In this issue of The Journal of Experimental Medicine, Hudson et al. 1 report on the identification of a cytokine, the macrophage migration inhibitory factor or MIF, that overcomes p53 function by suppressing its transcriptional activity, thus linking complex programs that govern proinflammatory response and cell fate. p53 has a key role in the regulation of cell growth and death, and its involvement in these regulatory systems may reflect its ability to respond to different cellular stress situations by inducing cell cycle arrest or apoptosis (see references 2–4 for reviews on the topic and below for more details on p53 function). When altered by germ line or somatic mutations, by aberrant patterns of expression, or through the inactivating potential of Mdm2 or certain viral oncoproteins, loss of wild-type p53 function is responsible for tumorigenesis and tumor progression 5 6 7. The relationship between certain chronic inflammatory conditions and cancer has been known for many years, and several postulates have been furnished to explain such an association (for recent reviews see references 8 and 9). The study of Hudson et al. provides a mechanistic connection between chronic inflammation and tumorigenesis.
The authors used two independent functional screens to isolate genes bypassing p53-mediated growth arrest or apoptosis, which both yielded cDNAs encoding MIF. Three different biological assays were then pursued to assess p53 status after treatment with MIF. MIF was either directly tested by transducing it into different cells or, as MIF was originally characterized as an extracellular cytokine, it was added as a soluble factor. These experiments revealed that p53 continues to possess the same microanatomical nuclear localization and maintain similar levels of expression in control and MIF-treated cells. However, MIF partially suppresses p53-dependent transcriptional activity, as evidenced by the low levels of p21, cyclin G1, and Mdm2 observed in treated versus untreated cells. In the presence of MIF, Hudson et al. also observed suppression of apoptosis. Finally, murine fibroblasts treated with MIF have an extended life span, supporting the concept that MIF may, at least in part, overcome cellular senescence.
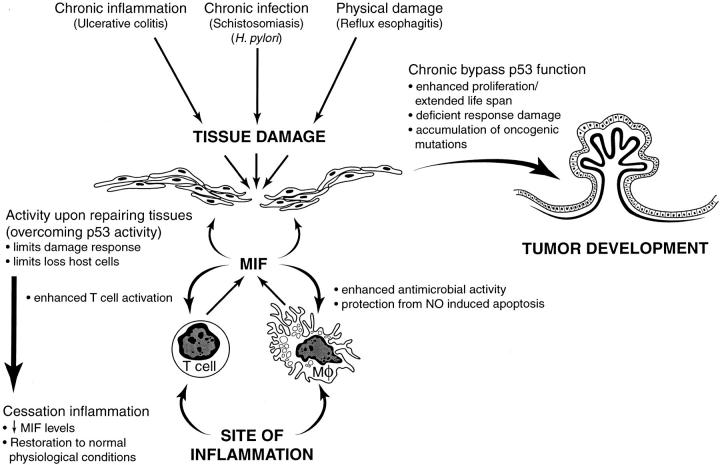
It is known that during tissue damage, lymphocytes and macrophages are recruited at sites of inflammation. MIF is released from T cells and macrophages, contributing to enhanced T cell activation and increased antimicrobial functions of macrophages. Besides these proinflammatory functions, activated macrophages also release reactive oxygen species. These, in turn, could induce macrophage apoptosis, which has been demonstrated to be p53 dependent. Hudson et al. propose that in the presence of active inflammatory cells, MIF would create a protective antiapoptosis feedback loop. In addition, overcoming p53 transactivation properties within repairing tissues would limit damage response and the loss of host cells. Cessation of inflammation will restore normal MIF levels and reinstate physiological conditions. However, chronic bypass of p53 key regulatory functions could enhance proliferation, extend life span, create a deficient response to genetic damage, and even allow accumulation of oncogenic mutations. Taken together, these phenomena could lead to cellular transformation and tumor development. Fig. 1 is a diagram illustrating this sequence of events.
Figure 1.
Diagram illustrating the working hypothesis of Hudson et al. relating chronic inflammation, release of MIF, and continuous bypass of p53 function as it may relate to tumorigenesis.
Based on the natural history of certain diseases and epidemiology studies, a strong association has been established between particular chronic inflammatory conditions and eventual tumor appearance 8 9 10 11. Fig. 1 summarizes some of these related processes, which include chronic infections (i.e., schistosomiasis and bladder cancer, Helicobacter pylori and gastric cancer, and hepatitis C and hepatocellular carcinoma); chronic tissue damage by chemical or physical agents (i.e., reflux esophagitis and esophageal cancer in Barrett's syndrome and chronic pancreatitis and pancreatic carcinoma); and disorders of uncertain etiology producing chronic inflammation (i.e., chronic nonspecific ulcerative colitis and Crohn's disease and colon cancer). For most of these processes, a sequence of histopathological events has been described 12. Continuous damage first appears to cause tissue atrophy. This is followed by increased proliferative activity, which is usually associated with “metaplasia,” a reversible change in which a differentiated cell type is replaced by another differentiated cell type. “Dysplasia” may then develop, characterized by irregular proliferative changes that yield atypical cells as part of a nonadaptive process. Dysplastic changes are often found adjacent to foci of cancer.
The morphological alterations described above have been related to unbalanced biological systems. A number of independent studies using clinical samples have shown that cell turnover is affected by the inflammatory response. This notion is supported by reports describing an increase in cell death by apoptosis as an early response to infection and continued inflammation. For example, H. pylori and certain inflammatory mediators, such as TNF and IFN-γ, have been shown to regulate apoptosis of gastric epithelial cells 13. As a consequence, atrophic changes could develop that may be reversible if the inflammatory stimuli cease. The continuous presence of MIF, together with the cell loss produced in the atrophic tissues, may then trigger a proliferative response by noncommitted epithelial cell precursors. There are multiple studies that have revealed the increased expression of the proliferative Ki-67 nuclear antigen in reactive, nonneoplastic epithelial cells at sites of chronic inflammation. For example, colorectal biopsies from patients with ulcerative colitis, undergoing colonoscopic surveillance for histopathological detection of dysplasia to select patients at high risk for prophylactic colectomy, showed increased Ki-67 staining in areas of reactive/regenerating epithelium 14. Moreover, certain studies have also disclosed the association between dysplasia and detection of mutations affecting critical genes, such as K-RAS in chronic pancreatitis tissues 15. Loss of heterozygosity (LOH) at tumor suppressor gene loci such as 3p, 9p21, and 17p11 (where p53 maps at 17p11.3) has also been documented in nonneoplastic mucosa from patients with chronic ulcerative colitis 16. Taken together, increased proliferation and the production of these molecular alterations may give rise to the morphological changes ascribed to dysplasia and lead to neoplastic transformation.
In addition to the studies referred to above, the direct association between MIF and cancer has also been documented in several reports. Protein expression profiles in breast ductal carcinoma versus normal breast tissue were analyzed by two-dimensional gels 17. There were 32 spots highly expressed in all carcinoma samples, one of them being MIF. In another study, differential display PCR was used to isolate genes that exhibited increased expression in prostatic adenocarcinoma metastases versus primary prostatic tumors 18. Three cDNA clones were identified, two corresponding to genes that showed no homology to known database sequences. The third cDNA was that of MIF. In a follow-up study, this group of investigators used immunohistochemistry, ELISA, and Northern blot analysis to assess MIF expression on a variety of normal prostatic tissues and primary and metastatic prostate tumors, as well as several prostate cancer cell lines 19. MIF was localized to the glandular epithelium, and the most intense expression was identified in metastatic carcinomas and in LNCaP cells.
Hudson et al. have now provided data showing that MIF treatment of cells inhibits the expression of endogenous downstream targets of p53, including p21 and bax 1. Furthermore, they found that MIF treatment blocked p53 transactivation in a reporter assay in transiently transfected cells 1. How does MIF cause this downregulation of p53? p53 possesses the domains and properties of many other transcriptional activators 2 3. Its NH2 terminus contains a strong acidic activation region that has been shown to interact in vitro with members of the general transcriptional machinery such as TBP, TAFs and p300/CBP, as well as with its negative regulator, Mdm2. The central “core” of the protein, where the vast majority of tumor-derived missense mutations are detected, contains its sequence-specific DNA binding domain, which recognizes sites located within the vicinity of its downstream target genes, including p21, cyclin G, and Mdm2. The COOH terminus contains a tetramerization region and a highly basic region that can negatively regulate DNA binding by the central core domain.
It is well documented that the p53 protein is extensively modified by phosphorylation and acetylation 4. Such modifications have been mapped at sites within the NH2- and COOH-terminal portions of the protein, and a number of studies have shown that certain modifications can affect DNA binding and protein interactions of p53. Importantly, there have been several reports in the past two years showing that many of these sites are targets of complex signaling pathways 4. Understanding the impact of modification of p53 after DNA damage is still in an early stage. However, although we now know that several sites can be inducibly modified, it has also been demonstrated that p53 protein in normal unstressed cells is phosphorylated at several sites 20. It has been shown that unmodified, bacterially expressed p53 protein binds DNA very poorly when compared with p53 isolated from eukaryotic cells 21. Furthermore, it is also clear that overexpression of p53 in mammalian cells, even in the absence of DNA damage–induced activation, results in a protein capable of inducing its downstream targets. Thus, constitutive or basal modification of p53 may be a necessary prerequisite for the protein to be functional in vivo. We can imagine a number of scenarios by which MIF could conceivably cause repression of p53, all of which are entirely speculative at this point. First, normal nonstress-induced modification of p53 protein may be affected by MIF treatment. MIF signaling might repress key basal phosphorylation events or, alternately, induce a new modification(s) in p53, which in either case somehow renders it unable to activate transcription. Second, p53 has been shown to be activated by noncovalent modifiers such as Ref-1 and HMG-1, which stimulate its DNA binding in vitro and transactivation in vivo 22. Perhaps MIF initiates a process that leads to inactivation or loss of these (or other as yet unidentified) p53 coactivators. Third, MIF may cause expression or activation of one or more factors that directly interact with p53 and prevent its binding to DNA or interactions with general transcription factors. Whatever the mode by which MIF represses the transactivation function of p53, elucidation of this process will undoubtedly shed new light onto both MIF and p53. It will also provide further knowledge regarding the link between complex programs such as those governing inflammatory responses and tumorigenesis.
References
- Hudson J.D., Shoaibi M.A., Maestro R., Carnero A., Hannon G.J., Beach D.H. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J. Exp. Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Ko L.J., Prives C. p53puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Giaccia A.J., Kastan M. The complexity of p53 modulationemerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C. Mutations of cell cycle regulators. Biological and clinical implications in human neoplasia. Am. J. Pathol. 1995;147:545–560. [PMC free article] [PubMed] [Google Scholar]
- Clurman B.E., Roberts J.M. Cell cycle and cancer. J. Natl. Cancer Inst. 1995;87:1499–1501. doi: 10.1093/jnci/87.20.1499. [DOI] [PubMed] [Google Scholar]
- Brown J.M., Wouters B.G. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- Wang J.M., Deng X., Gong W., Su S. Chemokines and their role in tumor growth and metastasis. J. Immunol. Methods. 1998;220:1–17. doi: 10.1016/s0022-1759(98)00128-8. [DOI] [PubMed] [Google Scholar]
- Frommel T.O., Zarling E.J. Chronic inflammation and cancerpotential role of Bcl-2 gene family members as regulators of cellular antioxidant status. Med. Hypotheses. 1999;52:27–30. doi: 10.1054/mehy.1997.0621. [DOI] [PubMed] [Google Scholar]
- Spechler S.J. The role of gastric carditis in metaplasia and neoplasia at the gastroesophageal junction. Gastroenterology. 1999;117:218–228. doi: 10.1016/s0016-5085(99)70571-8. [DOI] [PubMed] [Google Scholar]
- Talamini G., Falconi M., Bassi C., Sartori N., Salvia R., Caldiron E., Frulloni L., Di Francesco V., Vaona B., Bovo P. Incidence of cancer in the course of chronic pancreatitis. Am. J. Gastroenterol. 1999;94:1253–1260. doi: 10.1111/j.1572-0241.1999.01075.x. [DOI] [PubMed] [Google Scholar]
- Nardone G., Staibano S., Rocco A., Mezza E., D'armiento F.P., Insabato L., Coppola A., Salvatore G., Lucariello A., Figura N. Effect of Helicobacter pylori infection and its eradication on cell proliferation, DNA status, and oncogene expression in patients with chronic gastritis. Gut. 1999;44:789–799. doi: 10.1136/gut.44.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P. The role of inflammation in the pathogenesis of gastric cancer. Aliment. Pharmacol. Ther. 1999;13:13–18. doi: 10.1046/j.1365-2036.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- Sjoqvist U., Ost A., Lofberg R. Increased expression of proliferative Ki-67 nuclear antigen is correlated with dysplastic colorectal epithelium in ulcerative colitis. Int. J. Colorectal Dis. 1999;14:107–113. doi: 10.1007/s003840050194. [DOI] [PubMed] [Google Scholar]
- Wenger F.A., Zieren J., Peter F.J., Jacobi C.A., Muller J.M. K-ras mutations in tissue and stool samples from patients with pancreatic cancer and chronic pancreatitis. Langenbecks Arch. Surg. 1999;384:181–186. doi: 10.1007/s004230050189. [DOI] [PubMed] [Google Scholar]
- Park W.S., Pham T., Wang C., Pack S., Mueller E., Mueller J., Vortmeyer A., Zhuang Z., Fogt F. Loss of heterozygosity and microsatellite instability in non-neoplastic mucosa from patients with chronic ulcerative colitis. Int. J. Mol. Med. 1998;2:221–224. [PubMed] [Google Scholar]
- Bini L., Magi B., Marzocchi B., Arcuri F., Tripodi S., Cintorino M., Sanchez J.C., Frutiger S., Hughes G., Pallini V. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 1997;18:2832–2841. doi: 10.1002/elps.1150181519. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler K., Hudson P.B. Enhanced expression of macrophage migration inhibitory factor in prostatic adenocarcinoma metastases. Urology. 1996;48:448–452. doi: 10.1016/S0090-4295(96)00207-5. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler K., Fattor R.A., Hudson P.B. Expression of macrophage migration inhibitory factor in the human prostate. Diagn. Mol. Pathol. 1998;7:44–50. doi: 10.1097/00019606-199802000-00008. [DOI] [PubMed] [Google Scholar]
- Meek D.W. Post-translational modification of p53. Semin. Cancer Biol. 1994;5:203–210. [PubMed] [Google Scholar]
- Hupp T.R., Meek D.W., Midgley C.A., Lane D.P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Jayaraman L., Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell. Mol. Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]