases: expression analysis of the genes in vegetative growth. Microbiology 148:391-404. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff, H. I. M., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 6.Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein, and L. Thelander. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112:391-401. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quali, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, J. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, A. F., and C. L. Spreadbury. 1998. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J. Bacteriol. 180:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dannenberg, A. M., Jr. 1993. Immunopathogenesis of pulmonary tuberculosis. Hosp. Pract. 28:51-58. [DOI] [PubMed] [Google Scholar]
- 10.Desjardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 183:5311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engels, W. R. 1993. Contributing software to the Internet: the Amplify program. Trends Biochem. Sci. 18:448-450. [DOI] [PubMed] [Google Scholar]
- 12.Fagan, P. K., M. J. Walker, J. Chin, G. J. Eamens, and S. P. Djordjevic. 2001. Oral immunization of swine with attenuated Salmonella typhimurium aroA SL3261 expressing a recombinant antigen of Mycoplasma hyopneumoniae (NrdF) primes the immune system for a NrdF specific secretory IgA response in the lungs. Microb. Pathog. 30:101-110. [DOI] [PubMed] [Google Scholar]
- 13.Fritz, C., S. Maass, A. Kreft, and F. C. Bange. 2002. Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect. Immun. 70:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garriga, X., R. Eliasson, E. Torrents, A. Jordan, J. Barbe, I. Gibert, and P. Reichard. 1996. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229:189-192. [DOI] [PubMed] [Google Scholar]
- 15.Gordhan, B. G., and T. Parish. 2001. Gene replacement using pre-treated DNA. Methods Mol. Med. 54:77-92. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Y., and A. R. M. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, M., and S. J. Elledge. 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6105-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, A., E. Pontis, F. Aslund, U. Hellman, I. Gibert, and P. Reichard. 1996. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 271:8779-8785. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, A., E. Torrents, C. Jeanthon, R. Eliasson, U. Hellman, C. Wernstedt, J. Barbe, I. Gibert, and P. Reichard. 1997. B12-dependent ribonucleotide reductases from deeply rooted eubacteria are structurally related to the aerobic enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 94:13487-13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan, A., E. Torrents, I. Sala, U. Hellman, I. Gibert, and P. Reichard. 1999. Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181:3974-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karasseva, V., J. G. Weiszfeiler, and Z. Lengyel. 1977. Synthesis of vitamin B12 by various species of mycobacteria. Zentralbl. Bakteriol. 239:514-520. [PubMed] [Google Scholar]
- 23.Manganelli, R., S. Tyagi, and I. Smith. 2001. Real time PCR using molecular beacons. Methods Mol. Med. 54:295-310. [DOI] [PubMed] [Google Scholar]
- 24.Masalha, M., I. Borovok, R. Schreiber, Y. Aharonowitz, and G. Cohen. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J. Bacteriol. 183:7260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAdam, R. A., S. Quan, D. A. Smith, S. Bardarov, J. C. Betts, F. C. Cook, E. U. Hooker, A. P. Lewis, P. Woollard, M. J. Everett, P. T. Lukey, G. J. Bancroft, W. R. Jacobs, Jr., and K. Duncan. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148:2975-2986. [DOI] [PubMed] [Google Scholar]
- 26.Medina, E., and R. J. North. 1998. Resistance ranking of some common inbred mouse strains to Mycobacterium tuberculosis and relationship to major histocompatibility complex haplotype and Nramp1 genotype. Immunology 93:270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monje-Casas, F., J. Jurado, M. J. Prieto-Alamo, A. Holmgren, and C. Pueyo. 2001. Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J. Biol. Chem. 276:18031-18037. [DOI] [PubMed] [Google Scholar]
- 28.Moreira, A. L., L. Tsenova-Berkova, J. Wang, P. Laochumroonvorapong, S. Freeman, V. H. Freedman, and G. Kaplan. 1997. Effect of cytokine modulation by thalidomide on the granulomatous response in murine tuberculosis. Tuber. Lung Dis. 78:47-55. [DOI] [PubMed] [Google Scholar]
- 29.O'Gaora, P., S. Barnini, C. Hayward, E. Filley, G. Rook, D. Young, and J. Thole. 1997. Mycobacteria as immunogens. Development of expression vectors for use in multiple mycobacterial species. Med. Principles Pract. 6:91-96. [Google Scholar]
- 30.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked M. tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 31.Park, H.-D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole, A. M., D. T. Logan, and B. M. Sjoberg. 2002. The evolution of the ribonucleotide reductases: much ado about oxygen. J. Mol. Evol. 55:180-196. [DOI] [PubMed] [Google Scholar]
- 33.Rastogi, N., K. S. Goh, P. Ruiz, and M. Casal. 1995. In vitro activity of roxithromycin against the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 39:1162-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichard, P. 2002. Ribonucleotide reductases: the evolution of allosteric regulation. Arch. Biochem. Biophys. 397:149-155. [DOI] [PubMed] [Google Scholar]
- 35.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 36.SantaLucia, J., Jr. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi, L., Y. J. Jung, S. Tyagi, M. L. Gennaro, and R. J. North. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA 100:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smalley, D., E. R. Rocha, and C. J. Smith. 2002. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol. 184:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szekeres, T., M. Fritzer-Szekeres, and H. L. Elford. 1997. The enzyme ribonucleotide reductase: target for antitumor and anti-HIV therapy. Crit. Rev. Clin. Lab. Sci. 34:503-528. [DOI] [PubMed] [Google Scholar]
- 41.Torrents, E., I. Roca, and I. Gibert. 2003. Corynebacterium ammoniagenes class Ib ribonucleotide reductase: transcriptional regulation of an atypical genomic organization in the nrd cluster. Microbiology 149:1011-1020. [DOI] [PubMed] [Google Scholar]
- 42.Tsenova, L., A. L. Moreira, E. Party, V. H. Freedman, and G. Kaplan. 1997. Aerosol infection of mice with mycobacteria using a nose-only exposure device. J. Am. Biol. Safety Assoc. 2:20-31. [Google Scholar]
- 43.Vollack, K.-U., E. Härtig, H. Körner, and W. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 31:1681-1694. [DOI] [PubMed] [Google Scholar]
- 44.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 45.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for the sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, F., S. C. Curran, L. S. Li, D. Avarbock, J. D. Graf, M. M. Chua, G. Lu, J. Salem, and H. Rubin. 1997. Characterization of two genes encoding the Mycobacterium tuberculosis ribonucleotide reductase small subunit. J. Bacteriol. 179:6408-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]