Abstract
Certain peptides such as dynorphin A [dynA-(1–13)] enhance the release of antigenic peptides bound to class II MHC molecules at neutral pH. This enhanced release has been termed push off. Previous work has shown that the antigenic pigeon cytochrome c peptide PCC-(89–104) has at least two conformational isomers when bound to the class II MHC protein I-Ek. We have accordingly studied the push off of PCC-(89–104) from the complex PCC-(89–104)/I-Ek to see whether these isomeric conformations are distinguished by the push-off effect. A comparison of the association and dissociation kinetics of PCC-(89–104)/I-Ek in the presence of dynA-(1–13) shows that dynA-(1–13) does not simply replace PCC-(89–104) but rather acts catalytically. The major product is peptide-free I-Ek, which is receptive to further peptide binding. Evidence is presented that a two peptide—one MHC complex is formed in solution. This ternary complex represents the first step of the mechanism of push off. 19F NMR data are presented that indicate that dynA-(1–13) interacts specifically with only one of the two isomeric complexes of PCC-(89–104) and I-Ek. A push-off mechanism is proposed in which dynA-(1–13) binds outside the peptide binding groove. In a second step, the dissociation of one of the two isomers is specifically enhanced. Thus the push-off effect may be useful for identifying conformational isomers and for separating them.
Keywords: second peptide binding site, isomers, accelerated peptide release, 19F NMR
Class II major histocompatibility (MHC) molecules are αβ-heterodimeric, glycosylated membrane proteins. They present antigenic peptides to the T cell receptor (TCR) of CD4+-lymphocytes. The interaction between these molecules and the formation of a stable, ternary complex is one of the key events in the generation of a cellular immune response against a foreign pathogen (1). Class II MHC molecules are normally loaded with peptide in an acidic, endosomal compartment (2) with the aid of chaperones such as HLA-DM (3). However it is known that a direct binding of peptides to class II MHC molecules is also possible at the cell surface, but with low efficiency (4). For some applications, it would be desirable to bypass the acidic loading compartment and to load the MHC molecule with antigenic peptide under the neutral pH conditions of the cell surface with high efficiency.
A prerequisite for an efficient binding of peptides to MHC molecules at the cell surface is the accelerated release of already bound peptide and the subsequent binding of the second peptide. Over the past years, reports have described such an accelerated release of prebound peptide either at the cell surface or in vitro (5–7). de Kroon and McConnell (6, 7) have observed that different peptides are able to stimulate the release of another bound peptide from murine class II MHC-peptide complexes. The authors conclude that this replacement reaction, called push off, proceeds via a two peptide—one MHC intermediate. So far, such two peptide—one MHC complexes have been observed experimentally only in gels, not in solution (8).
In the course of studying kinetic and conformational isomers of an antigenic peptide derived from pigeon cytochrome c [PCC-(89–104)] complexed with a water-soluble version of the murine class II MHC molecule I-Ek (9), we observed a monophasic dissociation of this complex at neutral pH (10). On the other hand, 19F NMR studies revealed the presence of two conformational isomers at pH 7.0 (11). To further analyze these findings and to relate kinetic and structural isomers of class II MHC-peptide complexes, we examined the action of a push-off peptide, dynorphin A [dynA-(1–13)], on the dissociation kinetics of the PCC-(89–104)/I-Ek complex. We have combined kinetic analysis, fluorescence resonance energy transfer (FRET) (12), and 19F NMR to determine the molecular mechanism of push off. The results indicate that the first step of push off is the formation of a two peptide—one MHC complex in solution. Furthermore, the 19F NMR data indicate a specific interaction between dynA-(1–13) and only one of the observed two conformational isomers of the PCC-(89–104)/I-Ek complex. On the basis of our observation we propose a mechanism in which the major product is the empty class II MHC molecule, which is receptive for further peptide binding at neutral pH.
MATERIALS AND METHODS
Kinetic Experiments.
Peptide synthesis, purification, and protein isolation as well as complex preparation and dissociation experiments were performed as described in ref. 10. Sequences of the peptides used are PCC-(89–104), AERADLIAYLKQATAK, and dynA-(1–13), YGGFLRRIRPKLK. Association experiments were performed in phosphate-buffered saline (PBS, 10 mM sodium phosphate, pH 7.0/150 mM NaCl), at 25°C, 30 nM I-Ek in the presence of various concentrations (150 nM to 3 μM) of fluorescein-labeled peptides [F-PCC-(89–104) or F-dynA-(1–13)]. When indicated, a second, unlabeled, peptide was added to the association or dissociation reaction mixture. For the association reactions, 50-μl aliquots of the incubation solution were applied to a disposable 2-ml spin column (Sephadex G-50, equilibrated in PBS and blocked with 1 mg/ml lysozyme) at the indicated times. After washing with 230 μl of PBS, the class II MHC-peptide complex was eluted with 200 μl of PBS and immediately injected onto an HPLC size-exclusion chromatography column. To obtain quantitative information about the association process, the signal heights of the class II MHC-peptide complexes were standardized with 5(6)-carboxyfluorescein. In the case of F-dynA-(1–13), comparison of the concentration determined by absorption at 280 nm (Tyr) and 495 (F) showed that a quenching of a factor of 3.6 occurs. This quenching is due to the N-terminal Tyr as already reported by Watts and Voss (13). Further details of the experimental procedures, including the fitting procedures are given in ref. 10.
FRET Experiments.
All experiments were performed in a 1.8-ml volume in a Spex fluorometer under constant gentle stirring. Calculated aliquots of Texas red (TR)-dynA/dynA (1:50) stock solutions were diluted into 10 nM F-PCC/I-Ek in PBS at room temperature. Experimental parameters were: 492-nm excitation (2-nm width) and 612-nm emission (2-nm width), 20 scans over a time of 2 min with a delay of 3 s between each scan.
19F NMR Experiments.
Eighteen thousand scans were averaged at 4°C with a delay time of 3 s (>5 ⋅ T1) for each spectrum. For the push-off experiments shown in Fig. 5, a 0.08 mM solution of the fluorine-substituted peptide PCC-(89–104) A96A-19F, Y97Fp19F, A103A-19F/I-Ek in PBS was incubated with various amounts of dynA-(1–13). Calculated amounts of dynA-(1–13) in PBS were added to the NMR sample (added volume 10 μl of a stock solution) and incubated for 30 min at 25°C. The sample was immediately cooled to 4°C and subsequent spectra were acquired. Further experimental details as well as the processing procedures are given in ref. 11.
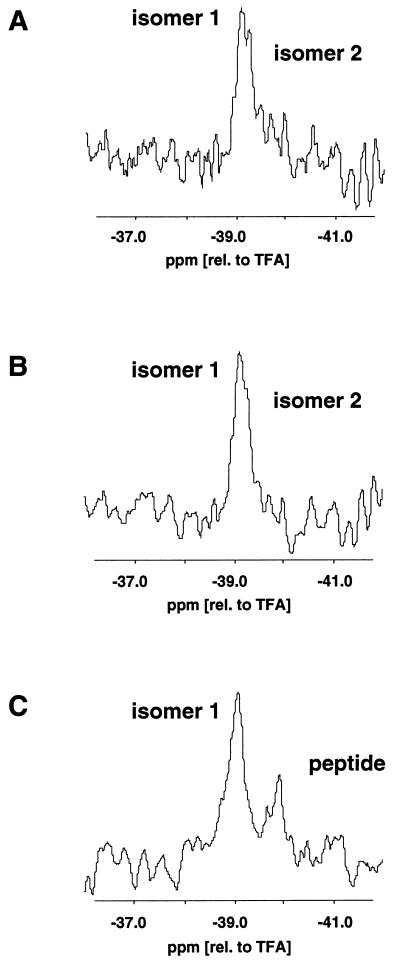
Figure 5.
19F NMR spectra of 0.08 mM F-PCC-(89–104) A96A-19F, Y97Fp19F, A103A-19F/I-Ek in the presence of various concentrations of dynA-(1–13). Shown are only the spectra of Y97Fp19F bound to the P3 pocket of I-Ek. (TFA, trifluoroacetic acid). (A) 0.8 μM dynA (molar ratio of MHC-peptide complex to dynA = 100:1). (B) 0.1 mM dynA (molar ratio of MHC-peptide complex to dynA = 1:1.25). (C) 0.8 mM dynA (molar ratio of MHC-peptide complex to dynA = 1:100). Based on a binding constant Kb of 15 μM−1 of dynA to the MHC-peptide complex a sigmoidal binding curve was calculated for a complex concentration of 0.08 mM. The concentrations of dynA in the 19F NMR experiments correspond to the lower asymptotic part of the sigmoidal binding curve (A), near the 50% value (B), and the upper asymptotic part of the sigmoidal binding curve (C). In A, two 19F NMR signals were observed for Y97Fp19F (labeled isomer 1 and 2). In B, the signal of isomer 2 is reduced to a shoulder. In C, only the signal of isomers 1 was detected in addition to the signal of unbound peptide. In all three spectra, the chemical shift of isomer 1 was identical within experimental error (±0.05 ppm).
RESULTS AND DISCUSSION
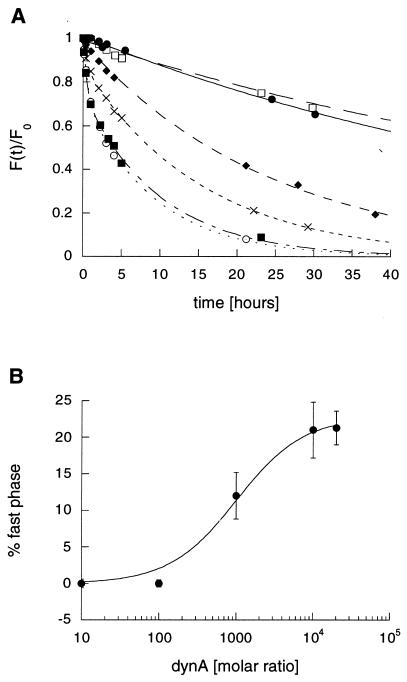
The influence of dynA-(1–13) on the dissociation kinetics of F-PCC-(89–104)/I-Ek in PBS, pH 7.0, at 25°C is shown in Fig. 1A. Increasing the concentration of dynA-(1–13) results in an accelerated dissociation of F-PCC-(89–104) and the appearance of a second, fast-dissociating, component. This indicates that two isomers are present in the F-PCC-(89–104)/I-Ek complex at pH 7.0. In the absence of dynA-(1–13) it is not possible to distinguish these complexes because of their apparently very similar off-rates. The fast-dissociating isomer is first observed at 15 μM dynA-(1–13), which corresponds to a 1,000-fold molar excess of this peptide. The observed magnitude follows a sigmoidal behavior (Fig. 1B). The maximum of fast-dissociating complex is generated in the presence of a 10,000-fold molar excess of dynA-(1–13) corresponding to 150 μM. Further increase of the dynA-(1–13) concentration results in no further increase of the generated fast-dissociating isomer. This saturation behavior suggests a second peptide binding site (14) outside the peptide binding groove of I-Ek. By assuming that the generation of a fast-dissociating complex of F-PCC-(89–104)/I-Ek is an indicator of push off by dynA-(1–13), one can extract an apparent binding constant from the data shown in Fig. 1B. Fitting a standard sigmoidal binding curve gives a binding constant Kb of 15 ± 1.4 μM−1. In addition to the generation of a fast-dissociating complex, dynA-(1–13) also accelerates the release of the slow-dissociating complex. The half-time of the slow-dissociating isomer F-PCC-(89–104)/I-Ek complex is reduced from 57 hr [1.5 μM dynA-(1–13)] to 7 hr [150 and 300 μM dynA-(1–13)]. This observation is consistent with a picture in which the kinetic isomers of the F-PCC-(89–104)/I-Ek complex interconvert, but with an interconversion rate that is slower than the off-rate of the fast-dissociating complex.
Figure 1.
(A) Dissociation kinetics of F-PCC-(89–104)/I-Ek in PBS at 25°C in the presence of various concentrations of dynA-(1–13). ●, No dynA; □, 1.5 μM dynA (100-fold molar excess); ♦, 15 μM dynA (1,000-fold molar excess); ×, 75 μM dynA (5,000-fold molar excess); ○, 150 μM dynA (10,000-fold molar excess); ■, 300 μM dynA (20,000-fold molar excess). Data were acquired until the fluorescence signal of each dissociation reaction [F(t)] was smaller than 5% of the fluorescence at time zero (F0). For clarity, data points are shown only up to 40 hr. (B) Percent observed fast-dissociating complex versus molar ratio of dynA-(1–13) to F-PCC-(89–104)/I-Ek. Values were taken from the corresponding dissociation curves in A. Error bars represent the standard deviation of two independent dissociation experiments.
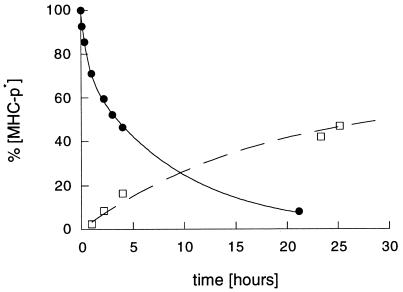
As suggested by de Kroon and McConnell (6, 7), these observations can be interpreted by a replacement reaction of F-PCC-(89–104) by dynA-(1–13). To investigate this hypothesis further, we examined the association kinetics of F-dynA-(1–13) to PCC-(89–104)/I-Ek under identical conditions (Fig. 2). Especially for the early data points, our data demonstrate that F-dynA-(1–13) does not simply replace PCC-(89–104). After 4 hr, around 50% of PCC-(89–104) is released, but only 20% of F-dynA-(1–13) forms a stable complex during this time period. For the association data in Fig. 2, a fluorescence quenching factor of 3.6 was included in the calculations. This factor was obtained by comparing the absorption of F-dynA-(1–13) at 280 nm (Tyr) and 495 nm (fluorescein). An identical quenching has been also reported for a Tyr-induced fluorescein quenching in IgG antibodies (13). Since we do not know whether Tyr-1 quenches the fluorescence of the fluorescein in F-dynA-(1–13) when bound to I-Ek, the values given in Fig. 2 for the F-dynA-(1–13)/I-Ek complex formed in the association reaction represent only the maximum values. Assuming that little or no quenching takes place within the F-dynA-(1–13)/I-Ek complex, the actual amount of complex formed is even less than the values given in Fig. 2. Nevertheless, the data show that dynA-(1–13) does not replace PCC-(89–104), but generates empty I-Ek molecules. These MHC molecules are highly receptive for further peptide binding. In experiments not shown here, we found that these I-Ek molecules bound PCC-(89–104) and Hb-(64–76) peptides faster and to an higher extent than I-Ek molecules that were not exposed to dynA-(1–13). This observation is in agreement with a report showing that class II MHC molecules have to be in an activated form to bind antigenic peptides efficiently (15). The data also suggest that there are at least two distinct steps in the mechanism of push off. In the first step, dynA-(1–13) binds to the F-PCC-(89–104)/I-Ek complex with an apparent binding constant of 15 ± 1.4 μM−1 and forms a two peptide—one MHC complex. In this ternary complex, one peptide [F-PCC-(89–104)] is bound inside the peptide binding cleft, while the other (dynA-(1–13)) is bound outside the groove at a second peptide binding side. At this stage the push-off event takes place. In the second step, dynA-(1–13), still outside the peptide binding cleft, either forms a stable class II MHC-peptide complex by occupying the groove (minor reaction product) or it dissociates from the protein (major reaction product).
Figure 2.
Comparison of the dissociation kinetics of 15 nM F-PCC-(89–104)/I-Ek in the presence of 150 μM dynA (10,000-fold molar excess) in PBS, pH 7.0, 25°C (●) and the association kinetics of 150 μM F-dynA-(1–13) to 15 nM PCC-(89–104)/I-Ek (□) under identical conditions. Plotted is the amount of class II MHC-peptide complex present at the indicated times.
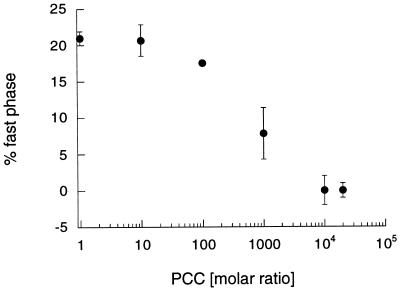
Since this proposed multistep mechanism includes an initial binding step of dynA-(1–13) to the class II MHC-peptide complex, one might be able to block this binding event. Fig. 3 summarizes the results of dissociation experiments of F-PCC-(89–104)/I-Ek in the presence of a 10,000-fold molar excess of dynA-(1–13) and various concentrations of unlabeled PCC-(89–104). The concentration of dynA-(1–13) used in these experiments corresponds to the conditions where the largest fraction of fast-dissociating isomer (Fig. 1B) is observed. Increasing the concentration of unlabeled PCC-(89–104) resulted in a sigmoidal decrease of detected fast-dissociating isomer. At a ratio of PCC-(89–104) to dynA-(1–13) of 1:1 and higher no fast-dissociating complex was observed. On the other hand, the monophasic dissociation curves of F-PCC-(89–104)/I-Ek have shorter half-times [t1/2 for 1:1 PCC-(89–104)/dynA-(1–13) of 22 hr] than the one measured in the absence of dynA-(1–13) (t1/2 = 51 hr) (data not shown). This result demonstrates that dynA-(1–13) and PCC-(89–104) compete for the same second peptide binding site. No fast-dissociating isomer is created because of this competition process, but peptide dissociation is still accelerated. Thus, the competition reduces the effective concentration of dynA-(1–13) bound to the class II MHC-peptide complex and the number of events yielding a push-off event. Nevertheless, the question arises why both peptides are able to bind to the same second peptide binding site, but only dynA-(1–13) is an effective push-off peptide. Dissociation experiments of F-PCC-(89–104)/I-Ek in the presence of a 10,000- and 20,000-fold molar excess of unlabeled PCC-(89–104) displayed no influence on the stability of the complex within experimental error (data not shown). One can speculate that the PCC-(89–104) peptide bound to the nonclassical binding site is oriented toward the peptide binding cleft in a way that there are no interactions between peptide side chains and the amino acid residues of the MHC protein. Consequently, PCC-(89–104) only occupies the second peptide site and dissociates immediately without accelerating peptide dissociation. On the hand, dynA-(1–13) might be bound to the second peptide binding site in an arrangement that orients peptide side chains toward the MHC protein in a favorable way. The resulting interaction between dynA-(1–13) and I-Ek destabilizes the peptide within the cleft and consequently accelerates the dissociation of the peptide inside the groove. NMR studies in solution (L.S. and H.M.M., unpublished results) indicate that dynA-(1–13) has a secondary structure in aqueous solution, whereas PCC-(89–104) has none. Assuming that dynA-(1–13) binds to I-Ek without losing its secondary structure, it might place one or more side chains toward the peptide binding cleft in a conformationally fixed orientation. As a result, one or more of the side chains of dynA-(1–13) could compete with prebound PCC-(89–104) for the binding pockets of I-Ek. Within this competition process only a small entropic penalty is paid due to the secondary structure of dynA-(1–13). In the case of PCC the same mode of action is entropically less favored. Consequently, only dynA-(1–13) can enhance peptide release.
Figure 3.
Inhibition of push off by PCC-(89–104). Shown is the amount of observed fast-dissociating complex of 15 nM F-PCC-(89–104)/I-Ek in the presence of 150 μM dynA-(1–13) (10,000-fold molar excess) and various concentrations of PCC-(89–104). Experiments were performed in PBS at 25°C.
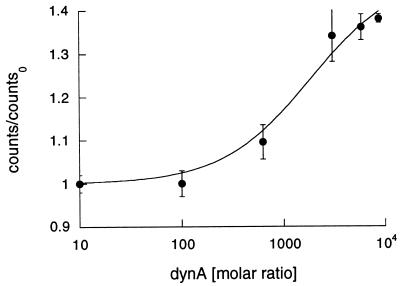
All our data indicate that push off proceeds by a multistep mechanism. Of central importance is the formation of a two peptide—one MHC complex in which dynA-(1–13) binds to a second peptide binding site and F-PCC-(89–104) is bound inside the peptide binding cleft. Such a ternary complex has been observed in a gel matrix but never in solution (8). To further investigate the formation of the proposed ternary complex we performed FRET experiments (Fig. 4). The dyes fluorescein [covalently coupled to the N terminus of PCC-(89–104), F-PCC-(89–104)] and TR (covalently bound to the N terminus of dynA-(1–13), TR-dynA-(1–13)) served as an energy transfer pair (16). The concentrations of dynA-(1–13) necessary to observe push off result in self-quenching of TR and fluorescence emission by excitation at 492 nm. The later finding is due to the long tail in the absorption spectrum of TR. Consequently, we diluted TR-dynA-(1–13) into dynA-(1–13) to yield a final molar ratio of 1:50. But even in this diluted solution, TR-dynA-(1–13) emitted after being excited at 492 nm. As a result, this emission was taken as background. The observed energy transfer between F-PCC-(89–104)/I-Ek and TR-dynA-(1–13) is the measured energy transfer divided by the background fluorescence of TR-dynA-(1–13) in the absence of F-PCC-(89–104)/I-Ek. As can be seen in Fig. 4, the observed Förster transfer is TR-dynA-(1–13) concentration dependent and levels off around 100 μM dynA-(1–13). The apparent binding constant Kb was determined to 18 ± 3.2 μM−1. This is in good agreement with the binding constant Kb (15 ± 1.4 μM−1) determined by kinetic analysis (Fig. 1A). In addition, the observed energy transfer enabled us to estimate the distance between the two fluorescent dyes. The Förster radius (17) of the fluorescein–TR pair varies between 41 and 56 Å (16). Taking a dilution of TR-dynA-(1–13) of 1:50 into account and assuming that both dyes tumble freely in solution (18), we obtain a rough estimate for the distance between the two dyes on the order of 10–15 Å. But more important, the FRET data demonstrate that the proposed two peptide—one MHC complex is formed in solution and represents a key intermediate in the mechanism of push off.
Figure 4.
FRET of 10 nM F-PCC-(89–104)/I-Ek in the presence of various concentrations of TR-dynA-(1–13)/dynA-(1–13) (1:50 molar ratio) in PBS at room temperature. To account for the emission of TR-dynA-(1–13) at 612 nm by exciting at 492 nm [excitation maximum of fluorescein (F)], measured counts were divided by the counts of TR-dynA-(1–13)/dynA-(1–13) (1:50) in the absence of F-PCC-(89–104)/I-Ek under identical conditions.
Some of our kinetic data indicate that the presence of kinetic isomers of the F-PCC-(89–04)/I-Ek complex may play a role in the push-off mechanism. Furthermore, the data imply that the observed accelerated dissociation of F-PCC-(89–104) in the presence of dynA-(1–13) is due to a specific interaction of dynA-(1–13) with only one of the two observed isomers. To investigate this hypothesis further, we employed 19F NMR measurements in solution (11). Here, three fluorine labels were introduced at specific positions of the PCC-(89–104) peptide. Fluorine replaced one proton in the methyl side chains of Ala-96 and Ala-103 (A96A-19F and A103A-19F), and fluorine replaced the hydroxyl group of Tyr-97 (Y97Fp19F). A103A-19F displayed only one 19F NMR signal. In the case of A96A-19F, two 19F NMR signals were observed, but one of them partially overlapped with the signal of A103A-19F. Therefore, only the 19F NMR signals of Y97Fp19F bound to the P3 pocket of I-Ek were monitored. In Fig. 5A, the 19F NMR signals of Y97Fp19F of the PCC A96A-19F, Y97Fp19F, A103A-19F/I-Ek complex are shown. Because of the very low concentration of protein (80 μM) used in this study, the signal-to-noise ratio is very low. Nevertheless, two signals arbitrarily labeled isomer 1 and isomer 2 were observed as already described in detail for this complex (11). In Fig. 5B, dynA-(1–13) was added and the solution was incubated at 25°C for 30 min before acquiring a subsequent spectrum at 4°C. The added concentration of dynA-(1–13) resulted in a final ratio of class II MHC-peptide complex to dynA-(1–13) of 1:1.25. Whereas the 19F NMR signal of isomer 1 is not affected by dynA-(1–13), the signal of isomer 2 is reduced and hardly visible as a shoulder. Increasing the concentration of dynA-(1–13) to a ratio of complex to dynA-(1–13) of 1:100 (Fig. 5C) resulted in a complete loss of the 19F NMR signal of isomer 2. Here again, the chemical shift as well as the signal height of the 19F NMR signal of isomer 1 is not affected. Removing dynA-(1–13) by ultrafiltration resulted in a reappearance of the 19F NMR signal of isomer 2 (spectra not shown). This result demonstrates again that the two isomers interconvert. More important, the 19F NMR data show that dynA-(1–13) interacts specifically with only one of the two isomers.
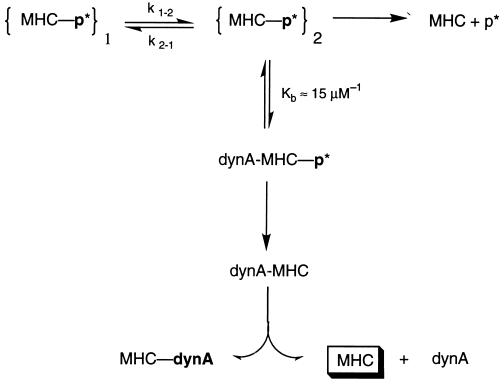
The data presented here are summarized in Fig. 6. As demonstrated by the reappearance of the 19F NMR signal of isomer 2 after removal of dynA-(1–13) and in a previous study (10), the two isomers of F-PCC-(89–104)/I-Ek interconvert. Although the dissociation kinetics of F-PCC-(89–104)/I-Ek are monophasic at pH 7.0 (10), two interconverting isomers with similar off-rates are present. This similarity in the off-rates is the reason why it is not possible to resolve their presence by the type of kinetic analysis employed here. In the presence of dynA-(1–13) however, biphasic dissociation curves were obtained (Fig. 1A). This indicates that dynA-(1–13) is destabilizing one of the two kinetic isomers. As a result, a push off of F-PCC-(89–104) bound to I-Ek by dynA-(1–13) is observed. In the first step of push off, dynA-(1–13) binds with an apparent binding constant Kb of about 15 μM−1 to the class II MHC-peptide complex. The binding constant was determined by the fraction of fast-dissociating component in the presence of dynA-(1–13) (Fig. 1B) as well as FRET experiments (Fig. 4). As shown by the 19F NMR experiments (Fig. 5), the interaction between dynA-(1–13) and the bound antigenic peptide [PCC-(89–104)] is specific for only one isomer. We assign this interaction arbitrarily to isomer 2. This binding process can be inhibited in the presence of unlabeled PCC-(89–104) (Fig. 3). Therefore, both peptides compete for the same second peptide binding site. While both peptides bind to this second site, only dynA-(1–13) effectively accelerates the dissociation reaction. It is not clear if this nonclassical binding site is identical to the one described for an insulin peptide (14). The general ability of peptides to bind to this second binding site indicates that push off is a multi step reaction, where binding and interaction are separate events. In the second step of push off, the dissociation of the antigenic peptide bound inside the peptide binding cleft of I-Ek is accelerated. This results in the observed fast-dissociating component in the dissociation kinetics. Because of interconversion of the two isomers, the isomer 2 state never completely depletes. The repopulation results in subsequent push off, the rate of which is now determined by the effective interconversion rate between the isomers. This repopulation and the dependence on the effective interconversion rate explain why the slow-dissociating component observed in the dissociation kinetics (Fig. 1A) has a smaller off-rate which is also dependent on the concentration of dynA-(1–13). At this stage, dynA is still bound to the second peptide binding site, which is outside the peptide binding cleft. In the third and final step, dynA either dissociates from the MHC molecules or occupies the peptide binding groove by itself. Association and dissociation data (Fig. 2) demonstrate that the major products formed in this reaction are empty class II MHC molecules. As has been described previously (15), these MHC molecules are highly receptive to further peptide binding. For example, the association rate and the final amount of PCC-(89–104) binding to empty I-Ek molecules generated by push off is roughly 8 times larger than the association to nonactivated I-Ek (data not shown). Additionally, the apparent binding constant of dynA-(1–13) to the class II MHC-peptide complex depends strongly on the antigenic peptide. As shown here, the highest push-off rate of F-PCC-(89–104) is observed around 150 μM dynA-(1–13) (10,000-fold molar excess). In case of an antigenic peptide derived from hemoglobin ([Hb-(64–76)] only 10 μM dynA is necessary to achieve the highest push-off rate. This suggests that the antigenic peptide bound inside the MHC peptide binding cleft influences the structure or architecture of the second peptide binding site. In summary, our data demonstrate that push off is not a simple replacement reaction. Rather it generates empty class II MHC molecules under neutral pH conditions by a two peptide—one MHC complex intermediate. The fact that push off is effective at neutral pH conditions opens up new possibilities for loading class II MHC molecules efficiently under the conditions that are encountered at the cell surface. Furthermore, our in vitro data show that some class II MHC-peptide complexes are susceptible to catalytic peptide release. This raises the questions of whether such an accelerated release happens in vivo and whether it might be involved in an alternative loading or recycling pathway of class II MHC molecules.
Figure 6.
Proposed mechanism of push off. The antigenic peptide [PCC-(89–104) or dynA-(1–13)] bound to the peptide binding cleft of I-Ek is shown in boldface, whereas the peptide bound to the second binding site is shown in lightface.
Acknowledgments
We thank Tom Anderson, Josh Rabinowitz, Marija Vrljic, Michael Belmares, and Peter Kasson for many stimulating discussions and critical reading of the manuscript and John Mumm for technical assistance in the protein purification. We are also indebted to Craig Beeson for many helpful suggestions and Elisabeth Mellins for helpful discussions and critical reading of the manuscript. L.S. was supported by the Deutsche Forschungsgemeinschaft (Grant Schm 1279/1–2). J.R.K. is a Stanford Undergraduate Fellow. This work was funded by the Howard Hughes Medical Institute (M.M.D.) and the National Institutes of Health (H.M.M.).
ABBREVIATIONS
- dynA
dynorphin A
- F
fluorescein
- FRET
fluorescence resonance energy transfer
- PCC
pigeon cytochrome c
- TR
Texas red
References
- 1.Hedrick S, Edelmann F J. Fundamental Immunology. New York: Raven; 1993. [Google Scholar]
- 2.Tulp A, Verwoerd D, Dobberstein B, Ploegh H, Pieters J. Nature (London) 1994;369:120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- 3.Sloan V S, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller D M. Nature (London) 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 4.Beeson C, Rabinowitz J, Tate K, Gütgemann I, Chien Y-H, Jones P P, Davis M M, McConnell H N. J Exp Med. 1996;184:777–782. doi: 10.1084/jem.184.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedrazzini T, Sette A, Albertson M, Grey H M. J Biol Chem. 1986;146:3496–3501. [PubMed] [Google Scholar]
- 6.de Kroon A I P M, McConnell H M. Proc Natl Acad Sci USA. 1993;90:8797–8801. doi: 10.1073/pnas.90.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Kroon A I P M, McConnell H M. J Immunol. 1994;152:609–619. [PubMed] [Google Scholar]
- 8.Tampé R, Clark B R, McConnell H M. Science. 1991;254:87–89. doi: 10.1126/science.1656526. [DOI] [PubMed] [Google Scholar]
- 9.Wettstein D, Boniface J J, Reay P A, Schild H, Davis M M. J Exp Med. 1991;174:219–228. doi: 10.1084/jem.174.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt L, Boniface J J, Davis M M, McConnell H M. Biochemistry. 1998;35:17371–17380. doi: 10.1021/bi9815593. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt L, Boniface J J, Davis M M, McConnell H N. J Mol Biol. 1999;286:2409–2420. doi: 10.1006/jmbi.1998.2463. [DOI] [PubMed] [Google Scholar]
- 12.Selvin P R. Methods Enzymol. 1995;246:300–335. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 13.Watt R M, Voss E W. Immunochemistry. 1977;14:533–541. doi: 10.1016/0019-2791(77)90308-1. [DOI] [PubMed] [Google Scholar]
- 14.Tompkins S M, Moore J C, Jensen P E. J Exp Med. 1996;183:857–866. doi: 10.1084/jem.183.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabinowitz J D, Vrljic M, Kasson P, Liang M N, Busch R, Boniface J J, Davis M M, McConnell H M. Immunity. 1998;9:699–708. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 16.Van Der Meer B W, Coker G, III, Chen S-Y S. Resonance Energy Transfer: Theory and Data. New York: VCH; 1994. [Google Scholar]
- 17.Förster T. Z Naturforsch A. 1949;4:321–347. [Google Scholar]
- 18.Epe B, Steinhaeuser K G, Wooley P. Proc Natl Acad Sci USA. 1983;80:2579–2583. doi: 10.1073/pnas.80.9.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]