Abstract
Two major receptors involved in human natural cytotoxicity, NKp46 and NKp44, have recently been identified. However, experimental evidence suggested the existence of additional such receptor(s). In this study, by the generation of monoclonal antibodies (mAbs), we identified NKp30, a novel 30-kD triggering receptor selectively expressed by all resting and activated human natural killer (NK) cells. Although mAb-mediated cross-linking of NKp30 induces strong NK cell activation, mAb-mediated masking inhibits the NK cytotoxicity against normal or tumor target cells. NKp30 cooperates with NKp46 and/or NKp44 in the induction of NK-mediated cytotoxicity against the majority of target cells, whereas it represents the major triggering receptor in the killing of certain tumors. This novel receptor is associated with CD3ζ chains that become tyrosine phosphorylated upon sodium pervanadate treatment of NK cells. Molecular cloning of NKp30 cDNA revealed a member of the immunoglobulin superfamily, characterized by a single V-type domain and a charged residue in the transmembrane portion. Moreover, we show that NKp30 is encoded by the previously identified 1C7 gene, for which the function and the cellular distribution of the putative product were not identified in previous studies.
Keywords: natural killer cells, triggering receptor, natural cytotoxicity
Natural killer (NK) cells provide an efficient effector mechanism by which immunosurveillance eliminates tumor or virally infected cells 1 2 3. A well-defined characteristic of NK cells is their ability to lyse target cells deficient in expression of MHC class I molecules 4. This observation has been the basis for identification of different inhibitory receptors expressed by NK cells. Upon binding to MHC class I molecules expressed on target cells, these receptors deliver inhibitory signals that down-regulate cytolytic functions 1 2 3 5 6. In humans, recognition of HLA class I molecules is mediated by two types of receptors: those belonging to the Ig superfamily (Ig-SF),1 which includes both killer inhibitory receptor (KIR; 7 8 9 10 11 12 13 14) and LIR-1/ILT-2 proteins 15 16 whose ligands are represented by various groups of HLA-A, -B, and -C alleles; and the lectin-like CD94/NKG2A receptor complex 17, which recognizes HLA-E molecules 18 19. The expression of these inhibitory receptors explains how NK cells can distinguish between HLA-deficient and normal cells 20 21. However, limited information existed on the activating NK receptors responsible for triggering natural cytotoxicity. Only recently, two distinct NK-specific receptors have been identified that play an important role in the NK cell–mediated recognition and killing of HLA class I–defective target cells. These receptors, termed NKp46 and NKp44 22 23, are members of the Ig-SF 24 25. Their cross-linking, induced by specific mAbs, leads to a strong NK cell activation resulting in increased intracellular Ca2+ levels, triggering of cytotoxicity, and lymphokine release 22 23. Importantly, mAb-mediated masking of NKp46 and/or NKp44 resulted in inhibition of NK cytotoxicity against most, but not all, target cells. These findings, while providing evidence for a central role of NKp46 and NKp44 in natural cytotoxicity, also implied the existence of additional receptors 23 24 26.
In this study, we identified and characterized NKp30, a third triggering receptor involved in NK cell–mediated recognition and killing of target cells. NKp30 is a member of the Ig-SF characterized by a single V-type domain that is selectively expressed on the surface of human NK cells.
Materials and Methods
mAbs.
The following mAbs were produced in our lab: JT3A (IgG2a, anti-CD3); BAB281 22 and KL247 (IgG1 and IgM, respectively, anti-NKp46); Z231 23 and KS38 (IgG1 and IgM, respectively, anti-NKp44); KD1 and c127 (IgG2a and IgG1, respectively, anti-CD16); c218 and GPR165 (IgG1 and IgG2a, respectively, anti-CD56); A6-136 (IgM, anti-HLA class I [6]); GL183 (IgG1, anti-p58.2 [7]); EB6 (IgG1, anti-p58.1 [8]); and Z199 (IgG2b, anti-NKG2A [17]).
D1.12 (IgG2a, anti–HLA-DR) mAb was provided by Dr. R.S. Accolla (University of Insubria, Varese, Italy). HP2.6 (IgG2a, anti-CD4) mAb was provided by Dr. P. Sanchez-Madrid (Hopital de la Princessa, Madrid, Spain).
The novel mAbs were derived by immunizing 5 wk–old Balb/C mice with activated (CD3−, CD56+, and CD16+) NK cells, either NK clones (EC1 and SA260 for A76 and Z25 mAbs, respectively) or a polyclonal NK cell population (for AZ20 mAb). After different cell fusions, the mAbs were selected for the ability to induce lysis in redirected killing assays against the FcγR+ P815 target cells.
Purification of PBLs and Generation of Polyclonal or Clonal NK Cell Populations.
PBLs were derived from healthy donors by Ficoll-Hypaque gradients and depletion of plastic-adherent cells. To obtain enriched NK cells, PBLs were incubated with anti-CD3 (JT3A), anti-CD4 (HP2.6), and anti–HLA-DR (D1.12) mAbs (30 min at 4°C), followed by goat anti–mouse-coated Dynabeads (Dynal; 30 min at 4°C) and immunomagnetic depletion 21 22 23. CD3−4−DR− cells were used in cytolytic assays or cultured on irradiated feeder cells in the presence of 100 U/ml rIL-2 (Proleukin; Chiron Corporation) and 1.5 ng/ml PHA (GIBCO BRL) to obtain polyclonal NK cell populations or, after limiting dilution 27, NK cell clones.
Flow Cytofluorimetric Analysis.
Cells were stained with the appropriate mAb followed by PE- or FITC-conjugated isotype-specific goat anti–mouse second reagent (Southern Biotechnology Associates Inc.). Samples were analyzed by one- or two-color cytofluorimetric analysis (FACScan™; Becton Dickinson), as described previously 7.
Cell Lines and Cytolytic Assays.
The following FcγR-negative targets were used: MEL15 (MEL15392, human melanoma [21]); M14 (human melanoma [24]); SMMC (human hepatocarcinoma [26]); A549 (human lung adenocarcinoma, no. CCL-185.1; American Type Culture Collection). FO-1 and 1174 mel (human melanomas) were provided by Dr. S. Ferrone (Roswell Park Cancer Center, Buffalo, NY); AUMA (human melanoma) was provided by Dr. P. Coulie (Catholic University of Louvain, Brussels, Belgium). The FcγR-positive target used was P815 (murine mastocytoma). PHA blasts, used as normal target cells, were obtained by culturing PBLs with 1.5 ng/ml PHA (GIBCO BRL).
Cells were tested for cytolytic activity in a 4-h 51Cr-release assay as described previously 8 26, either in the absence or presence of various mAbs. The concentrations of the various mAbs were 10 μg/ml for the masking experiments and 0.5 μg/ml for the redirected killing experiments. The E/T ratios are indicated in the text.
Determination of Intracellular Free Calcium [Ca2+]i Increase.
Determination of [Ca2+]i was performed as described previously 28. Fura-2–labeled NK cells were incubated for 30 min at 4°C with saturating amounts of anti-NKp30 mAb (AZ20) or medium alone. Cross-linking of this receptor was obtained by adding into the cuvette 20 μg/ml of affinity-purified goat anti–mouse antiserum (GAM; ICN Biomedicals, Inc.).
Biochemical Characterization of the NKp30 Molecules.
Integral NK cell membrane proteins 29 were prepared as follows: 25 × 106 cells were lysed in 100 μl TX buffer (20 mM sodium phosphate buffer, 1% Triton X-114, 10 mM EDTA, pH 8) for 30 min at 4°C, and centrifuged (5 min, 10,000 rpm). The supernatant was left for 10 min at 37°C, centrifuged, and the lower phase was resuspended 1:2 in TX buffer and left for 10 min at 4°C in order to clarify the lysates. The suspension was then left for 10 min at 37°C, centrifuged, and the lower phase was resuspended 1:3 in EB (0.0625 M Tris, pH 6.8, 10% glycerol, 2.3% SDS). Samples were analyzed in discontinuous SDS-PAGE, transferred to Immobilon P (Millipore Corp.), and probed with AZ20 mAb followed by rabbit anti–mouse horseradish peroxidase (HRPO; DAKO), or NKp30-specific antiserum followed by donkey anti–rabbit HRPO (Nycomed Amersham plc). The Renaissance Chemiluminescence kit (NEN Life Science Products) was used for detection.
NKp30 Polyclonal Antiserum.
A 2.5-kg HY/Cr male rabbit (Charles River Laboratories) was immunized with 100 μg/100 μl of the 15 amino acid peptide WVSQPPEIRTLEGSC (Eurogentec) conjugated with KLH 13. Four weekly treatments were performed, the first in association with 100 μl CFA and all others with 100 μl IFA. 1 wk after the last treatment, 10 ml of blood was drawn, and serum was tested and titered by ELISA against the immunizing and irrelevant peptides.
Analysis of the NKp30 Signal Transduction Complex.
NK cells (108) were stimulated or not with 100 μM sodium pervanadate 25, and 1% digitonin lysates were precleared five times with sepharose protein A–coupled KD1 (anti-CD16) mAb. Lysates were then immunoprecipitated with sepharose-CNBr–coupled Z231 and BAB281 mAbs, or with sepharose protein A–coupled NKp30-specific rabbit antiserum and preimmune rabbit serum. Samples were analyzed in a 15% SDS-PAGE under reducing conditions (5% 2-ME), transferred to Immobilon P (Millipore Corp.), and probed with antiphosphotyrosine mAb (PY20-HRPO; Transduction Laboratories) or anti-CD3ζ mAb (2H2; Immunotech), followed by rabbit anti–mouse HRPO (DAKO). The Renaissance Chemiluminescence kit (NEN Life Science Products) was used for detection.
Library Screening by cDNA Expression in COS-7 Cells.
The expression cDNA library was prepared in VR1012 plasmid (Vical Inc.) using RNA extracted from IL-2–activated polyclonal NK cells obtained from two healthy donors as described previously 24 25.
The library screening procedure was as described 24 25 30. In brief, cDNA library was transiently transfected in COS-7 cells, and selection of positive pools was performed by immunocytochemical staining using the specific anti-NKp30 mAb A76 and sib-selection.
DNA Sequencing.
DNA sequencing was performed using d-Rhodamine Terminator Cycle Sequencing kit and a 377 ABI Automatic Sequencer (Applied Biosystems/Perkin-Elmer).
Transient Transfections.
COS-7 cells (5 × 105/plate) were transfected with VR1012-NK-A1 (clone 5C) or with the vector alone by the DEAE-dextran or electroporation methods as described 13. After 48 h, transfected cells were used for cytofluorimetric analysis.
Analysis of NKp30 Transcript Expression by Northern Blotting.
To analyze NKp30 transcript expression in different cell lines of hemopoietic origin, RNA was size fractionated by denaturing agarose gel electrophoresis and transferred onto a positively charged nylon membrane (NEN Life Science Products). In particular, 10 μg of total RNA prepared using CsCl gradient, or 2 μg of poly A+ RNA prepared using oligo(dT) magnetic bead separation (Dynal AS), was loaded on each lane. Northern blots were performed under high stringency conditions as described 31. The NKp30 421-bp cDNA probe was obtained by PCR amplification performed with 25 pmol of each primer for 30 cycles (30 s at 94°C, 30 s at 60°C, 30 s at 72°C), followed by a 7-min incubation at 72°C. The sequences of the primers are: CAG GGC ATC TCG AGT TTC CGA CAT GGC CTG GAT GCT GTT G (NK-A1 up) and GAC TAG GAT CCG CAT GTG TAC CAG CCC CTA GCT GAG GAT G (NK-A1 down). The cDNA fragment was 32P-labeled by random priming 32.
Reverse Transcription PCR Amplification of NKp30 cDNA.
Total RNA extracted using RNAzol (Cinna/Biotecx) from polyclonal NK and T cell populations and clones and from different hemopoietic cell lines were reverse transcribed using oligo(dT) priming. Primers used for cDNA amplification of NKp30 (606 bp) were the following: 5′ CAG GGC ATC TCG AGT TTC CGA CAT GGC CTG GAT GCT GTT G (NK-A1 up) and 5′ GAT TTA TTG GGG TCT TTT GAA G (A76-38 reverse). Amplification was performed with 25 pmol of each primer for 30 cycles (30 s at 94°C, 30 s at 60°C, and 30 s at 72°C), followed by a 7-min incubation at 72°C. The amplification products were subcloned in pCR2.1 by TOPO-TA Cloning kit (Invitrogen), and subsequently sequenced.
Zoo-Blot Analysis.
Analysis of cross-species conservation of NKp30 gene was performed using a Zoo-Blot (CLONTECH). The Southern blot contained genomic DNA from humans, Rhesus monkey, Sprague–Dawley rat, BALB/c mouse, dog, cow, rabbit, chicken, and Saccharomyces cerevisiae yeast. The hybridization probe was the same 421-bp cDNA fragment used to hybridize the Northern blot. Washes were carried out at low stringency conditions as described 32.
Results
Identification of a Novel NK-specific Triggering Surface Molecule.
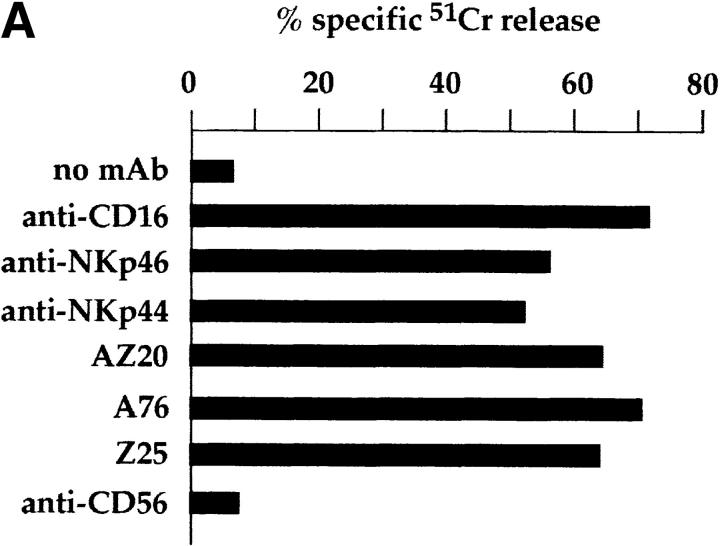
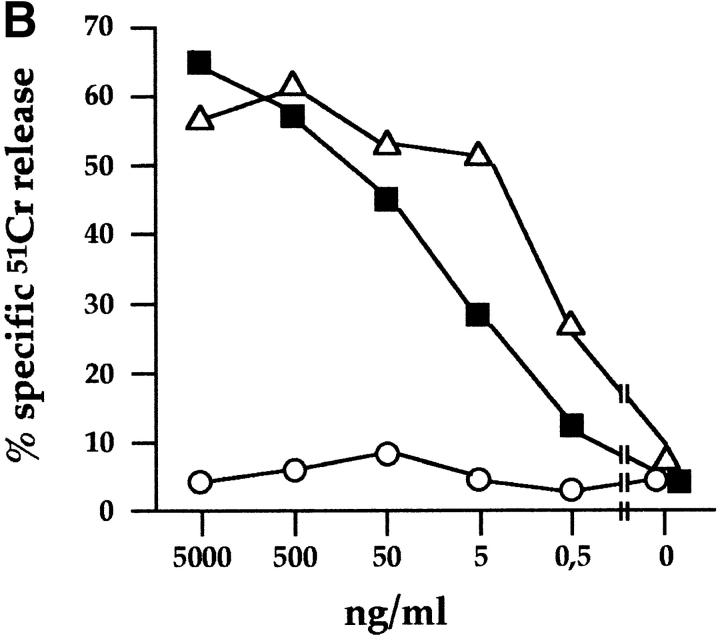
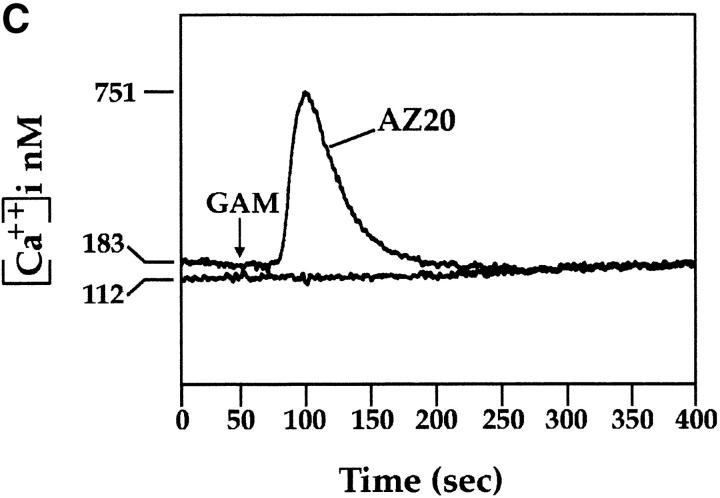
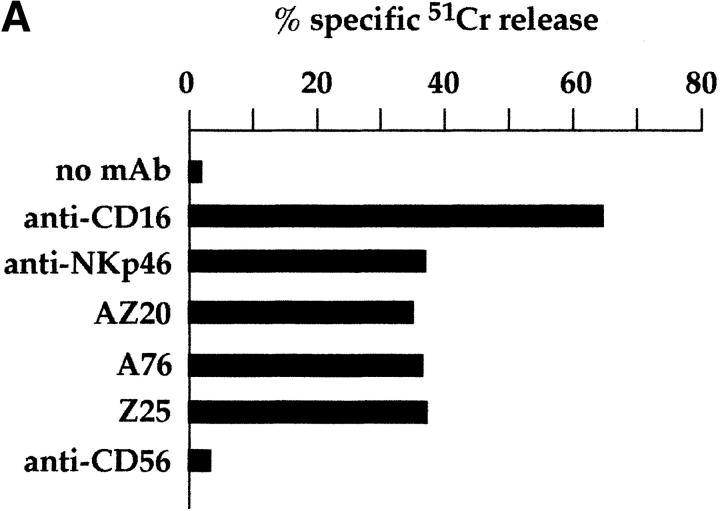
Mice were immunized with CD3−, CD16+, CD56+ NK cell clones or bulk populations. mAbs from different fusions were first selected according to their ability to induce lysis of the FcγR+ P815 target cells in a redirected killing assay using polyclonal NK cell populations or clones as effector cells. Three mAbs, A76, AZ20, and Z25 (all of IgG1 isotype), were selected that induced a strong cytolytic activity (Fig. 1 A) similar to that elicited by other mAbs specific for known triggering NK receptors, including CD16, NKp46, and NKp44 22 23 26. In Fig. 1 B, the NK cell cytotoxicity induced by graded amounts of AZ20 mAb is compared with that of isotype–matched anti-CD16 or anti-CD56 mAbs. The cytolytic response to AZ20 mAb paralleled that induced by anti-CD16 mAb, whereas anti-CD56 mAb had no effect. Moreover, as shown in Fig. 1 C, a sharp [Ca+]i increase was detected in the representative clone 3M16 after stimulation with AZ20 mAb. Notably, [Ca2+]i increments induced by this Ab occurred only in the presence of a goat anti–mouse second reagent, allowing efficient cross-linking of the activating receptor.
Figure 1.
Triggering of NK-mediated cytolytic activity induced by three new mAbs. (A) A representative polyclonal NK cell population was analyzed for cytolytic activity in a redirected killing assay against the FcγR-positive P815 target cell line in the absence or presence of c127 (anti-CD16), BAB281 (anti-NKp46), Z231 (anti-NKp44), AZ20, A76, Z25, and c218 (anti-CD56) mAbs. The E/T ratio used was 1:1. (B) The representative NK clone 3M16 was analyzed in a redirected killing assay against P815 target cells (E/T ratio 1:1) in the presence of graded amounts of AZ20 (▪), c127 (anti-CD16; ▵), or c218 (anti-CD56; ○) mAbs. All the mAbs used are of the IgG1 isotype. (C) Clone 3M16 was analyzed for [Ca2+]i mobilization in the presence of AZ20 mAb, followed by GAM. The negative control is represented by cells treated with GAM alone.
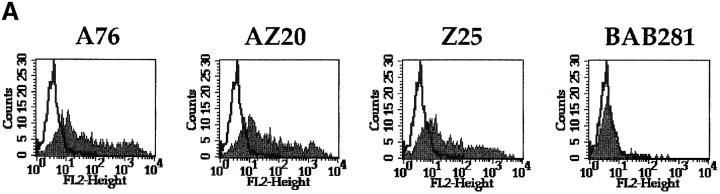
Analysis of the cell surface distribution of the molecule(s) recognized by A76, AZ20, and Z25 mAbs, performed by indirect immunofluorescence and FACS® analysis, revealed reactivity with various activated polyclonal or clonal NK cell populations derived from different donors (see below). These also included the infrequent CD16− NK cell clones. On the contrary, no mAb reactivity was detected with PHA-induced polyclonal T cell populations or TCR-α/β and -γ/δ T cell clones (derived from different donors). Neither was any reactivity detected with EBV-induced B cell lines, monocytic and dendritic cell lines, and different hemopoietic and nonhemopoietic tumor cell lines, including HL60, U937, Eo/A3, THP-1, Daudi, Jurkat, IGROV, and all the various tumor cell lines used as target cells in this study (data not shown).
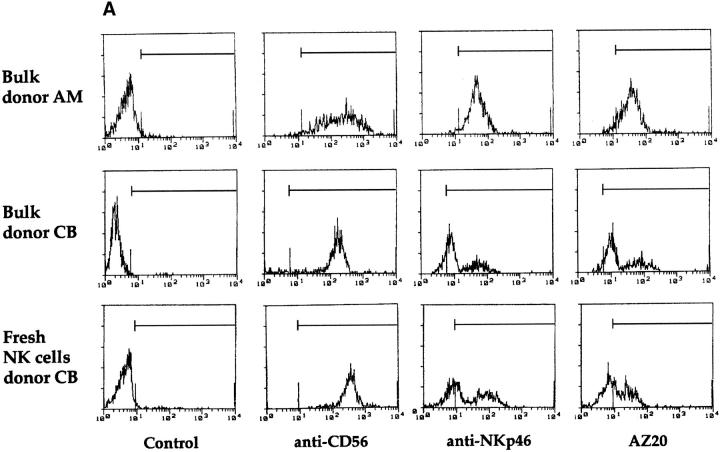
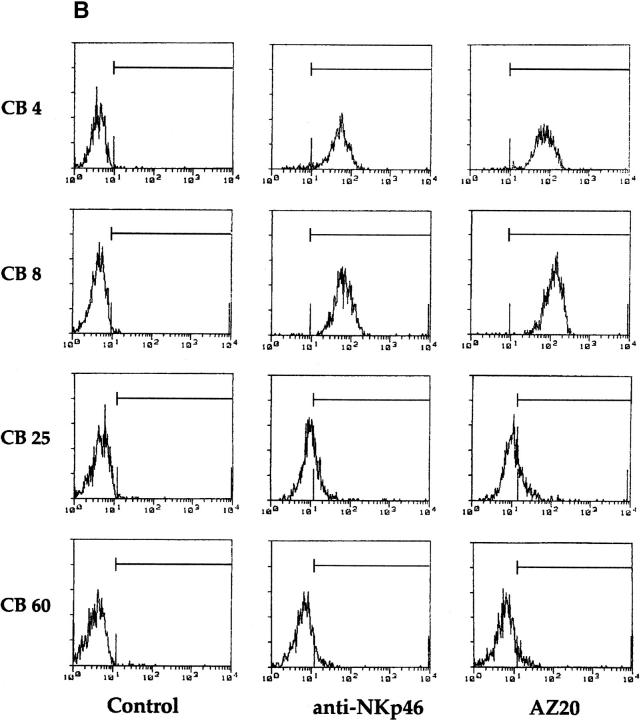
We recently showed that polyclonal NK cell populations from some donors were characterized by a bimodal distribution of fluorescence intensity of NKp46 molecules (NKp46bright and NKp46dull), and that NK clones derived from these individuals expressed a stable NKp46bright or NKp46dull phenotype 26. Importantly, the cytolytic activity of NK cell clones against NK-susceptible target cells strictly correlated with their NKp46 phenotype 26. We then analyzed the reactivity of the new mAbs on polyclonal NK cell populations and NK cell clones derived from individuals displaying different patterns of NKp46 expression. As shown in Fig. 2 A, the polyclonal NK cell population derived from the representative donor AM displayed a homogeneously bright phenotype when stained by either AZ20 or anti-NKp46 mAbs. On the contrary, in the polyclonal NK cells derived from donor CB, staining with the same mAbs resulted in a bimodal distribution of fluorescence. Notably, in donor CB, the same pattern of fluorescence intensity was also detectable in fresh purified NK cells (Fig. 2 A). Moreover, the analysis of several clones derived from donor CB revealed that NKp46bright clones were consistently AZ20bright, whereas NKp46dull clones always displayed an AZ20dull phenotype (Fig. 2 B).
Figure 2.
Cytofluorimetric analysis of resting or activated, polyclonal or clonal, NK cells. (A) Polyclonal NK cell populations, derived from donors AM and CB, and freshly isolated NK cells, derived from donor CB, were analyzed by immunofluorescence and FACS® analysis using c218 (anti-CD56), BAB281 (anti-NKp46), or AZ20 mAbs, followed by PE-conjugated goat anti–mouse IgG1. The control is represented by cells incubated with the second reagent alone. (B) NK cell clones, derived from donor CB, were analyzed by immunofluorescence and FACS® analysis using BAB281 (anti-NKp46) or AZ20 mAbs, followed by PE-conjugated goat anti–mouse IgG1.
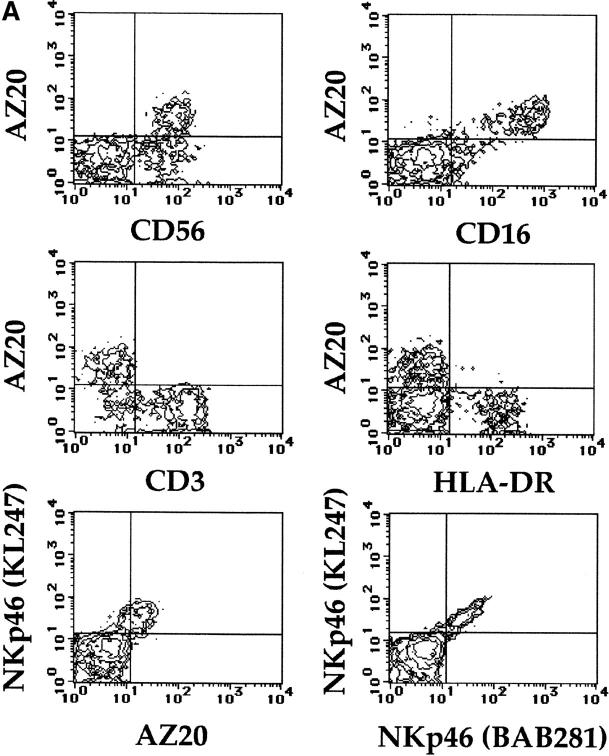
To further define the pattern of reactivity of the new mAbs in freshly isolated lymphocytes, PBLs derived from different individuals were assessed by double fluorescence analysis using informative mAbs. A representative donor is shown in Fig. 3 A: the surface molecule recognized by AZ20 mAb was selectively expressed on CD56+ cells. Moreover, most AZ20+ cells coexpressed CD16 molecules. However, AZ20 mAb did not stain CD3+ T lymphocytes or HLA-DR+ B lymphocytes. It is of note that the CD56+ AZ20− cell population detected in this donor also expressed surface CD3 molecules (not shown). Therefore, also in freshly derived lymphocytes, the reactivity of AZ20 mAb overlaps with that of anti-NKp46 mAb. A direct comparative analysis of the surface expression of NKp46 and AZ20 mAb–reactive molecules is shown in Fig. 3 A. The two molecules were clearly coexpressed by the same cell subset. However, no diagonal distribution could be detected in cells stained by AZ20 and anti-NKp46 mAbs, whereas this type of fluorescence distribution occurred when cells were stained simultaneously by two anti-NKp46 mAbs of different isotype.
Figure 3.
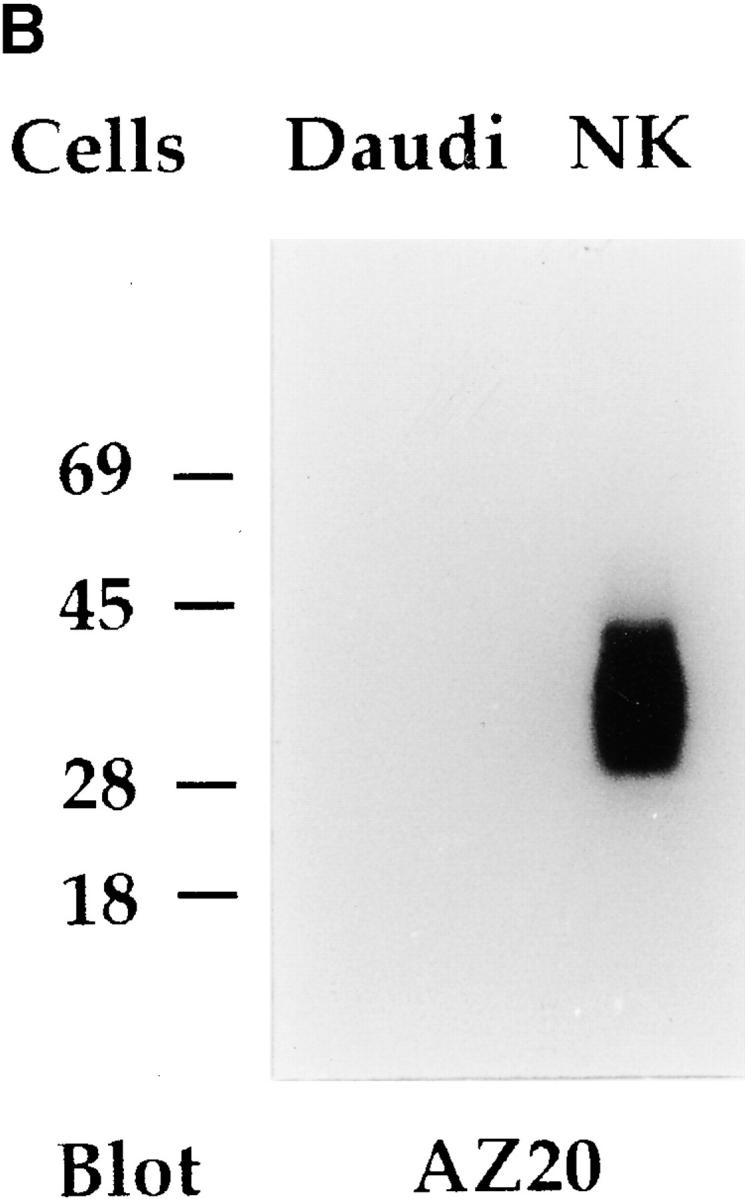
Pattern of expression of NKp30 in PBLs and Western blot analysis. (A) Freshly isolated PBLs, derived from a representative donor, were analyzed by two-color immunofluorescence and FACS® analysis with AZ20 mAb in combination with GPR 165 (IgG2a, anti-CD56), KD1 (IgG2a, anti-CD16), JT3A (IgG2a, anti-CD3), D1-12 (IgG2a, anti–HLA-DR), and KL247 (IgM, anti-NKp46) mAbs, followed by isotype-specific FITC- or PE-conjugated GAM second reagents. Double immunofluorescence with two anti-NKp46 mAbs of different isotype (KL247, IgM vs. BAB281, IgG1) is also shown. The contour plots were divided into quadrants representing unstained cells (lower left), cells with only red fluorescence (upper left), cells with red and green fluorescence (upper right), and cells with only green fluorescence (lower right). (B) Integral membrane proteins derived from Daudi (Burkitt lymphoma, negative control) and from a polyclonal NK cell population were analyzed in an 11% SDS-PAGE under nonreducing conditions and probed with AZ20 mAb. Molecular weight markers (in kD) are indicated on the left.
Notably, results identical to those described for AZ20 mAb were obtained with A76 and Z25 mAbs. These data suggested that the molecule recognized by the new mAbs may be distinct from NKp46. To directly evaluate this possibility, COS-7 cells transiently transfected with NKp46 cDNA 24 were analyzed for their reactivity with AZ20, A76, and Z25 mAbs. Cell transfectants, although reacting with different anti-NKp46 mAbs, were not stained by AZ20, A76, and Z25 mAbs (data not shown). Taken together, these data strongly suggest that A76, AZ20, and Z25 mAbs are specific for a novel surface molecule that defines all mature human NK cells, but is distinct from NKp46.
To analyze the biochemical characteristics of the surface molecules recognized by AZ20, A76, and Z25 mAbs, NK populations were surface labeled with 125I or biotin, immunoprecipitated with one or another mAb, and analyzed by SDS-PAGE. Under these conditions, no specific bands could be detected. Thus, integral membrane proteins were prepared from NK cells to further analyze a possible reactivity of the various mAbs in Western blot. As shown in Fig. 3 B, AZ20 mAb specifically reacted with an ∼30-kD molecule, thereafter termed NKp30. Under the same conditions, both A76 and Z25 mAbs displayed a poor reactivity (data not shown).
Cross-linking of NKp30 Induces Cytolytic Activity in Freshly Derived NK Cells.
Since NKp30 molecule, like NKp46, was expressed on fresh NK cells, we analyzed whether it could trigger the cytolytic activity of these cells as demonstrated previously for NKp46. As shown in Fig. 4 A, AZ20, A76, and Z25 mAbs induced a strong increase of cytolytic activity against P815 target cells, whereas the isotype–matched anti-CD56 mAb had no effect. This triggering effect was comparable to that obtained with anti-NKp46 mAb. Moreover, in these experiments, the use of AZ20 F(ab′)2 fragments did not induce triggering of cytolytic activity, indicating that mAb-dependent NKp30 stimulation requires efficient cross-linking mediated by FcγR on target cells (data not shown).
Figure 4.
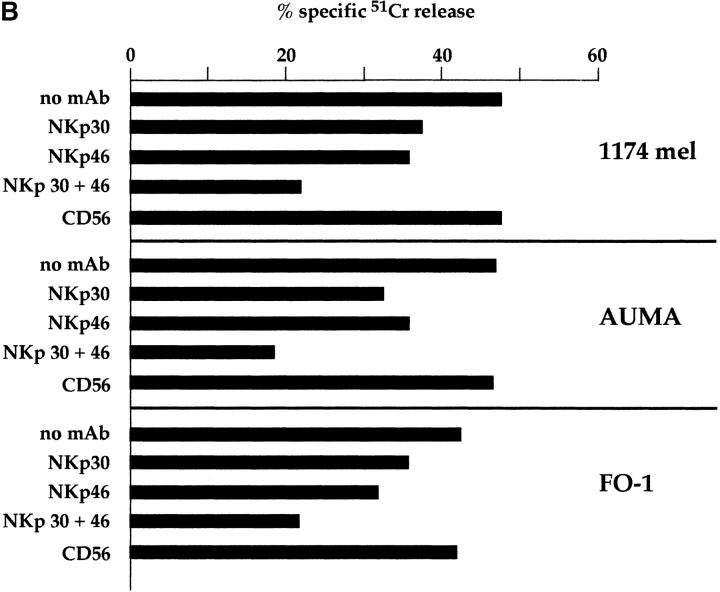
NKp30 functions as an activating receptor in fresh NK cells and is involved in their natural cytotoxicity. (A) Freshly isolated peripheral blood NK lymphocytes, derived from a representative donor, were analyzed for cytolytic activity in a redirected killing assay against the FcγR-positive P815 target cell line in the absence or presence of c127 (anti-CD16), BAB281 (anti-NKp46), AZ20, A76, Z25, and c218 (anti-CD56) mAbs. The E/T ratio used was 20:1. (B) Freshly isolated peripheral blood NK cells were analyzed for cytolytic activity against the indicated FcγR-negative/HLA class I–negative melanoma cell lines either in the absence or presence of mAbs to the indicated molecules. c218 (anti-CD56), AZ20 (anti-NKp30), and BAB281 (anti-NKp46) mAbs were used. The E/T ratio was 20:1.
Involvement of NKp30 in the Induction of Natural Cytotoxicity against Normal or Tumor Cells.
Previous data showed that mAb-mediated masking of NKp46 or NKp44 inhibited the non–MHC-restricted tumor cell lysis by activated NK cells 23 24 26. Moreover, masking of NKp46 also inhibited the natural cytotoxicity mediated by freshly isolated peripheral blood NK cells 26. We then evaluated whether masking of NKp30 could affect the cytolytic activity mediated by freshly derived NK cells or NK clones against a panel of FcγR-negative tumor target cells. As shown in Fig. 4 B, anti-NKp30 mAb, but not the isotype-matched anti-CD56 mAb, inhibited natural cytotoxicity mediated by fresh NK cells against the HLA class I–negative 1174 mel, AUMA, and FO-1 melanoma cell lines. In addition, a greater inhibitory effect occurred when anti-NKp30 mAb was used in combination with anti-NKp46 mAb. This suggests that NKp30 and NKp46 may represent receptors that act synergistically in triggering the natural cytotoxicity of fresh NK cells.
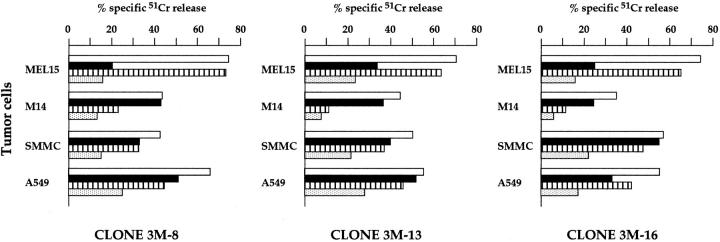
In view of these data, we further analyzed the effect of mAb-mediated masking of NKp30 on the tumor cell killing by activated NK cells. Fig. 5 shows three representative NK cell clones analyzed in a cytolytic assay against different tumor targets, including two melanomas (MEL15 and M14), a hepatocarcinoma (SMMC), and a lung adenocarcinoma (A549). In previous studies, we showed that the cytolytic activity against the M14 melanoma was confined to NK clones displaying the NKp46bright phenotype and could be inhibited by mAb-mediated masking of NKp46 receptor. On the other hand, NKp46bright clones also killed MEL15. However, neither masking of NKp46 nor of NKp44 significantly inhibited their cytolytic activity 26. These data strongly suggested the existence in these clones of additional triggering receptors responsible for the cytotoxicity against MEL15 target cells. As illustrated above, NKp30 is brightly expressed in NKp46bright clones. Therefore, it is conceivable that it may play a role in the killing of MEL15 target cells. Indeed, as shown in Fig. 5, anti-NKp30 mAb sharply inhibited the NK-mediated lysis of MEL15 cells (>50% of inhibition). Anti-NKp46 mAb exerted a minor effect, whereas an isotype-matched anti-CD56 mAb had no effect (data not shown). On the contrary, lysis of M14 melanoma was inhibited by anti-NKp46 mAb, whereas anti-NKp30 mAb had virtually no effect. Thus, while NKp46 appears as the major receptor involved in lysis of M14, NKp30 plays a central role in the killing of MEL15.
Figure 5.
Involvement of NKp30 and NKp46 in the tumor cell lysis mediated by NK cell clones. Three NK cell clones were analyzed for cytolytic activity against MEL15, M14, SMMC, and A549 FcγR-negative target cell lines either in the absence (white bars) or presence of AZ20 (anti-NKp30; black bars), BAB281 (anti-NKp46; striped bars) or both AZ20 and BAB281 (stippled bars) mAbs. The E/T ratio was 4:1.
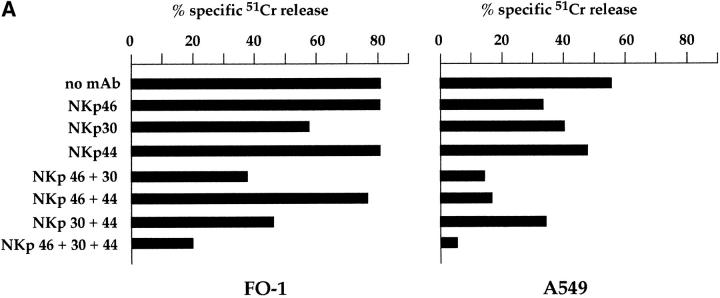
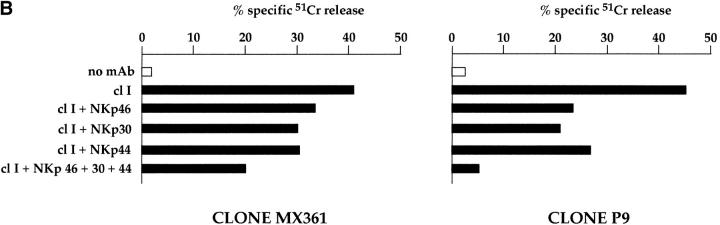
Analysis of the same NK clones in cytolytic assays against other tumor target cells such as SMMC and A549 (Fig. 5) revealed a balanced contribution of NKp46 and NKp30 to the induction of cytotoxicity. Indeed, while mAb-mediated masking of NKp46 or NKp30 alone had a moderate inhibitory effect, the simultaneous masking of the two molecules resulted in a significant inhibition. These results indicate that the two receptors may exert an additive effect in the induction of cytotoxicity against certain target cells. Cooperation in NK cell triggering was previously demonstrated for NKp46 and NKp44 23. Further analysis revealed that NKp30 could exert an additive effect in the induction of NK-mediated cytotoxicity, not only with NKp46, but also with NKp44. Fig. 6 A shows the cytolytic activity of the representative NK clone MIL69 against FO-1 or A549 tumor cells. Target cell lysis was only partially inhibited by mAb-mediated masking of NKp30, NKp44, or NKp46 receptors. However, the combined masking of two receptors resulted in a higher inhibitory effect, whereas the simultaneous masking of the three receptors gave the maximal inhibition. Isotype-matched anti-CD56 mAb had no inhibitory effect either when used alone or in combination with other mAbs (data not shown).
Figure 6.
NKp30 cooperates with NKp46 and NKp44 in the induction of NK-mediated cytotoxicity against tumor or normal autologous target cells. (A) The representative NK clone MIL69 was analyzed for cytolytic activity against FO-1 or A549 FcγR-negative target cell lines either in the absence or presence of mAbs to the indicated molecules. The following mAbs were used: KL247 (anti-NKp46), AZ20 (anti-NKp30), and KS38 (anti-NKp44). The E/T ratios were 2:1 (FO-1) and 3:1 (A549). (B) Two NK cell clones (MX361 and P9) were analyzed for cytolytic activity against autologous PHA blasts either in the absence (white bars) or presence of mAbs to the indicated molecules (black bars). The mAbs used were A6-136 (anti–HLA class I [cl I]), KL247 (anti-NKp46), KS38 (anti-NKp44), and AZ20 (anti-NKp30). The E/T ratio was 10:1.
We further analyzed the role of NKp30 alone or in combination with other receptors in cytolytic assay using PHA-induced T cell blasts as a source of normal target cells. In these experiments, lysis of autologous cells by NK cell clones was obtained by mAb-mediated masking of HLA class I molecules on target cells to disrupt the interaction with the HLA class I–specific inhibitory receptors expressed on NK cells. Also under these experimental conditions, the mAb-mediated masking of single receptors had only a partial inhibitory effect on cytotoxicity (Fig. 6 B). On the other hand, the simultaneous masking of NKp30, NKp46, and NKp44 receptors strongly reduced (or virtually abrogated) target cell lysis (see the representative clones MX361 and P9). These data support the notion that the ligands recognized by these receptors are expressed not only in tumor, but also in normal cells.
Finally, we analyzed the possible involvement of NKp30 in the recognition of murine target cells. Previous studies indicated that the NK-mediated killing of murine cells was abrogated by the mAb-mediated masking of NKp46 receptors, thus suggesting that NKp46 may represent the unique human NK receptor able to recognize ligand(s) expressed on murine cells 24 26. This hypothesis was substantiated by the recent identification of a murine homologue that shared a high degree of identity with the human NKp46 receptor 33. Although not shown, the mAb-mediated masking of NKp30 had no effect on the lysis of both BW1502 and YAC-1 murine thymoma target cells.
Altogether, the above data indicate that NKp30 functions as a major receptor involved in the NK-mediated cytotoxicity against normal target cells and most, but not all, tumor cells. In addition, NKp30 may cooperate with NKp46 and NKp44, most likely depending on the expression of specific ligands by the target cell analyzed.
Molecular Cloning of the cDNA Encoding the NKp30 Molecule.
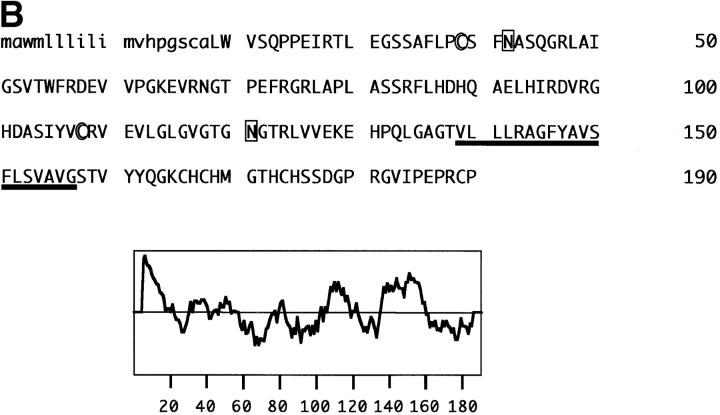
In an attempt to identify the cDNA encoding the NKp30 molecule, a cDNA expression library was generated from the mRNA of human polyclonal NK cells 24. COS-7 cells transfected with different cDNA library pools were stained with A76 mAb by an immunocytochemical detection method. A 674-bp cDNA (NK-A1 clone 5C; sequence data available from EMBL/GenBank/DDBJ under accession no. AJ223153) was isolated that contained a single open reading frame (ORF) of 573 bp. Transfection of COS-7 cells with clone 5C cDNA construct resulted in the surface expression of a molecule that was recognized by all the various anti-NKp30 mAbs (Fig. 7 A), but not by anti-NKp46 mAbs as assessed by cytofluorimetric analysis. As shown in Fig. 7 B, clone 5C ORF encoded a putative 190 amino acid polypeptide belonging to the Ig-SF, characterized by a signal peptide of 18 amino acids and by an extracellular region of 120 amino acids forming an Ig-like domain of the V type. The extracellular portion contains two potential N-linked glycosylation sites and no consensus sequences for O-linked glycosylation. A region rich in hydrophobic amino acids, potentially involved in protein–protein interactions, is connecting the Ig V-like domain with the transmembrane region. The 19 amino acid transmembrane region contains the positively charged amino acid, Arg, and the 33 amino acid cytoplasmic portion lacks typical immunoreceptor tyrosine-based activating motif (ITAM) consensus sequences. The presence of a charged amino acid in the transmembrane domain is a feature common to other triggering receptors expressed on NK cells 24 25 34 35 36 37. These charged residues are usually thought to be involved in the association with ITAM-containing signaling polypeptides.
Figure 7.
Cytofluorimetric analysis of the NKp30 molecules expressed in COS-7 cell transfectants; amino acid sequence and hydrophobicity plot of NKp30. (A) COS-7 cells transfected with clone 5C cDNA construct were stained with anti-NKp30 (A76, AZ20, and Z25) or with anti-NKp46 (BAB281) mAbs followed by PE-conjugated goat anti–mouse IgG1, and analyzed by flow cytometry. White profiles represent cells incubated with the second reagent alone (i.e., negative controls). (B) The putative signal peptide is indicated in lowercase letters, and the transmembrane region is underlined. Cysteines involved in the Ig-like fold are circled, and putative N-glycosylation sites are boxed. A Kyte-Doolittle hydrophobicity plot is shown at the bottom. DNA and protein sequence analyses were performed using GeneWorks, MacVector suites, NetOGlyc version 2.0 (available at http://www.cbs.dtu.dk/services/NetOGlyc), and PSORT Prediction Servers (available at http://psort.nibb.ac.jp:8800/). The sequence data are available from EMBL/GenBank/DDBJ under accession no. AJ223153.
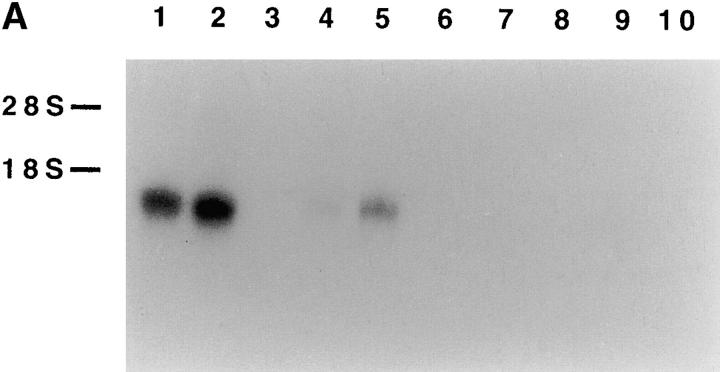
Searching EMBL/GenBank/DDBJ databases revealed that the clone 5C cDNA was identical to a previously identified alternatively spliced form of the 1C7 gene (available under accession no. AF031138). This gene has been mapped on human chromosome 6, in the TNF cluster of the MHC gene complex 38. To date, however, owing to the lack of specific mAb, neither the function nor the surface distribution of the putative product of 1C7 gene could be identified. Moreover, the 1C7 transcript could not be revealed by Northern blot on different tissues and cell lines. On the other hand, by reverse transcriptase (RT)-PCR, the 1C7 transcript could be amplified by RNA isolated from spleen (but not from other tissues) or certain lymphoid and myeloid cell lines. These data suggested that 1C7 transcripts could be poorly represented, or could be expressed at substantial levels only in a narrow range of cell types 39. Our present analysis of NKp30 expression by Northern blotting revealed a mRNA of ∼1 kb in polyclonal NK cell populations and NK cell lines, including NKL and NK3.3. On the contrary, consistent with the lack of reactivity with anti-NKp30 mAbs, no NKp30 mRNA could be detected in human monocytes or cell lines of different histotype, including U937, Jurkat, HL60, and LCL 721.221 cells (Fig. 8 A). In some of these cell lines that were negative for mRNA expression by Northern blot (and for anti-NKp30 mAb surface staining), it has been possible to detect transcripts when analyzed by the RT-PCR technique. This finding is likely to reflect a low level of NKp30 transcription resulting in lack of NKp30 surface expression. Moreover, Northern blot analysis of multiple human tissues showed selective expression of NKp30 transcript only in spleen cells (data not shown). Altogether, these data are consistent with the notion that NKp30 expression is largely NK specific.
Figure 8.
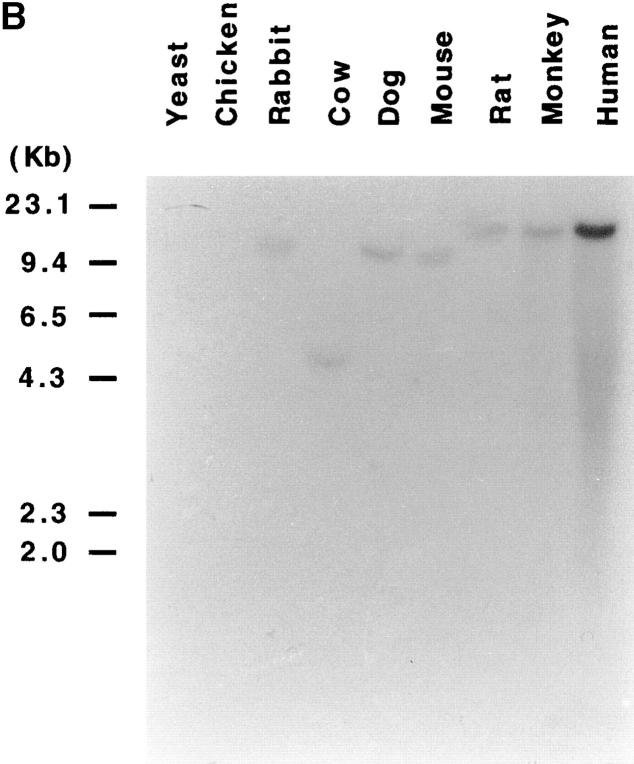
Northern blot analysis of NKp30 transcript expression and Zoo-Blot analysis. (A) Total RNA was isolated from cells of different origin. Lanes 1 and 2, polyclonal NK cell populations; lane 3, blank; lane 4, an NK cell line (NKL); lane 5, an NK cell line (NK3.3); lane 6, human monocytes; lane 7, a histiocytic lymphoma cell line (U937); lane 8, a T lymphoma cell line (Jurkat); lane 9, an acute promyelocytic leukemia cell line (HL60); and lane 10, an EBV-transformed B cell line (LCL721.221). 10 μg of each RNA preparation (2 μg of poly A+ RNA from polyclonal NK cell populations, lanes 1 and 2) were hybridized with the 421-bp NKp30 cDNA probe. The positions of 28S and 18S ribosomal RNA subunits are indicated on the left. (B) A Southern blot containing genomic DNA from humans, Rhesus monkey, Sprague-Dawley rat, BALB/c mouse, dog, cow, rabbit, chicken, and S. cerevisiae yeast was hybridized under low stringency condition with the 421-bp NKp30 cDNA probe.
Finally, the human NKp30 cDNA probe hybridized with genomic DNA from monkey, rat, mouse, dog, cow, and rabbit. These data suggest that the NKp30-encoding gene may be conserved in different species (Fig. 8 B).
Biochemical Characterization of the NKp30 Complex.
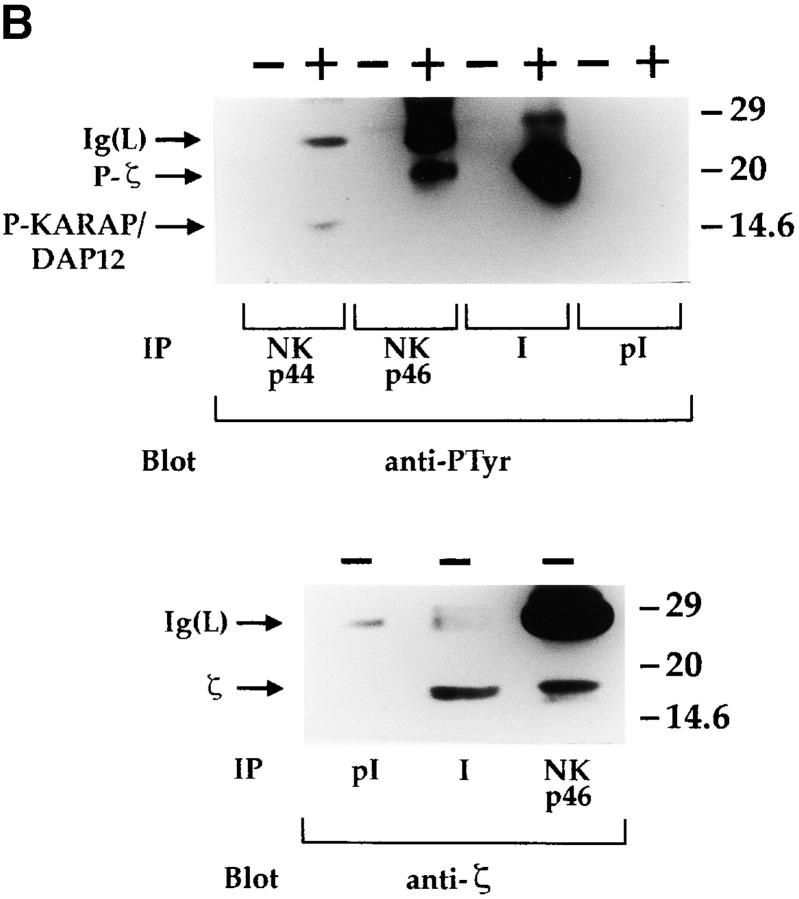
An NKp30-specific antiserum was generated by immunizing rabbits with an NH2-terminal NKp30 peptide. As shown in Fig. 9 A, the antiserum recognized in Western blot a molecule identical to that previously detected by AZ20 mAb. Unlike the AZ20 mAb, the antiserum immunoprecipitates NKp30 molecules from polyclonal NK cell populations labeled with biotin (data not shown). Thus, a polyclonal NK cell population, treated or not with sodium pervanadate, was immunoprecipitated with the NKp30-specific antiserum and probed with antiphosphotyrosine mAb. To avoid nonspecific binding of rabbit Ig to CD16 molecules, cell lysates were extensively precleared with anti-CD16 mAb. Moreover, in all experiments, preimmune rabbit serum was used as negative control. In these experiments, no tyrosine phosphorylation of NKp30 receptor could be detected (data not shown). On the other hand, NKp30 receptor associated with a molecule that became tyrosine phosphorylated upon sodium pervanadate treatment (Fig. 9 B) and comigrated with the NKp46-associated CD3ζ chain. The identity between the NKp30-associated molecule and CD3ζ polypeptides was directly demonstrated by its reactivity with anti-CD3ζ mAb (Fig. 9 B).
Figure 9.
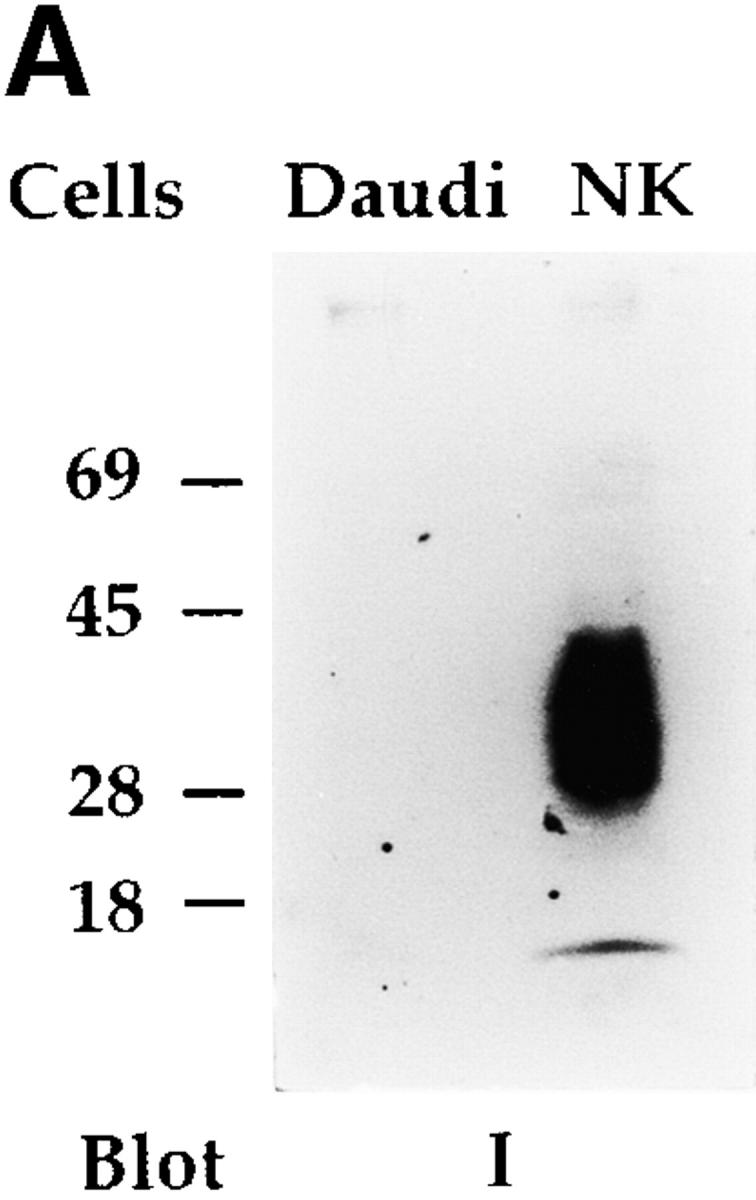
Biochemical analysis of the NKp30 receptor complex by the use of a specific antiserum. (A) Integral membrane proteins derived from Daudi (as negative control) and from a polyclonal NK cell population were analyzed in an 11% SDS-PAGE under nonreducing conditions and probed with NKp30-specific rabbit antiserum (I). Molecular weight markers (in kD) are indicated on the left. (B) 1% digitonin cell lysates derived from a polyclonal NK cell population, untreated (−) or treated (+) with sodium pervanadate, were immunoprecipitated with Z231 mAb (anti-NKp44), BAB281 mAb (anti-NKp46), NKp30-specific rabbit antiserum (I), and preimmune rabbit serum (pI). Samples were analyzed in a 15% SDS-PAGE under reducing conditions and probed with either antiphosphotyrosine (anti-PTyr) or anti-CD3ζ (anti-ζ) mAbs. Ig light chains (Ig(L)), tyrosine-phosphorylated CD3ζ (P-ζ), tyrosine-phosphorylated KARAP/DAP12 (P-KARAP/DAP12), and the nonphosphorylated form of CD3ζ (ζ) are indicated by arrows. Molecular weight markers (in kD) are indicated on the right.
Thus, NKp30, similar to other NK triggering receptors including CD16 34 35 and NKp46 23, can transduce activating signals via association with the ITAM-containing CD3ζ polypeptides. These data are in agreement with the lack of ITAM in the NKp30 cytoplasmic tail, and with the presence of a charged residue in its transmembrane portion.
Discussion
In this study, because of the generation of specific mAbs, we identified and characterized NKp30, a novel triggering receptor that plays an important role in the natural cytotoxicity of both resting and activated human NK cells. Similar to NKp46, NKp30 is selectively expressed by all NK cells, both freshly isolated and cultured in IL-2, thus representing an optimal marker for NK cell identification. Although it belongs to the Ig-SF, NKp30 does not display any substantial homology with previously identified NK receptors.
In many respects, NKp30 appeared similar to NKp46. Indeed, their parallel expression on all NK cells (including the rare CD16− cells), the existence for both of a high or low density pattern of surface expression, together with their similar functional characteristics, led to the thought that the surface molecule recognized by the new mAbs could be identical or strictly related to NKp46. However, NKp30 and NKp46 displayed different molecular masses and, functionally, appeared to play a complementary role in the induction of natural cytotoxicity. Moreover, molecular cloning revealed that NKp30 is a protein with very limited homology with NKp46, as the two molecules display only 13% identity and 15% similarity, and are encoded by genes located on different chromosomes.
The receptors responsible for the NK cell triggering during natural cytotoxicity and tumor cell lysis have remained elusive until recently. Available data were consistent with the hypothesis of the existence of multiple triggering NK receptors involved in natural cytotoxicity. In this context, we recently identified NKp46 and NKp44, two receptors involved in recognition and lysis of a variety of tumor targets. Both belong to the Ig-SF, but neither displays significant identity. They associate to different signal transducing polypeptides (CD3ζ/Fc∈RIγ and KARAP/DAP12, respectively) that become tyrosine phosphorylated upon NK cell activation. NKp46 and NKp44 were shown to cooperate in the process of tumor cell lysis by human NK cells. However, lysis of certain target cells was only marginally NKp46 and/or NKp44 dependent, since mAb-mediated masking of these molecules did not significantly interfere with cytotoxicity 26. This finding strongly suggested the existence of additional triggering receptors that could induce cytotoxicity against these target cells. Moreover, although clearly NKp46 and/or NKp44 dependent, the cytolytic activity against other tumor cell lines could not be abrogated by mAb-mediated masking of both molecules, suggesting again the existence of additional receptor(s) cooperating with NKp46 and NKp44. Indeed, we show here that NKp30 represents a receptor that may cooperate with NKp46 and NKp44 in the induction of cytotoxicity against a variety of target cells. Perhaps more importantly, NKp30 represents the major receptor in inducing NK-mediated killing of certain tumor target cells, the lysis of which is largely NKp46/NKp44 independent. Remarkably, NKp30, similar to NKp46, is also involved in NK cell activation and target cell killing by fresh NK cells.
As discussed above, the surface expression of NKp30 parallels that of NKp46. Indeed, NK cells displaying an NKp46dull or an NKp46bright phenotype were also characterized by NKp30dull or NKp30bright fluorescence. We showed previously that NK cell clones characterized by an NKp46dull phenotype consistently express low amounts of NKp44 26. The finding that NK cells express parallel densities of different triggering receptors may explain the existence of NK cell subsets displaying different “natural” cytolytic activity. For example, it was difficult to understand why the cytolytic activity against some target cells (such as MEL15), although largely NKp46 independent, was essentially confined to NK clones expressing the NKp46bright phenotype. These results can now be explained by the finding that only NKp46bright cells express a high density of NKp30 receptor. Thus, the previous demonstration of major differences in cytolytic activity of NKp46dull and NKp46bright cells can now be applied also to NK cells displaying different NKp30 phenotypes. Along this line, the cytolytic activity of NKp30dull NK cell clones was markedly reduced compared with NKp30bright clones (data not shown).
NKp30, similar to NKp46, associates with CD3ζ, which is most likely involved in signaling via the receptor complex. However, CD3ζ does not appear to be necessary for the surface expression of both receptors, at least in COS-7 cells (this report, and reference 24). Molecular cloning revealed that NKp30 is the product of 1C7, a gene previously mapped on human chromosome 6 in the HLA class III region 38 39. However, neither the function nor the cellular distribution of the putative product of 1C7 gene was known, and no indications existed of its role in natural cytotoxicity. In addition, the analysis of 1C7 transcript expression was limited to RT-PCR, whereas no detection had been possible by Northern blot analysis 39. It should also be stressed that no correlation between transcript and surface expression could be established because of the lack of specific mAbs. In this study, we show that a precise correlation exists between the surface expression of NKp30, as determined by staining with three different mAbs, and mRNA expression, as assessed by Northern blot. On the contrary, the detection of 1C7 transcripts by RT-PCR does not allow prediction of the surface expression of the 1C7/NKp30 molecule.
In conclusion, the NKp30 molecule represents a third member of an emerging family of receptors, termed natural cytotoxicity receptors (NCRs; 40), that are involved in NK cell triggering upon recognition of non-HLA ligands. These receptors appear to complement each other in the induction of target cell lysis by NK cells. The relative contribution of each receptor is likely to reflect the expression/density of their specific ligands on target cells. It has recently been shown that CD16 is also involved in natural cytotoxicity, thus suggesting that in addition to Fc binding and antibody-dependent cell-mediated cytotoxicity, CD16 may play a role in the regulation of NK cell function 41. Besides CD16 and the different NCRs, several other surface molecules that can mediate NK cell triggering have been identified in humans and rodents. These include CD2 42 43, CD69 44, CD28 45, 2B4 46, and NKR-P1 47. However, their actual role in natural cytotoxicity has yet to be clarified, since in most instances these activating structures are not NK restricted.
Finally, although the identification of different NCRs constitutes a major step forward in our understanding of the NK cell physiology, both the nature and the distribution of the NCR ligands on target cells remain to be determined. Based on the available data, it is possible to envisage a novel mechanism of tumor escape consisting in the downregulation (on tumor cells) of ligand molecules specifically recognized by NK-specific triggering receptors. Thus, the identification of such ligands will allow the analysis of their distribution in normal versus tumor cells, and define whether a correlation exists between ligand expression and susceptibility to NK-mediated lysis by different tumor cells.
Acknowledgments
This work was supported by grants awarded by Associazione Italiana per la Ricerca sul Cancro, Istituto Superiore di Sanità, Ministero della Sanità, Ministero dell'Università e della Ricerca Scientifica e Tecnologica, and Consiglio Nazionale delle Ricerche, Progetto Finalizzato Biotecnologie.
Footnotes
1used in this paper: GAM, goat anti–mouse antiserum; HRPO, horseradish peroxidase; Ig-SF, Ig superfamily; ITAM, immunoreceptor tyrosine-based activating motif; KIR, killer inhibitory receptor; NCR, natural cytotoxicity receptor; ORF, open reading frame; RT, reverse transcriptase
References
- Moretta A., Biassoni R., Bottino C., Pende D., Vitale M., Poggi A., Mingari M.C., Moretta L. Major histocompatibility complex class I-specific receptors on human natural killer and T lymphocytes. Immunol. Rev. 1997;155:105–117. doi: 10.1111/j.1600-065x.1997.tb00943.x. [DOI] [PubMed] [Google Scholar]
- Lanier L.L. NK cell receptors. Annu. Rev. Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- Long E.O. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 1998;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- Ljunggren H.-G., Karre K. In search of the missing ‘self’MHC molecules and NK cell recognition. Immunol. Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- Renard V., Cambiaggi A., Vély F., Bléry M., Olcese L., Olivero S., Bouchet M., Vivier E. Transduction of cytotoxic signals in natural killer cellsa general model of fine tuning between activatory and inhibitory pathways in lymphocytes. Immunol. Rev. 1997;155:205–221. doi: 10.1111/j.1600-065x.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]
- Ciccone E., Pende D., Vitale M., Nanni L., Di Donato C., Bottino C., Morelli L., Viale O., Amoroso A., Moretta A., Moretta L. Self class I molecules protect normal cells from lysis mediated by autologous natural killer cells. Eur. J. Immunol. 1994;24:1003–1006. doi: 10.1002/eji.1830240434. [DOI] [PubMed] [Google Scholar]
- Moretta A., Tambussi G., Bottino C., Tripodi G., Merli A., Ciccone E., Pantaleo G., Moretta L. A novel surface antigen expressed by a subset of human CD3−CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J. Exp. Med. 1990;171:695–714. doi: 10.1084/jem.171.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A., Bottino C., Pende D., Tripodi G., Tambussi G., Viale O., Orengo A.M., Barbaresi M., Merli A., Ciccone E., Moretta L. Identification of four subset of human CD3−CD16+ NK cells by the expression of clonally distributed functional surface moleculescorrelation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J. Exp. Med. 1990;172:1589–1598. doi: 10.1084/jem.172.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagtmann N., Biassoni R., Cantoni C., Verdiani S., Malnati M., Vitale M., Bottino C., Moretta L., Moretta A., Long E.O. Molecular clones of the p58 NK cell receptor reveal Ig-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- Colonna M., Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- Vitale M., Sivori S., Pende D., Augugliaro R., Di Donato C., Amoroso A., Malnati M., Bottino C., Moretta L., Moretta A. Physical and functional independency of p70 and p58 NK cell receptors for HLA class Itheir role in the definition of different groups of alloreactive NK cell clones. Proc. Natl. Acad. Sci. USA. 1996;93:1453–1457. doi: 10.1073/pnas.93.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A., Chang C., Franz-Bacon K., McClanahan T., Phillips J.H., Lanier L.L. Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J. Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- Pende D., Biassoni R., Cantoni C., Verdiani S., Falco M., Di Donato C., Accame L., Bottino C., Moretta A., Moretta L. The natural killer cell receptor specific for HLA-A allotypesa novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140 kD disulphide-linked dimer. J. Exp. Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohring C., Scheidegger D., Samaridis J., Cella M., Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- Cosman D., Fanger N., Borges L., Kubin M., Chin W., Peterson L., Hus M.L. A novel immunoglobilin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- Samaridis J., Colonna M. Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cellsstructural evidence for new stimulatory and inhibitory pathways. Eur. J. Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- Sivori S., Vitale M., Bottino C., Marcenaro E., Sanseverino L., Parolini S., Moretta L., Moretta A. CD94 functions as a natural killer cell inhibitory receptor for different HLA class I allelesidentification of the inhibitory form of CD94 by the use of novel monoclonal antibodies. Eur. J. Immunol. 1996;26:2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- Braud V.M., Jones E.Y., McMichael A.J. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- Lee N., Llano M., Carretero M., Ishitani A., Navarro F., Lopez-Botet M., Geraghty D. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido F., Ruiz-Cabello F., Cabrera R., Pérez-Villar J.J., López-Botet M., Duggan-Keen M., Stern P.L. Implications for immunosurveillance of altered HLA-class I phenotypes in human tumours. Immunol. Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
- Pende D., Accame L., Pareti L., Mazzocchi A., Moretta A., Parmiani G., Moretta L. The susceptibility to natural killer cell-mediated lysis of HLA class I-positive melanomas reflects the expression of insufficient amounts of HLA class I alleles. Eur. J. Immunol. 1998;28:2384–2394. doi: 10.1002/(SICI)1521-4141(199808)28:08<2384::AID-IMMU2384>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Sivori S., Vitale M., Morelli L., Sanseverino L., Augugliaro R., Bottino C., Moretta L., Moretta A. p46, a novel natural killer cell–specific surface molecule that mediates cell activation. J. Exp. Med. 1997;186:1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale M., Bottino C., Sivori S., Sanseverino L., Castriconi R., Marcenaro R., Augugliaro R., Moretta L., Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells is involved in non–MHC restricted tumor cell lysis. J. Exp. Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessino A., Sivori S., Bottino C., Malaspina A., Morelli L., Moretta L., Biassoni R., Moretta A. Molecular cloning of NKp46a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni C., Bottino C., Vitale M., Pessino A., Augugliaro R., Malaspina A., Parolini S., Moretta L., Moretta A., Biassoni R. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J. Exp. Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S., Pende D., Bottino C., Marcenaro E., Pessino A., Biassoni R., Moretta L., Moretta A. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur. J. Immunol. 1999;29:1656–1666. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Moretta A. Frequency and surface phenotype of human T lymphocytes producing interleukin 2. Analysis by limiting dilution and cell cloning. Eur. J. Immunol. 1985;151:148–155. doi: 10.1002/eji.1830150208. [DOI] [PubMed] [Google Scholar]
- Poggi A., Pardi R., Pella N., Morelli L., Sivori S., Vitale M., Revello V., Moretta A., Moretta L. CD45-mediated regulation of LFA1 function in human natural killer cells. Anti-CD45 monoclonal antibodies inhibit the calcium mobilization induced via LFA1 molecules. Eur. J. Immunol. 1993;23:2445–2463. doi: 10.1002/eji.1830231012. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Brakenhoff R.H., Gerretsen M., Knippels E.M.C., van Dijk M., van Essen H., Weghuis D.O., Sinke R.J., Snow G.B., van Dongen G.A.M.S. The human E48 antigen, highly homologous to the murine Ly-6 antigen ThB, is a GPI-anchored molecule apparently involved in keratinocyte cell–cell adhesion. J. Cell. Biol. 1995;129:1677–1689. doi: 10.1083/jcb.129.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biassoni R., Ferrini S., Prigione I., Moretta A., Long E.O. CD3-negative lymphokine-activated cytotoxic cells express the CD3 epsilon-gene. J. Immunol. 1988;140:1685–1689. [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. Molecular cloningA Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1982. [Google Scholar]
- Biassoni R., Pessino A., Bottino C., Pende D., Moretta L., Moretta A. The murine homologue of the human NKp46, a triggering receptor involved in the induction of natural cytotoxicity. Eur. J. Immunol. 1999;29:1014–1020. doi: 10.1002/(SICI)1521-4141(199903)29:03<1014::AID-IMMU1014>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lanier L.L., Yu G., Phillips J.H. Co-association of CD3ζ with a receptor (CD16) for IgG Fc on human natural killer cells. Nature. 1989;342:803–805. doi: 10.1038/342803a0. [DOI] [PubMed] [Google Scholar]
- Vivier E., Rochet N., Kochan J.P., Presky D.H., Schlossman S.F., Anderson P. Structural similarity between Fc receptors and T cell receptors. Expression of the γ subunit of the Fc∈RI in human T cells, natural killer cells and thymocytes. J. Immunol. 1991;147:4263–4270. [PubMed] [Google Scholar]
- Olcese L., Cambiaggi A., Semenzato G., Bottino C., Moretta A., Vivier E. Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells. J. Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- Lanier L.L., Corliss B., Wu J., Phillips J.H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- Nalabolu S.R., Shukla H., Nallur G., Parimoo S., Weissman S.M. Genes in a 220-kb region spanning the TNF cluster in human MHC. Genomics. 1996;31:215–222. doi: 10.1006/geno.1996.0034. [DOI] [PubMed] [Google Scholar]
- Neville M.J., Campbell R.D. A new member of the Ig superfamily and a V-ATPase G subunit are among the predicted products of novel genes close to the TNF locus in the human MHC. J. Immunol. 1999;162:4745–4754. [PubMed] [Google Scholar]
- Moretta A., Biassoni R., Bottino C., Mingari M.C., Moretta L. The “natural cytotoxicity receptors” which trigger the NK-mediated cytolysiselusive no more. Immunol. Today. 1999;In press doi: 10.1016/s0167-5699(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Mandelboim O., Malik P., Davis D.M., Jo C.H., Boyson J.E., Strominger J.L. Human CD16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc. Natl. Acad. Sci. USA. 1999;96:5640–5644. doi: 10.1073/pnas.96.10.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis R.L., Roozemond R.C., van de Griend R.J. Induction and blocking of cytolysis in CD2+, CD3− NK and CD2+, CD3+ cytotoxic T lymphocytes via the CD2 50kD sheep erythrocyte receptor. J. Immunol. 1986;136:3939–3944. [PubMed] [Google Scholar]
- Lanier L.L., Corliss B., Phillips J.H. Arousal and inhibition of human NK cells. Immunol. Rev. 1997;155:145–154. doi: 10.1111/j.1600-065x.1997.tb00947.x. [DOI] [PubMed] [Google Scholar]
- Moretta A., Poggi A., Pende D., Tripodi G., Orengo A.M., Pella N., Augugliaro R., Bottino C., Ciccone E., Moretta L. CD69-mediated pathway of lymphocyte activationanti-CD69 monoclonal antibodies trigger the cytolytic activity of different lymphoid effector cells, with the exception of cytolytic T lymphocytes expressing TCR α/β. J. Exp. Med. 1991;174:1393–1398. doi: 10.1084/jem.174.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma M., Cayabyab M., Buck D., Phillips J.H., Lanier L.L. Involvement of CD28 in MHC-unrestricted cytotoxicity mediated by a human NK leukemia cell line. J. Immunol. 1992;149:1115–1123. [PubMed] [Google Scholar]
- Mathew P.A., Garni-Wagner B.A., Land K., Takashima A., Stoneman E., Bennett M., Kumar V. Cloning and characterization of the 2B4 gene encoding a molecule associated with non-MHC-restricted killing mediated by activated natural killer cells and T cells. J. Immunol. 1993;151:5328–5337. [PubMed] [Google Scholar]
- Bezouska K., Yuen C., O'Brien J., Childs R.A., Chai W., Lawson A.M., Drbal K., Fiserova A., Pospisil M., Feizi T. Oligosaccharide ligands for NKR-P1 protein activate NK cells and cytotoxicity. Nature. 1994;372:150–157. doi: 10.1038/372150a0. [DOI] [PubMed] [Google Scholar]