Abstract
By stimulating blood lymphocytes from a renal cell carcinoma patient in vitro with the autologous tumor cells, we obtained cytolytic T lymphocyte (CTL) clones that killed several autologous and allogeneic histocompatibility leukocyte antigen (HLA)-B7 renal carcinoma cell lines. We identified the target antigen of these CTLs by screening COS cells transfected with the HLA-B7 cDNA and with a cDNA library prepared with RNA from the tumor cells. The antigenic peptide recognized by the CTLs has the sequence LPRWPPPQL and is encoded by a new gene, which we named RU2. This gene is transcribed in both directions. The antigenic peptide is not encoded by the sense transcript, RU2S, which is expressed ubiquitously. It is encoded by an antisense transcript, RU2AS, which starts from a cryptic promoter located on the reverse strand of the first intron and ends up on the reverse strand of the RU2S promoter, which contains a polyadenylation signal. This mechanism of antigen expression is unprecedented and further illustrates the notion that many peptides recognized by T cells cannot be predicted from the primary structure of the major product of the encoding gene. Antisense transcript RU2AS is expressed in a high proportion of tumors of various histological types. It is absent in most normal tissues, but is expressed in testis and kidney, and, at lower levels, in urinary bladder and liver. Short-term cultures of normal epithelial cells from the renal proximal tubule expressed significant levels of RU2AS message and were recognized by the CTLs. Therefore, this antigen is not tumor specific, but corresponds to a self-antigen with restricted tissue distribution.
Keywords: renal cell carcinoma, cytolytic T lymphocytes, antisense, peptides
Several antigens recognized by cytolytic T cells on human tumors have been defined 1. Although some of them are expressed in various tumor types, most have been characterized using antitumor CTLs isolated from the lymphocytes of melanoma patients. They can be classified according to four groups: (a) shared tumor-specific antigens, which are encoded by MAGE-type genes that are silent in most normal tissues and expressed in many tumors; (b) melanocyte differentiation antigens, such as tyrosinase, gp100, and Melan-AMART1, which are also expressed in normal melanocytes; (c) antigens resulting from tumor-specific point mutations in genes expressed ubiquitously; and (d) antigens that are overexpressed in tumor cells (for a review, see reference 1).
Because isolating CTLs against tumors other than melanoma is more difficult, little is known about the antigens targeted by the immune system on such tumors. Three antigens have been defined on renal cell carcinoma (RCC). One corresponds to the first group of tumor antigens and is encoded by gene RAGE, which is silent in most normal tissues but expressed only in a limited number of RCC samples 2. The two other examples belong to the group of mutated antigens and result from a point mutation in the HLA-A2 or in the hsp70-2 gene 3 4. Some CTLs against RCC were also found to recognize normal kidney cells, suggesting the recognition of kidney differentiation antigens 5 6. These antigens would be to kidney cancer what melanocytic differentiation antigens are to melanoma. However, their molecular nature has not been defined. Here, we provide the first definition of such a kidney differentiation antigen. The mechanism of expression of this antigen is very unusual: it is encoded by the reverse strand of a housekeeping gene.
Materials and Methods
Cell Lines.
Cell lines LE9211-RCC, LE9211-EBV, and LB23-SAR have been described previously 2. Lines LB2043-PTEC and LB2046-PTEC were derived from surgical samples of normal kidney cortex and grown in Iscove's medium supplemented with ACL4 7 and epithelial growth factor (10 ng/ml) 8. They were confirmed to derive from the proximal tubule by testing the expression of aquaporin-1 by immunohistochemistry 9. Stable transfectant LB23-SAR-4.1 was obtained by transfection of cDNA 4.1 cloned into plasmid pEF-BOSpuro-PL3 10 using the calcium phosphate precipitation method, selection with puromycin, and cloning by limiting dilution. CTL clones were obtained exactly as described 2.
CTL Assays.
Chromium-release and TNF stimulation assays were performed as described previously 2. Before being used as stimulators for a TNF assay, cells from LB2043- and LB2046-PTEC were transiently transfected with the HLA-B7 cDNA using DMRIE-C (GIBCO BRL) according to the instructions of the manufacturer. IFN-γ production was measured by ELISA using antibodies from Biosource.
Cloning of Gene RU2 and RU2 cDNAs.
We used the same cDNA library as that described previously 2, using the procedure detailed elsewhere 11. In brief, mRNA was converted to cDNA with the Superscript Choice System (GIBCO BRL) using an oligo-dT primer containing a NotI site at its 5′ end. cDNAs were then ligated to BstXI adaptors and digested with NotI. After size fractionation, the cDNAs were unidirectionally cloned into the BstXI and NotI sites of plasmid pcDNAI/Amp (Invitrogen). Plasmid DNA was purified from pools of ∼100 independent cDNA clones and transfected together with DNA from an HLA-B7 cDNA construct into COS cells by the DEAE-dextran-chloroquine method. Transfected COS cells were then screened for their ability to stimulate the release of TNF by CTL 361A/21. The 5-terminal sequence was obtained by rapid amplification of cDNA ends (RACE) PCR using the 5′ RACE System (GIBCO BRL). The genomic library was prepared in phage vector lambdaGEM-11 (Promega Corp.) using standard techniques. Sequencing reactions were performed by PCR with the dideoxy-termination method and analyzed either manually or on an ABI310 Genetic Analyzer (Perkin-Elmer Applied Biosystems).
PCR Assay.
Expression of RU2S and RU2AS was measured by reverse transcription (RT)-PCR on cDNA produced from total RNA as described previously 2. Primers used were VDE87 and VDE93 for RU2S, and VDE119/VDE120 for RU2AS. PCR conditions were: 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 54°C for primers VDE87/VDE93 or at 62°C for primers VDE119/VDE120, and 1 min at 72°C, and a final elongation step of 15 min at 72°C. Primer sequences were: 5′-CCGTCAGGAACATCTACA-3′ (VDE87), 5′-CCAACAGCCACATAAAAC-5′ (VDE93), 5′-TAAATGGGTGGGCGGTGGGGGAGAC-3′ (VDE119), and 5′-TAGGCTGTTTGGAAAGGGT-AGCACA-3′ (VDE120). Because primers VDE119 and VDE120 also amplify genomic DNA, positive samples were retested after DNase treatment, to exclude any false positives due to DNA contamination of the RNA samples. Control RT-PCRs were also performed in which the addition of reverse transcriptase was omitted during cDNA synthesis. For the quantitative PCR shown in Fig. 8, the PCR conditions were first optimized to ensure linearity of the reaction according to De Plaen et al. 12. Optimized conditions for primers VDE119/VDE120 were: 33 cycles with 1 min at 94°C, 2 min at 68°C, and 3 min at 72°C. Serial twofold dilutions of each sample were then tested together with dilutions of positive control sample LE9211-RCC, which was chosen as a reference. PCR products were then visualized on agarose gels stained with ethidium bromide, and the level of expression of each sample was inferred from the comparison with the reference control. The results were then normalized for RNA integrity by quantifying the actin message in the same way 12. Results were expressed as percentages of the expression level measured in reference sample LE9211-RCC.
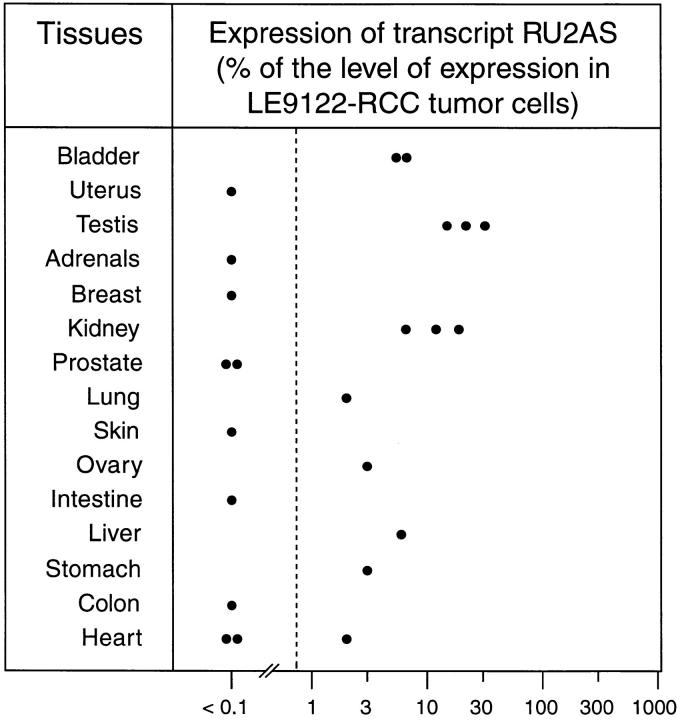
Figure 8.
Quantification of transcript RU2AS by RT-PCR. The level of RU2AS transcript in the indicated normal tissues was measured by RT-PCR as described in Materials and Methods and is expressed as percentage of the level of RU2AS found in LE9211-RCC tumor cells.
Results
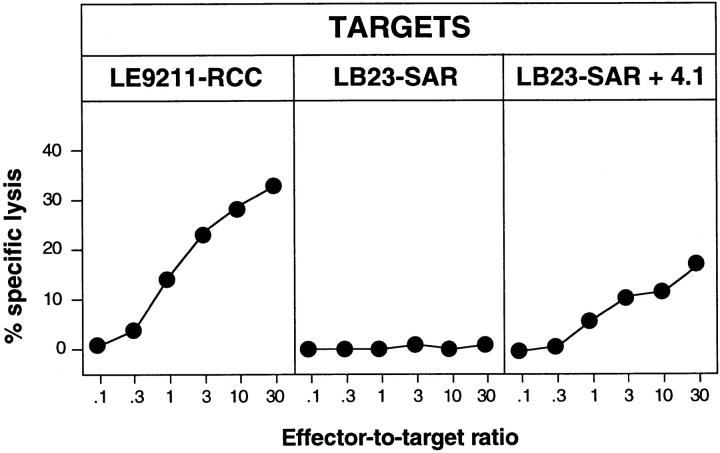
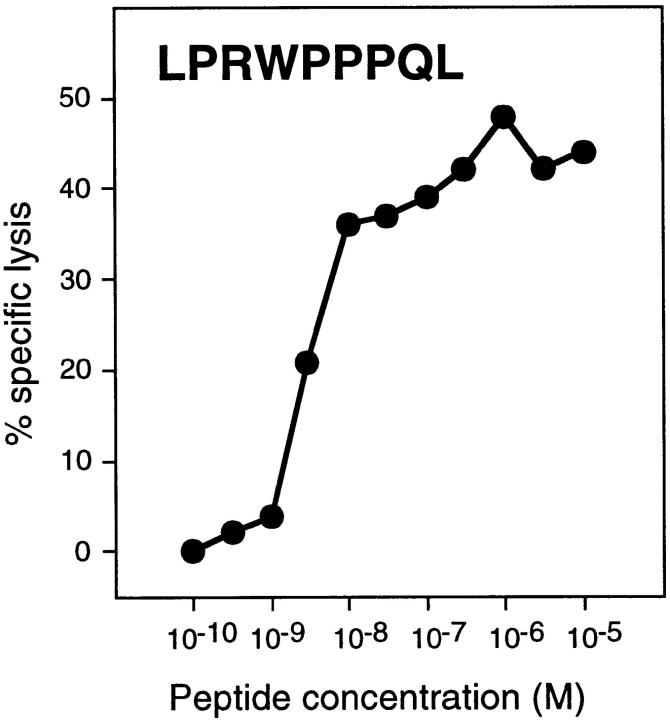
CTL clone 361/A21 was isolated by stimulating blood lymphocytes of a kidney cancer patient in vitro with autologous tumor cells LE9211-RCC, and by cloning the responding cells by limiting dilution 13. This clone was able to lyse both autologous and allogeneic tumor cells sharing the HLA-B7 specificity, but it did not lyse autologous EBV-transformed B cells or NK target K562 (Fig. 1). The HLA-B7 restriction of this CTL clone was confirmed by the observation that an anti–HLA-B7 antibody blocked the recognition of the tumor cells (Fig. 2 A). To identify the target antigen of CTL 361A/21, we prepared an oligo-dT–based unidirectional cDNA library with RNA from LE9211-RCC cells, and we transfected DNA from this library into COS cells together with DNA from an HLA-B7 cDNA plasmid construct. The transfected cells were screened by adding the CTLs to the microcultures and measuring the production of TNF. We isolated cDNA 4.1, which was able to stimulate the CTLs when transfected into COS cells together with the HLA-B7 cDNA (Fig. 2 B). This cDNA clone was 924 bp long. Its sequence was new and contained an open reading frame coding for a protein of 84 amino acids (Fig. 3). To exclude that recognition of the transfected COS cells was an artifact of high expression levels after transient transfection, we transfected an HLA-B7–positive sarcoma line with cDNA 4.1. A stable transfectant was obtained, and it was recognized by the CTLs (Fig. 4). To identify the antigenic epitope recognized by CTL 361A/21, we tested several synthetic peptides encoded by cDNA 4.1 and bearing the binding motif for HLA-B7, which is P in position 2 and L/F in position 9 14. We found that peptide LPRWPPPQL was able to sensitize autologous EBV-B cells to lysis by the CTLs (Fig. 5).
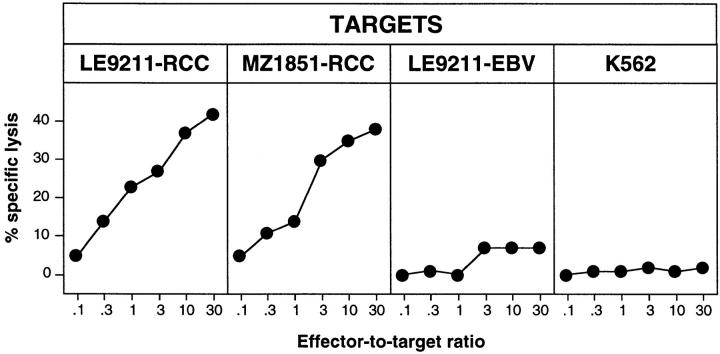
Figure 1.
Lytic activity of CTL clone 361A/21. The following target cells were tested in a standard 4-h chromium-release assay: LE9211-RCC, the autologous tumor line; MZ1851-RCC, an allogeneic HLA-B7–positive renal cell carcinoma line; LE9211-EBV, autologous B cells transformed with EBV; and K562, NK target cells. Results of a representative experiment are shown. CTL 361A/21 is representative of 12 CTL clones isolated from the lymphocytes of patient LE9211 and recognizing the same antigen.
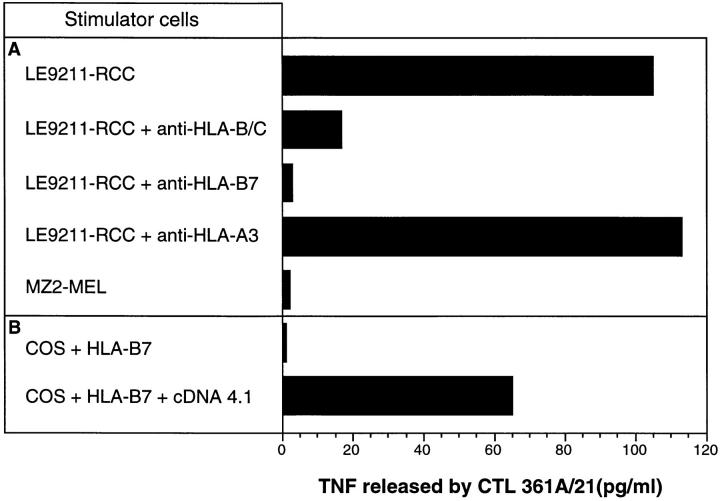
Figure 2.
Isolation of a cDNA coding for the antigen recognized by CTL 361A/21. (A) MHC restriction of CTL 361A/21. CTL activation was followed by measuring the production of TNF after stimulation with autologous tumor cells in the presence of the indicated anti-HLA mAbs. The HLA typing of patient LE9211 is HLA-A3, -B7, -B35, -Cw4, and -Cw7. MZ2-MEL is an allogeneic melanoma line that was used as a negative control. Antibodies W6/32 (anti–HLA class I), 4E (anti–HLA-B/C), ME1 (anti–HLA-B7), and GAPA3 (anti–HLA-A3) were used as described (reference 13). 3,000 CTLs were mixed with 30,000 tumor cells and IL-2 (25 U/ml), and TNF production was measured 18 h later as described elsewhere (reference 11). (B) cDNA 4.1 cloned into plasmid pcDNAI/Amp was transfected into COS cells together with the HLA-B*0702 cDNA cloned into plasmid pcDSRalpha. CTL 361A/21 (3,000 cells) was added after 24 h, and TNF production was measured 18 h later.
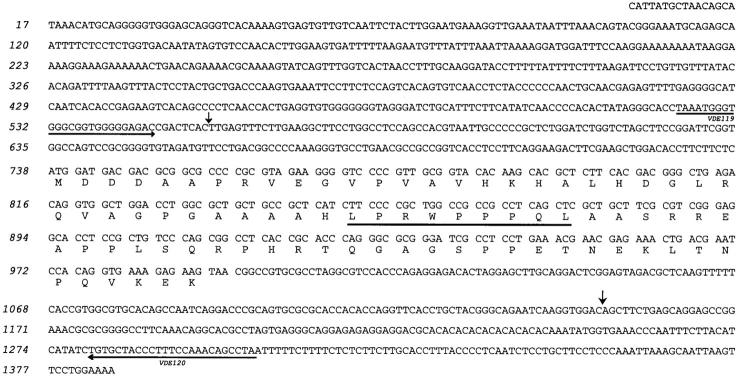
Figure 3.
Sequence of transcript RU2AS and its predicted protein product. The antigenic peptide recognized by CTL 361A/21 is underlined. The sequence shown is compiled from the sequence of cDNA 4.1 and the 5′ sequence obtained by RACE PCR. The vertical arrows indicate the limits of the fragment corresponding to the reverse strand of the first exon of the gene (see Fig. 6). The horizontal arrows indicate primers VDE119 (forward) and VDE120 (reverse), which were used for testing the expression of RU2AS by RT-PCR. The RU2AS sequence is available from EMBL/GenBank/DDBJ under accession no. AF181722.
Figure 4.
Lysis by CTL 361A/21 of LB23-SAR sarcoma cells transfected with cDNA 4.1. A representative clone obtained after stable transfection of cDNA 4.1 into HLA-B7 LB23-SAR sarcoma cells was used as the target in a standard chromium-release assay with CTL 361A/21 as the effector. Chromium release was measured after 4 h.
Figure 5.
Lysis by CTL 361A/21 of LE9211-EBV cells pulsed with synthetic peptide LPRWPPPQL. Chromium-labeled LE9211-EBV cells were incubated with the indicated peptide concentrations for 30 min before addition of CTLs at an E/T ratio of 10. Chromium release was measured after 4 h.
To obtain the full-length sequence of the transcript corresponding to cDNA 4.1, we amplified its 5′ end by PCR and sequenced the product. This extended the 4.1 sequence by 458 nucleotides (Fig. 3). The 1,382-bp sequence appeared to represent the full-length message, since it corresponded to a unique band observed by PCR amplification of the 5′ end. However, when cDNA 4.1 was used as a double-stranded probe on a Northern blot prepared with RNA from LE9211-RCC cells, it hybridized with a band of 2.2 kb, substantially larger than our putative full-length sequence. To solve this paradox, we screened our cDNA library by hybridization using cDNA 4.1 as a double-stranded probe. We obtained three identical clones of 2.2 kb, represented by clone I.1. Remarkably, the coding strand of clone I.1 did not contain the sequence of the coding strand of cDNA 4.1, but contained a 595-bp sequence that was complementary to the coding strand of cDNA 4.1. Since both cDNAs contained a poly(A) tail at their 3′ end and derived from an unidirectional library, their coding strands appeared to correspond to two genuinely distinct messages, one read in the sense and the other in the antisense direction of the same gene.
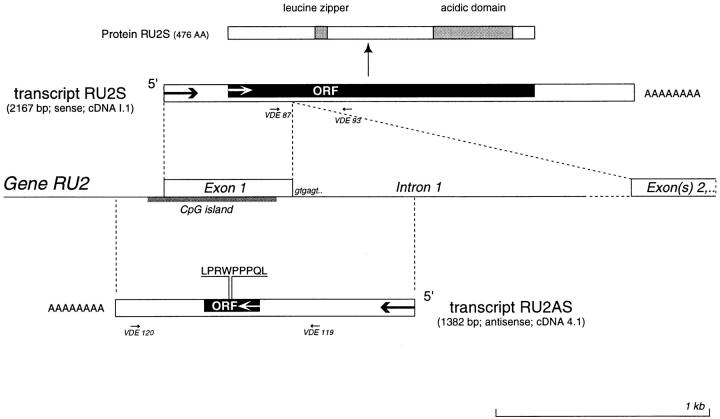
We cloned the genomic sequence by screening a phage library constructed with genomic DNA from LE9211-RCC cells, using cDNA 4.1 as a probe. We isolated a positive phage containing a 14-kb insert, from which we sequenced the relevant part. The comparison of the three sequences revealed that cDNA I.1 corresponds to the fully spliced transcript of the gene, whereas cDNA 4.1 corresponds to an aberrant message that starts on the antisense strand of the first intron, is transcribed backwards on the antiparallel strand of exon 1, and ends with a polyadenylation site that is located on the reverse strand of the promoter (Fig. 6). The gene, which we called RU2, is therefore transcribed in both directions, producing a message of 2,167 bp on one strand and another message of 1,382 bp on the other strand. The longer message, which corresponds to cDNA I.1, appears to be the “normal” RU2 message, since it encodes a longer protein with potentially functional domains (Fig. 6 and Fig. 7) and is expressed at higher levels and in a wider range of tissues (see below). We compared the sequence of the RU2 gene present in tumor cells LE9211-RCC to that of the RU2 gene present in autologous EBV-B cells and found no mutation. By fluorescent in situ hybridization (FISH) analysis, we mapped gene RU2 to chromosome 6p22.1 (data not shown).
Figure 6.
Partial structure of gene RU2. Black boxes indicate the open reading frames (ORF). The donor splice site of intron 1 is shown. Potentially functional domains of the RU2S protein are indicated. The small arrows indicate the primers used for testing the expression of the two opposite transcripts by RT-PCR: primers VDE87 and VDE93 were used to detect expression of sense transcript RU2S, whereas primers VDE119 and VDE120 were used to detect antisense transcript RU2AS. The 3′ part of intron 1 (dotted line) and the exons that follow it were not sequenced. The genomic sequence of RU2 is available from EMBL/GenBank/DDBJ under accession no. AF181720.
Figure 7.

Sequence of transcript RU2S (cDNA I.1) and its predicted protein product. The vertical arrow indicates the end of the first exon. The horizontal arrows indicate primers VDE87 (forward) and VDE93 (reverse), which were used for testing the expression of RU2S by RT-PCR. The RU2S sequence is available from EMBL/GenBank/DDBJ under accession no. AF181721.
We tested the expression of the two opposite RU2 transcripts by RT-PCR. For the sense message, which we named RU2S and which is represented by cDNA I.1, we used primers located in different exons, as indicated in Fig. 6. We found that RU2S was expressed in all of the tissues tested (Table ). This suggests a housekeeping function for this gene, which is consistent with the presence of a CpG island in its promoter and in most of exon 1 (Fig. 6). We then tested the expression of the antisense transcript, named RU2AS and represented by cDNA 4.1, using a primer located in the promoter region and a primer located in the first intron (Fig. 6). As opposed to the sense transcript, the antisense was only found in normal kidney, bladder, liver, and testis (Table ). However, a very high proportion of tumors of various histological origins also express RU2AS, including tumors derived from tissues that are negative for RU2AS, such as melanomas, sarcomas, and colorectal carcinomas (Table ).
Table 1.
Expression of the Sense (RU2S) and Antisense (RU2AS) Transcripts of Gene RU2
| Expression of transcript | No. of tumors expressing RU2AS/ No. of tumors tested | |||
|---|---|---|---|---|
| Normal tissues | RU2S | RU2AS | Tumor samples | |
| Adrenals | + | − | Renal carcinomas | 10/10 |
| Breast | + | − | Colorectal carcinomas | 14/15 |
| Colon | + | − | Melanomas | 15/24 |
| Heart | ± | − | Sarcomas | 7/9 |
| Kidney | + | + | Leukemias | 12/18 |
| Liver | + | + | Brain tumors | 6/9 |
| Lung | + | − | Thyroid carcinomas | 4/5 |
| Ovary | + | − | Mammary carcinomas | 5/10 |
| Prostate | + | − | Prostatic carcinomas | 4/10 |
| Skin | + | − | Esophageal tumors | 3/9 |
| Stomach | + | − | Bladder tumors | 3/10 |
| Testis | + | + | Lung carcinomas | 4/20 |
| Urinary bladder | + | ± | Head and neck tumors | 1/10 |
| Uterus | + | − | Mesotheliomas | 0/4 |
Expression was measured by RT-PCR as indicated in Materials and Methods.
To determine the level of expression of RU2AS in the normal tissues that were found to be positive, we repeated the RT-PCR using a limiting number of cycles and an optimized annealing temperature, to allow a quantitative amplification of the messenger RNA 15. The results shown in Fig. 8 are expressed as percentages of the level of expression found in cell line LE9211-RCC, which is arbitrarily considered as expressing 100% of RU2AS. We found that testis expresses 16–31%, kidney 7–19%, and bladder and liver 6%.
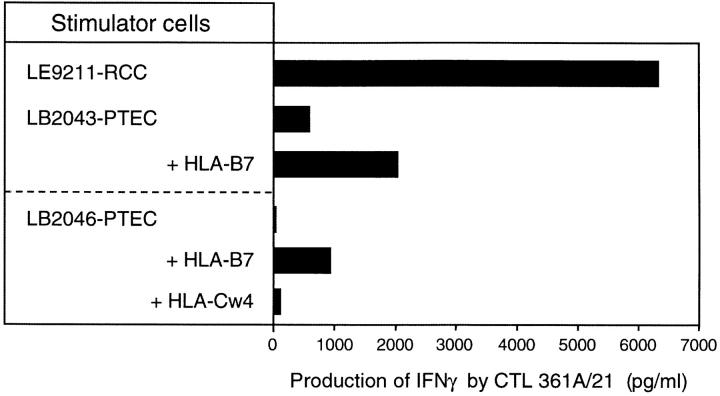
We previously observed, with a CTL clone directed against a MAGE-1 peptide, that cell lines expressing the MAGE-1 mRNA below a threshold of ∼10% were not recognized by the CTLs 15. Therefore, to determine whether the level of RU2AS in normal kidney is above the threshold for CTL recognition, we derived short-term cell lines from two samples of normal kidney. These two lines appeared to derive from the proximal tubule epithelium, since they expressed aquaporin-1, a specific marker of those tubules (9; data not shown). We measured the level of RU2AS in those lines by RT-PCR, and found that line LB2043-PTEC expressed 50% of the control level, whereas LB2046-PTEC expressed 25%. We then tested those cell lines for recognition by CTL 361A/21, after transient transfection of the HLA-B7 cDNA. As shown in Fig. 9, both lines were able to stimulate the production of IFN-γ by the CTLs. Line LB2043-PTEC was also recognized without transfection of HLA-B7, suggesting that it already expressed the HLA-B7 molecule. We conclude that this antigen is not tumor specific and is expressed in at least a subtype of normal kidney cells.
Figure 9.
Recognition of normal kidney cell lines by CTL 361A/21. LB2043-PTEC and LB2046-PTEC are two short-term cell lines established from normal kidney samples and corresponding to epithelial cells of the proximal tubule. They were transiently transfected with the indicated HLA cDNAs before the addition of 10,000 CTL 361A/21 in the presence of 25 U/ml IL-2. The presence of IFN-γ in the supernatant was measured by ELISA after an overnight incubation.
Expression of antisense transcripts is well known in prokaryotes, where it constitutes an important mechanism of gene regulation 16. Recently, several natural antisense RNA were also described in eukaryotes. They are believed to control gene expression, either by targeting the sense transcript for rapid degradation or by preventing its translation (for a review, see reference 17). To find out whether the two opposite RU2 transcripts exert a similar regulatory role, we transfected COS cells with the HLA-B7 cDNA and with both the sense and the antisense cDNAs at various ratios. We found that a 200-fold excess of RU2S over RU2AS did not prevent recognition of the transfected cells by our CTLs directed against the RU2AS-encoded peptide, indicating that an excess of sense transcript does not significantly affect stability and translation of the antisense transcript (data not shown).
Discussion
Our results contribute to the dissection of the immune response against renal cancer cells and provide the first molecular definition of the antigenic target of CTLs recognizing both normal and tumoral kidney cells. Such CTLs have been isolated from several patients by different groups, and they represent an important part of the T cell response against kidney cancer, at least in vitro 5 6. In one case, such CTLs were isolated from the lymphocytes of a patient who had been successfully treated with IL-2/LAK therapy, suggesting a possible in vivo relevance of this type of immune response 6. Interestingly, this patient did not show clinically detectable signs of autoimmunity despite the complete tumor response and the presence in his blood of CTLs against normal kidney cells. However, whether kidney differentiation antigens can be successfully and safely used for cancer immunotherapy is as yet uncertain.
The mechanism of expression of the RU2AS antigen is unprecedented and further supports the notion that many antigens recognized by T cells cannot be predicted from the primary structure of the major product of the encoding gene, but rather result from nonclassical mechanisms acting at the level of transcription, translation, or processing 18. One of the first such examples was a melanoma antigenic peptide, which was found to be encoded by the intronic part of an aberrant transcript starting on a cryptic promoter in an intron of the gene coding GnTV, a ubiquitous enzyme involved in protein glycosylation 19. This mechanism resembles that of RU2AS, except that the cryptic promoter is located on the sense and not the antisense strand of the intron. Other antigens have been found to be encoded by retained introns resulting from splicing defects in the MUM-1, gp100, and TRP2 genes 20 21 22. Translation of alternative open reading frames of genes TRP1 and NY-ESO-1 was also found to provide antigenic peptides recognized by CTLs on melanoma 23 24 25. Finally, a tyrosinase peptide was found to be modified after translation, with the asparagine residue of a glycosylation site changed into an aspartic acid, apparently after removal of the glucid moiety before processing of the peptide by the proteasome 26.
Altogether, these findings indicate that the variety of peptides presented to T lymphocytes is larger than expected, and that some potentially useful antigens cannot be predicted from the sequence of the cellular protein content.
Acknowledgments
We thank V. Ha Thi and L. Pilotte for technical assistance, and S. Depelchin and S. Mapp for editorial assistance.
This work was partially supported by the Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Office for Science, Technology and Culture; La Fédération contre le Cancer, Brussels, Belgium; and Caisse Générale d'Epargne et de Retraite-Assurances and VIVA, Brussels, Belgium. B. Gaugler was supported by a fellowship from the European Community. M. Probst-Kepper was supported by the Deutsche Forschungsgemeinschaft and by the Schering Forschungsgesellschaft, Germany.
Footnotes
B. Gaugler's present address is Laboratoire d'Immunologie des Tumeurs, Institut Paoli-Calmettes, 232 Bd. de Ste. Marguerite, F-13009 Marseille, France.
Abbreviations used in this paper: RACE, rapid amplification of cDNA ends; RCC, renal cell carcinoma; RT, reverse transcription.
References
- Van den Eynde B., van der Bruggen P. T cell-defined tumor antigens. Curr. Opin. Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- Gaugler B., Brouwenstijn N., Vantomme V., Szikora J.-P., Van der Spek C.W., Patard J.-J., Boon T., Schrier P., Van den Eynde B.J. A new gene coding for an antigen recognized by autologous cytolytic T lymphocytes on a human renal carcinoma. Immunogenetics. 1996;44:323–330. doi: 10.1007/BF02602776. [DOI] [PubMed] [Google Scholar]
- Brändle D., Brasseur F., Weynants P., Boon T., Van den Eynde B. A mutated HLA-A2 molecule recognized by autologous cytotoxic T lymphocytes on a human renal cell carcinoma. J. Exp. Med. 1996;183:2501–2508. doi: 10.1084/jem.183.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin C., Kremer F., Angevin E., Scott V., Triebel F. A hsp70-2 mutation recognized by CTL on a human renal cell carcinoma. J. Immunol. 1999;162:1730–1738. [PubMed] [Google Scholar]
- Bernhard H., Karbach J., Wölfel T., Busch P., Störkel S., Stöckle M., Wölfel C., Seliger B., Huber C., Meyer zum Büschenfelde K.-H., Knuth A. Cellular immune response to human renal-cell carcinomasdefinition of a common antigen recognized by HLA-A2-restricted cytotoxic T-lymphocyte (CTL) clones. Int. J. Cancer. 1994;59:837–842. doi: 10.1002/ijc.2910590621. [DOI] [PubMed] [Google Scholar]
- Brouwenstijn N., Hoogstraten C., Verdegaal E.M.E., Van der Spek C.W., Deckers J.G., Mulder A., Osanto S., Schrier P.I. Definition of unique and shared T-cell defined tumor antigens in human renal cell carcinoma. J. Immunother. 1998;21:427–434. doi: 10.1097/00002371-199811000-00004. [DOI] [PubMed] [Google Scholar]
- Gazdar A.F., Oie H.K. ReGrowth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:6011–6012. [PubMed] [Google Scholar]
- Detrisac C.J., Sens M.A., Garvin J., Spicer S.S., Sens D.A. Tissue culture of human kidney epithelial cells of proximal tubule origin. Kidney Int. 1984;25:383–390. doi: 10.1038/ki.1984.28. [DOI] [PubMed] [Google Scholar]
- Deen P.M.T., van Os C.H. Epithelial aquaporins. Curr. Opin. Cell Biol. 1998;10:435–442. doi: 10.1016/s0955-0674(98)80055-0. [DOI] [PubMed] [Google Scholar]
- Guéguen M., Patard J.-J., Gaugler B., Brasseur F., Renauld J.-C., Van Cangh P.J., Boon T., Van den Eynde B. An antigen recognized by autologous CTL on a human bladder carcinoma. J. Immunol. 1998;160:6188–6194. [PubMed] [Google Scholar]
- De Plaen E., Lurquin C., Brichard V., van der Bruggen P., Renauld J.-C., Coulie P., Szikora J.-P., Wölfel T., Van Pel A., Boon T. Cloning of genes coding for antigens recognized by cytolytic T lymphocytes. In: Lefkovits I., editor. The Immunology Methods Manual. Academic Press Ltd; London: 1997. pp. 692–718. [DOI] [PubMed] [Google Scholar]
- De Plaen E., Lurquin C., Lethé B., van der Bruggen P., Brichard V., Renauld J.-C., Coulie P., Van Pel A., Boon T. Identification of genes coding for tumor antigens recognized by cytolytic T lymphocytes. Methods. 1997;12:125–142. doi: 10.1006/meth.1997.0462. [DOI] [PubMed] [Google Scholar]
- Brouwenstijn N., Gaugler B., Krüse K.M., Van der Spek C.W., Mulder A., Osanto S., Van den Eynde B.J., Schrier P.I. Renal cell carcinoma-specific lysis by cytotoxic T lymphocyte clones isolated from peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Int. J. Cancer. 1996;68:177–182. doi: 10.1002/(SICI)1097-0215(19961009)68:2<177::AID-IJC6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Rammensee H.-G., Bachmann J., Stevanovic S. MHC Ligands and Peptide Motifs. Molecular Biology Intelligence Unit 1997. Springer-Verlag; Heidelberg: pp. 462 [Google Scholar]
- Lethé B., van der Bruggen P., Brasseur F., Boon T. MAGE-1 expression threshold for the lysis of melanoma cell lines by a specific CTL Melanoma Res. 7Suppl. 21997. S83 S88 [PubMed] [Google Scholar]
- Simons R.W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu. Rev. Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Vanhée-Brossollet C., Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- Mayrand S.-M., Green W.R. Non-traditionally derived CTL epitopesexceptions that prove the rules? Immunol. Today. 1998;19:551–556. doi: 10.1016/s0167-5699(98)01342-5. [DOI] [PubMed] [Google Scholar]
- Guilloux Y., Lucas S., Brichard V.G., Van Pel A., Viret C., De Plaen E., Brasseur F., Lethé B., Jotereau F., Boon T. A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J. Exp. Med. 1996;183:1173–1183. doi: 10.1084/jem.183.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulie P.G., Lehmann F., Lethé B., Herman J., Lurquin C., Andrawiss M., Boon T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. USA. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupetti R., Pisarra P., Verrecchia A., Farina C., Nicolini G., Anichini A., Bordignon C., Sensi M., Parmiani G., Traversari C. Translation of a retained intron in tyrosinase-related protein (TRP)-2 mRNA generates a new cytotoxic T lymphocyte (CTL)-defined and shared human melanoma antigen not expressed in normal cells of the melanocytic lineage. J. Exp. Med. 1998;188:1005–1016. doi: 10.1084/jem.188.6.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P.F., El-Gamil M., Li Y.F., Fitzgerald E.B., Kawakami Y., Rosenberg S.A. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J. Immunol. 1997;159:303–308. [PubMed] [Google Scholar]
- Wang R.-F., Johnston S.L., Zeng G., Topalian S.L., Schwartzentruber D.J., Rosenberg S.A. A breast and melanoma-shared tumor antigenT cell responses to antigenic peptides translated from different open reading frames. J. Immunol. 1998;161:3596–3606. [PubMed] [Google Scholar]
- Wang R.-F., Parkhurst M.R., Kawakami Y., Robbins P.F., Rosenberg S.A. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J. Exp. Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarnoudse C.A., van den Doel P.B., Heemskerk B., Schrier P.I. Interleukin-2-induced, melanoma-specific T cells recognize camel, an unexpected translation product of LAGE-1. Int. J. Cancer. 1999;82:442–448. doi: 10.1002/(sici)1097-0215(19990730)82:3<442::aid-ijc19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Skipper J.C.A., Hendrickson R.C., Gulden P.H., Brichard V., Van Pel A., Chen Y., Shabanowitz J., Wölfel T., Slingluff C.L., Jr., Boon T. An HLA-A2–restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]