Abstract
Interleukin (IL)-4 is an immunoregulatory cytokine that exerts distinct biological activities on different cell types. Our studies indicate that interferon regulatory factor (IRF)-4 is both a target and a modulator of the IL-4 signaling cascade. IRF-4 expression is strongly upregulated upon costimulation of B cells with CD40 and IL-4. Furthermore, we find that IRF-4 can interact with signal transducer and activator of transcription (Stat)6 and drive the expression of IL-4–inducible genes. The transactivating ability of IRF-4 is blocked by the repressor factor BCL-6. Since expression of IRF-4 is mostly confined to lymphoid cells, these data provide a potential mechanism by which IL-4–inducible genes can be regulated in a lineage-specific manner.
Keywords: CD40, interleukin 4, signal transducer and activator of transcription 6, BCL-6, interferon regulatory factor 4
Interleukin (IL)-4 is an immunoregulatory cytokine that exerts a wide variety of effects on many different cell types 1 2 3. IL-4 plays a central role in the regulation of immune responses by promoting the differentiation of T helper cells toward the Th2 subset. Furthermore, IL-4 synergizes with stimuli provided by CD40 to drive B cell activation, proliferation, and differentiation, thus strongly enhancing humoral immunity 4.
IL-4 mediates its biological actions by activating a signaling cascade, which follows the paradigm originally described for the IFN signaling pathway 5 6 7. Binding of IL-4 to its receptor leads to the activation, via Janus kinases (JAKs), of a latent cytoplasmic protein, signal transducer and activator of transcription (STAT) 6 (Stat6) 8. Tyrosine phosphorylation of Stat6 allows it to homodimerize and rapidly translocate into the nucleus where it modulates gene transcription by binding to distinct cis-elements termed gamma-activated sites (GAS). Although studies in Stat6-deficient mice have demonstrated that Stat6 plays a key role in IL-4 signaling 9 10 11, additional factors are likely to participate in this cascade. Indeed, the rapidity of Stat6 activation in response to IL-4 cannot solely explain the inducibility of IL-4 target genes like CD23, whose activation displays a delayed kinetics 12. Moreover, tyrosine phosphorylation of Stat6 in response to IL-4 has been detected in a wide variety of cells 13 14 15 16 17. Thus, the differential gene expression triggered by IL-4 in diverse cell types must involve the presence and/or recruitment of additional, possibly cell type–specific, cofactors.
Consistent with the requirements for additional protein–protein interactions in the regulation of IL-4–responsive genes, the activity of Stat6 can be downregulated by B cell lymphomas 6 (BCL-6). BCL-6 is a Krüppel zinc finger transcriptional repressor that is highly expressed in germinal center B cells and is frequently altered in non-Hodgkin lymphoma 18 19 20 21. BCL-6 has been shown to bind to the GAS element in the CD23b promoter and repress Stat6-mediated promoter function 22. Mice deficient in BCL-6 display lack of germinal centers as well as an inflammatory disease characterized by exaggerated Th2 responses including elevated levels of serum IgG1 and IgE, supporting a role for BCL-6 as a repressor of the IL-4 signaling pathway in vivo 22 23. However, generation of mice deficient in both BCL-6 and Stat6 failed to abrogate the Th2 inflammatory disease, suggesting that not all of the defects present in the BCL-6–deficient mice can be simply ascribed to its repression of Stat6 function 24.
Studies of the IFN signaling pathway have demonstrated that another family of transcription factors, the IFN regulatory factor (IRF) family, is both a target and a modulator of this cytokine signaling cascade 25 26. Genetic studies have demonstrated a role for IRF proteins in the proliferation, differentiation, and apoptosis of a variety of immune effector cells, suggesting the involvement of IRFs in pathways in addition to those triggered by the IFNs 27 28. In particular, lack of one of these newly cloned IRF proteins, IRF-4 (also termed Pip, LSIRF, ICSAT, or MUM1 [29–32]), results in striking defects in the function of mature B and T cells 33. Consistent with these results, expression of both murine and human IRF-4 is mostly confined to the lymphoid compartment and is induced in response to mitogenic stimuli 31 32 34. Functional studies have shown that IRF-4 is involved in the transactivation of both the κ and λ Ig light chain enhancers as well as the CD20 promoter 29 35 36 37. However, the effect of IRF-4 on these enhancers is unlikely to explain the profound immunological impairments demonstrated by the gene targeting studies. Additional IRF-4 targets and the physiological stimuli that activate IRF-4 are unknown.
We are interested in understanding the downstream effectors of the IL-4 signaling pathways in B cells as well as the mechanisms underlying the cross-talk between the CD40 and IL-4 signaling pathways. We focused our attention on the regulation of the CD23 gene 38 39 40 41. CD23 induction in response to IL-4 and CD40 occurs with delayed kinetics, and displays a synergistic response when the two stimuli are combined 12 42 43. In humans, CD23 exists as two alternatively spliced isoforms, termed CD23a and CD23b, which differ only in the intracytoplasmic domain. Expression of CD23a and CD23b is controlled by two separate promoters 44. IL-4 inducibility of the CD23b isoform has been functionally mapped to a Stat6 binding element within the CD23b promoter termed the CD23b GAS 13 45. We have found that the recently cloned IRF-4 can physically interact with Stat6 and act as a transactivator of the CD23b GAS. Presence of BCL-6 blocks the transactivating potential of IRF-4. While BCL-6 is known to be downregulated by CD40 46, IRF-4 expression is markedly induced in response to IL-4 and CD40. Since IRF-4 expression is largely restricted to lymphoid cells, these data provide a potential mechanism by which expression of IL-4–inducible genes can be modulated in a cell type–specific manner. Furthermore, the ability of CD40 and IL-4 to target the expression of multiple components of this nucleoprotein complex may underlie the synergistic interaction between these two pathways.
Materials and Methods
Cell Lines and Cultures.
The human B cell lines Ramos (obtained from Dr. Seth Lederman, Columbia University) and BL-41 (obtained from Dr. Riccardo Dalla-Favera, Columbia University) are EBV-negative Burkitt's lymphomas. JY (obtained from Dr. Riccardo Dalla-Favera) is an EBV-transformed lymphoblastoid B cell line. U937 (obtained from Dr. Kathryne Calame, Columbia University) is a monocytic cell line. All cells were grown in IMDM supplemented with 10% fetal calf serum (Atlanta Biologicals, Inc.).
Ramos B cells (10–20 × 106) were stimulated with 0.1 μg/ml of either anti-CD40 or an isotype-matched control Ab in a final volume of 10 ml at 37°C for 24 h. IL-4 treatment (100 U/ml; PeproTech) was carried out simultaneously in separate plates in a final volume of 10 ml. Tonsillar mononuclear cells were obtained from surgical specimens after routine tonsillectomies and were isolated as described previously 47.
Abs.
The rabbit polyclonal antiserum against IRF-4 used in electrophoretic mobility shift assay (EMSA) experiments was a gift of Dr. Hirai, Tokoyo Universty, Tokyo, Japan 32. We subsequently generated our own rabbit polyclonal anti–IRF-4 antiserum using a similar glutathione S-transferase (GST)–IRF-4 (nucleotides 441–924) fusion protein as the immunogen (Babco). As previously indicated, this GST fusion protein contains a portion of the IRF-4 protein that is specific to IRF-4 and thus avoids cross-reactivity with other IRFs 32. This antiserum was used for immunoprecipitations and immunoblot analyses. Blots were also reprobed with a commercially available anti–IRF-4 antiserum (Santa Cruz Biotechnology), which gave identical results. Rabbit polyclonal antisera against human Stat6, Stat3, p65, IRF-2, IFN consensus sequence binding protein (ICSBP), or BCL-6, were purchased from Santa Cruz Biotechnology, Inc. The hybridomas secreting the anti-CD40 mAb G28-5 (IgG1) or an isotype-matched control mAb were obtained from American Type Culture Collection.
DNA Constructs.
Full-length human IRF-4 cDNA cloned into pBluescript vector (pBSKS-myc-IRF-4) was a gift of Dr. Riccardo Dalla-Favera 30. The IRF-4 expression plasmid (pCEP4-IRF-4) was constructed by cloning the entire coding region of the IRF-4 cDNA into the NotI and XhoI sites of the mammalian expression vector pCEP4 (Invitrogen). For construction of the GST–IRF-4 expression plasmid, the entire coding region of the IRF-4 cDNA was cloned, in frame, into the filled EcoRI site of pGEX-3X vector (Amersham Pharmacia Biotech). The in frame junctions in the GST–IRF-4 fusion construct were confirmed by DNA sequencing in an automated cycle sequencer (Perkin Elmer). The full-length human BCL-6 cDNA cloned into pMT2T mammalian expression vector was a gift of Dr. Riccardo Dalla-Favera 19.
The CD23a cDNA was a gift of Dr. Kikutani, Osaka University, Osaka, Japan 44 and Dr. Wang, Harvard University, Boston, MA 48. To generate an antisense riboprobe of CD23a/b, a 600-bp 5′-EcoRI-HindIII-3′ fragment from the 5′ end of CD23a cDNA was cloned in opposite orientation, into the HindIII and EcoRI sites of pGEM1 in vitro transcription vector (Promega). Transcription of the antisense RNA was driven by SP6 RNA polymerase promoter.
To prepare the CD23b GAS firefly luciferase reporter construct, a trimer of the CD23b GAS element was synthesized with flanking BamHI sites (GIBCO BRL) and then cloned into the BamHI site (immediately upstream of minimal TK promoter) of the TK200 luciferase reporter vector (a gift of Dr. Kathryne Calame). The CD23b promoter firefly luciferase reporter construct in the pGL3-enhancer vector (Promega) was a gift of Dr. Seth Lederman. The pRL-TK reporter plasmid encoding renilla luciferase to determine the transfection efficiency was purchased from Promega.
DNA Binding Assays and Cell Extracts.
The preparation and employment of DNA oligonucleotide probes for EMSAs have been described previously 49 with the following modifications: 2% glycerol was added to the polyacrylamide gel, and a reduced poly(dI-dC) concentration (1 μg/reaction) was used in the binding buffer. The CD23b GAS oligonucleotides used in these studies were as follows: CD23b GAS wild-type (wt), 5′-gatcGGGTGAATTTCTAAGAAAGGGAC-3′; CD23b GAS M1, 5′-gatcGGGTGAATTTCTAAGGTCGGGAC-3′; CD23b GAS M2, 5′-gatcGGGTGGTCTTCTAAGAAAGGGAC-3′; and CD23b GAS M3, 5′-gatcGGGTGAATGCTGAAGAAAGGGAC-3′. Oligonucleotide competition and Ab interference assays were performed as described previously 49. Nuclear and whole cell extracts (WCEs) were prepared as described previously 49 50.
Immunoprecipitations, Western Blot Analysis, and UV Cross-linking Assays.
Cell extracts were immunoprecipitated with an anti–IRF-4, anti–BCL-6, or anti-Stat6 antiserum as described previously 49. The immunoprecipitates were resolved by 7% SDS-PAGE. The gel was transferred to a nitrocellulose membrane, and then immunoblotted with a Stat6, BCL-6, or IRF-4 Ab. The bands were visualized by ECL (Amersham Pharmacia Biotech). UV cross-linking was performed as described previously 49.
GST Pull-down Assays.
GST fusion proteins were expressed in Escherichia coli DH5α and affinity-purified on glutathione (GSH)-agarose beads (Sigma Chemical Co.), as described previously 51. The concentration of each fusion protein was determined by SDS-PAGE/Coomassie staining. For pull-down assays, lysates (∼2 mg of total cell proteins) from control or stimulated Ramos or JY cells were incubated with ∼100 μg of GST alone or GST–IRF-4 fusion protein immobilized onto GSH-agarose beads in 400 μl of 1× lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1 mM EDTA, 0.05% NP-40, 10% glycerol, 1 mM dithiothreitol, 1 mM Na3VO4, 1 mM PMSF, 1 μg/ml leupeptin, 3 μg/ml aprotinin) for 4 h at 4°C with constant agitation. The complexes were extensively washed with 1× lysis buffer (containing 0.5% NP-40). The bound proteins were eluted from the beads by boiling them in SDS-PAGE sample buffer, fractionated on a 7% SDS-polyacrylamide gel, and then blotted onto a nitrocellulose membrane. The blot was probed with either a Stat6, Stat3, or BCL-6 Ab.
Northern Analysis and RNase Protection Assays.
Total RNA was extracted by using the RNA-STAT60 kit (TelTest, Inc.). Northern blot analysis was performed with 10 μg of total RNA according to standard protocols. The blot was probed with either a human IRF-4 cDNA, a BCL-6 cDNA, or a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA radiolabeled by the DNA Labeling beads (−dCTP) kit (Amersham Pharmacia Biotech). RNase protection analysis was performed as described previously 52. 10 μg of total RNA was hybridized simultaneously to an antisense riboprobe of CD23a/b as well as of β-actin (Ambion) transcribed by SP6 RNA Polymerase in vitro Transcription System (Promega) using [α-32P]UTP. The annealed products were digested with ribonuclease T2 (GIBCO BRL), and then analyzed on a 6% polyacrylamide-urea denaturing gel.
Transient Transfections.
For the transient transfection assays, 6 × 106 U937 cells were cotransfected with 25 μg of a CD23b GAS wt or a CD23b GAS M2 firefly luciferase reporter plasmid and 25 μg of various expression plasmids (as indicated in the figure legend) by electroporation at 300 V and 960 μF with a BTX electroporator as described previously 53. 5 μg of the pRL-TK reporter plasmid expressing renilla luciferase under the control of the thymidine kinase promoter was added to each transfection as a transfection efficiency control. JY cells (10 × 106) were cotransfected by the DEAE-dextran method 54 with 10 μg of the CD23b promoter firefly luciferase reporter vector and 5 μg of pRL-TK plasmid in the presence of 15 μg of a BCL-6 expression plasmid (pMT2T-BCL6) or the equivalent amount of empty pMT2T vector. After transfection, the cells were equally split into two 2-ml cultures and then incubated for 16–24 h in the presence or absence of IL-4 (10 ng/ml). The transfected cells were then harvested, lysed, and assayed for luciferase activities with the Dual Luciferase Assay System (Promega) according to the manufacturer's instructions. The firefly luciferase activity was normalized on the basis of renilla luciferase activity.
Results
CD40 and IL-4 Synergistically Upregulate both CD23a and CD23b.
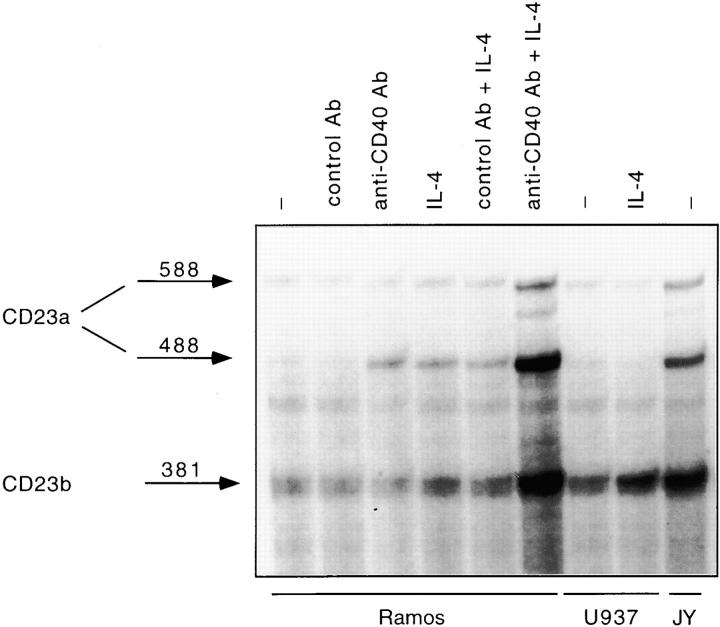
To identify and characterize the downstream effectors of the IL-4 signaling cascade, we focused our attention on CD23, a B cell activation molecule whose regulation displays lineage- and stage-specific features 41 55. Although IL-4 and CD40 can individually induce modest levels of CD23, costimulation of human tonsillar B cells or purified centroblasts with both CD40 and IL-4 leads to a synergistic induction of surface CD23 expression 42 43. We found that Ramos, an EBV-negative Burkitt's lymphoma cell line extensively used to study the CD40 and/or IL-4 signaling pathways 56 57 58 59, mimics the physiological regulation of CD23 in normal B cells (data not shown). Since no study had previously explored the CD23 isoform(s) induced in response to CD40 or the CD40 and IL-4 combination, we performed RNase protection assays. As shown in Fig. 1, exposure of Ramos cells to either anti-CD40 Ab or IL-4 alone preferentially upregulated the CD23a isoform. A slight effect of IL-4 on the expression of CD23b could also be detected. Coculturing Ramos cells with both stimuli led to a strong induction of both CD23a and CD23b transcripts. Consistent with previous reports, constitutively high levels of both CD23a and CD23b were detected in an EBV-transformed B cell line (JY), whereas IL-4 stimulation of a monocytic cell line, U937, only upregulated the CD23b isoform 44 48. β-actin levels were equivalent in all lanes (data not shown). Thus, costimulation with CD40 and IL-4 leads to a synergistic induction of both CD23 isoforms.
Figure 1.
The synergistic upregulation of CD23 in response to CD40 and IL-4 reflects an induction of both CD23a and CD23b isoforms. Ramos cells were cultured for 24 h either in the absence of any stimulus or in the presence of an anti-CD40 Ab (0.1 μg/ml), an isotype-matched control Ab (0.1 μg/ml), human IL-4 (100 U/ml), a combination of anti-CD40 Ab plus IL-4, or a control Ab plus IL-4. U937 cells were either unstimulated or stimulated with IL-4. JY cells were left unstimulated. After harvesting the cells, total RNA was extracted and subjected to RNase protection analysis. The antisense riboprobe corresponds to a 600-bp 5′ EcoRI-HindIII fragment of the CD23a cDNA. The protected fragments of 588 and 488 nucleotides correspond to two isoforms of CD23a transcript which differ by one base change at position 96 in the 5′ untranslated region, while the 381-nucleotide protected fragment corresponds to the CD23b transcript (reference 44).
IRF-4 Binds to the CD23b GAS.
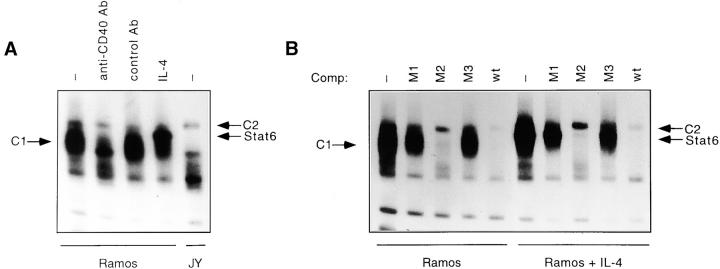
IL-4 stimulation has been shown to lead to the rapid binding of Stat6 to a functional element within the CD23b promoter termed the CD23b GAS 13 45. However, our RNase protection assay studies had revealed only a minimal induction of CD23b in response to IL-4 alone. To determine whether additional factors could target the CD23b GAS and modulate Stat6 activity, we first performed EMSA experiments on Ramos cells stimulated with either IL-4, anti-CD40, or a control Ab. As shown in Fig. 2 A, Ramos cells contain multiple CD23b GAS binding complexes, most noticeably a broad constitutive (C1) and a slower mobility constitutive complex (C2). CD40 stimulation of Ramos cells led to a decrease in the intensity of the C1 complex (Fig. 2 A). Stimulation of Ramos cells with IL-4 activates an additional superimposed complex that contains Stat6 (Fig. 2 C). In contrast to Ramos cells, the appearance of the CD23b GAS binding complexes in JY, an EBV-transformed B cell line expressing constitutively high levels of CD23b, was strikingly different. The C1 complex was absent, while the C2 complex became clearly visible (Fig. 2 A).
Figure 2.
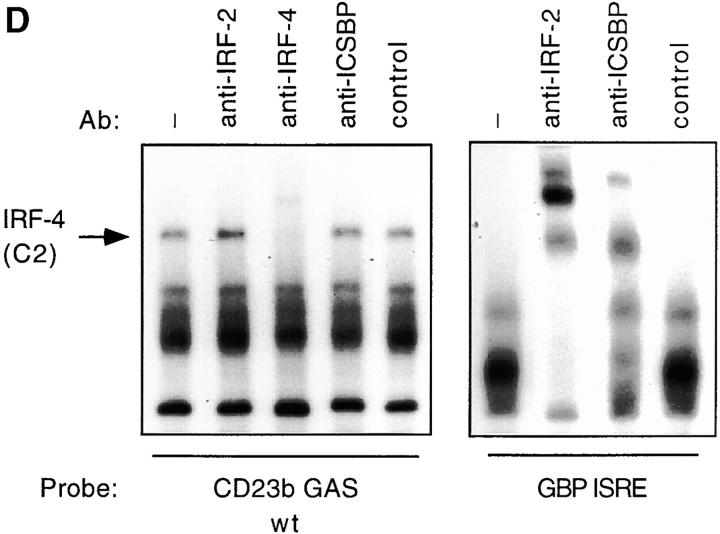
The CD23b GAS is targeted by Stat6, BCL-6, and IRF-4. (A) Ramos cells were either unstimulated or stimulated with anti-CD40 Ab (0.1 μg/ml), an isotype-matched control Ab (0.1 μg/ml), or human IL-4 (100 U/ml) for 24 h. JY cells were left unstimulated. Nuclear extracts were then prepared and analyzed by EMSA using a 32P-labeled CD23b GAS wt probe. (B) Ramos cells were cultured and assayed as described in A. Oligonucleotide competition assays were performed either in the absence or presence of a 100-fold molar excess of cold GAS oligonucleotides (see Table ) added to the shift reaction as indicated. (C) Ramos and JY cells were cultured as described in A. Nuclear extracts were then prepared and analyzed by EMSA using either a 32P-labeled CD23b GAS wt probe or a 32P-labeled CD23b GAS M1 probe (Table ) as indicated. Ab interference mobility shift assays were carried out by addition of antisera against Stat6, BCL-6, IRF-4, or control. All antisera were added at a final dilution of 1:20 for 30 min at 4°C before incubation with the probe for 20 min at 25°C. (D) JY cells were unstimulated. Nuclear extracts were then prepared and analyzed by EMSA using a 32P-labeled CD23b GAS wt probe (left). Ab interference mobility shift assays were carried out by addition of antisera against IRF-2, IRF-4, ICSBP, or control as indicated. All antisera were added as outlined above. As a control for the IRF-2 and ICSBP antisera, Ab interference analysis using a 32P-labeled GBP-ISRE probe (reference 32) was simultaneously performed on Ramos extracts (right).
To dissect the CD23b GAS binding complexes, we then carried out EMSA experiments using a panel of mutated CD23b GAS elements as cold competitors of the radiolabeled CD23b GAS wt probe. Interestingly, a survey of the CD23b GAS had revealed that it contains two potential core sequences for IRF binding (GAAT and GAAA; Table ) 60. We thus mutated each of these two sites (M2 and M1, respectively) as well as a region between them (M3). These oligonucleotide competition experiments revealed that the C1 and C2 complexes displayed a differential pattern of competition (Fig. 2 B). In particular, the CD23b GAS M2 mutant failed to compete the C2 complex, suggesting that this complex targets the potential 5′ IRF recognition sequence.
Table 1.
Sequence Comparison between the Wt and Mutant CD23b GAS Oligonucleotides
| CD23b GAS wt | 5′-GGGTGAATTTCTAAGAAAGGGAC-3′ |
|---|---|
| CD23b GAS M1 | 5′-GGGTGAATTTCTAAGGTCGGGAC-3′ |
| CD23b GAS M2 | 5′-GGGTGGTCTTCTAAGAAAGGGAC-3′ |
| CD23b GAS M3 | 5′-GGGTGAATGCTGAAGAAAGGGAC-3′ |
| Stat6 consensus | TTNNNNNNAA |
| IRF response element core sequence | GAAA |
| BCL-6 binding site | GAAAATTCCTAGAAAGCATA |
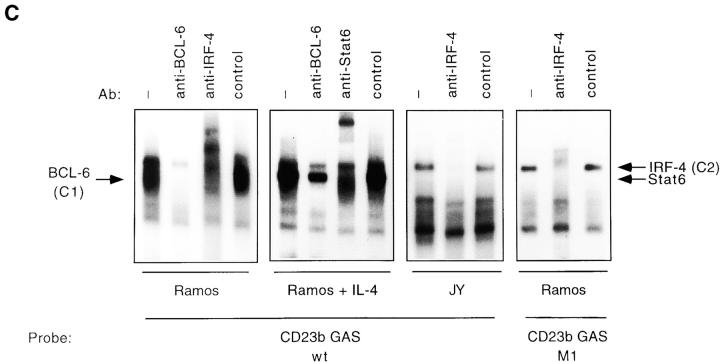
Previous studies have shown that purified BCL-6 can bind the CD23b GAS 22. Furthermore, expression of BCL-6 is downregulated by CD40 stimulation of B cells, and undetectable in EBV-transformed B cell lines 46 61. These observations suggested that the C1 complex, which we had detected in Ramos but not in JY cells, might contain BCL-6. Indeed, incubation of nuclear extracts from Ramos cells with a BCL-6 antiserum, but not with a control antiserum, led to the disappearance of the C1 complex (Fig. 2 C). These experiments also clearly revealed the presence of the C2 complex in both untreated and IL-4–treated cells. In IL-4–stimulated cells, an additional Stat6 complex could also be observed.
Our cold competition experiments suggested that binding of the C2 complex to the CD23b GAS requires the presence of a potential IRF binding site. To directly assess whether the lymphoid-specific IRF-4 participated in the CD23b GAS binding complexes, we then performed Ab interference EMSA assays with an anti–IRF-4 antiserum 32. Incubation of extracts from untreated Ramos cells with anti–IRF-4 antiserum, but not with a control antiserum, supershifted the C2 complex (Fig. 2 C). Addition of the IRF-4 antiserum completely blocked the appearance of the C2 complex in JY extracts. Since detection of the C2 complex in Ramos cells is hindered by the presence of the BCL-6 complex, we also performed Ab interference assays using as a probe the CD23b GAS M1 mutant, which has lost the ability to bind BCL-6. As shown in Fig. 2 C, this probe clearly detected the C2 complex in Ramos cells, and addition of the anti–IRF-4, but not a control, antiserum led to its disappearance. Additional Ab interference assays demonstrated that antisera against another IRF family member, IRF-2 or ICSBP, failed to affect the appearance of the CD23b GAS binding complexes in either JY or Ramos despite appropriately supershifting complexes binding to the guanylate binding protein (GBP)–IFN-stimulated regulatory element (ISRE) (Fig. 2 D, and data not shown). Furthermore, no effect of the anti–IRF-4 antiserum on the IL-4–inducible Stat6 complex or on the IFN-γ–inducible Stat1 complex binding to the IRF-1 GAS was noted, indicating that this effect was specific for the complex binding to the CD23b GAS (data not shown). We have also detected IRF-4 binding to a cis-element adjacent to the Stat6 binding site in the CD23a promoter (data not shown).
Thus, these data indicate that IRF-4, BCL-6, and Stat6 can all target the CD23b GAS element. Interestingly, these experiments also suggest that binding of IRF-4 and Stat6 to the CD23b GAS is not cooperative and can occur independently of each other.
IRF-4 Physically Interacts with Stat6.
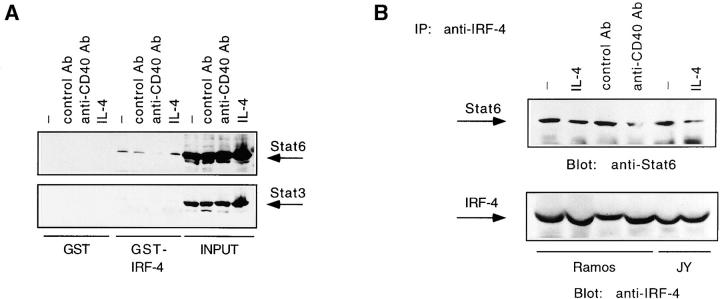
Interactions between an IRF and STATs are critical for the formation of the IFN-α–inducible complex, IFN-stimulated gene factor 3 (ISGF3), which contains Stat1 and Stat2 as well as p48, a member of the IRF family 62. Since Stat6 and IRF-4 can bind to adjacent DNA elements, we then proceeded to test whether IRF-4 can physically interact with Stat6 by performing GST fusion protein binding assays (Fig. 3 A, top). An association of Stat6 with IRF-4 could indeed be detected by incubating a GST–IRF-4 fusion protein with extracts from Ramos cells stimulated with anti-CD40, a control Ab, or human IL-4. No interaction of Stat6 with the GST moiety alone was observed. Furthermore, reprobing with anti-Stat3, anti-Stat1, anti-Stat2, and anti-Stat5 antisera revealed that none of these STATs were able to complex with IRF-4 (Fig. 3 A, bottom, and data not shown). To confirm that the IRF-4–Stat6 interaction could occur in vivo, we used an anti–IRF-4 antiserum to immunoprecipitate extracts from Ramos cells stimulated with either anti-CD40 Ab, a control Ab, or IL-4. Consistent with our previous observations, presence of the Stat6 protein could be detected in the IRF-4 immunoprecipitates from both stimulated and unstimulated Ramos lysates (Fig. 3 B, top). Similar results were also observed in JY cells. Stripping and reprobing of the filter with an anti–IRF-4 Ab ensured for equal loading of the immunoprecipitates (Fig. 3 B, bottom). Consistent with recent studies, which have detected IRF-4 in both the nuclear and cytoplasmic compartments (Riccardo Dalla-Favera, personal communication), the IRF-4–Stat6 association can occur in the absence of IL-4 stimulation. Interestingly, in both GST and coimmunoprecipitation experiments the IRF-4–Stat6 interaction appears to decrease upon stimulation of Ramos cells with the CD40 Ab.
Figure 3.
IRF-4 interacts with Stat6. (A) Ramos cells were stimulated as described in the legend to Fig. 2. WCEs were then prepared and incubated with immobilized GST–IRF-4 fusion protein. Bound proteins were eluted, resolved by 7% SDS-PAGE, blotted onto a nitrocellulose membrane, and then probed with a Stat6 (top) or Stat3 Ab (bottom). Binding to immobilized GST alone is shown as a control. (B) Ramos cells were stimulated as described in the legend to Fig. 2. JY cells were either unstimulated or stimulated with IL-4 (100 U/ml). WCEs were then prepared and immunoprecipitated with an IRF-4 antiserum. The immunoprecipitated proteins were resolved by 7% SDS-PAGE and then analyzed by Western blotting using a Stat6 Ab (top). The blot was later stripped and reprobed with an anti–IRF-4 antiserum (bottom) using protein A–horseradish peroxidase conjugate as a secondary detection reagent to probe the primary Ab binding.
Thus, our data indicate that IRF-4 is capable of specifically interacting with Stat6 both in vitro and in vivo.
Expression of IRF-4 Is Stimulated by CD40 and IL-4.
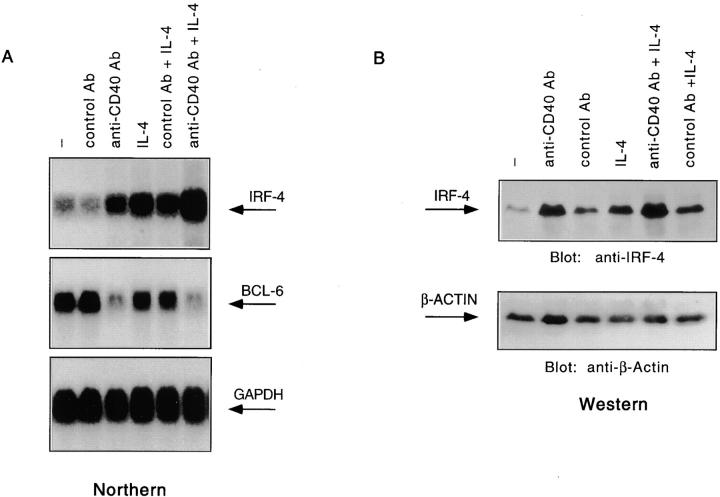
The preceding experiments suggested that IRF-4 could bind a functional element of the CD23b promoter. Since CD40 and IL-4 synergistically induce CD23b, we proceeded to determine whether CD23b upregulation by these stimuli was accompanied by changes in IRF-4 expression. Therefore, we cultured Ramos cells with an anti-CD40 Ab, IL-4, or a combination of anti-CD40 Ab and IL-4. Simultaneous cultures with a control Ab were also included. Northern analysis of total RNA derived from this experiment revealed that IRF-4 expression was upregulated in response to either CD40 or IL-4 stimulation (Fig. 4 A, top). A much stronger induction was noted when the anti-CD40 Ab was used in conjunction with IL-4. Consistent with previous reports 46 61, CD40 stimulation of Ramos cells also led to the downregulation of BCL-6 (Fig. 4 A, middle). The same pattern of IRF-4 upregulation in response to CD40 and/or IL-4 was also detected in another EBV-negative Burkitt's lymphoma cell line, BL-41 (data not shown), and by Western analysis, in human tonsillar cells (Fig. 4 B). Strong induction of IRF-4 was also observed in response to transfectants expressing CD40 ligand (CD40L) but not to control transfectants, indicating that IRF-4 expression is a physiological target of the CD40–CD40L interaction (59; data not shown). Kinetic studies showed that induction of IRF-4 is first noted at 2 h and can still be detected at 24 h (data not shown). No effect on IRF-4 expression was noted by culturing Ramos cells with either IFN-γ or IFN-α (data not shown). Reprobing of this Northern blot with a probe for ICSBP, an IRF family member closely related to IRF-4 63, revealed that the CD40 and IL-4 signaling cascades do not upregulate ICSBP expression (data not shown). Consistent with previous reports 34, we also found that IRF-4 levels are highly increased in EBV-transformed B cell lines (data not shown). Thus, expression of IRF-4 in B cells is specifically targeted by the CD40 and IL-4 signaling cascades as well as by EBV transformation.
Figure 4.
IRF-4 expression is upregulated in response to CD40 and/or IL-4. (A) Ramos cells were stimulated as described in the legend to Fig. 1. Total RNA was then extracted, and 10 μg of the RNA was assayed by Northern blotting as per standard protocols. The blot was then probed with either an IRF-4 cDNA (top), a BCL-6 cDNA (middle), or a GAPDH cDNA (bottom) radiolabeled by random hexamer priming. (B) Tonsillar mononuclear cells were stimulated as described in the legend to Fig. 1. WCEs were prepared, electrophoresed on a 7% SDS-polyacrylamide gel, and then analyzed by Western blotting using an anti–IRF-4 Ab (top). The blot was later stripped and reprobed with a β-actin Ab to ensure for equal loading.
IRF-4 Interacts with the Krüppel Zinc Finger Transcriptional Repressor, BCL-6.
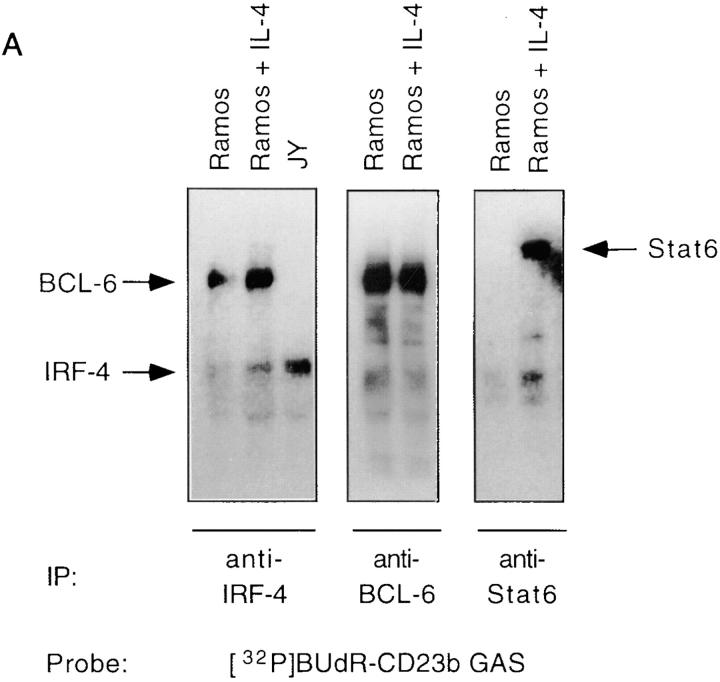
The induction of IRF-4 expression by CD40 and IL-4 as well as by EBV suggested that IRF-4 might function as a transactivator of CD23 gene expression. However, we had detected constitutive binding of IRF-4 to the CD23b GAS in unstimulated Ramos cells, which only express low levels of CD23b. We then reasoned that, in this setting, IRF-4 function might be modulated by interaction with a repressor. Consistent with this hypothesis, our EMSA experiments had revealed binding of BCL-6 to the CD23b GAS in Ramos, but not in JY cells, which express high constitutive levels of CD23b. To assess whether IRF-4 could interact with BCL-6, we then performed UV cross-linking experiments with a bromodeoxyuridine-substituted CD23b GAS probe. This was followed by immunoprecipitations with an IRF-4 antiserum, a BCL-6 antiserum, or a Stat6 antiserum as control (Fig. 5 A). The immunoprecipitates were then resolved by SDS-PAGE and Western blotted. Consistent with our EMSA results (Fig. 2), an ∼62-kD protein was recognized by the IRF-4 antiserum. This corresponds to the molecular mass of IRF-4 plus the DNA probe. An additional cross-linked protein was also immunoprecipitated by the IRF-4 antiserum in Ramos, but not in JY cells. The size of this additional protein, after correction for the probe contribution, was ∼90 kD, and was identical to that of the BCL-6 protein cross-linked to the CD23b GAS as determined by simultaneous immunoprecipitation with an anti–BCL-6 antiserum. Indeed, reprobing of the Western blot with an anti–BCL-6 antiserum confirmed that the p90 protein could be recognized by this antiserum (data not shown). Thus, the anti–IRF-4 antiserum can coprecipitate IRF-4 and BCL-6 bound to the CD23b GAS.
Figure 5.
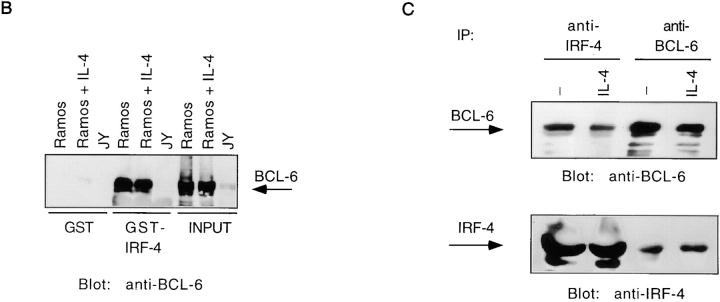
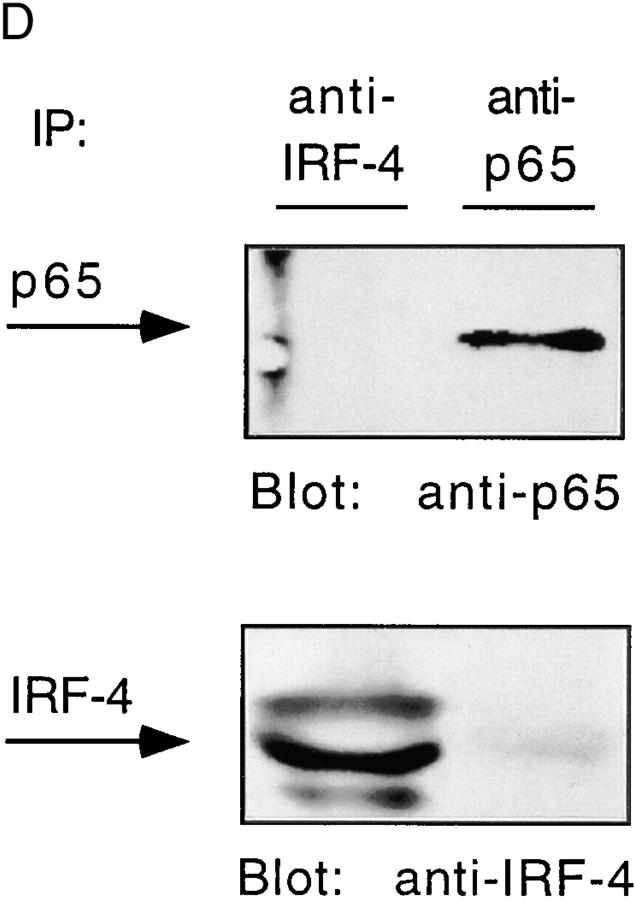
IRF-4 interacts with BCL-6. (A) Ramos cells were either unstimulated or stimulated with IL-4 (100 U/ml). JY cells were unstimulated. Nuclear extracts were then prepared as described in the legend to Fig. 2, and incubated with a radiolabeled 5-bromodeoxyuridine–substituted CD23b GAS wt probe (BUdR-CD23b GAS wt) in a standard shift reaction. DNA–protein complexes were irradiated twice with 1,000 mJ of UV, and then immunoprecipitated (IP) with either an IRF-4, a BCL-6, or a Stat6 antiserum. The immunoprecipitates were resolved by 7% SDS-PAGE, transferred to a nitrocellulose membrane, and then exposed to an x-ray film. No cross-linking was detected in the absence of UV irradiation. (B) Ramos and JY cells were cultured as described in A. Nuclear extracts were then prepared and incubated with immobilized GST–IRF-4 fusion protein. Bound proteins were eluted, fractionated by 7% SDS-PAGE, transferred to a nitrocellulose membrane, and then probed with a BCL-6 antiserum. Binding to immobilized GST alone is shown as a control. (C) Ramos cells were either unstimulated or stimulated with IL-4 (100 U/ml). Nuclear extracts were prepared and immunoprecipitated with either an anti–IRF-4 or an anti–BCL-6 antiserum. The immune complexes were resolved by 7% SDS-PAGE, and then analyzed by Western blotting using an anti–BCL-6 antiserum (top). The blot was later stripped and reprobed with an anti–IRF-4 antiserum (bottom), as described in the legend to Fig. 3. (D) JY cells were unstimulated. Nuclear extracts were prepared and immunoprecipitated with either an anti–IRF-4 or an anti-p65 antiserum. The immune complexes were resolved by 7% SDS-PAGE, and then analyzed by Western blotting using an anti-p65 antiserum (top). The blot was later stripped and reprobed with an anti–IRF-4 antiserum (bottom), as described above.
To determine whether interaction of IRF-4 and BCL-6 could occur independently of the presence of the CD23b GAS, we performed pull-down assays with a GST–IRF-4 fusion protein. As shown in Fig. 5 B, incubation of a GST–IRF-4 fusion protein with extracts from Ramos cells revealed a very strong association of IRF-4 with BCL-6. No interaction was observed with the GST moiety alone or upon incubation of the GST–IRF-4 with JY extracts, consistent with the lack of BCL-6 expression in EBV-transformed B cells. We also subjected extracts from Ramos cells cultured with or without IL-4 to immunoprecipitation assays with either an anti–IRF-4 or anti–BCL-6 antiserum (Fig. 5 C). This experiment demonstrated that IRF-4 and BCL-6 coimmunoprecipitated. Association of BCL-6 and IRF-4 was not affected by IL-4 treatment. Surprisingly, we did not detect increased levels of IRF-4 in our immunoprecipitations of IL-4– or CD40-treated extracts (Fig. 3 B and Fig. 5 C), despite an induction of IRF-4 levels by these stimulations (Fig. 4). We suspect that this may be due either to a limited ability of this antiserum to immunoprecipitate increasing amounts of IRF-4 or to the ability of IRF-4 upon IL-4–CD40 treatment to form alternative complexes that cannot be recognized by this antiserum.
To further corroborate the specificity of the association of IRF-4 with BCL-6 and Stat6, we also performed coimmunoprecipitation experiments with either an anti–IRF-4 or an antiserum against a nuclear factor κB (NF-κB) family member, p65. These assays were conducted on extracts from JY cells, which constitutively express p65 in the nucleus (49; Fig. 5 D). These experiments failed to detect an association of IRF-4 with p65.
These studies thus indicate that IRF-4 can interact with the Krüppel zinc finger transcriptional repressor, BCL-6, both in vitro and in vivo. Furthermore, no such interaction is detected in cells (JY) that express high levels of CD23b, suggesting that the presence of BCL-6 affects the functional ability of IRF-4 to modulate CD23b transcription.
The Ability of IRF-4 to Transactivate CD23b Can Be Blocked by BCL-6.
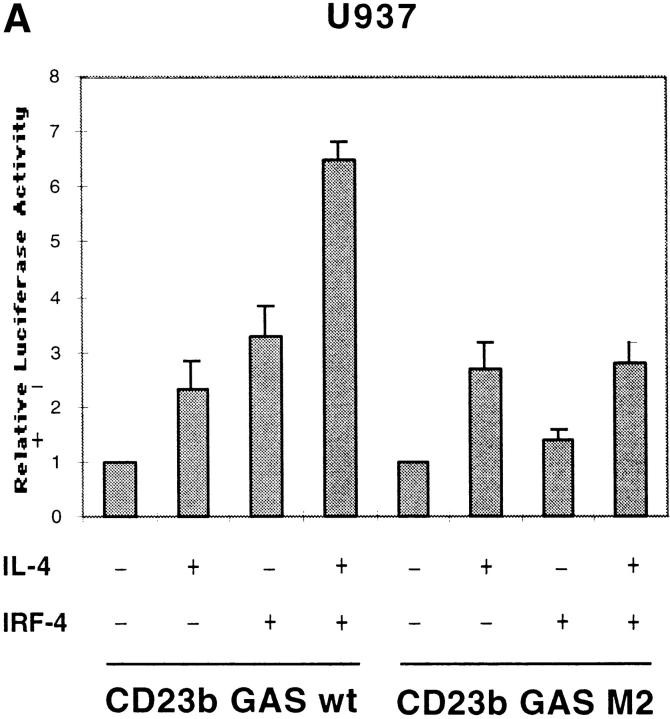
To directly assess whether IRF-4, in the absence of BCL-6, could function as a transactivator of CD23b, we performed transient transfection assays in U937 cells, a monocytic cell line that is capable of activating Stat6 in response to IL-4, but lacks both IRF-4 and BCL-6. Cotransfection of an IRF-4 expression vector with a luciferase reporter construct driven by an oligomerized CD23b GAS wt element resulted in a threefold induction in the luciferase activity, suggesting that IRF-4 can act as a transactivator of CD23b in the absence of Stat6 activation (Fig. 6 A). This level of induction was similar to that observed upon stimulation of U937 with IL-4. Interestingly, cotransfection of IRF-4 augmented, albeit not in a synergistic manner, the induction of the CD23b GAS wt reporter construct in response to IL-4. Consistent with the results of our EMSA experiments, a reporter construct driven by a mutant CD23b GAS element, which binds Stat6 but not IRF-4 (CD23b GAS M2, Fig. 2 B), displayed a normal IL-4 inducibility but could not be activated by IRF-4 cotransfection (Fig. 6 A). Furthermore, overexpression of IRF-4 was unable to enhance the IL-4–mediated activation of this CD23b GAS M2 reporter construct. Thus, these data suggest that optimal transactivation of CD23b requires the presence of both Stat6 and IRF-4.
Figure 6.
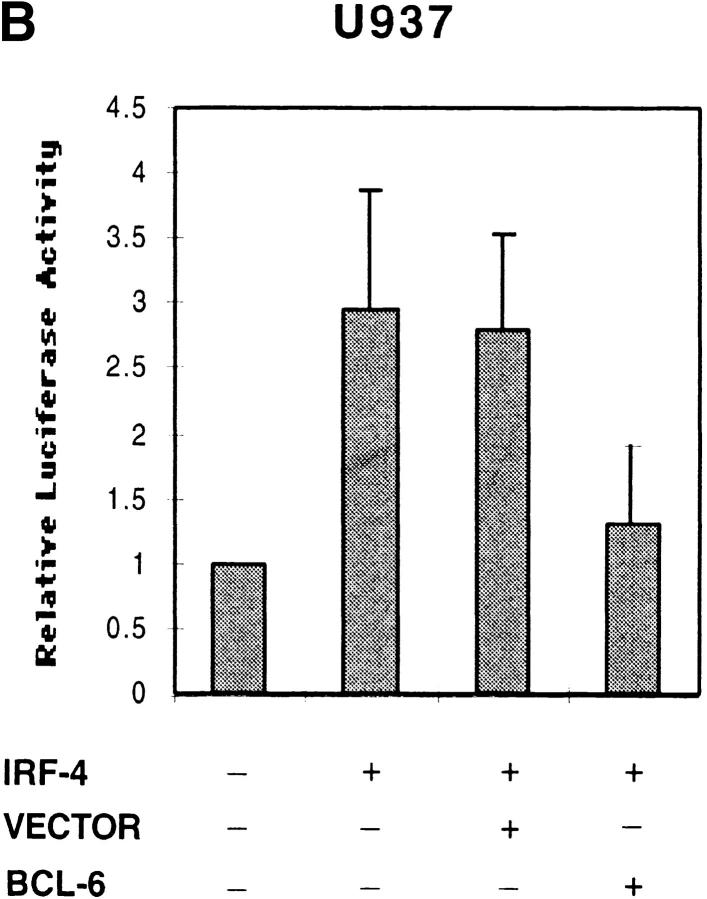
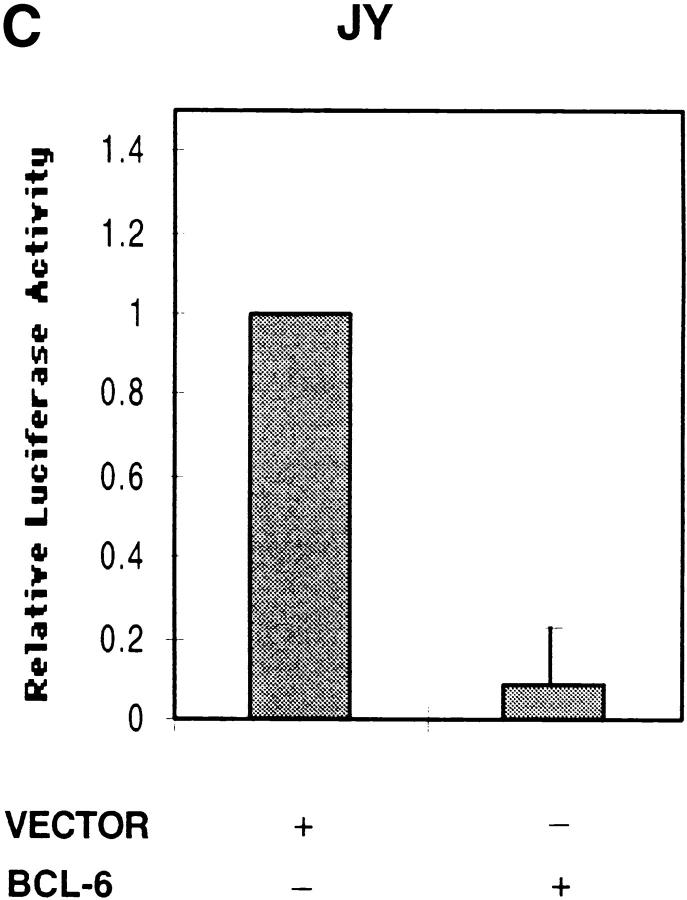
Effects of IRF-4 on the transactivation of CD23b. (A) U937 cells were cotransfected with a luciferase reporter construct driven by either an oligomerized CD23b GAS wt or an oligomerized CD23b GAS M2 element, and either an IRF-4 expression plasmid (pCEP4-IRF4) or equivalent amounts of empty vector. The transfected cells were equally split into two 2-ml aliquots and then incubated for 24 h in the presence or absence of IL-4 (10 ng/ml). The data are presented relative to the activity of the reporter construct in control U937 cells, which was set to 1.0, as indicated, in each experiment. Results show the mean ± SE of four independent experiments. (B) U937 cells were cotransfected with a luciferase reporter construct driven by an oligomerized CD23b GAS wt element, and an IRF-4 expression plasmid (pCEP4-IRF4) in the presence of either an BCL-6 expression vector (pMT2T-BCL6) or equivalent amounts of empty pMT2T vector, as indicated. The transfected cells were left unstimulated for 16 h. The data are presented relative to the activity of the reporter construct in control U937 cells, which was set to 1.0, as indicated, in each experiment. Results show the mean ± SE of three independent experiments. (C) JY cells were cotransfected with a CD23b promoter luciferase reporter construct in the presence of an expression plasmid for BCL-6 (pMT2T-BCL6) or of equivalent amounts of empty pMT2T vector. The transfected cells were left unstimulated for 16 h. The data are presented relative to the activity of the reporter construct in control JY cells, which was set to 1.0, as indicated, in each experiment. Results show the mean ± SE of three independent experiments.
Since BCL-6 is known to repress Stat6 function, we then proceeded to determine whether BCL-6 could block IRF-4 function. Indeed, cotransfection of BCL-6, but not of an empty vector, with IRF-4 was able to repress the ability of IRF-4 to induce the activity of the CD23b GAS reporter construct (Fig. 6 B). A similar inhibitory effect was also detected when BCL-6 was cotransfected with a reporter construct driven by the CD23b promoter in JY, an EBV-transformed B cell line, which constitutively expresses high levels of IRF-4 but no activated Stat6 (Fig. 6 C). These studies thus indicate that IRF-4 can transactivate the CD23b GAS and that BCL-6 can block IRF-4 function independently of its inhibitory effects on Stat6.
Discussion
IRF-4, a Lineage-specific Effector of the IL-4 and CD40 Signaling Pathways.
Biochemical and genetic studies have demonstrated that Stat6 plays a key role in IL-4 signaling 10. However, the rapid activation of Stat6 cannot solely account for the complex biological activities of IL-4 1 3. The IL-4 signaling pathway must thus use additional effector molecules. Our studies demonstrate that IRF-4 is both a target and a modulator of the IL-4 signaling pathway. This dual role of IRF-4 is thus reminiscent of that of IRF-1 in the IFN signaling cascade 26. In contrast to the IL-4 induction of Stat6, which has been detected in a wide variety of cells 13 14 15 16 17, IRF-4 expression is largely restricted to the lymphoid compartment 31 32 34. Thus, the recruitment of IRF-4 by the IL-4 signaling pathway provides a potential mechanism by which this cytokine can regulate the expression of target genes in a lineage-specific manner.
Similarly to IRF-1, whose induction is not restricted to the IFN pathway 64 65, we found that the expression of IRF-4 in B cells can be independently upregulated by stimulation via the CD40 receptor. CD40 engagement is also known to lead to the rapid activation of the NF-κB/rel family of transcription factors 4. Although other IRFs have been shown to interact with NF-κB/rel proteins 66 67 68, we did not detect a physical interaction of IRF-4 with p65 (Fig. 5 D). However, this finding does not exclude that IRF-4 may be able to associate with other NF-κB family members and/or functionally modulate the activity of CD40-inducible NF-κB complexes. Such an interaction may allow IRF-4 to participate in the regulation of genes that are activated in response to CD40 alone.
Our studies indicate that costimulation of B cells with CD40 and IL-4 leads to maximal IRF-4 induction. This is accompanied by the simultaneous CD40-mediated downregulation of BCL-6 (Fig. 4 A), consistent with previous reports 46 61. Since germinal center T cells have been shown to express CD40L as well as IL-4 69, the transcriptional events triggered by activation of B cells with both stimuli are physiologically relevant. Thus, one may predict that B cells that have been successfully selected in the germinal center and have received appropriate T cell help should express high levels of IRF-4 in addition to low or absent BCL-6. In support of this notion, a small subpopulation of centrocytes with such a phenotype has been identified in close apposition to follicular dendritic cells. These cells have been postulated to represent “surviving centrocytes” (Riccardo Dalla-Favera, personal communication). The coordinated induction of IRF-4 and downregulation of BCL-6 may thus be an important step in the progression of B cells toward the terminal stages of differentiation.
Interplay between IRF-4, Stat6, and BCL-6.
Our studies demonstrate that Stat6 and IRF-4 can physically interact. Preliminary experiments suggest that association of IRF-4 with Stat6 involves the COOH-terminal region of IRF-4. This portion of IRF-4 contains a putative α-helical region, which displays strong homology to the STAT-interacting domain of p48, the IRF component of the ISGF3 complex 63 70. As in the case of the p48–Stat2 complex 71, tyrosine phosphorylation of Stat6 does not appear to be needed for its association with IRF-4. However, unlike the ISGF3 complex 70 72, IRF-4 does not appear to be involved in recruiting Stat6 to the CD23b GAS. Our inability to detect a cooperative complex containing both IRF-4 and Stat6 also contrasts with the strong phosphorylation-dependent cooperative interaction between IRF-4 and the Ets protein, PU.1 29 35 36.
Our transient transfection experiments indicate that IRF-4 can act as a transactivator and augment the Stat6 inducibility of CD23b. Our studies have revealed an additive rather than a synergistic interaction between IRF-4 and Stat6, suggesting that recapitulation of the synergistic induction of CD23b in vitro may require regions flanking the CD23b GAS and/or additional components. For example, studies of the IFN-β enhanceosome, a model system for transcriptional synergism, have demonstrated that this regulatory region (−110 to −53) contains multiple IRF-1 binding sites 73. Consistent with this notion, our survey of the CD23b promoter has revealed that the regions flanking the CD23b GAS may contain additional IRF-4 binding sites. Our inability to detect enhanced IRF-4 DNA binding in EMSAs upon IL-4 treatment (Fig. 2 A) may thus be due to the lack of these additional sites in the probes used. Although, in the CD23 system, IRF-4 acts as a positive regulator, IRF-4 contains multiple regions with transactivation and/or repressing potential 74 75 76 77. Therefore, the precise outcome of the Stat6–IRF-4 interaction is likely to be dictated by the specific arrangement of their DNA binding sites as well as by the presence of additional cofactors.
Indeed, our studies suggest that the function of IRF-4 can be profoundly affected by the presence of the Krüppel zinc finger transcriptional repressor, BCL-6 19 20 21 78 79. BCL-6 is able to repress IRF-4 function in the absence of Stat6. This may allow BCL-6 to modulate the expression of Stat6-independent target genes. Indeed, we have found that BCL-6 binds to the IRF-4 binding site present in the Igκ 3′ enhancer (35 36; data not shown). In contrast to the CD23b GAS, this DNA element does not bind Stat6. Thus, the BCL-6–IRF-4 interaction may underlie some of the defects exhibited by the BCL-6–deficient mice that are not corrected by the lack of Stat6 24.
Various mechanisms may account for the repressive effects of BCL-6 on IRF-4 function. In addition to the known ability of BCL-6 to recruit the corepressor machinery 80 81 82 83, BCL-6 may prevent high-affinity DNA binding by IRF-4 as suggested by our UV cross-linking experiments, which detected stronger CD23b GAS–IRF-4 complexes in the absence of BCL-6. Furthermore, previous studies have indicated that IRF-4 contains an inhibitory region (amino acids 207–300) that can mask its own transactivation domain 84. Thus, BCL-6 may maintain IRF-4 in an autoinhibitory state. IRFs have previously been shown to be critical components of the virally induced IFN-β enhanceosome 73. Interestingly, another member of the Krüppel zinc finger family of transcription factors, PRDI-BF/Blimp1 85 86 87, can bind to one of the IRF binding sites within the IFN-β enhanceosome and act as a repressor of IFN-β gene expression 85. Thus, interaction between IRFs and Krüppel proteins may be a conserved feature of the transcriptional regulation of a variety of genes.
The ability of the CD23b GAS to bind an IRF family member as well as its organizational features are indeed reminiscent of regulatory DNA elements targeted by enhanceosomes 88. This suggests that the CD23b GAS and its flanking regions may function in the assembly of higher-order transcriptional complexes. The synergistic induction of CD23b by the CD40 and IL-4 signaling pathways may thus result from their ability to simultaneously target the expression/function of Stat6, IRF-4, and BCL-6, leading to a profound remodeling of the architecture of this nucleoprotein complex. Although we have been unable to clearly demonstrate formation of a trimolecular complex consisting of Stat6, BCL-6, and IRF-4, prolonged exposures of our UV cross-linking experiments (Fig. 5 A) have revealed that the IRF-4 antiserum can immunoprecipitate, in addition to IRF-4 and BCL-6, a faint band of mobility identical to that of Stat6. Assembly of this complex may thus require specific three-dimensional contacts, which we are unable to fully reproduce with the techniques available to us. Alternatively, some of the interactions between Stat6, IRF-4, and BCL-6 could be mediated by additional cofactors. These findings thus lend support to the notion that assembly of these enhanceosome-like complexes may represent ideal targets for the final integration of signaling pathways 73. Furthermore, presence of lineage-specific components like IRF-4 and stage-specific repressors like BCL-6 within these complexes would endow cells with the ability to regulate gene expression in response to signals such as CD40 and IL-4 in a context-appropriate manner.
Acknowledgments
We thank Dr. Hirai for the anti–IRF-4 antiserum and Drs. Wang and Kikutani for the CD23 cDNA. We are very thankful to Dr. Dalla-Favera and members of his laboratory for providing us with cell lines and the IRF-4 and BCL-6 constructs. We are grateful to Yang Xu for her technical assistance. We also thank Drs. J. Siu and C. Schindler for reading this manuscript as well as for their constant support. We thank Drs. Dalla-Favera and Thanos for their critical reading of this manuscript.
This work was supported by the New York City Council Speaker's Fund for Biomedical Research: Toward the Science of Patient Care, and a Grant-in-Aid from the American Heart Association.
Footnotes
Abbreviations used in this paper: BCL-6, B cell lymphomas 6; EMSA, electrophoretic mobility shift assay; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GAS, IFN-γ–activated site(s); GBP, guanylate binding protein; GSH, glutathione; GST, GSH S-transferase; ICSBP, IFN consensus sequence binding protein; IRF, IFN regulatory factor; ISGF, IFN-stimulated gene factor; ISRE, IFN-stimulated regulatory element; NF-κB, nuclear factor κB; STAT, signal transducer and activator of transcription; WCE, whole cell extract; wt, wild-type.
References
- Brown M., Hurai J. Functions of IL-4 and control of its expression. Crit. Rev. Immunol. 1997;17:1–32. doi: 10.1615/critrevimmunol.v17.i1.10. [DOI] [PubMed] [Google Scholar]
- Chomarat P., Banchereau J. An update on interleukin-4 and its receptor. Eur. Cytokine Netw. 1997;8:333–344. [PubMed] [Google Scholar]
- Paul W.E. Interleukin-4a prototypic immunoregulatory lymphokine. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- Laman J., Claassen E., Noelle R. Functions of CD40 and its ligand, gp39 (CD40L) Crit. Rev. Immunol. 1996;16:59–108. doi: 10.1615/critrevimmunol.v16.i1.40. [DOI] [PubMed] [Google Scholar]
- Darnell J., Kerr I., Stark G. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1420. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Ihle J., Kerr I. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- Leonard W.J., O'Shea J.J. JAKS and STATSbiological implications. Annu. Rev. Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Nelms K., Huang H., Ryan J., Keegan A., Paul W.E. Interleukin-4 receptor signalling mechanisms and their biological significance. Adv. Exp. Med. Biol. 1998;452:37–43. doi: 10.1007/978-1-4615-5355-7_5. [DOI] [PubMed] [Google Scholar]
- Kaplan M., Schindler U., Smiley S., Grusby M. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Takeda K., Tanaka T., Shi W., Matsumoto M., Minami M., Kashiwamura S., Nakanishi K., Yoshida N., Kishimoto T., Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- Shimoda K., van Deursen J., Sangster M.Y., Sarawar S.R., Carson R.T., Tripp R.A., Chu C., Quelle F.W., Nosaka T., Vignali D.A. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- Rousset F., de Malefijt R., Slierendregt B., Aubry J.-P., Bonnefoy J.-Y., Defrance T., Banchereau J., de Vries J. Regulation of Fc receptor for IgE (CD23) and class II MHC antigen expression on Burkitt's lymphoma cell lines by human IL-4 and IFN-γ. J. Immunol. 1988;140:2625–2632. [PubMed] [Google Scholar]
- Kohler I., Rieber E. Allergy-associated I∈ and Fc∈ receptor II (CD23b) genes activated via binding of an interleukin-4-induced transcription factor to a novel responsive element. Eur. J. Immunol. 1993;23:3066–3071. doi: 10.1002/eji.1830231204. [DOI] [PubMed] [Google Scholar]
- Schindler C., Kashleva H., Pernis A., Pine R., Rothman P. STF-IL4a novel IL-4 induced signal transducing factor. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:1350–1356. doi: 10.1002/j.1460-2075.1994.tb06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriggl R., Berchtold S., Friedrich K., Standke G.J., Kammer W., Heim M., Wissler M., Stocklin E., Gouilleux F., Groner B. Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells. Mol. Cell. Biol. 1997;17:3663–3678. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.H., Uetani K., Haque S.J., Williams B.R., Dweik R.A., Thunnissen F.B., Calhoun W., Erzurum S.C. Interferon-γ and interleukin-4 stimulate prolonged expression of inducible nitric oxide synthase in human airway epithelium through synthesis of soluble mediators. J. Clin. Invest. 1997;100:829–838. doi: 10.1172/JCI119598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay C., Porter B., Guenette D., Billir B., Arend W.P. Interleukin-4 (IL-4) and IL-13 enhance the effect of IL-1β on production of IL-1 receptor antagonist by human primary hepatocytes and hepatoma HepG2 cellsdifferential effect on C-reactive protein production. Blood. 1999;93:1299–1307. [PubMed] [Google Scholar]
- Cattoretti G., Chang C.-C., Cechova K., Zhang J., Ye B.H., Falini B., Louie D.C., Offit K., Chaganti R.S.K., Dalla-Favera R. BCL-6 protein is expressed in germinal center B cells. Blood. 1995;1:45–53. [PubMed] [Google Scholar]
- Chang C., Ye B., Chaganti R., Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfert V., Allman D., He Y., Staudt L. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- Ye B., Lista F., Coco F.L., Knowles D., Offit K., Chaganti R., Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- Dent A.L., Shaffer A.L., Yu X., Allman D., Staudt L.M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Ye B., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S.K. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997;16:161–169. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Dent A., Hu-Li J., Paul W.E., Staudt L.M. T helper type 2 inflammatory disease in the absence of interleukin 4 and transcription factor STAT6. Proc. Natl. Acad. Sci. USA. 1998;95:13823–13828. doi: 10.1073/pnas.95.23.13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Darnell J. Transcriptional responses to polypeptide ligandsthe JAK-STAT pathway. Annu. Rev. Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- Harada H., Taniguchi T., Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- Vaughan P., van Wijnen A., Stein J., Stein G. Interferon regulatory factorsgrowth control and histone gene regulation—it's not just interferon anymore. J. Mol. Med. 1997;75:348–359. doi: 10.1007/s001090050120. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Tanaka N., Taki S. Regulation of the interferon system, immune response and oncogenesis by the transcription factor interferon regulatory factor-1 Eur. Cytokine Netw. 9Suppl. 31998. 43 48 [PubMed] [Google Scholar]
- Eisenbeis C., Singh H., Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- Iida S., Rao P., Butler M., Corradini P., Boccadoro M., Klein B., Chaganti R., Dalla-Favera R. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat. Genet. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- Matsuyama T., Grossman A., Mittrucker H., Siderovski D., Kiefer F., Kawakami T., Richardson C., Taniguchi T., Yoshinaga S., Mak T. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T., Nishida J., Tanaka T., Sakai R., Mitani K., Taniguchi T., Yazaki Y., Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol. Cell. Biol. 1996;16:1283–1294. doi: 10.1128/mcb.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittrucker H., Matsuyama T., Grossman A., Kundig T., Potter J., Shahinian A., Wakeham A., Patterson B., Ohashi P., Mak T. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- Grossman A., Mittrucker H., Nicholl J., Suzuki A., Chung S., Antonio L., Suggs S., Sutherland G., Siderovski D., Mak T. Cloning of human lymphocyte-specific interferon regulatory factor (hLSIRF/hIRF4) and mapping of the gene to 6p23-25. Genomics. 1996;37:229–233. doi: 10.1006/geno.1996.0547. [DOI] [PubMed] [Google Scholar]
- Pongubala J.M.R., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., Atchinson M.L. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol. Cell. Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala J.M.R., Beveren C.V., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., Atchinson M.L. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- Himmelmann A., Riva A., Wilson G.L., Thevenin B.P.L.C., Kehrl J.H. PU.1/Pip and basic helix loop helix zipper transcription factors interact with binding sites in the CD20 promoter to help confer lineage- and stage-specific expression of CD20 in B lymphocytes. Blood. 1997;90:3984–3995. [PubMed] [Google Scholar]
- Kikutani H., Kishimoto T. Molecular genetics and biology of two different species of Fc∈RII. Res. Immunol. 1990;141:249–258. doi: 10.1016/0923-2494(90)90116-g. [DOI] [PubMed] [Google Scholar]
- Conrad D. FceRII/CD23the low affinity receptor for IgE. Annu. Rev. Immunol. 1990;8:623–645. doi: 10.1146/annurev.iy.08.040190.003203. [DOI] [PubMed] [Google Scholar]
- Richards M., Katz D. Biology and chemistry of the low affinity IgE receptor (Fc∈RII/CD23) Crit. Rev. Immunol. 1991;11:65–86. [PubMed] [Google Scholar]
- Bonnefoy J.-Y., Lecoanet-Henchoz S., Aubry J.-P., Gauchat J.-F., Graber P. CD23 and B-cell activation. Curr. Opin. Immunol. 1995;7:355–359. doi: 10.1016/0952-7915(95)80110-3. [DOI] [PubMed] [Google Scholar]
- Paterson R., Lack G., Domenico J.M., Delespesse G., Leung D.Y., Finkel T.H., Gelfand E.W. Triggering through CD40 promotes interleukin-4-induced CD23 production and enhanced soluble CD23 release in atopic disease. Eur. J. Immunol. 1996;26:1979–1984. doi: 10.1002/eji.1830260902. [DOI] [PubMed] [Google Scholar]
- Choe J., Kim H.-S., Armitage R., Choi Y.S. The functional role of B cell antigen receptor stimulation and IL-4 in the generation of human memory B cells from germinal center B cells. J. Immunol. 1997;159:3757–3766. [PubMed] [Google Scholar]
- Yokota A., Kikutani H., Tanaka T., Sato R., Barsumian E., Suemura M., Kishimoto T. Two species of human Fc∈ receptor II (Fc∈RII/CD23)tissue-specific and IL-4-specific regulation of gene expression. Cell. 1988;55:611–618. doi: 10.1016/0092-8674(88)90219-x. [DOI] [PubMed] [Google Scholar]
- Kotanides H., Reich N.C. Requirement of tyrosine phosphorylation for rapid activation of a DNA binding factor by IL-4. Science. 1993;262:1265–1267. doi: 10.1126/science.7694370. [DOI] [PubMed] [Google Scholar]
- Allman D., Jain A., Dent A., Maile R., Selvaggi T., Kehry M., Staudt L. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- Cleary A.M., Fortune S., Yellin M.J., Chess L., Lederman S. Opposing roles of CD95 (Fas/APO-1) and CD40 in the death and rescue of human low density tonsillar B cells. J. Immunol. 1995;155:3329–3337. [PubMed] [Google Scholar]
- Wang F., Gregory C., Sample C., Rowe M., Liebowitz D., Murray R., Rickinson A., Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytesEBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Xia D., Jiang M., Lee S., Pernis A. Signaling pathways mediated by the TNF- and cytokine-receptor families target a common cis-element of the IRF-1 promoter. J. Immunol. 1998;161:5997–6004. [PubMed] [Google Scholar]
- Grumont R., Gerondakis S. The subunit composition of NF-κB complexes changes during B-cell development. Cell Growth Differ. 1994;5:1321–1331. [PubMed] [Google Scholar]
- Frangioni J.V., Neel B.G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Azam M., Lee C., Strehlow I., Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- Berrier A., Siu G., Calame K. Transcription of a minimal promoter from the NF-IL6 gene is regulated by CREB/ATF and SP1 proteins in U937 promonocytic cells. J. Immunol. 1998;161:2267–2275. [PubMed] [Google Scholar]
- Thienes C.P., Monte L.D., Monticelli S., Busslinger M., Gould H.J., Vercelli D. The transcription factor B cell-specific activator protein (BSAP) enhances both IL-4- and CD40-mediated activation of the human ∈ germline promoter. J. Immunol. 1997;158:5874–5882. [PubMed] [Google Scholar]
- Gordon J. CD23a novel multifunctional regulator of the immune system that binds IgE. Monogr. Allergy. 1991;29:1–49. [PubMed] [Google Scholar]
- Lederman S., Yellin M., Cleary A., Pernis A., Inghirami G., Cohn L., Covey L., Lee J., Rothman P., Chess L. T-BAM/CD40-L on helper T lymphocytes augments lymphokine-induced B cell Ig isotype switch recombination and rescues B cells from programmed cell death. J. Immunol. 1994;152:2163–2171. [PubMed] [Google Scholar]
- Ning Z., Norton J., Li J., Murphy J. Distinct mechanisms for rescue from apoptosis in Ramos human B cells by signaling through CD40 and interleukin-4 receptorrole for inhibition of an early response gene, Berg36. Eur. J. Immunol. 1996;26:2356–2363. doi: 10.1002/eji.1830261013. [DOI] [PubMed] [Google Scholar]
- Lens S., Tesselaar K., de Drijver B.F., van Oers M., van Lier R. A dual role for both CD40-ligand and TNF-α in controlling human B cell death. J. Immunol. 1996;156:507–514. [PubMed] [Google Scholar]
- Cheng G., Cleary A., Ye Z., Hong D., Lederman S., Baltimore D. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- Escalante C., Yie J., Thanos D., Aggarwal A. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- Cattoretti, G., C. Chang, K. Cechova, J. Zhang, B. Ye, B. Falini, D. Louie, K. Offit, R. Chaganti, and R. Dalla-Favera. 1997. Downregulation of BCL-6 gene expression by CD40 and EBV latent membrane protein-1 (LMP1) and its block in lymphoma carrying BCL-6 rearrangements. Blood. 90(Suppl. 1):175a. (Abstr.)
- Darnell J.E. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Brass A., Kehrli E., Eisenbeis C., Storb U., Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- Fujita T., Reis L., Watanabe N., Kimura Y., Taniguchi T., Vilcek J. Induction of the transcription factor IRF-1 and interferon-β mRNAs by cytokines and activators of second-messenger pathways. Proc. Natl. Acad. Sci. USA. 1989;86:9936–9940. doi: 10.1073/pnas.86.24.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harroch S., Revel M., Chebath J. Induction by interleukin-6 of interferon regulatory factor-1 (IRF-1) gene expression through the palindromic interferon response element pIRE and cell type-dependent control of IRF-1 binding to DNA. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:1942–1949. doi: 10.1002/j.1460-2075.1994.tb06463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew P., Franzoso G., Becker K., Bours V., Carlson L., Siebenlist U., Ozato K. NFκB and interferon regulatory factor 1 physically interact and synergistically induce major histocompatibility class I gene expression. J. Interferon Cytokine Res. 1995;15:1037–1045. doi: 10.1089/jir.1995.15.1037. [DOI] [PubMed] [Google Scholar]
- Drew P., Franzoso G., Carlson L., Biddison W., Siebenlist U., Ozato K. Interferon regulatory factor-2 physically interacts with NF-κB in vitro and inhibits NF-κB induction of major histocompatibility class I and β2-microglobulin gene expression in transfected human neuroblastoma cells. J. Neuroimmunol. 1995;63:157–162. doi: 10.1016/0165-5728(95)00140-9. [DOI] [PubMed] [Google Scholar]
- Neish A.S., Read M.A., Thanos D., Pine R., Maniatis T., Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol. Cell. Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiagbe V.K., Inghirami G., Thorbecke G.J. The physiology of germinal centers. Crit. Rev. Immunol. 1996;16:381–421. [PubMed] [Google Scholar]
- Horvath C., Stark G., Kerr I., Darnell J.J. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moczygemba M., Gutch M.J., French D.L., Reich N.C. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-α-stimulated transcription factor ISGF3. J. Biol. Chem. 1997;272:20070–20076. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- Qureshi S.A., Salditt-Georgieff M., Darnell J.E. Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. USA. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D. Mechanisms of transcriptional synergism of eukaryotic genes. The interferon-beta paradigm. Hypertension. 1996;27:1025–1029. doi: 10.1161/01.hyp.27.4.1025. [DOI] [PubMed] [Google Scholar]
- Renkawitz R. Transcriptional repression in eukaryotes. Trends Genet. 1990;6:192–197. doi: 10.1016/0168-9525(90)90176-7. [DOI] [PubMed] [Google Scholar]
- Cowell I. Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Johnson A. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- Herschbach B., Johnson A. Transcriptional repression in eukaryotes. Annu. Rev. Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- Kawamata N., Miki T., Ohashi K., Suzuki K., Fukuda T., Hirosawa S., Aoki N. Recognition DNA sequence of a novel putative transcription factor, BCL6. Biochem. Biophys. Res. Commun. 1994;204:366–374. doi: 10.1006/bbrc.1994.2468. [DOI] [PubMed] [Google Scholar]
- Baron B.W., Stanger R.R., Hume E., Sadhu A., Mick R., Kerckaert J.-P., Deweindt C., Bastard C., Nucifora G., Zeleznick-Le N., McKeithan T. BCL6 encodes a sequence-specific DNA-binding protein. Genes Chromosomes Cancer. 1995;13:221–224. doi: 10.1002/gcc.2870130314. [DOI] [PubMed] [Google Scholar]
- David G., Alland L., Hong S.H., Wong C.W., DePinho R.A., Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- Wong C.W., Privalsky M.L. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARα and BCL-6. J. Biol. Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- Dhordain P., Albagli O., Lin R.J., Ansieu S., Quief S., Leutz A., Kerckaert J.P., Evans R.M., Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K.D., Bardwell V.J. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Nagulapalli S., Atchinson M.L. Transcription factor Pip enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol. Cell. Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A.D., Maniatis T. Identification and characterization of a novel repressor of β-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Turner C.A., Jr., Mack D., Davis M.M. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Huang S. Blimp-1 is the murine homolog of the human transcriptional repressor PRDI-BF. Cell. 1994;78:9. doi: 10.1016/0092-8674(94)90565-7. [DOI] [PubMed] [Google Scholar]
- Collins T., Read M., Neish A., Whitley M., Thanos D., Maniatis T. Transcriptional regulation of endothelial cell adhesion moleculesNF-κB and cytokine-inducible enhancers. FASEB J. 1995;9:899–909. [PubMed] [Google Scholar]