Abstract
T helper (Th) lymphocytes, when reactivated, recall expression of those cytokines they had been instructed to express in earlier activations, even in the absence of specific cytokine-inducing factors. In cells that memorize their expression, the cytokine genes are modified by chromatin rearrangement and demethylation, suggesting that they have been somatically imprinted. Here we show, by using inhibitors blocking the cell cycle in various stages, that for the instruction of a Th cell to express interleukin (IL)-4 or IL-10 upon restimulation, entry of the cell into the S phase of the first cell cycle after initial activation is required. Separation of the IL-4 receptor (IL-4R) and T cell antigen receptor (TCR) signals in time, demonstrates that this instruction is dependent on concomitant signaling from both receptors. In Th cells, inhibited to progress into the first S phase after activation, the IL-4R and TCR signals can be memorized for at least 1 d, priming the T cell to become instructed for expression of IL-4 upon restimulation, when entering the S phase after release of the cell cycle block. The requirement of the initial S phase of T cell activation, for instruction of Th cells to express IL-4 or IL-10 upon restimulation points to the decisive role of epigenetic modification of cytokine genes as a molecular correlate of the memory to express particular cytokines.
Keywords: cytokine memory, T cell differentiation, interleukin 4, interleukin 10, CFSE
Thelper (Th) lymphocytes, by selective expression of cytokines, control immune reactions and call up distinct immune effector functions (for a review, see reference 1). Upon primary activation, naive T cells become instructed for expression of cytokines by costimulatory signals, such as CD28 for IL-2 (for a review, see reference 2), IL-4 for IL-4 3, and IL-12 for IFN-γ expression 4 5. In Th cells, expression of cytokines is transient. After primary activation of Th cells, cytokines are expressed for several days 6 7 8 9. Later, the expression of cytokines is memorized by Th cells. When restimulated, they recall expression of those cytokines they had been instructed to express earlier, without requirement for the original costimulator, and to some extent even in the presence of adverse costimulators 10 11 12 13 14. The molecular basis for cytokine memory in Th lymphocytes is not clear. Recently, several groups have provided evidence for somatic imprinting of cytokine genes in T cells expressing them. Analysis of DNA methylation and DNase I hypersensitivity of the genomic regions of various cytokine genes 15 16 17 18 19 20 has shown that those cytokine genes that the T cells had originally expressed remained “accessible” later, and that accessibility is correlated with the ability for expression of the cytokines upon recall stimulation.
The mechanism of epigenetic imprinting of cytokine genes remains obscure. Chromatin remodeling of the IL-4 locus is dependent not only on IL-4R signaling, but also on concomitant TCR signaling 20. It is not yet clear how the TCR and IL-4R signaling pathways are connected to mechanisms for epigenetic modification. Recently, several groups have suggested that the instruction of Th cells to express cytokines upon restimulation may relate to proliferation of Th cells after activation. Using carboxyfluorescein to analyze cytokine expression in correlation with the proliferative history of activated Th cells, Gett and Hodgkin 21 have claimed that distinct cytokines are expressed in Th cells that had undergone distinct numbers of cell cycles, independent of the duration of Th cell activation. Bird et al. 19, basically using the same approach, have claimed that for the instruction to express IFN-γ upon restimulation, a Th cell must have entered the S phase of the first cell cycle after activation, whereas for the instruction to express IL-4, the cell would have to progress through at least three full cell cycles. Thus, cytokine genes would differ in their ability to count cell cycles and convert this information into a stable imprint for expression.
We show here that naive Th cells, when activated, are able to express IL-4 and IL-10 upon restimulation before they have divided. The instruction for expression of IL-4 requires the entry of the cell into the S phase of the first cell cycle after onset of primary activation. Instruction to express IL-10 requires progression of the activated T cell into the M phase. Priming of Th cells to express IL-4 upon restimulation is induced by concomitant signaling through the TCR and IL-4R and does not require entry of the activated cell into the cell cycle.
Materials and Methods
Mice.
Mice homozygously transgenic (tg)1 for the DO11.10 α/β-TCR (OVA-TCRtg/tg 22) on BALB/c background (gift from Dennis Y. Loh, Washington University School of Medicine, St. Louis, MO) and BALB/c mice were bred under specific pathogen-free conditions in our animal facility. All animal experiments were performed in accordance with institutional, state, and federal guidelines.
Isolation of Naive CD62L+CD4+ Cells and Labeling with CFSE.
Magnetic isolation of naive CD62L+CD4+ T cells was performed using two-parameter high-gradient magnetic cell separation (MACS [11]). Splenic cells from OVA-TCRtg/tg mice were stained with FITC-conjugated anti-CD4 mAb (GK1.5 23) and MultiSort anti-FITC microbeads (Miltenyi Biotec). CD4+ cells were isolated by positive selection on VS+ columns using the MidiMACS system (Miltenyi Biotec 24). After release of MultiSort microbeads, the CD4+ cells were stained with anti-CD62L MACS microbeads (Miltenyi Biotec). CD62L+CD4+ cells were positively selected on VS+ columns to a purity of >99%, as determined by cytometric analysis.
Labeling of naive CD62L+CD4+ cells with 5-(and 6-)carboxyfluorescein diacetate, succinimidyl ester (CFSE; Molecular Probes) was performed as described 25. In brief, cells were washed and resuspended at a concentration of 107/ml in PBS. CFSE was added at a final concentration of 5 μM and incubated for 5 min at room temperature. The reaction was stopped by washing the cells with RPMI 1640 (Life Technologies) containing 10% FCS (Sigma Chemical Co.).
Cell Culture.
Cell cultures were set up with 2 × 106 cells/ml in complete RPMI 1640, containing 10% FCS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.3 mg/ml glutamine, and 10 μM 2-ME. The antigenic peptide OVA323–339 (Neosystem) was used at 0.5 μM. Irradiated spleen cells (3,000 rad) from BALB/c mice were used as APCs for OVA-TCRtg/tg T cells at a 5:1 ratio. Th cell cultures were split on day 2 or day 3.
Recombinant murine IL-12 (gift from Maurice Gately, Hoffmann-La Roche, Nutley, NJ) was used at 1 ng/ml and neutralizing anti–IL-4 mAb (11B11 26) at 6 μg/ml. IL-4 was added at 30 ng/ml (culture supernatant of murine myeloma cell line P3-X63 Ag.8.653 transfected with murine IL-4 cDNA 27), anti–IFN-γ mAb (AN18.17.24 28) at 5 μg/ml, and anti–IL-12 mAb (C17.8.6 29) at 6 μg/ml. Recombinant human IL-2 (Hoffmann-La Roche) was used at 50 U/ml where indicated.
Progression of the cell cycle was inhibited by supplement of the following drugs: 300 μM l-mimosine (ICN 30), 2 μg/ml aphidicolin (Sigma Chemical Co. 31), 1 μg/ml nocodazole (Sigma Chemical Co. 32), or 200 nM paclitaxel (ICN 33). In the presence of the inhibitors, 86–93% of the Th cells were viable on day 2 and 62–83% on day 3, with 83% for l-mimosine. Without inhibitors, 95% viable Th cells were detectable on day 2 and 90% on day 3 according to staining with propidium iodide, in the experiment shown in Fig. 4. Calculating back from the numbers of Th cells in the various generations to the number of naive Th cells that had proliferated in culture, an estimated >50% of originally seeded cells were represented even after 4 d of culture. Also in the cultures with cell cycle inhibitors, 40–60% of the starting Th cells were still alive on day 2, and at least 20% on day 4, in the presence of l-mimosine. In all experiments using cell cycle inhibitors, they were also added to washing buffers and during the restimulation with PMA/ionomycin.
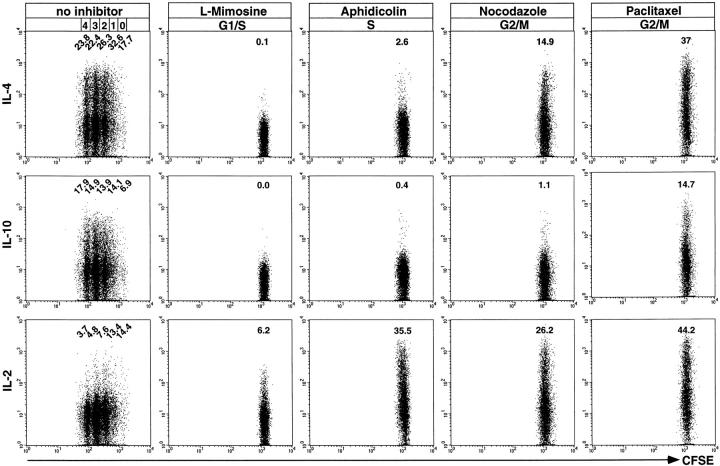
Figure 4.
Instruction for IL-4 and IL-10 expression during the first cell cycle after primary activation. CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were activated with OVA323–339 and APCs in the presence of IL-4, anti–IL-12, and anti–IFN-γ for 3 d either with or without inhibitors of cell cycle progression. After restimulation with PMA/ionomycin in the presence or absence of cell cycle inhibitors, cells were analyzed for the expression of IL-4, IL-10, and IL-2. Intracellular stainings for IL-4 and IL-10 were controlled by blocking the digoxigenized anticytokine detection mAbs with an excess of the respective unconjugated anticytokine mAbs (data not shown). Frequencies of cytokine-positive OVA-TCR+ cells in the indicated generations are corrected for background frequencies.
Activation of Th cells was controlled by staining with anti-CD25 (PharMingen), anti-CD44 (IM7.8.1 34), anti-CD62L (MEL14 35), and anti-CD69 (PharMingen). In the experiment described in Fig. 7, activation of naive Th cells immediately after isolation resulted in 76% CD25+ Th cells 36 h after onset of activation. Without TCR triggering, 17–30% and 8–25% of the originally seeded naive Th cells survived for 72 and 96 h, respectively. Cells treated with IL-4 showed the higher and cells cultivated without IL-4 the lower survival rate. Of the remaining Th cells, 50–62.5% and 41.9–59%, cultivated for 72 and 96 h respectively, reacted to TCR stimulation with upregulation of CD25, analyzed 36 h after activation by antigen and APCs.
Figure 7.
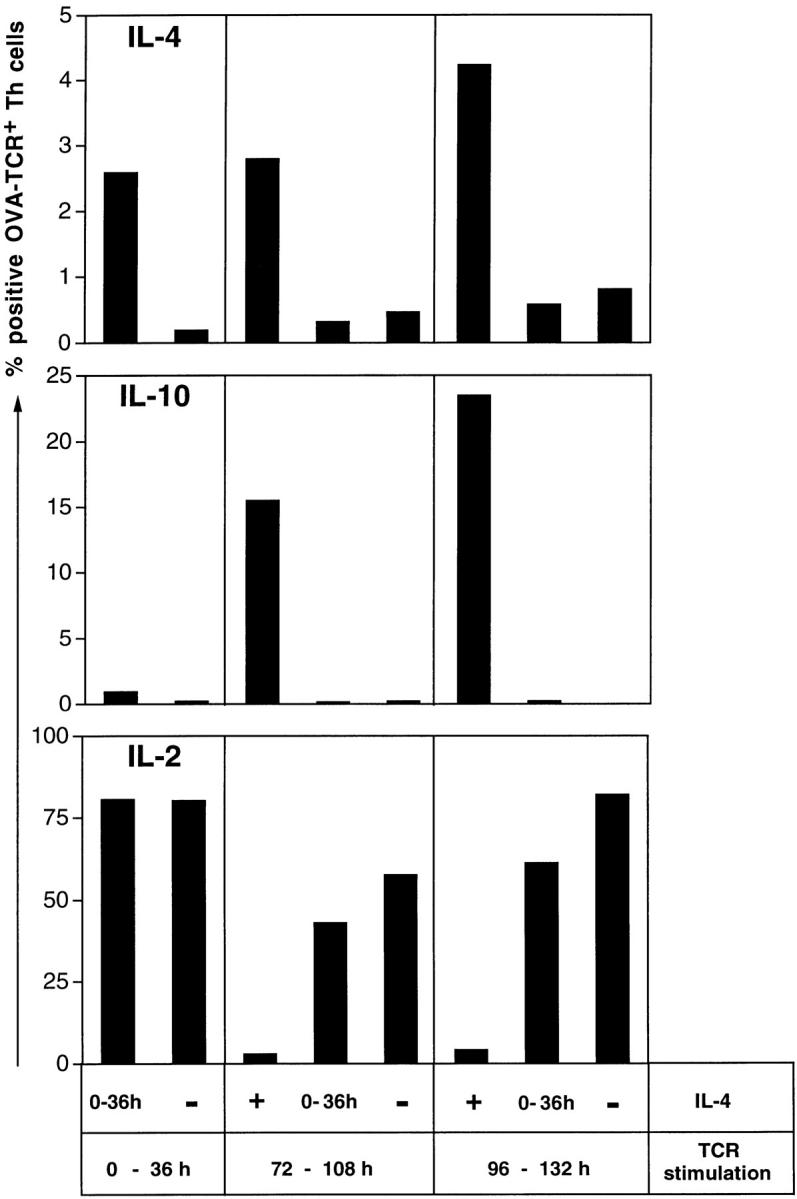
Priming for IL-4 instruction is dependent on both IL-4R and TCR signaling. CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were cultivated either in the presence of IL-4 (+) or with anti–IL-4 (−) for the entire culture period, or with IL-4 for the first 36 h (0–36 h). OVA323–339 and APCs were either added directly after isolation of the naive Th cells (0 – 36 h TCR stimulation), or after 72 h (72 – 108 h TCR stimulation) or 96 h (96 – 132 h TCR stimulation). Anti–IL-12 and anti–IFN-γ were added to all cultures. In all cultures, the antigen was withdrawn 36 h after it was added. On day 4 after stimulation with OVA323–339 and APCs, cells were restimulated with PMA/ionomycin and examined for expression of IL-4, IL-10, and IL-2. The specificities of the intracellular stainings for IL-4 and IL-10 were controlled by blocking the digoxigenized anticytokine detection mAbs with an excess of the respective unconjugated anticytokine mAbs (data not shown). Indicated percentages among the OVA-TCR+ cells are corrected for the respective control values.
Four-Color Cytometric Analysis of Expression of Intracellular Cytokines and Surface Markers versus Proliferation.
Cells (2 × 106/ml) were restimulated with 10 ng/ml PMA and 1 μg/ml ionomycin (both Sigma Chemical Co.) for 5 h. Brefeldin A (Sigma Chemical Co.) was added at 5 μg/ml for the last 3 h of stimulation. Cells were then fixed with 2% formaldehyde 6. For intracellular cytokine staining, cells were permeabilized with 0.5% saponin (Sigma Chemical Co.) in PBS/BSA/azide and incubated with the following cytokine-specific mAbs: PE-coupled anti–IL-4 (11B11), anti–IL-5 (TRFK5), and anti–IL-10 (JES5-16E3) at 3 μg/ml (PharMingen); digoxigenin (Dig)-conjugated anti–IL-2 (S4B6 36) at 3 μg/ml, anti–IL-4 (11B11) at 1 μg/ml, anti–IL-10 (JES5-2A5 [37, 38]) at 1.5 μg/ml, anti–IFN-γ (AN18.17.24) at 1 μg/ml, and anti–TNF-α (MP6-XT22; PharMingen) at 1 μg/ml. Digoxigenized primary mAbs were detected with anti-Dig Fab fragments (Boehringer Mannheim) conjugated to Cy5. Anti-Dig–Cy5 and PE-labeled isotype control mAbs (3 μg/ml; PharMingen) were used as controls. In parallel, the specificity of the staining for intracellular IL-4 and IL-10 was controlled by blocking the staining of digoxigenized anticytokine mAbs by incubating the cells 15 min before and during the staining of IL-4 and IL-10, with an excess of the respective unconjugated anticytokine mAbs. OVA-TCRtg/tg cells were identified according to staining with the clonotypic mAb KJ1-26.1 39. Four-color cytometric analysis of intracellular cytokines and cell proliferation was performed by gating on KJ1-26.1+ lymphocytes and on individual cell generations, identified by CFSE staining. In cells of these subpopulations, coexpression of two cytokines was analyzed as indicated. The population of undivided Th cells was identified according to CFSE staining intensity of nonactivated, KJ1-26.1− Th cells in the same cultures.
Cytometric analysis was performed on a FACSCalibur™ using CELLQuest™ research software (Becton Dickinson). Dead cells were excluded according to forward and side scatter and staining with propidium iodide (0.4 μg/ml).
Statistical Analysis of Cytokine Coexpression.
The observed (obs.) frequencies of cytokine-coexpressing cells were compared with the expected (exp.) values calculated for random coincidence of two independent variables using the test for phi (φ) correlation coefficients (values ranging from −1 to 1) 40. A φ value of −1 indicates that two analyzed cytokines are never coexpressed, a value of 0 identifies random coexpression, and a φ coefficient of 1 marks a pair of cytokines that is always coexpressed in individual cells. We observed only φ values ≥ 0 41.
Results
Instruction of Th Cells to Express Cytokines upon Restimulation Is Correlated with But Not Dependent on Cell Proliferation.
Here we analyze whether cellular proliferation is involved in the regulation of cytokine expression during Th cell differentiation from naive precursors into cytokine-producing cells. Naive CD62L+CD4+ cells were purified to >99% from the spleens of OVA-TCRtg/tg BALB/c mice by two-parameter MACS. The isolated cells were labeled with CFSE, allowing for the cytometric discrimination of individual generations of proliferating cells. CFSE-labeled, naive, OVA-specific Th cells were stimulated with the antigenic peptide OVA323–339 and congenic APCs, i.e., irradiated BALB/c splenocytes, either in the presence of IL-12 and anti–IL-4, to analyze expression of IFN-γ, IL-2, and TNF-α, or in the presence of IL-4, anti–IL-12, and anti–IFN-γ, to analyze expression of IL-4, IL-5, and IL-10. Expression of the cytokines was evaluated by intracellular immunofluorescence, after recall stimulation of the cells with PMA/ionomycin, at various time points after primary activation. The frequencies of cytokine-expressing Th cells were determined for each T cell population that had undergone a defined number of cell divisions, according to CFSE staining, as shown in Fig. 1 A. In Fig. 1 B, the frequencies of cells capable of expressing the various cytokines at different time points of stimulation are compared for cells of different generations. Cells that had not divided could be analyzed up to day 2 in IL-4–supplemented cultures and up to day 3 in IL-12–supplemented cultures. In those cells, expression of IFN-γ, IL-2, TNF-α, IL-4, and IL-10 was clearly detectable, revealing that the expression of these cytokines in Th cells does not require cell division. For IL-5, the low frequency of expressing cells did not allow us to make a reliable statement.
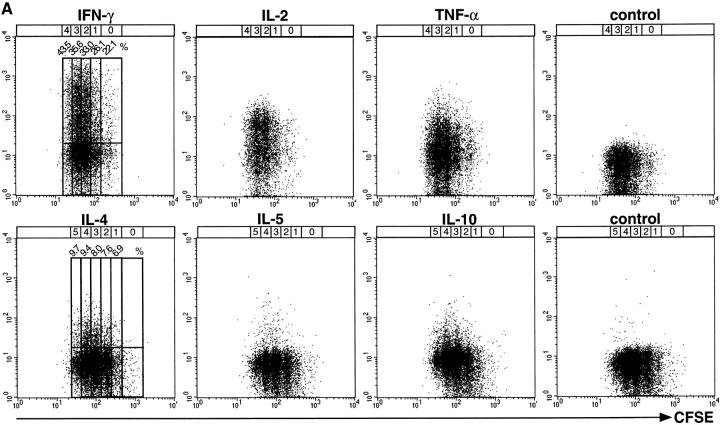
Figure 1.
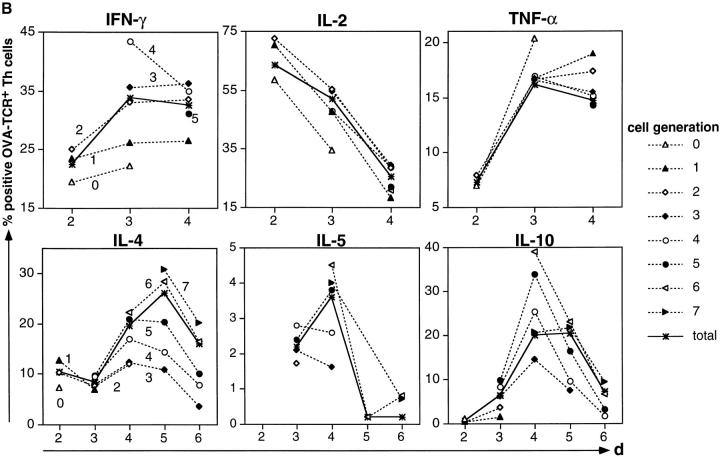
Correlation of cytokine expression with proliferation of activated Th lymphocytes. CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were stimulated with OVA323–339 and APCs either in the presence of IL-12 and anti–IL-4 for the analysis of IFN-γ, IL-2, and TNF-α expression, or with IL-4, anti–IL-12, and anti–IFN-γ for the analysis of IL-4, IL-5, and IL-10 expression. Cells were restimulated with PMA/ionomycin after 2–6 d. After fixation and permeabilization, the cells were intracellularly stained for cytokines. (A) Flow-cytometric analysis of cytokine expression in OVA-TCR+ Th cells of different generations, restimulated at day 3 after primary activation. For analysis, gates were set on OVA-TCR+ Th cells, identified by the clonotype-specific mAb KJ1-26. To determine the frequencies of cytokine-expressing OVA-TCR+ Th cells, regions were set according to the control stainings, using the secondary detection antibody or PE-conjugated isotype control mAb. The percentages of IFN-γ– and IL-4–expressing Th cells are plotted for the individual cell generations. The number and position of individual cell generations are indicated above the dot plots according to CFSE intensity of nondivided and nonstimulated OVA-TCR− Th cells. (B) Percentages of cytokine-positive OVA-TCR+ Th cells at different time points after priming within the total cell populations (solid line), and in cells of different generations (dashed lines). Indicated frequencies include subtraction of the respective control staining values (background frequencies).
In cells that had divided from one up to seven times, we observed expression of all cytokines analyzed, generally at higher frequencies in populations that had divided more often. For example, IL-4 was expressed on day 5 by 10.8% of the cells that had divided three times but by 30.7% of the cells that had divided seven times. IL-10 was expressed on day 5 by 7.4% of cells divided three times and by 23.1 and 21.8% of cells divided six and seven times. Within a given generation, the frequencies of cytokine-producing cells were not constant but varied over time of stimulation. For cells divided five times, the frequencies of IL-4–expressing cells rose from 9.7% on day 3 to 20.9 and 20.4% on days 4 and 5 and then dropped to 10.1% on day 6. In summary, the ability of primarily activated Th cells to produce IL-4, IL-10, and IFN-γ upon restimulation is correlated with but not dependent on cell proliferation.
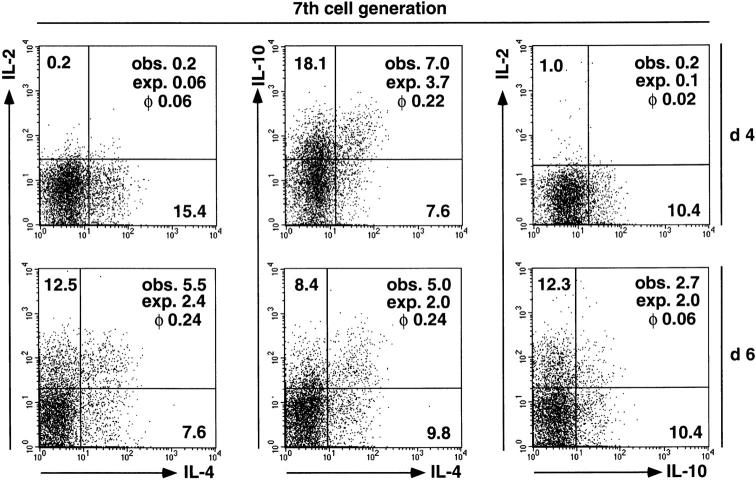
The degree of coexpression of the cytokines IL-2, IL-4, and IL-10 is not correlated with the proliferative activity of Th cells, showing that the Th cells are not sequentially instructed to express the various cytokine genes upon restimulation. φ correlation coefficients for the simultaneous expression of two cytokines in individual Th cells were calculated for the total population and separately for cells of each generation at various time points after initial activation and polarization with IL-4 (Table ). In the total cell population, the cytokines IL-2 and IL-4 were expressed independently of each other until day 4, with φ values ranging from 0.03 to 0.04. On day 5 and day 6, expression was correlated with φ = 0.13 and φ = 0.23. φ correlation coefficients for IL-4 and IL-10 rose from 0.11 on day 3 to 0.34 on day 5. φ values for IL-2 and IL-10 coexpression ranged from 0.02 on day 4 to 0.06 on days 5 and 6. The correlation of cytokine coexpression in cells of different generations at each time point analyzed was similar to that of the total populations at that time point. As shown in Fig. 2 for cells of the seventh generation, analyzed on days 4 and 6 after priming, cells with a given record of cell divisions showed an increased coexpression of IL-2 and IL-4, whereas coefficients of IL-4 and IL-10 coexpression and IL-2 and IL-10 coexpression were similar at both time points. Thus, a certain number of cell cycles is not linked to a certain degree of cytokine coexpression nor does higher division number necessarily imply a higher probability of cytokine coexpression, arguing against the idea of a cell cycle counting mechanism being involved in the instruction of Th cells to express cytokines upon restimulation, with different cell cycle numbers required for each cytokine 21.
Table 1.
Coexpression of Cytokines in Individual Cells in Correlation with Proliferation
| Cell generation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Total | ||
| IL-2/IL-4 | Day 2 | 0.07 | 0.06 | 0.14 | 0.04 | |||||||
| Day 3 | 0.01 | 0.07 | 0.07 | 0.07 | 0.03 | |||||||
| Day 4 | 0.08 | 0.10 | 0.04 | 0.06 | 0.04 | |||||||
| Day 5 | 0.24 | 0.18 | 0.18 | 0.15 | 0.13 | |||||||
| Day 6 | 0.28 | 0.24 | 0.27 | 0.23 | 0.23 | |||||||
| IL-4/IL-10 | Day 3 | 0.10 | 0.11 | 0.15 | 0.08 | 0.11 | ||||||
| Day 4 | 0.24 | 0.25 | 0.24 | 0.22 | 0.23 | |||||||
| Day 5 | 0.30 | 0.39 | 0.38 | 0.37 | 0.34 | |||||||
| Day 6 | 0.21 | 0.24 | 0.27 | 0.22 | 0.26 | |||||||
| IL-2/IL-10 | Day 4 | 0.04 | 0.00 | 0.04 | 0.02 | 0.02 | ||||||
| Day 5 | 0.00 | 0.12 | 0.14 | 0.09 | 0.06 | |||||||
| Day 6 | 0.06 | 0.06 | 0.08 | 0.04 | 0.06 | |||||||
CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were activated with OVA323–339 and APCs in the presence of IL-4, anti–IL-12, and anti–IFN-γ. From day 2 to day 6, samples were restimulated and analyzed for the simultaneous expression of IL-2 and IL-4, IL-4 and IL-10, and IL-2 and IL-10 in individual cell generations and in the total cell population of OVA-TCR+ cells. Phi (φ) correlation coefficients are given.
Figure 2.
Cytokine coexpression of seven times divided Th cells on days 4 and 6 after activation. CFSE-labeled, naive CD62L+ CD4+ OVA-TCRtg/tg cells were activated with OVA323–339 and APCs in the presence of IL-4, anti–IL-12, and anti–IFN-γ. Cells were restimulated on days 4 and 6 after priming. By gating on OVA-TCR+ cells and on cells of the seventh generation, cells were analyzed for the simultaneous expression of two of the cytokines IL-2, IL-4, and IL-10. The observed percentages (obs.) of cytokine-positive cells are shown in each quadrant, set according to the control stainings. In addition, the expected frequencies (exp.) calculated for random coincidence of two independent variables, and the correlation coefficient (φ) of the respective cytokine pair are indicated.
Instruction for IL-4 and IL-10 Expression Requires Progression of Th Cells into the S Phase.
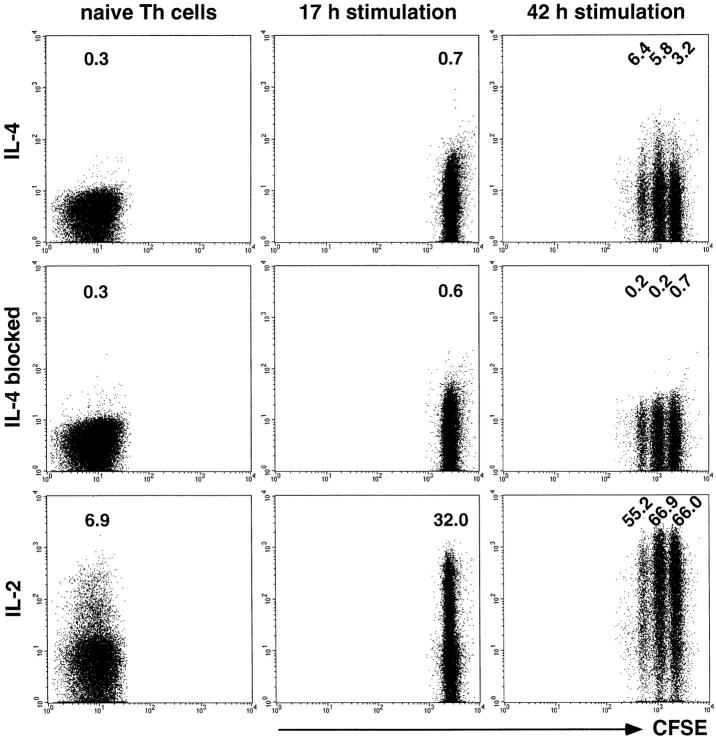
We have analyzed the time point when Th cells become first instructed to express IL-4 upon later restimulation. CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were activated with antigen, APCs, IL-4, anti–IL-12, and anti–IFN-γ, and analyzed for expression of IL-4 and IL-2 after recall stimulation with PMA/ionomycin (Fig. 3). Although IL-2 production was inducible in naive Th cells and those activated for 17 and 42 h, IL-4 was expressed neither by naive Th cells nor by cells stimulated for 17 h. 42 h after onset of primary activation, IL-4 expression was readily detectable after restimulation both in cells that had divided once or twice, and in cells that had not divided, at frequencies ranging from 2.5 to 6.2%.
Figure 3.
Instruction for IL-4 expression after primary activation of Th lymphocytes. Naive CD62L+CD4+ OVA-TCRtg/tg cells were either directly stimulated with PMA/ionomycin or they were labeled with CFSE and activated for 17 or 42 h with OVA323–339 and APCs in the presence of IL-4, anti–IL-12, and anti–IFN-γ before PMA/ionomycin recall. Intracellular IL-4 and IL-2 expression were examined among OVA-TCR+ cells. The specificity of the intracellular IL-4 staining was controlled by blocking the digoxigenized anti–IL-4 detection mAb with an excess of the unconjugated anti–IL-4 mAb. Indicated frequencies of IL-2–producing cells have been corrected for the background frequencies of the respective control stainings.
Since instruction of Th cells to express IL-4 upon restimulation is not dependent on completion of the first cell division after initial activation, as shown above, we analyzed next whether entry of the cells into the first cell cycle may be required. Using inhibitors to arrest activated Th cells in different phases of the first cell cycle, we determined the frequencies of Th cells that were induced to express IL-4, IL-10, and IL-2 after PMA/ionomycin restimulation (Fig. 4). The analysis was performed 3 d after onset of primary activation, when a high frequency of cells was expected to express IL-4 and/or IL-10 upon restimulation (Fig. 1 B). At that time point, nonarrested cells had divided up to four times with 17.7–32.6% of them expressing IL-4, 6.9–17.9% IL-10, and 3.7–14.4% IL-2. In contrast, cells arrested with l-mimosine in the late G1 phase of the first cell cycle expressed neither IL-4 nor IL-10. IL-2 expression was clearly detectable in 6.2% of these cells. For cells that had been allowed to progress into the early S phase and had been arrested by aphidicolin, blocking the elongation process during DNA replication, 2.6% expressed IL-4 and 0.4% IL-10. When cells were arrested in the G2/M phase of the first cell cycle by nocodazole, inhibiting the formation of metaphase microtubules, i.e., the initiation of metaphase, 14.9% produced IL-4 and 1.1% IL-10. When arrested with paclitaxel, which prevents depolymerization of microtubules in metaphase, i.e., metaphase/anaphase transition, 37% of the cells expressed IL-4 and 14.7% IL-10.
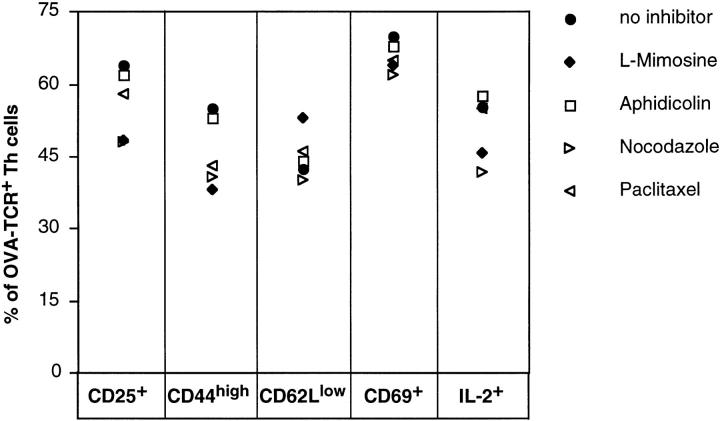
To exclude that the observed differences in the abilities of Th cells to express the various cytokines upon restimulation were caused by decreased overall Th cell activation in cultures with cell cycle inhibitors, expression of the markers of Th cell activation CD25, CD44, CD62L, and CD69 was analyzed in comparison with expression of IL-2 (Fig. 5). None of the cell cycle inhibitors could block expression of any of the activation markers CD25, CD44, and CD69, or prevent downregulation of expression of CD62L, which occurs after Th cell activation, or block expression of IL-2.
Figure 5.
Expression of activation markers after primary activation of Th cells, inhibited for cell cycling. CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were activated with OVA323–339, APCs, IL-4, anti–IL-12, and anti–IFN-γ in the presence or absence of cell cycle inhibitors, as indicated. 1 d after priming, IL-4 and the antigen were removed and anti–IL-4, anti–IL-12, anti–IFN-γ, IL-2, and fresh inhibitors were added. On day 2 after onset of stimulation, live cells were analyzed for expression of CD25, CD44, CD62L, and CD69. Induction of IL-2 expression was determined by intracellular staining after PMA/ionomycin restimulation in the presence of the indicated inhibitors at that time point. The frequencies of positive cells were determined by gating on OVA-TCR+ cells.
Priming of Th Cells to Become Instructed for IL-4 Expression Is Independent of DNA Synthesis.
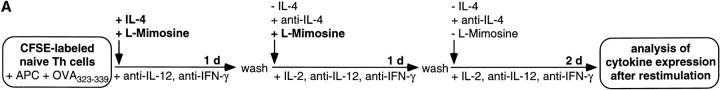
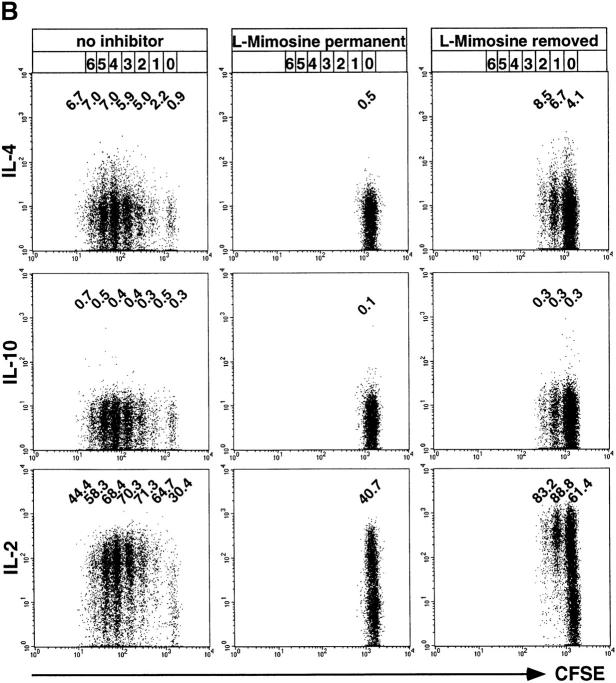
The above results show that processes of the early S phase of the first cell cycle after initial activation are involved in instructing Th lymphocytes for IL-4 expression upon restimulation. We now analyzed whether Th cells can be primed to become instructed for IL-4 expression, independently of entry into cell cycling. As illustrated in Fig. 6 A, we activated CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg Th cells in the presence of l-mimosine for 2 d, to prevent the S phase–dependent instruction of the cells for IL-4 expression. IL-4 and antigen were removed from the culture after 1 d, and anti–IL-4 was added. After 2 d, l-mimosine was also removed, and the cells were further cultivated for 2 d in the absence of antigen and IL-4. At that time, some cells had undergone one or two cell divisions, and all cells were examined for their ability to express IL-4, IL-10, and IL-2 upon recall stimulation with PMA/ionomycin (Fig. 6 B). Compared with the previous experiments, the overall frequencies of IL-4–expressing cells were reduced and IL-2 levels were enhanced in the control cell population, which had not been blocked by l-mimosine. This was probably because IL-4 had only been added for the first day of stimulation, and had been blocked throughout the last 3 d. However, IL-4–expressing cells were clearly detectable in cells of all generations, at frequencies ranging from 0.9 to 7.0%. As expected, control cultures with permanent inhibition of DNA synthesis by l-mimosine showed no IL-4–expressing cells.
Figure 6.
Priming for IL-4 instruction is independent of cell cycling. (A) CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg cells were stimulated with OVA323–339, APCs, IL-4, anti–IL-12, and anti–IFN-γ in the presence of l-mimosine. After 1 d of priming, IL-4 and the antigen were removed and anti–IL-4 and IL-2 added, while l-mimosine was added for an additional 1 d. 2 d after onset of stimulation, l-mimosine was withdrawn and cells were cultured for an additional 2 d. Cells were examined for IL-4, IL-10, and IL-2 expression after recall stimulation with PMA/ionomycin on day 4. (B) The frequencies of IL-4–, IL-10–, and IL-2–expressing OVA-TCR+ cells are shown for cells activated as described in A, compared with cells stimulated and restimulated in the continuous presence or absence of l-mimosine for 4 d. The specificities of the intracellular stainings for IL-4 and IL-10 were controlled by blocking the digoxigenized anticytokine detection mAbs with an excess of the respective unconjugated anticytokine mAbs (data not shown). Indicated percentages among the OVA-TCR+ cells are corrected for the respective control staining values. More cells than depicted were analyzed, but not plotted, to illustrate the separation of the individual generations.
Stimulating naive Th cells with antigen and IL-4, while inhibiting instruction of the Th cells for IL-4 expression with l-mimosine, led to priming of the cells to become instructed later, when relieved from l-mimosine, and allowed to enter S phase. After 2 d of culture in the absence of l-mimosine, 6.7, 8.5, and 4.1% of IL-4–expressing cells could be readily detected among cells that had divided once or twice or not at all. This priming of Th cells to become instructed for expression of cytokines upon restimulation, as induced by IL-4, was restricted to IL-4. Expression of IL-10 was not detectable on day 4 at significant levels in any of the cell populations initially activated in the presence of antigen and IL-4 for only the first day.
Priming for IL-4 Instruction Requires Simultaneous IL-4R and TCR Triggering.
Inhibiting the entry of an activated T cell into the S phase of the first cell cycle by l-mimosine had shown that Th cells can maintain the signal(s) for instruction of IL-4 expression for at least 1 d. Is this priming due to signaling via the IL-4R alone, or is signaling of the TCR also required? To analyze this, naive Th cells were first stimulated with IL-4 for 36 h, then with antigen at various time points after removal of IL-4, before they were analyzed for expression of IL-4 upon restimulation.
CFSE-labeled, naive CD62L+CD4+ OVA-TCRtg/tg Th cells were stimulated with IL-4, but not with antigen, in the presence of anti–IL-12 and anti–IFN-γ. After 36 h, IL-4 was removed and anti–IL-4 was added to the cultures. After 72 or 96 h, the Th cells were stimulated with antigen and APCs. Induction of IL-4 expression was compared in these cells with Th cells that had been activated as described above, but either in the presence or absence of IL-4 for the entire culture period, and with Th cells that had been stimulated directly after isolation with or without IL-4, antigen, and APCs for 36 h. In all cultures, the antigen was withdrawn 36 h after it was added. On day 4 after onset of stimulation with antigen and APCs, the cells were restimulated with PMA/ionomycin and analyzed for expression of IL-4, IL-10, and IL-2 by intracellular immunofluorescence.
2.6% of the Th cells that had been activated simultaneously and directly after isolation with antigen, APCs, and IL-4 for 36 h expressed IL-4, compared with 0.2% of Th cells activated alike but without IL-4 (Fig. 7). Of the Th cells that had been activated 72 or 96 h after isolation with antigen and APCs for 36 h, 2.8 and 4.3% expressed IL-4, if they had been exposed to IL-4 for the entire culture period, and only 0.5 and 0.8% if not. Cells that had been stimulated with IL-4 for 36 h, but IL-4 then having been withdrawn for another 36 or 60 h before TCR stimulation, did not express IL-4 at higher frequencies than those cells that had never been exposed to IL-4 in vitro (0.3 and 0.6% vs. 0.5 and 0.8%). Thus, the initial stimulation with IL-4 for 36 h in the absence of TCR triggering did not result in priming for IL-4 instruction. According to CFSE staining, the various cell populations showed similar proliferation activities under all culture conditions after antigen stimulation (data not shown).
IL-10 was hardly detectable in cell populations that had been activated with IL-4 for only 36 h. However, in those cultures containing IL-4 for the entire culture period, IL-10 was expressed by 15.5 and 23.5% of the cells. IL-2 was expressed in all Th cell populations at high frequencies, except those that had been incubated with IL-4 all the time and that expressed IL-10 at high frequencies. IL-2 and IL-10 expression and the analysis of proliferation using CFSE staining indicate that the Th cells were still able to respond to TCR stimulation, even 72 and 96 h after isolation, and that their failure to react with expression of IL-4 is due to a lack of priming for IL-4 expression. From this experiment, it is clear that simultaneous IL-4R and TCR stimulation is required to prime a Th cell to become instructed for IL-4 recall expression.
Discussion
Cytokine expression in Th lymphocytes is transient, but can be memorized by the cells upon neutral or even adverse restimulation 6 7 8 9 10 11 12 13 14. Here we show that in primarily activated Th lymphocytes, entry into the first cell cycle is required and sufficient to instruct the Th cells to produce IL-4 and IL-10 upon restimulation, whereas IL-2 expression is independent of cell cycling. Moreover, independently of DNA synthesis, Th cells are primed to become instructed for IL-4 recall expression, by simultaneous signaling via IL-4R and TCR but not IL-4R alone. This coordinate signaling can be maintained for at least 1 d.
Two groups have recently used a similar technical approach to analyze the dependency of cytokine expression on proliferation in activated T cells 19 21. Gett and Hodgkin 21 claim that cell cycling may act as an intrinsic clock, allowing expression of distinct cytokines time-independently only after a certain number of cell divisions. We find a similar correlation between proliferative activity of the cells and their capacity to express cytokines. However, already in activated but nondivided Th cell populations, cells expressing IL-2, IL-4, IL-10, IFN-γ, and TNF-α are clearly detectable. Our results rule out the possibility that even completion of the first cell cycle after activation is required to allow expression of any of these cytokines upon restimulation. Moreover, if cell division controlled the successive expression of various cytokine genes, the frequencies of cells coexpressing cytokines, which are switched on early and later, would increase in correlation to proliferation rather than time. We find here that coexpression of any two of the cytokines IL-2, IL-4, and IL-10 is not correlated with the number of cell divisions a cell has performed after activation.
However, in cells that had divided a given number of times, the capacity for cytokine expression was correlated to the time of stimulation. We did not observe differentiation of Th cells for IL-4 expression as early as 17 h after primary stimulation. After 42 h, IL-4–producing cells were evident upon restimulation, also in the nondivided population. The time required to become instructed for IL-4 expression after recall stimulation could reflect the time needed by a naive Th cell to progress into the S phase of the first cell cycle after activation. The correlative increase in frequencies of cells expressing cytokines with increasing number of cell cycles performed may rather be explained by assuming that the cells get instructed for cytokine expression not only in the first but also in later cell generations, increasing the frequencies of cytokine-expressing cells among those cell populations that have divided most often.
Bird et al. 19 report that instruction of an activated Th cell to express IFN-γ requires entry of the cells into the S phase of the first cell cycle, whereas cells would have to complete three cell cycles to become instructed for IL-4 expression. In contrast, we find Th cells instructed for expression of IL-4 already in the nondivided population, but only among those cells that had entered the S phase of the first cell cycle. We can exclude that the IL-4–expressing Th cells we observe are preactivated Th cells, since none of the purified naive Th cells or Th cells stimulated for 17 h with antigen expressed IL-4 upon restimulation. The reason for the discrepancy between the present data and those of Bird et al. 19 could be the overall lower frequency of IL-4–expressing cells in the cultures of Bird et al., making it difficult to identify them in the nondivided cell population. Gett and Hodgkin 21, analyzing the capacity for IL-4 expression in Th cells once divided, apparently detected a low frequency of IL-4–expressing cells but did not check expression of IL-4 in nondivided cells. Taking the work of Bird et al. 19 and our results together, instruction of Th cells for IFN-γ, IL-4, or IL-10 expression requires the cells to enter the S phase of the first cell cycle after activation, pointing to a common molecular mechanism for instruction of the cells to express these “effector” cytokines upon restimulation. For IL-10 instruction, the cells apparently have to complete the S phase. IL-2 expression in naive cells does not seem to require entry into the cell cycle. It is likely that those cells have already been instructed for IL-2 expression in the thymus 20 42.
The molecular basis for instruction of Th cells to express particular cytokines after restimulation is less clear, as is its stability and plasticity. Apart from cytokine-dependent modulations of the signaling and transcription repertoire of Th cells expressing particular cytokines, restricting their potential to respond to signals inducing expression of other cytokines 43 44 45 46 47 48 49, modifications of the cytokine gene loci themselves have been demonstrated. Demethylation has been described for the DNA comprising the IL-3, IL-4, IL-5, or IFN-γ genes of Th cell lines, clones, and ex vivo–activated T cell populations containing cells expressing the respective cytokines upon restimulation 15 16 17 19. Furthermore, it has been shown for such T cell populations that transcription of the IL-4, IL-13, and IFN-γ genes parallels the appearance of particular DNase I hypersensitive sites within the expressed cytokine gene loci 18 20. These results suggest that the instruction for expression of particular cytokines might be maintained in Th cells by epigenetic somatic imprinting of the respective cytokine gene loci.
The initiation of the somatic imprinting of cytokine genes remains enigmatic. This is even more so in light of the monoallelic expression of cytokine genes in many Th cells, suggesting that the instruction of a Th cell to express a particular cytokine is a stochastic event for each allele in a given population of Th cells 50 51 52 53. We describe here that instruction of a Th cell for IL-4 and IL-10 recall expression is dependent on progression of the activated Th cell through the S phase of the first cell cycle, while Bird et al. 19 have shown the same for IFN-γ. The most obvious characteristic of the S phase of the first cell cycle is that the first DNA synthesis after activation of the naive T cell occurs. One might speculate that an epigenetic modification of the DNA, most likely demethylation, may be the molecular correlate of cytokine gene instruction. How would demethylation be targeted to the cytokine gene locus? The presence of sequence- or site-specific demethylating enzymes has not been demonstrated to date 54. “Unspecific” demethylation machineries would have to be directed to a particular area of DNA, most likely by flagging of the DNA through sequence-specific proteins, which may or may not belong to the transcription machinery. We here provide indirect evidence for such a flagging of cytokine genes for molecular modification. Th cells that had been activated via TCR and IL-4R, but inhibited to enter the cell cycle, are primed to become instructed for IL-4 expression later, when allowed to progress through S phase, even in the absence of the inductive stimuli. For the priming for IL-4 instruction, signaling by IL-4 alone or sequential stimulation by IL-4 and antigen is not sufficient. The IL-4R and TCR have to be stimulated simultaneously, although it cannot be excluded at present that activation by antigen, followed by IL-4 stimulation, would not work as well. This is in accordance with the results of Agarwal and Rao 20, who showed that chromatin remodeling of the IL-4 gene is detectable only after stimulation of naive Th cells with IL-4 and anti-CD3 and not with IL-4 alone.
Default induction of IL-4 expression is dependent on IL-4R/signal transducer and activator of transcription (STAT)6 signaling 55 56 57. However, IL-4R– or STAT6-independent IL-4 induction has also been observed. An IL-4–independent pathway for the induction of IL-4 has been demonstrated in IL-4R–deficient NK T cells 58. IL-4 expression is also inducible in CD3+ T cells deficient for STAT6 and BCL-6 59, suggesting a role for BCL-6 as an inhibitor of IL-4 expression, probably by competing with STAT6 for DNA binding motifs 60. According to the present results, in wild-type cells components of both the IL-4R and TCR signaling pathways are required to prime Th cells for IL-4 instruction. Evidence has been obtained that STAT6 is crucial for chromatin remodeling and expression of IL-4, using STAT6-deficient cells 19. In those cells, expression of IL-4 could be induced by trichostatin A, an inhibitor of histone deacetylation (i.e., an activator of nucleosome displacement), and azacytidine, preventing DNA methylation. For STAT6, as well as for nuclear factor of activated T cells (NFAT)1 and activating protein 1 (AP-1), involved in TCR signaling, binding to and activation by the coactivators p300/cAMP response element–binding protein (CREB)–binding protein (CBP), which exhibit histone acetyltransferase activity, have been described 61 62 63 64. In line with this, BCL-6, the inhibitor of STAT6, binds to silencing mediator of retinoid and thyroid receptor (SMRT), a protein able to recruit histone deacetylase activity 65 66.
The speculative sequence of events for induction of molecular memory of cytokine expression appears to be IL-4R– and TCR-mediated chromatin rearrangement by specific acetylation of histones, perhaps involving STAT6 and displacement of BCL-6, followed by binding of specific, yet unidentified flagging factors to the DNA of the cytokine gene locus of the primed cell, in the G0/G1 phase preceding the first cell cycle of the activated cell. Finally, imprinting of the cytokine gene locus by demethylation during the S phase of the first cell cycle after activation would provide the molecular basis for cytokine memory.
Acknowledgments
We are grateful to Dr. D.Y. Loh for providing OVA-TCR transgenic DO11.10 mice, and to Drs. A. O'Garra, M. Gately, E. Schmitt, and G. Trinchieri for generous gifts of mAbs and reagents. We thank T. Geske and M. Steinbach for expert technical support; Drs. A. Hamann, T. Kamradt, and A. Scheffold for critical reading of the manuscript; and T. Stamm and Dr. O. Sözeri for helpful discussion.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 421), the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BEO/21/11340), the Senatsverwaltung für Wissenschaft, Forschung und Kultur des Landes Berlin, and the Studienstiftung des deutschen Volkes (M. Löhning).
Footnotes
1used in this paper: CFSE, 5-(and 6-)carboxyfluorescein diacetate, succinimidyl ester; Dig, digoxigenin; MACS, high-gradient magnetic cell separation; STAT, signal transducer and activator of transcription; tg, transgenic
References
- Abbas A.K., Murphy K.M., Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- Bluestone J.A. New perspectives of CD28-B7-mediated T cell costimulation. Immunity. 1995;2:555–559. doi: 10.1016/1074-7613(95)90000-4. [DOI] [PubMed] [Google Scholar]
- Le Gros G., Ben-Sasson S.Z., Seder R., Finkelman F.D., Paul W.E. Generation of interleukin 4 (IL-4)–producing cells in vivo and in vitroIL-2 and IL-4 are required for in vitro generation of IL-4–producing cells. J. Exp. Med. 1990;172:921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.S., Macatonia S.E., Tripp C.S., Wolf S.F., O'Garra A., Murphy K.M. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- Seder R.A., Gazzinelli R., Sher A., Paul W.E. Interleukin-12 acts directly on CD4+ T cells to enhance priming for interferon γ production and diminishes interleukin 4 inhibition of such priming. Proc. Natl. Acad. Sci. USA. 1993;90:10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenmacher M., Schmitz J., Radbruch A. Flow cytometric determination of cytokines in activated murine T helper lymphocytesexpression of interleukin 10 in interferon γ and in interleukin 4-expressing cells. Eur. J. Immunol. 1994;24:1097–1101. doi: 10.1002/eji.1830240513. [DOI] [PubMed] [Google Scholar]
- Assenmacher M., Löhning M., Scheffold A., Manz R.A., Schmitz J., Radbruch A. Sequential production of IL-2, IFN-γ and IL-10 by individual staphylococcal enterotoxin B-activated T helper lymphocytes. Eur. J. Immunol. 1998;28:1534–1543. doi: 10.1002/(SICI)1521-4141(199805)28:05<1534::AID-IMMU1534>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Cardell S., Sander B. Interleukin 2, 4 and 5 are sequentially produced in mitogen-stimulated murine spleen cell cultures. Eur. J. Immunol. 1990;20:389–395. doi: 10.1002/eji.1830200223. [DOI] [PubMed] [Google Scholar]
- Lederer J.A., Perez V.L., DesRoches L., Kim S.M., Abbas A.K., Lichtman A.H. Cytokine transcriptional events during helper T cell subset differentiation. J. Exp. Med. 1996;184:397–406. doi: 10.1084/jem.184.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Shibuya K., Hosken N., Openshaw P., Maino V., Davis K., Murphy K., O'Garra A. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J. Exp. Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenmacher M., Löhning M., Scheffold A., Richter A., Miltenyi S., Schmitz J., Radbruch A. Commitment of individual Th1-like lymphocytes to expression of IFN-γ versus IL-4 and IL-10selective induction of IL-10 by sequential stimulation of naive Th cells with IL-12 and IL-4. J. Immunol. 1998;161:2825–2832. [PubMed] [Google Scholar]
- Sornasse T., Larenas P.V., Davis K.A., de Vries J.E., Yssel H. Differentiation and stability of T helper 1 and 2 cells derived from naive human neonatal CD4+ T cells, analyzed at the single-cell level. J. Exp. Med. 1996;184:473–483. doi: 10.1084/jem.184.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo S.J., Jacobson N.G., Dighe A.S., Gubler U., Murphy K.M. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity. 1995;2:665–675. doi: 10.1016/1074-7613(95)90011-x. [DOI] [PubMed] [Google Scholar]
- Perez V.L., Lederer J.A., Lichtman A.H., Abbas A.K. Stability of Th1 and Th2 populations. Int. Immunol. 1995;7:869–875. doi: 10.1093/intimm/7.5.869. [DOI] [PubMed] [Google Scholar]
- Young H.A., Ghosh P., Ye J., Lederer J., Lichtman A., Gerard J.R., Penix L., Wilson C.B., Melvin A.J., McGurn M.E. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-γ gene. J. Immunol. 1994;153:3603–3610. [PubMed] [Google Scholar]
- Melvin A.J., McGurn M.E., Bort S.J., Gibson C., Lewis D.B. Hypomethylation of the interferon γ gene correlates with its expression by primary T-lineage cells. Eur. J. Immunol. 1995;25:426–430. doi: 10.1002/eji.1830250218. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D.R., Shirley K.M., McDonald L.E., Bielefeldt-Ohmann H., Kay G.F., Kelso A. Distinct methylation of the interferon γ (IFN-γ) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytesregional IFN-γ promoter demethylation and mRNA expression are heritable in CD44highCD8+ T cells. J. Exp. Med. 1998;188:103–117. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto N., Koyano-Nakagawa N., Yokota T., Arai N., Miyatake S., Arai K. Th2-specific DNase I hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int. Immunol. 1998;10:1981–1985. doi: 10.1093/intimm/10.12.1981. [DOI] [PubMed] [Google Scholar]
- Bird J.J., Brown D.R., Mullen A.C., Moskowitz N.H., Mahowald M.A., Sider J.R., Gajewski T.F., Wang C.R., Reiner S.L. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- Agarwal S., Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Gett A.V., Hodgkin P.D. Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc. Natl. Acad. Sci. USA. 1998;95:9488–9493. doi: 10.1073/pnas.95.16.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M., Heimberger A.B., Loh D.Y. Induction by antigen of intrathymic apoptosis of CD4+CD8+ TCRlo thymocytes in vivo. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- Dialynas D.P., Quan Z.S., Wall K.A., Pierres A., Quintans J., Loken M.R., Pierres M., Fitch F.W. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5similarity of L3T4 to the human Leu-3/T4 molecule. J. Immunol. 1983;131:2445–2451. [PubMed] [Google Scholar]
- Miltenyi S., Müller W., Weichel W., Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Lyons A.B., Parish C.R. Determination of lymphocyte division by flow cytometry. J. Immunol. Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Ohara J., Paul W.E. Production of a monoclonal antibody to and molecular characterization of B-cell stimulatory factor-1. Nature. 1985;315:333–336. doi: 10.1038/315333a0. [DOI] [PubMed] [Google Scholar]
- Karasuyama H., Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur. J. Immunol. 1988;18:97–104. doi: 10.1002/eji.1830180115. [DOI] [PubMed] [Google Scholar]
- Prat M., Gribaudo G., Comoglio P.M., Cavallo G., Landolfo S. Monoclonal antibodies against murine γ interferon. Proc. Natl. Acad. Sci. USA. 1984;81:4515–4519. doi: 10.1073/pnas.81.14.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka M., Kubin M., Vieira L.Q., Ozmen L., Garotta G., Scott P., Trinchieri G. Interleukin 12 is required for interferon γ production and lethality in lipopolysaccharide-induced shock in mice. Eur. J. Immunol. 1995;25:672–676. doi: 10.1002/eji.1830250307. [DOI] [PubMed] [Google Scholar]
- Lalande M. A reversible arrest point in the late G1 phase of the mammalian cell cycle. Exp. Cell Res. 1990;186:332–339. doi: 10.1016/0014-4827(90)90313-y. [DOI] [PubMed] [Google Scholar]
- Ikegami S., Taguchi T., Ohashi M., Oguro M., Nagano H., Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-α. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- Zieve G.W., Turnbull D., Mullins J.M., McIntosh J.R. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- Schiff P.B., Horwitz S.B. Taxol stabilizes microtubules in mouse fibroblast cells. Proc. Natl. Acad. Sci. USA. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge I.S., Lesley J., Schulte R., Hyman R., Trotter J. Biochemical characterization and cellular distribution of a polymorphic, murine cell-surface glycoprotein expressed on lymphoid tissues. Immunogenetics. 1982;15:299–312. doi: 10.1007/BF00364338. [DOI] [PubMed] [Google Scholar]
- Gallatin W.M., Weissman I.L., Butcher E.C. A cell-surface molecule involved in organ-specific homing of lymphocytes. Nature. 1983;304:30–34. doi: 10.1038/304030a0. [DOI] [PubMed] [Google Scholar]
- Mosmann T.R., Cherwinski H., Bond M.W., Giedlin M.A., Coffman R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- Abrams J.S., Roncarolo M.G., Yssel H., Andersson U., Gleich G.J., Silver J.E. Strategies of anti-cytokine monoclonal antibody developmentimmunoassay of IL-10 and IL-5 in clinical samples. Immunol. Rev. 1992;127:5–24. doi: 10.1111/j.1600-065x.1992.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sander B., Hoiden I., Andersson U., Möller E., Abrams J.S. Similar frequencies and kinetics of cytokine producing cells in murine peripheral blood and spleen. Cytokine detection by immunoassay and intracellular immunostaining. J. Immunol. Methods. 1993;166:201–214. doi: 10.1016/0022-1759(93)90361-a. [DOI] [PubMed] [Google Scholar]
- Haskins K., Kubo R., White J., Pigeon M., Kappler J., Marrack P. The major histocompatibility complex–restricted antigen receptor on T cells. I. Isolation with a monoclonal antibody. J. Exp. Med. 1983;157:1149–1169. doi: 10.1084/jem.157.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop Y.M.M., Fienberg S.E., Holland P.W. Discrete Multivariate Analysis. Theory and Practice 1975. MIT Press; Cambridge, MA: pp. 557 [Google Scholar]
- Löhning M., Grogan J.L., Coyle A.J., Yazdanbakhsh M., Meisel C., Gutierrez-Ramos J.C., Radbruch A., Kamradt T. T1/ST2 expression is enhanced on CD4+ T cells from schistosome egg-induced granulomasanalysis of Th cell cytokine coexpression ex vivo. J. Immunol. 1999;162:3882–3889. [PubMed] [Google Scholar]
- Rothenberg E.V., Ward S.B. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin-2 gene regulation. Proc. Natl. Acad. Sci. USA. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Paul W.E. Impaired interleukin-4 signaling in T helper type 1 cells. J. Exp. Med. 1998;187:1305–1313. doi: 10.1084/jem.187.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Ranganath S.H., Weindel K., Bhattacharya D., Murphy T.L., Sha W.C., Murphy K.M. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- Ho I.C., Lo D., Glimcher L.H. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4–dependent and –independent mechanisms. J. Exp. Med. 1998;188:1859–1866. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo S.J., Dighe A.S., Gubler U., Murphy K.M. Regulation of the interleukin (IL)-12R β2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge L., Barberis-Maino L., Biffi M., Passini N., Presky D.H., Gubler U., Sinigaglia F. Selective expression of an interleukin-12 receptor component by human T helper 1 cells. J. Exp. Med. 1997;185:825–831. doi: 10.1084/jem.185.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach E.A., Szabo S.J., Dighe A.S., Ashkenazi A., Aguet M., Murphy K.M., Schreiber R.D. Ligand-induced autoregulation of IFN-γ receptor β chain expression in T helper cell subsets. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- Pernis A., Gupta S., Gollob K.J., Garfein E., Coffman R.L., Schindler C., Rothman P. Lack of interferon-γ receptor β chain and the prevention of interferon-γ signaling in TH1 cells. Science. 1995;269:245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- Holländer G.A., Zuklys S., Morel C., Mizoguchi E., Mobisson K., Simpson S., Terhorst C., Wishart W., Golan D.E., Bhan A.K., Burakoff S.J. Monoallelic expression of the interleukin-2 locus. Science. 1998;279:2118–2121. doi: 10.1126/science.279.5359.2118. [DOI] [PubMed] [Google Scholar]
- Bix M., Locksley R.M. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- Naramura M., Hu R.J., Gu H. Mice with a fluorescent marker for interleukin 2 gene activation. Immunity. 1998;9:209–216. doi: 10.1016/s1074-7613(00)80603-2. [DOI] [PubMed] [Google Scholar]
- Riviere I., Sunshine M.J., Littman D.R. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–228. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. DNA methylation and imprintingwhy bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Takeda K., Tanaka T., Shi W., Matsumoto M., Minami M., Kashiwamura S., Nakanishi K., Yoshida N., Kishimoto T., Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- Shimoda K., van Deursen J., Sangster M.Y., Sarawar S.R., Carson R.T., Tripp R.A., Chu C., Quelle F.W., Nosaka T., Vignali D.A. Lack of IL-4–induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- Kaplan M.H., Schindler U., Smiley S.T., Grusby M.J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Noben-Trauth N., Shultz L.D., Brombacher F., Urban J.F., Jr., Gu H., Paul W.E. An interleukin-4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:10838–10843. doi: 10.1073/pnas.94.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent A.L., Hu-Li J., Paul W.E., Staudt L.M. T helper type 2 inflammatory disease in the absence of interleukin-4 and transcription factor STAT6. Proc. Natl. Acad. Sci. USA. 1998;95:13823–13828. doi: 10.1073/pnas.95.23.13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent A.L., Shaffer A.L., Yu X., Allman D., Staudt L.M. Control of inflammation, cytokine expression, and germinal center formation by Bcl-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Ohmori Y., Hamilton T.A. STAT6 is required for the anti-inflammatory activity of interleukin-4 in mouse peritoneal macrophages. J. Biol. Chem. 1998;273:29202–29209. doi: 10.1074/jbc.273.44.29202. [DOI] [PubMed] [Google Scholar]
- Gingras S., Simard J., Groner B., Pfitzner E. p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res. 1999;27:2722–2729. doi: 10.1093/nar/27.13.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez C., Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP) J. Exp. Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias J., Alberts A.S., Brindle P., Claret F.X., Smeal T., Karin M., Feramisco J., Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- Dhordain P., Albagli O., Lin R.J., Ansieau S., Quief S., Leutz A., Kerckaert J.P., Evans R.M., Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L., Kao H.Y., Chakravarti D., Lin R.J., Hassig C.A., Ayer D.E., Schreiber S.L., Evans R.M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]