Abstract
A novel T cell–specific adaptor protein, RIBP, was identified based on its ability to bind Rlk/Txk in a yeast two-hybrid screen of a mouse T cell lymphoma library. RIBP was also found to interact with a related member of the Tec family of tyrosine kinases, Itk. Expression of RIBP is restricted to T and natural killer cells and is upregulated substantially after T cell activation. RIBP-disrupted knockout mice displayed apparently normal T cell development. However, proliferation of RIBP-deficient T cells in response to T cell receptor (TCR)-mediated activation was significantly impaired. Furthermore, these activated T cells were defective in the production of interleukin (IL)-2 and interferon γ, but not IL-4. These data suggest that RIBP plays an important role in TCR-mediated signal transduction pathways and that its binding to Itk and Rlk/Txk may regulate T cell differentiation.
Keywords: T cell activation, signal transduction, adaptor protein, Tec tyrosine kinases, T helper type 1/T helper type 2 cells
It is well established that T cell activation critically depends on signal transduction pathways mediated by protein tyrosine kinases (PTKs) of the Src (Fyn and Lck) and Syk (Syk and ZAP-70) families 1 2 3 4. In contrast, the biological functions of members of the Tec family of tyrosine kinases are not well understood, nor have the biochemical mechanisms by which they operate been elucidated. These kinases contain a Src homology (SH)3 domain capable of binding proline-rich sequences, an SH2 domain capable of binding phosphotyrosine-containing sequences, and a tyrosine kinase domain 5 6. However, unlike other Src family kinases, members of the Tec family characteristically lack a negative regulatory tyrosine residue at the COOH terminus 7 8 9, whose dephosphorylation is normally required for kinase activity.
Multiple members of the Tec subfamily have been identified in T cells and B cells, including Itk, Rlk/Txk, Btk, and Tec. Itk, a 72-kD IL-2–inducible PTK expressed in T cells and NK cells 9 10, has been reported to be involved in T cell activation. Mice deficient in Itk display deficits in both T cell development and peripheral T cell activation 11; these T cells exhibit a diminished intracellular Ca2+ flux in response to TCR/CD3 signaling, and consistent with this, reduced phospholipase Cγ-1 phosphorylation and inositol triphosphate production 12. Additionally, Itk has been implicated in CD28 signal transduction, although whether it functions to positively or negatively regulate this pathway is unclear 13 14 15. Tec is expressed in multiple hematopoietic and nonhematopoietic cell types, including T cells 16 17. Although a recent report suggests that it may function in antigen receptor signal transduction in T cells 18, other evidence suggests that Tec is involved in signaling through receptors for the cytokines IL-3, IL-6, and GM-CSF (19 20 21, respectively). Finally, we identified previously a novel member of the Tec subfamily of Src-related PTKs, Rlk (also called Txk), that is expressed primarily in T cells, mast cells, and testes 22 23. Unlike other Tec subfamily members, Rlk (a 62-kD protein) lacks an NH2-terminal pleckstrin homology domain, and its expression is downregulated in naive T cells after activation. Moreover, there is substantially more Rlk expressed in Th1-type than Th2-type cells 22, suggesting that it may be involved in T cell differentiation. Although one recent study implicates Rlk in the binding of the cytoplasmic tail of the counter-costimulatory receptor CTLA-4 24, little is known about the biochemical targets of this PTK or its possible functions in T cell activation.
To begin to elucidate the function(s) of Rlk and related Tec kinases, we sought to identify associated proteins using the yeast two-hybrid system for analyzing interprotein interactions. A unique cDNA clone was identified that interacted with both Rlk and Itk. This cDNA encodes a putative adaptor protein, termed RIBP (for Rlk/Itk-binding protein). RIBP mRNA is expressed in T cells and NK cells and is significantly upregulated after T cell activation. Finally, targeted disruption of the RIBP gene via homologous recombination was shown to inhibit TCR/CD3-induced T cell proliferation, and IL-2 and IFN-γ production, but not IL-4 production. Together these data suggest that this novel molecule may provide an important adaptor function for Tec subfamily kinases critically involved in T cell activation and differentiation.
Materials and Methods
Bait cDNA Cloning and Yeast Two-Hybrid cDNA Library Screen.
The Y153 Saccharomyces cerevisiae strain (auxotrophic for adenine, uracil, leucine, and tryptophan) as well as the bait and trap expression vectors, pAS1 and pACT, respectively, have been described 25. The Rlk and Itk chimeric bait constructs were generated by cloning full-length cDNA, PCR-amplified using primers encoding NcoI and BamHI sites, into the NcoI and BamHI sites in the multiple cloning site of pAS1. The Itk cDNA in pcDNA1 (Invitrogen) was a gift of Dan Littman (New York University, New York, NY). A mouse T cell lymphoma MATCHMAKER cDNA library in pACT (Clontech) was screened using the Rlk bait as per the manufacturer's protocol. Full-length clones were identified by screening a custom-made Uni-ZAP mouse day 16 fetal thymic cDNA library (Stratagene) using a cDNA probe derived from the positive yeast clone, 2.3.2.
Cell Culture and Transfections.
Human embryonic kidney HEK293 cells were maintained in DMEM (GIBCO BRL) supplemented with 10% FCS, penicillin/streptomycin (100 μg/ml), and 2-ME. RIBP-HA cDNA was generated by cloning full-length (the short form) RIBP cDNA into pcDNA3 (Invitrogen) modified by ligation of a double-stranded oligonucleotide encoding a Kozak sequence and an HA tag between its HindIII and NotI sites (a gift from the laboratory of Harinder Singh, University of Chicago), between NotI and XbaI sites. Lck cDNA in the PEF expression vector was obtained from the laboratory of David Straus (University of Chicago). Transient transfections were carried out using a standard calcium phosphate precipitation method. Medium was exchanged 24 h after transfection, and cells were harvested 48 h after transfection.
Immunoprecipitation and Immunoblotting Analysis.
HEK293 cells were harvested and lysed in 1 ml of lysis buffer (0.5% Triton X-100, 50 mM Tris [pH 7.4], 150 mM NaCl, 5 mM EDTA [pH 8.0], 1 mM sodium vanadate, 10 μg/ml leupeptin, 10 μM aprotinin, 1 mM PMSF). Aliquots of the lysates (400 μg) were precleared at 4°C with 50 μl of protein G–agarose beads (GIBCO BRL) coated with 2.5 μg of rat IgG (Southern Biotechnology Associates). Immunoprecipitations were performed by incubating the precleared lysate with 50 μl of protein G–agarose beads precoated with 2.5 μg of rat high-affinity anti-HA Ab (Roche Diagnostics). Preclears and immunoprecipitates were washed four times in lysis buffer. All steps were performed at 4°C. The preclears and immunoprecipitates were boiled in 50 μl of 2× reducing SDS sample buffer and subjected to SDS-PAGE on a 10% gel. Proteins were subsequently transferred to a polyvinyldifluoride membrane preblocked in a 10% nonfat milk/1× TBST (Tris-buffered saline/Tween: 10 mM Tris, 150 mM NaCl, 0.05% Tween-20) solution. Blots were probed with a rabbit polyclonal anti-Itk antiserum (a gift from the laboratory of Dan Littman) solution (1:5,000 dilution in 5% nonfat milk/TBST) and developed with a 1:8,000 diluted anti–rabbit Ig (Amersham Pharmacia Biotech). For anti-RIBP immunoblotting, a rabbit polyclonal anti-RIBP antiserum solution (1:1,000 dilution in 2% nonfat milk/TBST) was used to probe blots.
Chromosomal Localization of the Mouse RIBP Gene.
The RIBP gene was mapped by Southern blot analyses of two sets of multilocus crosses: (NFS/N or C58/J × Mus musculus musculus) × M. m. musculus and (NFS/N × Mus spretus) × M. spretus or C58/J. Offspring of these crosses have been previously typed for >1,200 loci distributed over the 19 autosomes and the X chromosome 26 27. Recombinational distances were calculated according to Green 28, and gene order was established by minimizing the number of recombinants.
Northern and Southern Blot Analysis.
For Northern blot analysis, total RNA was isolated from the indicated tissues and cell types using TRIzol (GIBCO BRL) according to the manufacturer's protocol or as described previously 23. Northern blots were probed for the presence of RIBP mRNA using 32P-labeled cDNA probes derived from either full-length RIBP or RIBP 1–208 (encoding RIBP from the NH2 terminus to the end of the SH2 domain) by random priming, using a Prime-It II kit (Stratagene). The RIBP transcript was detected as a band ∼1.7 kb in size, migrating faster than the 18S rRNA band.
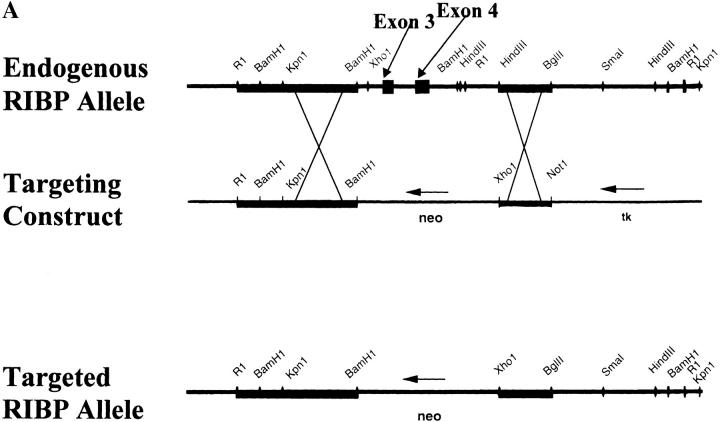
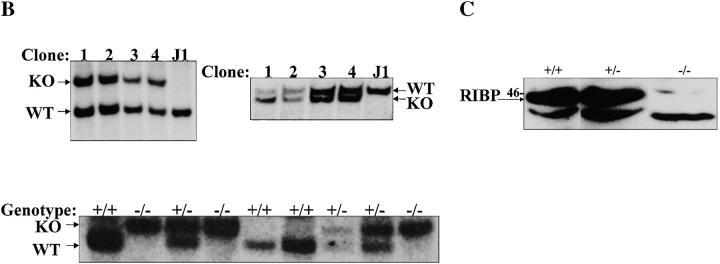
For Southern blot analysis, genomic DNA was prepared from embryonic stem (ES) cell clones by standard methods, and digested with the indicated restriction endonucleases. The DNA was denatured and, after gel electrophoresis and membrane transfer, hybridized to either a 5′ or 3′ 32P-labeled RIBP cDNA probe (as indicated in the text, and see Fig. 4 B). Bands representing either the endogenous or targeted RIBP allele were detected based on size, as described in the text and in the legend to Fig. 4 B.
Figure 4.
Targeted disruption of the RIBP gene. (A) Partial restriction endonuclease maps of the endogenous RIBP locus, targeting construct, and the targeted RIBP locus. Top: Restriction endonuclease map of the endogenous RIBP locus. Exons 3 and 4, deleted by homologous recombination in the KO mice, are indicated. Middle: Restriction endonuclease map of the targeting construct. Bottom: Restriction endonuclease map of the targeted RIBP locus. (B) Southern blot analyses of genomic DNA from targeted ES cell clones and genotyping of a litter from an intercross of RIBP heterozygous parents. Top left: Genomic DNA from ES cell clones was digested with EcoRI and XhoI, subjected to gel electrophoresis, and subsequently hybridized to a 5′ RIBP cDNA probe (a 1.8-kb fragment spanning the 5′-most EcoRI site to the KpnI/Asp718 site). The wild-type RIBP allele is represented by a 3.8-kb band, and the targeted allele is represented by a 5.5-kb band. Targeted ES cell clones are lanes 1–4, and a wild-type ES cell clone is denoted J1. Top right: ES cell genomic DNA was digested with Asp718 and XhoI, subjected to gel electrophoresis, and subsequently hybridized to a 3′ RIBP cDNA probe (a 1-kb fragment contained within the SmaI to HindIII fragment, near the 3′ end of RIBP, as shown in A). The wild-type RIBP allele is represented by a 20-kb band, and the targeted allele is represented by a 15-kb band. ES cell clones are as indicated previously. Bottom: Mouse tail genomic DNA samples from progeny of a mating of RIBP heterozygous parents were digested, subjected to gel electrophoresis, and subsequently hybridized to a 5′ RIBP cDNA probe as described previously for ES cell clone samples (top left). (C) Verification of absence of RIBP expression in RIBP KO mice. Lysates were prepared from activated splenocytes from wild-type, heterozygous, and homozygous KO mice and immunoblotted with an anti-RIBP polyclonal antiserum.
Targeted Deletion of the RIBP Gene and Generation of RIBP-deficient Mice.
The targeting construct was electroporated into the R1 ES cell line, derived from the 129/Sv mouse strain, using standard techniques 29. Homologous recombination at the RIBP locus was detected by Southern blot analysis using 5′ and 3′ probes binding outside the RIBP regions used in the targeting construct (see above, and Fig. 4 B). Targeted ES cell clones were used for blastocyst injection, and the resulting chimeric animals were backcrossed to C57BL/6 mice. Agouti offspring carrying the targeted RIBP allele were interbred to generate progeny homozygous for the targeted RIBP allele. Progeny were typed by Southern blot analysis of tail genomic DNA (see Fig. 4 B), and subsequently, by PCR analysis, using primer pairs which either amplified endogenous sequence between exons 2 and 3 or sequences in the neo cassette.
Flow Cytofluorimetric Analyses.
Single cell suspensions from thymi, lymph nodes, and spleens were prepared according to standard methods. Cells were then blocked with an anti-FcR mAb, 2.4G2, and stained with the Abs indicated in the text. Abs used in these experiments were conjugated to biotin or one of the following fluorochromes: FITC, PE, or Cy-Chrome. Ab-stained cells were analyzed on a FACScan™ (Becton Dickinson) flow cytometer.
Proliferation Assays.
Whole lymph node cells, splenocytes, or a 1:1 mixture of lymph node or splenic T cells cultured with T cell–depleted, irradiated syngeneic splenic APCs, from wild-type (either progeny from heterozygous intercrosses or [C57BL/6 × 129] F2 mice; The Jackson Laboratory), heterozygous, or homozygous knockout (KO) mice were used in these experiments. Soluble anti-CD3∈ mAb (145-2C11) with or without soluble anti-CD28 mAb (PV-1) was added to wells of round-bottomed 96-well plates (Costar) at the concentrations indicated in the text. When used, final concentrations of PMA and ionomycin were 10 ng/ml and 0.5 μM, respectively. Cells (2 × 105) were added to each well at a final concentration of 1 × 106/ml and cultured at 37°C in DMEM supplemented with 10% FCS, penicillin/streptomycin (100 μg/ml), and 2-ME. After 40 h, individual wells were pulsed with [3H]thymidine (1 μCi/well) for 8 h, for a total duration of 48 h of stimulation. Cell contents were then harvested onto fiberglass filters, scintillation fluid was added (25 μl/well), and counts were determined using a TopCount microplate scintillation counter (Packard).
IL-2 ELISAs.
IL-2 ELISAs were performed on culture supernatants of activated cell cultures (described above). Nunc-Immuno plates (Nalge Nunc) were used for these assays. Plates were coated with an anti–IL-2 coating Ab overnight at 4°C, washed, and blocked in 2% BSA/PBS for 1 h at room temperature. Supernatants, diluted in 2% BSA/PBS, were incubated on the coated plates overnight at 4°C. The next day, plates were washed, then incubated with an anti–IL-2 detecting Ab for 1 h at room temperature. Plates were washed and incubated with a 1:5,000 dilution of streptavidin-horseradish peroxidase (Zymed), after which they were developed using TMB One-Step Substrate (Dako). Abs and IL-2 standards were all purchased from Endogen.
RNase Protection Assays.
Splenocytes from wild-type (either [C57BL/6 × 129] F2 described above, or when indicated, C57BL/6 mice), heterozygous, and homozygous KO mice were activated with the indicated concentrations of soluble anti-CD3 mAb with or without anti-CD28. At the indicated times, cells were harvested, and RNA was prepared as described above. Cytokine RNase protection assays were performed using the RiboQuant Multi-Probe RNase Protection Assay System (PharMingen) according to the manufacturer's protocol. In brief, a 32P-labeled RNA probe was synthesized via in vitro transcription from the mCK-1 plasmid template. Aliquots of 2.5 μg of total RNA per sample were hybridized overnight at 56°C to the probe. Samples were then digested with RNase A, after which they were treated with Proteinase K and phenol/chloroform extracted. Samples were then precipitated, resuspended in 1× loading buffer, heated briefly at 90°C, and subjected to electrophoresis on a 5% denaturing polyacrylamide gel. Gels were dried, and autoradiography was performed. Densitometry on the cytokine and housekeeping gene bands was performed using a Molecular Dynamics ImageQuant® densitometer.
Results
RIBP Is a Novel Adaptor Protein That Interacts with Rlk and Itk.
The yeast two-hybrid system of detecting interprotein interactions was used to identify potential Rlk-binding proteins that might mediate or regulate Tec family kinase function. A cDNA encoding full-length Rlk was cloned into a yeast vector encoding the GAL4 DNA-binding domain, thus yielding a “bait” fusion protein consisting of Rlk physically linked to this DNA-binding protein. A mouse T cell lymphoma cDNA library was screened using the Rlk bait. This library consisted of “trap” cDNAs cloned into a yeast vector encoding the GAL4 transactivation domain. The T cell cDNA library was chosen as Rlk is highly expressed both in the thymus and in peripheral T cells. Two specificity criteria were used as measures of GAL4-mediated transcriptional activation: (a) histidine synthesis by the yeast, and (b) β-galactosidase synthesis. A total of 750,000 yeast colonies were screened. 86 of these colonies were identified as positive for bait–trap interaction by the first pass criterion of growth on histidine-deficient (His−) medium supplemented with 3-aminotriazole, an inhibitor of weak endogenous histidine synthesis. Among the 86 colonies, 56 were positive for interaction with the Rlk bait fusion protein based on the second criterion of β-galactosidase synthesis (blue/white selection). The cDNA library plasmids isolated from the β-galactosidase–positive yeast colonies were subsequently transformed into bacteria to distinguish multiple trap cDNAs present within the same yeast colony during the original screening. A total of 65 unique cDNA clones were identified and subsequently retransformed into yeast. One clone, 2.3.2, was found to interact with Rlk with high specificity, and was chosen for further study.
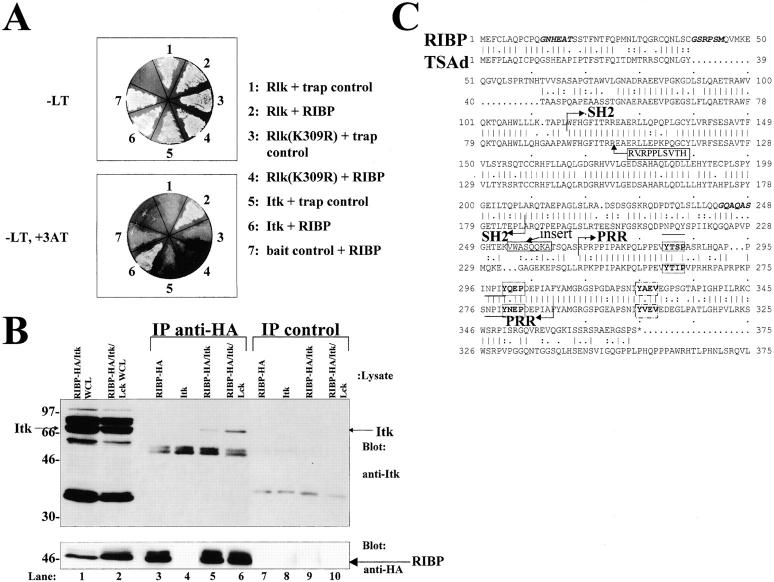
Initial sequence analysis demonstrated that clone 2.3.2 did not represent a full-length cDNA; consequently, a mouse fetal thymic cDNA library was screened using a probe derived from the positive yeast cDNA clone. This screen yielded a full-length cDNA that was used to examine interactions with Rlk and Itk. Yeast transformed with the full-length cDNA, termed RIBP, interacted specifically with both Rlk and Itk (Fig. 1 A, bottom; nos. 2 and 6, respectively). We further examined whether the interaction of RIBP with Rlk depended on the kinase function of Rlk. An altered Rlk that contained a point mutation in the kinase domain (K309→R, denoted K309R) previously shown to abolish kinase activity (our unpublished observations) was incapable of binding RIBP (Fig. 1 A, bottom; no. 4). This suggests that the bait–trap interaction is critically dependent on the functionality and activity of the tyrosine kinase domain of Rlk.
Figure 1.
Identification of an Rlk and Itk interacting protein, RIBP. (A) Yeast two-hybrid system interactions between RIBP and Itk, Rlk, or kinase-mutant Rlk (K309R). Top: Growth of bait plus trap–transformed yeast on leucine and tryptophan–deficient (−LT) medium (positive control for transformation with both constructs). Bottom: Assay for a specific bait–trap interaction. Growth of bait plus trap–transformed yeast on –LT, +3-aminotriazole (3AT, endogenous histidine synthesis inhibitor) medium. For interactions with Rlk, results are representative of 11 independent experiments. For interactions with Itk, results are representative of two independent experiments. For interactions with Rlk(K309R), results are representative of four independent experiments. (B) Western blot demonstrating association of RIBP with Itk in transfected HEK293 cells. Top: Lysates prepared from HEK293 cells transfected with the indicated constructs (top of the blot) were precleared with rat IgG on protein G beads, and subsequently, the supernatants were immunoprecipitated with rat anti-HA mAb. Immunoprecipitates and preclears were resolved by SDS-PAGE, transferred to a polyvinyldifluoride membrane, and immunoblotted with an anti-Itk antiserum. Bottom: Immunoblot from B was stripped and reprobed with mouse anti-HA to assess the amount of RIBP in the immunoprecipitates. Results are representative of three experiments. WCL, whole cell lysates. (C) Amino acid sequence homology between RIBP and TSAd. NH2 and COOH termini of a putative SH2 domain and PRR are indicated by arrows. Dots between amino acids in RIBP and TSAd indicate homology, with two dots indicating greater homology. The amino acid insertions present in the alternatively spliced forms of RIBP and TSAd are boxed with a solid line; NPXY PTB domain sequences are underlined, and YXXP and YXXV sequences, in boldface, are in boxes with dashed lines or dot-dashed lines, respectively. Putative N-myristoylation sites are in boldface and italics. The RIBP nucleotide sequence is available from EMBL/GenBank/DDBJ under accession no. AF203343.
Human epithelial kidney (HEK293) cells were transfected transiently with constructs encoding HA-tagged RIBP and/or Itk to directly examine protein–protein interactions in mammalian cells. Cell lysates were immunoprecipitated with an anti-HA Ab, resolved by SDS-PAGE, and subjected to immunoblotting using an anti-Itk antiserum. As shown in Fig. 1 B, a specific interaction between RIBP and Itk was detected (top panel, lane 5). Coexpression of the Src family kinase, Lck, increased the amount of Itk coprecipitated with RIBP (Fig. 1 B, top; compare lane 6 with lane 5). This Lck-dependent augmentation of RIBP–Itk complex formation was not due to increased levels of Itk in the cell lysates or RIBP-HA in the immunoprecipitates (Fig. 1 B, top, lane 2 vs. lane 1; and bottom, lane 6 vs. lane 5, respectively). The results suggested that the interaction of RIBP with Itk is regulated, at least in part, by Src family kinase–specific phosphorylation of one or both proteins.
Structural Analysis and Chromosomal Location of RIBP.
Sequence and genomic analyses were performed on the RIBP gene product. RIBP contains an SH2 domain (highly homologous to the SH2 domain of Shc, 47% [data not shown]), a proline-rich region (PRR) capable of binding SH3 domains, and an NPIY substrate sequence for binding by phosphotyrosine-binding (PTB) domains 30 present within the PRR (Fig. 1 C). Further analysis of the cDNA library identified a second RIBP transcript that represented an alternatively spliced form with an 8 amino acid sequence insert (VWASQQKA). Other potentially significant amino acid sequences within RIBP include two YXXP motifs, implicated in binding the SH2 domains of rasGAP, Abl, and Crk 31, and one YXXV motif, potentially capable of binding the SH2 domain of the tyrosine phosphatase, SHP-2 32. Additionally, RIBP contains three potential sites for N-myristoylation (Fig. 1 C). These structural features and the absence of a catalytic domain suggest that RIBP functions as an adaptor protein. One adaptor protein, TSAd 33, showed extensive sequence homology to RIBP. The human TSAd protein exhibits a 68% sequence identity and a 76% sequence similarity with RIBP. The sequence homology between RIBP and TSAd was greatest within the SH2 domain, where the two proteins share an 87% sequence identity (80/92 amino acids; Fig. 1 C). Furthermore, TSAd exists in two alternatively spliced forms, similar to RIBP, with a 10 amino acid insert present in the longer form. Although these results suggested that RIBP and TSAd may be species homologues, the sequence insertions in RIBP and TSAd are in different locations within the molecules. The 10 amino acid insertion in TSAd is located within the SH2 domain, whereas the 8 amino acid insertion in RIBP is located within one of the exons in the region between the SH2 domain and the PRR (Fig. 1 C).
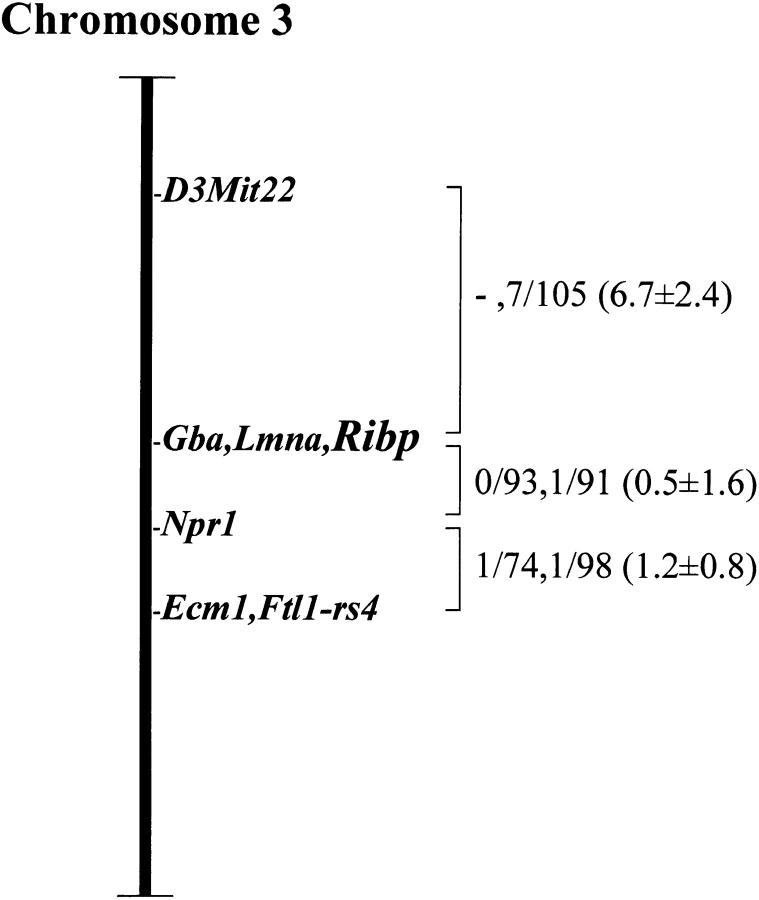
Genomic linkage studies of the RIBP gene were performed by Southern blot analyses of two sets of multilocus crosses (described in Materials and Methods). Digestion of DNA samples from parental mice with HindIII produced RIBP fragments of 8.6 and 2.9 kb in M. spretus and 11.0 and 10.8 kb in NFS/N. BamHI produced fragments of 12.4 and 4.0 kb in NFS/N and 16.5 kb in M. m. musculus. Inheritance of the variant fragments was compared with that of other markers previously typed in these crosses. The RIBP locus was mapped to mouse chromosome 3 (Fig. 2). Closest linkage was detected with Gba, for which no recombinants were detected in 171 mice, indicating that the genes are within 1.7 cM of one another at the upper limit of the 95% confidence interval. Similarly, no recombination was detected with Lmna in 103 mice, placing RIBP within 2.9 cM of Lmna. The RIBP gene maps to a region of mouse chromosome 3 with conserved synteny to human chromosome 1q21 34 35, where both human GBA 36 and LMNA 37, as well as TSAd 33 and Shc 38, are located. Thus, the RIBP gene has a genomic localization congruent with that of the TSAd and Shc genes, supporting the possibility of gene duplication events in this region.
Figure 2.
Chromosomal localization of mouse RIBP. To the right of the map are recombination fractions between adjacent loci, with the first fraction representing data from the M. m. musculus crosses and the second from the M. spretus crosses. Numbers in parentheses are recombinational distances ± SE. The M. m. musculus crosses were not typed for D3Mit22.
RIBP Is Restricted in Expression to T Lymphocytes and NK Cells.
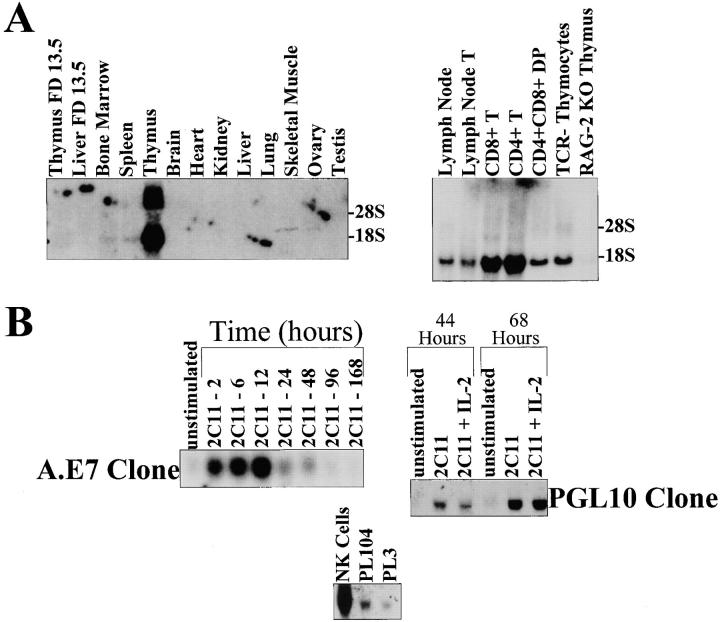
Tec family members have been shown to have restricted tissue expression. In the cases of Rlk and Itk, expression is largely restricted to T cells, with Itk also expressed in NK cells and Rlk also expressed in mast cells. Northern blot analyses were performed on RNA samples isolated from various tissues in order to examine the tissue expression of RIBP. A 1.7-kb transcript representing RIBP was expressed in the thymus, lymph node, and to a lesser extent, in the spleen and bone marrow (Fig. 3 A, left panel, lane 5; right panel, lanes 1 and 2; and left panel, lanes 4 and 3, respectively). In addition, analysis of multiple hematopoietic cell lines revealed that RIBP was expressed in T cell lines (e.g., BW.P and C57), but not expressed in a B cell lymphoma, A20, or a mastocytoma, P815 (data not shown). Northern blot analyses of thymocyte subpopulations demonstrated that RIBP mRNA was expressed in CD4+CD8+ thymocytes (Fig. 3 A; right panel, lane 5), and in both CD4+ and CD8+ peripheral T cells (Fig. 3 A, right panel, lanes 4 and 3, respectively). Additionally, although RIBP was expressed in TCR− thymocytes (Fig. 3 A, right panel, lane 6), little expression was detected in recombinase-activating gene (RAG)-2–deficient thymus (Fig. 3 A, right panel, lane 7). These data suggest that RIBP expression may be dependent on signal transduction through the pre-TCR, and consequently, that its expression levels vary during the course of T cell development.
Figure 3.
Northern blot analysis of RIBP tissue and T cell subset expression and its induction by T cell activation. (A) Northern blot analysis of RIBP gene expression in various tissues, and in different thymocyte subsets and peripheral T cells. A blot with RNA from various tissues was probed with an oligonucleotide derived from RIBP cDNA (left). Reprobing of the blot with an oligonucleotide specific for the housekeeping gene EF demonstrated equivalent RNA loading (not shown). RIBP expression in lymph node and thymocyte subsets was assessed similarly (right). Ethidium bromide staining of the gel revealed equivalent RNA loading except for lanes 3 and 4 (not shown), which had greater amounts of RNA present. (B) Northern blot analysis of regulation of RIBP gene expression by T cell activation. Top: RIBP expression in A.E7 Th1 T cell clones (left) and PGL10 Th1 T cell clones (right) stimulated with anti-CD3 (0.1 μg/ml for A.E7, and 5.0 μg/ml for PGL10) for the indicated times. Total RNA per sample was assessed by ethidium bromide staining, and demonstrated equivalent loading (not shown). Results for regulation of RIBP expression in T cell clones are representative of a total of four experiments. Bottom: Expression of RIBP in NK cells and Th2 T cell clones.
RIBP mRNA was found to be rapidly induced after T cell activation (Fig. 3 B, top panel). RIBP expression was upregulated in the Th1 T cell clone, A.E7, in as little as 2 h after exposure to anti-CD3 (Fig. 3 B, top left panel). Similarly, RIBP mRNA was induced in another Th1 T cell clone, pGL10, when activated with anti-CD3 (Fig. 3 B, top right panel) or ovalbumin in the presence of appropriate APCs (data not shown). The addition of exogenous IL-2 had no effect on RIBP expression, although it significantly increased [3H]thymidine incorporation (Fig. 3 B, top right panel, and data not shown). In addition to T lymphocytes, NK cells stimulated with IL-2 were observed to express high levels of RIBP mRNA (Fig. 3 B, bottom panel). Furthermore, RIBP was also expressed in the Th2 clones PL104 and PL3, as also shown in Fig. 3 B (bottom panel).
Generation of RIBP-deficient Mice.
RIBP KO mice were generated by disrupting the RIBP gene via homologous recombination. A targeting construct was generated that replaced exons 3 and 4 (encoding the SH2 domain) with a DNA segment encoding a neo gene. The targeting construct, depicted in Fig. 4 A, was introduced into the R1 ES cell line derived from the 129/Sv mouse strain. Southern blot analyses of drug-resistant ES cells demonstrated successful targeted disruption at the RIBP locus (Fig. 4 B, top). Genomic DNA samples from control and targeted ES cells were digested with EcoRI and XhoI, resolved by gel electrophoresis, and hybridized to a 5′ RIBP cDNA probe. As seen in Fig. 4 B (top left panel), probing of control digested ES cell DNA revealed bands of the predicted size (3.8 kb). In contrast, DNA from targeted ES cell clones revealed bands migrating at 5.5 kb, consistent with the replacement of the third and fourth exons with the neo cassette. Similar results were observed using a 3′ RIBP cDNA probe of control versus targeted ES cell genomic DNA samples digested with Asp718 (an isoschizomer of KpnI) and XhoI (Fig. 4 B, top right panel). The neomycin-resistant ES cells (clone 2) were injected into C57BL/6 blastocysts using standard techniques 29. Chimeric mice were bred to C57BL/6 mice to establish germline transmission, and heterozygous mice were interbred to obtain homozygous KO progeny. Southern blot analyses performed on tail genomic DNA samples from the progeny demonstrated successful targeted disruption at the RIBP locus (Fig. 4 B, bottom panel). Initial studies revealed that homozygous KO mice lacked RIBP mRNA expression based on Northern blot analyses (data not shown). The lack of RIBP expression in the RIBP KO offspring was confirmed by immunoblot analyses using an RIBP-specific polyclonal antiserum. As seen in Fig. 4 C, lysates from wild-type C57BL/6 and heterozygous splenocytes activated with anti-CD3 contained significant amounts of RIBP protein migrating at ∼46 kD. In contrast, no RIBP protein was detected in activated T cells from homozygous RIBP-deficient animals.
Analyses of Thymic and Peripheral T Cell Populations in RIBP-deficient Mice.
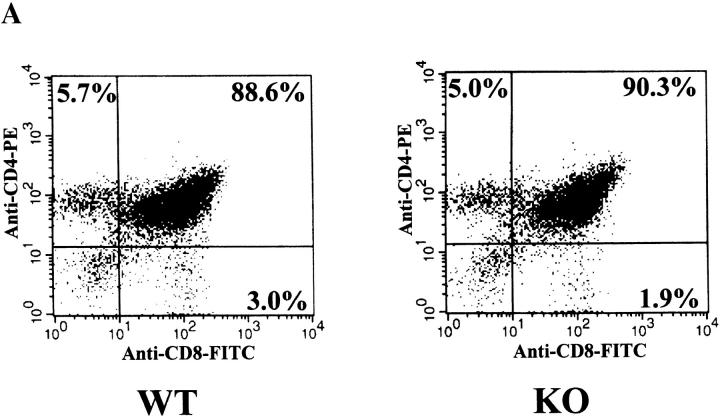
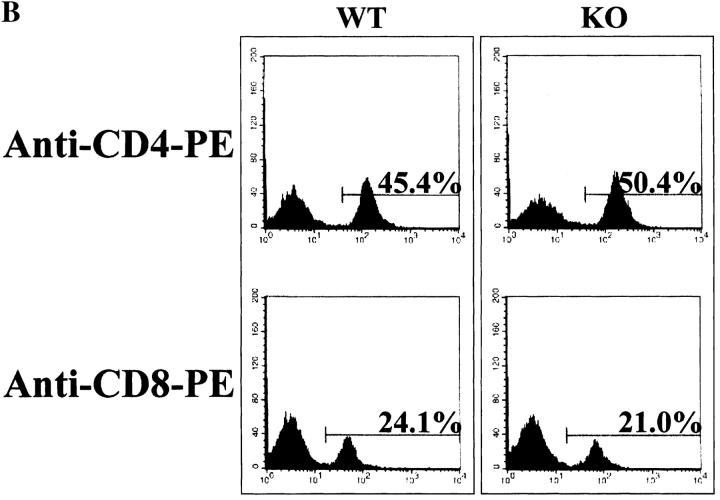
RIBP KO mice did not display any gross developmental abnormalities, and a normal Mendelian distribution of RIBP KO progeny was observed. The percentages of CD4−CD8−, CD4+CD8+, CD4+CD8−, and CD4−CD8+ thymocytes did not differ between RIBP KO and wild-type (Fig. 5 A) or heterozygous (data not shown) control mice, and no significant qualitative or quantitative differences in lymph node and splenic T cell populations were evident (Fig. 5 B, and data not shown, respectively). In addition, the CD4+/CD8+ ratio in both lymph node (Fig. 5 B) and spleen (data not shown) was similar among the various groups. Moreover, no obvious differences in the sizes or cell yields of thymus, spleen, and lymph nodes isolated from RIBP KO mice were apparent compared with wild-type control mice (data not shown). These data suggest that RIBP does not have an essential role in T cell development or homeostasis of peripheral T cell populations.
Figure 5.
Flow cytofluorimetric analysis of T cell development and peripheral T cell subsets in RIBP KO mice. (A) Thymocyte development in RIBP KO mice. Mice used were 7–10 wk of age. Cells were stained with FITC-conjugated anti-CD8 mAb and PE-conjugated anti-CD4 mAb to distinguish double negative, double positive, and single positive populations. Staining of thymocytes pooled from three wild-type mice (WT, left) compared with thymocytes pooled from three RIBP KO mice (right). (B) Lymph node T cell subsets in RIBP KO mice. Lymph node cells from 7–10 wk old wild-type (left) and KO (right) mice were stained with the same Abs as in A, as described in Materials and Methods. Results are representative of four independent experiments.
Proliferative Responses of RIBP-deficient T Cells in Response to TCR Ligation Are Impaired.
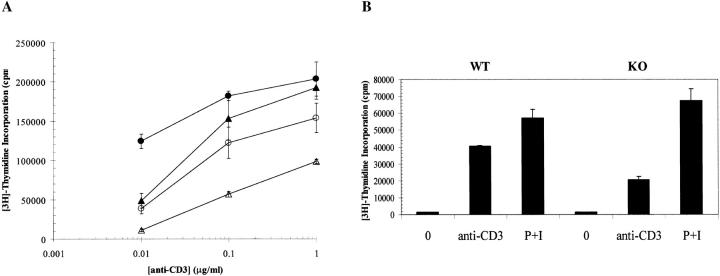
Given the findings that (a) RIBP expression is augmented by T cell activation, and (b) its binding partners Itk and Rlk have been shown to be requisite for T cell responses to activation stimuli 11 39, studies to evaluate the functional status of RIBP KO T cells were undertaken. In response to TCR/CD3-mediated signals delivered by an anti-CD3 mAb, proliferation of RIBP KO T cells was considerably reduced relative to control T cell proliferation (Fig. 6 A). This proliferative impairment was observed across the range of anti-CD3 concentrations tested, and ranged from an 80% reduction in proliferation at the lowest (anti-CD3 concentration) to a 50% reduction in proliferation at the highest (anti-CD3 concentrations). Although RIBP KO T cells displayed an apparent defect in TCR-mediated T cell activation, they were capable of receiving CD28-mediated costimulation (Fig. 6 A). These data indicate that RIBP KO T cells are impaired in their ability to respond to TCR-mediated signals, whereas responsiveness to CD28 signals is less affected.
Figure 6.
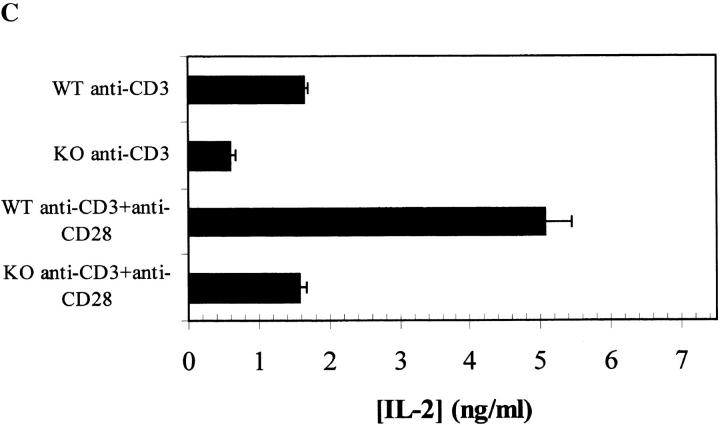
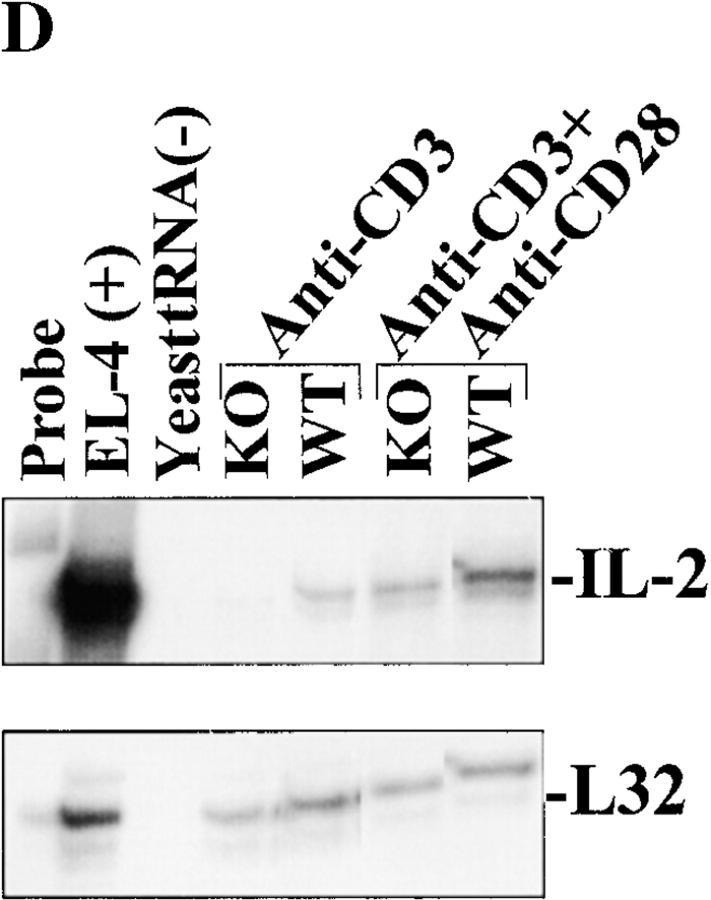
Impairment of RIBP KO T cell proliferation and IL-2 production in response to TCR-triggered activation. (A) Proliferation of T cells from RIBP heterozygous and RIBP KO mice as a function of TCR stimulus (anti-CD3). Splenocytes from heterozygous (filled symbols) or KO (open symbols) mice were stimulated with the indicated concentrations of soluble anti-CD3 alone (triangles), with or without soluble anti-CD28 added at 1.0 μg/ml (circles). Proliferation was assessed at 48 h, with a [3H]thymidine pulse added after 40 h, for an 8-h pulse duration. Results are representative of four independent experiments performed using splenocytes, and three independent experiments using purified T cells and irradiated, T cell–depleted APCs. (B) Proliferation of T cells from wild-type (WT) and RIBP KO mice in response to either anti-CD3 or PMA plus ionomycin (P+I). Proliferation was assessed as above. (C) Splenocytes from wild-type and RIBP KO mice were stimulated with 1 μg/ml of soluble anti-CD3, with or without 1 μg/ml of soluble anti-CD28. Supernatants were collected at either 6, 24, or 48 h, and the concentration of IL-2 was determined by ELISA; values listed above were obtained at 24 h. Results are representative of 11 independent experiments. (D) Splenocytes from RIBP KO or wild-type mice were activated for the indicated times with anti-CD3 with or without anti-CD28. Ab concentrations were 1 μg/ml for both mAbs. RNA prepared from cells was subsequently subjected to RNase protection assays, as described in Materials and Methods. 32P autoradiogram of RNase protection assay polyacrylamide gel. Top: IL-2 mRNA levels in wild-type and RIBP KO T cells activated for the indicated times with the stimuli listed. EL4 mRNA is a positive control for cytokine gene expression, and yeast tRNA is a negative control. Bottom: mRNA levels for the L32 housekeeping gene. Results are representative of three independent experiments.
To determine whether the RIBP KO T cells were defective in either proximal or distal components of the TCR signaling pathway, RIBP KO T cells were treated with either anti-CD3 or PMA and ionomycin, and proliferation was compared with that of wild-type T cells activated with the same stimuli. As shown in Fig. 6 B, RIBP KO T cell proliferation in response to anti-CD3 was 50% reduced compared with wild-type T cells. However, wild-type and RIBP KO T cells stimulated with PMA and ionomycin proliferated equivalently, suggesting that the inadequacy in TCR-mediated signaling in RIBP KO T cells lies upstream of pathways triggered by protein kinase C activation or intracellular Ca2+ flux. These findings are consistent with those reported for T cells deficient in either Itk 11 or both Itk and Rlk 39.
Reduced Cytokine Production by Activated RIBP KO T Cells.
RIBP KO splenocytes were stimulated with anti-CD3 and examined for cytokine production. Consistent with the reduced proliferative response (Fig. 6A and Fig. B), a significant reduction in the amount of IL-2 in the culture supernatant was observed (Fig. 6 C). Furthermore, although CD28-mediated costimulation increased IL-2 production by both wild-type and KO T cells, the relative differences in IL-2 levels were maintained. These results were confirmed using an RNase protection assay to determine relative levels of IL-2 transcripts in activated RIBP KO versus wild-type T cells. As shown in Fig. 6 D, IL-2 mRNA was detected at 6 h in (C57BL/6 × 129) F2 wild-type T cells stimulated with anti-CD3. In contrast, little IL-2 mRNA was apparent in activated RIBP KO T cells at this time point (Fig. 6 D, lane 5 versus lane 4). Densitometry analyses revealed that IL-2 mRNA levels in RIBP KO T cells were reduced by ∼80% relative to wild-type cells (data not shown). Also, consistent with the reduction in supernatant levels of IL-2 (Fig. 6 C), the relative amounts of IL-2 mRNA from RIBP KO T cells activated with anti-CD3 and anti-CD28 were still significantly reduced compared with control T cells (Fig. 6 D, lane 7 versus lane 6).
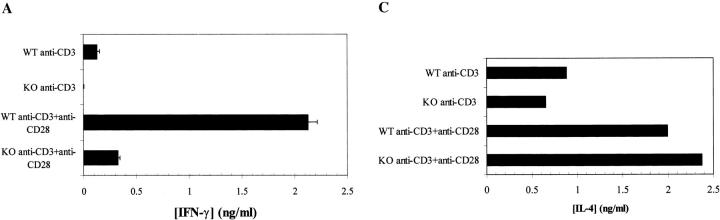
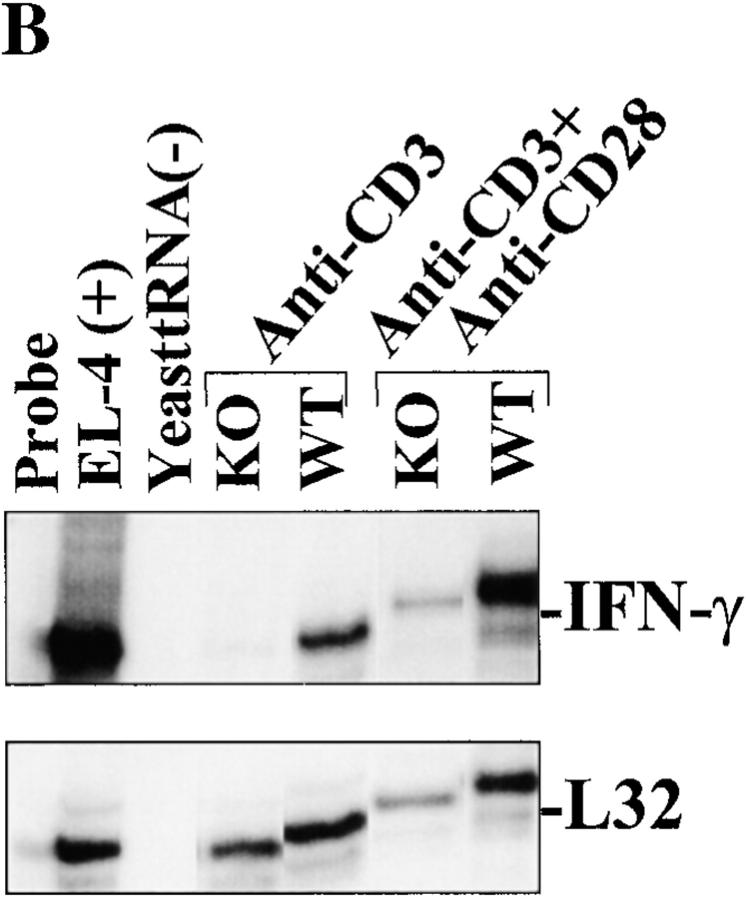
In addition to a reduction in IL-2 levels, a pronounced decrease in IFN-γ production was observed in RIBP KO T cells (Fig. 7 A). As was the case for IL-2, this reduction in IFN-γ production correlated with decreased IFN-γ mRNA levels (Fig. 7 B), suggesting that RIBP affects lymphokine production at the level of transcription and/or mRNA stabilization. Little IFN-γ mRNA was observed in anti-CD3–activated RIBP KO T cells relative to wild-type T cells; densitometry revealed that this was a >90% reduction (Fig. 7 B; densitometry data not shown). However, anti-CD28 costimulation did restore the IFN-γ decrease observed in RIBP KO T cells, suggesting that this defect may be an indirect consequence of the reduced activation and IL-2 production. Finally, despite the reductions in IL-2 and IFN-γ production by RIBP KO T cells, no significant reduction in IL-4 production was observed (Fig. 7 C). In fact, in some experiments, activated RIBP KO splenocytes produced more IL-4 than did control splenocytes. Thus, the lack of RIBP appears to more profoundly affect the ability of T cells to produce Th1-type cytokines than Th2-type cytokines.
Figure 7.
Demonstration of defective IFN-γ induction and normal IL-4 induction by activated RIBP KO T cells. (A) ELISA determination (see Fig. 6 C) of IFN-γ production by RIBP KO T cells. Results are representative of at least three independent experiments. (B) 32P autoradiogram of RNase protection assay polyacrylamide gel, as described in the legend to Fig. 6 D. Stimulation conditions are also as described for Fig. 6 D. Top: IFN-γ mRNA levels in wild-type (WT) and RIBP KO T cells activated for the indicated times with the stimuli listed. EL4 mRNA is a positive control for cytokine gene expression, and yeast tRNA is a negative control. Bottom: mRNA levels for the L32 housekeeping gene. Results are representative of three independent experiments. (C) IL-4 production by activated control and RIBP KO T cells. Supernatants (see Fig. 6 C and 7 A) were collected at 48 h and assessed for IL-4 by ELISA. Results are representative of at least three independent experiments.
Discussion
In this study, a yeast two-hybrid T cell cDNA library screen was used to search for proteins that could associate with the T cell–specific members of the Tec family of tyrosine kinases, Rlk and Itk. A novel adaptor molecule, termed RIBP, was identified that interacts with a site in Rlk and Itk regulated by tyrosine phosphorylation. RIBP is expressed in thymocytes, and at low levels in resting peripheral T cells. Expression of RIBP is upregulated after T cell activation, coinciding with the expression levels of one of its binding partners, Itk, and high expression levels are observed in activated NK cells. The restriction of RIBP expression to T cells and NK cells mirrors the expression patterns of several signaling molecules, including Itk 9, linker for activation of T cells (LAT 40), which is also expressed in mast cells, and ZAP-70 41. These results are consistent with the proposed common lineage of these cells 42. Finally, genetic disruption of RIBP was found to significantly impair TCR/CD3-mediated proliferation, and IL-2 and IFN-γ production, although IL-4 production was unaffected. Furthermore, the TCR-mediated signal transduction defects observed in RIBP KO T cells were bypassed using the downstream mitogens, PMA and ionomycin. These results suggest that RIBP positively regulates TCR signaling by acting as a molecular link between Itk/Rlk and other components of the TCR signaling pathway. The consequences of this interaction include the regulation of TCR-mediated signal transduction, and consequently, lymphokine production.
RIBP is a complex adaptor protein. The gene encodes a PRR that may bind SH3 domain–containing proteins such as other Src family kinases and other adaptor proteins. In addition, a tyrosine-phosphorylated NPIY sequence present in RIBP may bind PTB domain–containing proteins such as Shc. Proteins such as Shc have been implicated in TCR signal transduction via interactions with ZAP-70 43 and Cbl, an adaptor which is thought to negatively regulate ZAP-70 activation 44 45 and binds to the SH3 domain of Itk. The tyrosine-based protein binding motifs present in RIBP (YXXP and YXXV) may bind the SH2 domains of rasGAP, Abl, Crk, and SHP-2, molecules previously shown to regulate T cell signal transduction. Finally, RIBP contains an SH2 domain that may be involved in binding to other phosphotyrosine-containing proteins. In this regard, preliminary results indicate that the SH2 domain of RIBP is required for interaction with Rlk, whereas the PRR and tyrosine-based protein binding motifs are unnecessary (our unpublished observations).
The molecular mechanisms that link RIBP, Rlk, and Itk, to the proximal event of TCR ligation are unclear. The finding that Lck coexpression augments the association of RIBP with Itk is consistent with a role for RIBP in the regulation of TCR signal transduction. Tyrosine phosphorylation of Itk, catalyzed by Lck or Fyn, is required for optimal Itk kinase activity 46 47, and consequently, T cell activation. Phosphorylation of Itk occurs at Y511, a residue in the activation loop of the kinase domain of Itk 48. Thus, RIBP could be functionally coupled to TCR engagement, as its binding to Itk is enhanced by the activity of this CD4/CD8 coreceptor–associated Src family kinase in vitro.
RIBP may bind to both Itk and Rlk, raising the question as to its physiological partner and the relationship of its binding specificity to the functional deficit evident in RIBP KO T cells. The expression pattern of RIBP suggests that its physiological binding partner may be Itk in the periphery. Previous studies have shown that Rlk is highly expressed in resting T cells, but downregulated after T cell activation. In contrast, Itk gene expression is upregulated after T cell activation, similar to RIBP. Furthermore, the phenotype observed in RIBP KO T cells more closely resembles the phenotype of Itk-deficient T cells, which display diminished proliferation and IL-2 production in response to TCR ligation. In contrast, Rlk KO T cells have only minor deficits in TCR-mediated T cell activation 39. However, there are circumstances when the binding of RIBP to Rlk may be physiologically relevant. For instance, both RIBP and Rlk are expressed in thymocytes. Although we have not identified any profound changes in T cell development in the RIBP KO or Rlk KO mice (our unpublished observations, and reference 39, respectively), there may be subtle, as yet unidentified effects of this interaction in the thymus. Moreover, both Rlk and RIBP are expressed in NK cells, and may play a role in this lymphocyte subset. In fact, recent evidence indicates that Itk and Rlk have overlapping functions in T cell activation, as mice deficient in both kinases have more severely diminished peripheral T cell function than mice deficient in either kinase alone 39. Thus, further biochemical and breeding studies to generate RIBP/Itk and RIBP/Rlk double-KO mice will be needed in order to determine the differential effects of RIBP on the functions of Itk versus Rlk. It is important to emphasize that the functional impairment present in RIBP KO T cells was not as severe as that observed in Rlk/Itk double-KO T cells. There are two possible reasons for these findings. First, there may be functional homologues of RIBP that can partially compensate for the absence of RIBP in KO T cells. Second, not all of the intracellular effects on TCR signaling that Rlk and Itk mediate may require RIBP or any putative homologues.
The fact that CD28-mediated costimulation partially compensates for the severely diminished proliferative response to TCR/CD3-mediated T cell activation in RIBP KO mice (Fig. 6 A) suggests that the RIBP KO T cells are capable of responding to the CD28-mediated costimulatory signal. Interestingly, Itk has been reported to be both a positive and negative regulator of CD28-mediated T cell activation. On the one hand, Itk is physically associated with CD28 after T cell activation 49, and Jurkat T cells expressing CD28 cytoplasmic tail mutants incapable of recruiting Itk are suppressed in their ability to produce IL-2. However, Littman and colleagues demonstrated that T cells from Itk-deficient mice were hyperresponsive to CD28 costimulation 15. Therefore, it remains to be determined in what context, if any, RIBP is involved in CD28 signal transduction.
Finally, the results of this study support a role for RIBP in the regulation of Th cell subset differentiation. IL-2 and IFN-γ production were both significantly reduced in activated RIBP KO T cells (Fig. 6 and Fig. 7), whereas IL-4 production was unaffected (Fig. 7 C). Such a selective impairment in IFN-γ and IL-2 production suggests that RIBP may act in signaling pathways that promote Th1 differentiation. The lack of RIBP may reduce TCR-mediated signal strength via impairment of Itk and/or Rlk function, resulting in a selective loss of Th1 differentiation, as suggested by Bottomly and colleagues 50. This may occur by generating decreased TCR signals that drive Th2 skewing. In fact, both Itk and Rlk have been implicated in T cell differentiation (Schaeffer, E., personal communication). Also, Rlk is selectively expressed in Th1-type T cells. These results may explain, at least in part, the finding that RIBP selectively affects Th1-type cytokine production.
In conclusion, we have identified and functionally characterized a novel T cell adaptor molecule involved in the regulation of responses to T cell activation stimuli, specifically proliferation and lymphokine production. This adaptor, RIBP, appears to function in TCR/CD3-mediated signal transduction, consistent with previously reported data regarding its binding partners, the Tec tyrosine kinases Rlk and Itk. The binding of RIBP to Itk and Rlk may provide important biochemical links of these two important kinases with other components in the T cell activation machinery. Further molecular studies will be needed to determine the specific sites of action of RIBP within various T cell signal transduction pathways.
Acknowledgments
We thank Colin Duckett for assistance provided in yeast two-hybrid techniques, Roli Khattri and Kyung-Mi Lee for technical expertise provided in biochemistry experiments, Edward Schaeffer and Pamela Schwartzberg for information on Rlk KO and Rlk/Itk KO mice, Dan Littman for Itk constructs and anti-Itk antisera, and John Ortaldo for NK cells. We also thank Sean O'Herrin, Ingrid Rulifson, Benoit Salomon, Qizhi Tang, and Patrick Fields for valuable discussions.
K. Rajagopal is supported by the Medical Scientist Training Program. This work was funded by a National Institutes of Health Program Project Grant to J.A. Bluestone (PO1 AI35294-6).
Footnotes
Abbreviations used in this paper: ES, embryonic stem; KO, knockout; PRR, proline-rich region; PTB, phosphotyrosine-binding; PTK, protein tyrosine kinase; RAG, recombinase-activating gene; RIBP, Rlk/Itk-binding protein; SH, Src homology; TBST, Tris-buffered saline/Tween.
References
- Chan A.C., Shaw A.S. Regulation of antigen receptor signal transduction by protein tyrosine kinases. Curr. Opin. Immunol. 1996;8:394–401. doi: 10.1016/s0952-7915(96)80130-0. [DOI] [PubMed] [Google Scholar]
- Bolen J.B. Protein tyrosine kinases in the initiation of antigen receptor signaling. Curr. Opin. Immunol. 1995;7:306–311. doi: 10.1016/0952-7915(95)80103-0. [DOI] [PubMed] [Google Scholar]
- DeFranco A.L. Transmembrane signaling by antigen receptors of B and T lymphocytes. Curr. Opin. Cell Biol. 1995;7:163–175. doi: 10.1016/0955-0674(95)80024-7. [DOI] [PubMed] [Google Scholar]
- Weiss A., Littman D.R. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- Neet K., Hunter T. Vertebrate non-receptor protein-tyrosine kinase families. Genes Cells. 1996;1:147–169. doi: 10.1046/j.1365-2443.1996.d01-234.x. [DOI] [PubMed] [Google Scholar]
- Rawlings D.J., Witte O.N. The Btk subfamily of cytoplasmic tyrosine kinasesstructure, regulation and function. Semin. Immunol. 1995;7:237–246. doi: 10.1006/smim.1995.0028. [DOI] [PubMed] [Google Scholar]
- Andreotti A.H., Bunnell S.C., Feng S., Berg L.J., Schreiber S.L. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- Haire R.N., Ohta Y., Lewis J.E., Fu S.M., Kroisel P., Litman G.W. TXK, a novel human tyrosine kinase expressed in T cells shares sequence identity with Tec family kinases and maps to 4p12. Hum. Mol. Genet. 1994;3:897–901. doi: 10.1093/hmg/3.6.897. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Asao H., Ohtani K., Nakamura M., Sugamura K. A novel human tyrosine kinase gene inducible in T cells by interleukin 2. FEBS Lett. 1993;324:1–5. doi: 10.1016/0014-5793(93)81520-a. [DOI] [PubMed] [Google Scholar]
- Siliciano J.D., Morrow T.A., Desiderio S.V. itk, a T-cell-specific tyrosine kinase gene inducible by interleukin 2. Proc. Natl. Acad. Sci. USA. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.C., Littman D.R. Altered T cell receptor signaling and disrupted T cell development in mice lacking Itk. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- Liu K.Q., Bunnell S.C., Gurniak C.B., Berg L.J. T cell receptor–initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J. Exp. Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S., Truitt K., Lu Y., Lapushin R., Khan H., Imboden J.B., Mills G.B. Efficient CD28 signalling leads to increases in the kinase activities of the TEC family tyrosine kinase EMT/ITK/TSK and the SRC family tyrosine kinase LCK. Biochem. J. 1998;330:1123–1128. doi: 10.1042/bj3301123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Abe H., Yagita H., Okumura K., Nakamura M., Sugamura K. Itk, a T cell-specific tyrosine kinase, is required for CD2-mediated interleukin-2 promoter activation in the human T cell line Jurkat. Eur. J. Immunol. 1997;27:834–841. [PubMed] [Google Scholar]
- Liao X.C., Fournier S., Killeen N., Weiss A., Allison J.P., Littman D.R. Itk negatively regulates induction of T cell proliferation by CD28 costimulation. J. Exp. Med. 1997;186:221–228. doi: 10.1084/jem.186.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano H., Mano K., Tang B., Koehler M., Yi T., Gilbert D.J., Jenkins N.A., Copeland N.G., Ihle J.N. Expression of a novel form of Tec kinase in hematopoietic cells and mapping of the gene to chromosome 5 near Kit. Oncogene. 1993;8:417–424. [PubMed] [Google Scholar]
- Mano H., Ishikawa F., Nishida J., Hirai H., Takaku F. A novel protein-tyrosine kinase, tec, is preferentially expressed in liver. Oncogene. 1990;5:1781–1786. [PubMed] [Google Scholar]
- Yang W.C., Ghiotto M., Barbarat B., Olive D. The role of tec protein-tyrosine kinase in T cell signaling. J. Biol. Chem. 1999;274:607–617. doi: 10.1074/jbc.274.2.607. [DOI] [PubMed] [Google Scholar]
- Mano H., Yamashita Y., Sato K., Yazaki Y., Hirai H. Tec protein-tyrosine kinase is involved in interleukin-3 signaling pathway. Blood. 1995;85:343–350. [PubMed] [Google Scholar]
- Takahashi-Tezuka M., Hibi M., Fujitani Y., Fukada T., Yamaguchi T., Hirano T. Tec tyrosine kinase links the cytokine receptors to PI-3 kinase probably through JAK. Oncogene. 1997;14:2273–2282. doi: 10.1038/sj.onc.1201071. [DOI] [PubMed] [Google Scholar]
- Yamashita Y., Watanabe S., Miyazato A., Ohya K., Ikeda U., Shimada K., Komatsu N., Hatake K., Miura Y., Ozawa K., Mano H. Tec and Jak2 kinases cooperate to mediate cytokine-driven activation of c-fos transcription. Blood. 1998;91:1496–1507. [PubMed] [Google Scholar]
- Hu Q., Davidson D., Schwartzberg P.L., Macchiarini F., Lenardo M.J., Bluestone J.A., Matis L.A. Identification of Rlk, a novel protein tyrosine kinase with predominant expression in the T cell lineage. J. Biol. Chem. 1995;270:1928–1934. doi: 10.1074/jbc.270.4.1928. [DOI] [PubMed] [Google Scholar]
- Sommers C.L., Huang K., Shores E.W., Grinberg A., Charlick D.A., Kozak C.A., Love P.E. Murine txka protein tyrosine kinase gene regulated by T cell activation. Oncogene. 1995;11:245–251. [PubMed] [Google Scholar]
- Schneider H., Schwartzberg P.L., Rudd C.E. Resting lymphocyte kinase (Rlk/Txk) phosphorylates the YVKM motif and regulates PI 3-kinase binding to T-cell antigen CTLA-4. Biochem. Biophys. Res. Commun. 1998;252:14–19. doi: 10.1006/bbrc.1998.9559. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Adamson M.C., Silver J., Kozak C.A. The mouse homolog of the gibbon ape leukemia virus receptorgenetic mapping and a possible receptor function in rodents. Virology. 1991;183:778–781. doi: 10.1016/0042-6822(91)91010-e. [DOI] [PubMed] [Google Scholar]
- Kozak C.A., Peyser M., Krall M., Mariano T.M., Kumar C.S., Pestka S., Mock B.A. Molecular genetic markers spanning mouse chromosome 10. Genomics. 1990;8:519–524. doi: 10.1016/0888-7543(90)90039-w. [DOI] [PubMed] [Google Scholar]
- Green E.L. Genetics and Probability in Animal Breeding Experiments. Oxford University Press; New York: 1981. [Google Scholar]
- Love P.E., Shores E.W., Johnson M.D., Tremblay M.L., Lee E.J., Grinberg A., Huang S.P., Singer A., Westphal H. T cell development in mice that lack the zeta chain of the T cell antigen receptor complex. Science. 1993;261:918–921. doi: 10.1126/science.7688481. [DOI] [PubMed] [Google Scholar]
- Zhou S., Margolis B., Chaudhuri M., Shoelson S.E., Cantley L.C. The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J. Biol. Chem. 1995;270:14863–14866. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Shoelson S.E., Chaudhuri M., Gish G., Pawson T., Haser W.G., King F., Roberts T., Ratnofsky S., Lechleider R.J. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- Fuhrer D.K., Feng G.S., Yang Y.C. Syp associates with gp130 and Janus kinase 2 in response to interleukin-11 in 3T3-L1 mouse preadipocytes. J. Biol. Chem. 1995;270:24826–24830. doi: 10.1074/jbc.270.42.24826. [DOI] [PubMed] [Google Scholar]
- Spurkland A., Brinchmann J.E., Markussen G., Pedeutour F., Munthe E., Lea T., Vartdal F., Aasheim H.C. Molecular cloning of a T cell-specific adapter protein (TSAd) containing an Src homology (SH) 2 domain and putative SH3 and phosphotyrosine binding sites. J. Biol. Chem. 1998;273:4539–4546. doi: 10.1074/jbc.273.8.4539. [DOI] [PubMed] [Google Scholar]
- Prins J.B., Meisler M.H., Seldin M.F. Mouse chromosome 3. Mamm. Genome. 1994;5:S40–S50. [PubMed] [Google Scholar]
- Moseley W.S., Seldin M.F. Definition of mouse chromosome 1 and 3 gene linkage groups that are conserved on human chromosome 1evidence that a conserved linkage group spans the centromere of human chromosome 1. Genomics. 1989;5:899–905. doi: 10.1016/0888-7543(89)90132-8. [DOI] [PubMed] [Google Scholar]
- Ginns E.I., Choudary P.V., Tsuji S., Martin B., Stubblefield B., Sawyer J., Hozier J., Barranger J.A. Gene mapping and leader polypeptide sequence of human glucocerebrosidaseimplications for Gaucher disease. Proc. Natl. Acad. Sci. USA. 1985;82:7101–7105. doi: 10.1073/pnas.82.20.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydner K.L., McNeil J.A., Lin F., Worman H.J., Lawrence J.B. Chromosomal assignment of human nuclear envelope protein genes LMNA, LMNB1, and LBR by fluorescence in situ hybridization. Genomics. 1996;32:474–478. doi: 10.1006/geno.1996.0146. [DOI] [PubMed] [Google Scholar]
- Huebner K., Kastury K., Druck T., Salcini A.E., Lanfrancone L., Pelicci G., Lowenstein E., Li W., Park S.H., Cannizzaro L. Chromosome locations of genes encoding human signal transduction adapter proteins, Nck (NCK), Shc (SHC1), and Grb2 (GRB2) Genomics. 1994;22:281–287. doi: 10.1006/geno.1994.1385. [DOI] [PubMed] [Google Scholar]
- Schaeffer E.M., Debnath J., Yap G., McVicar D., Liao X.C., Littman D.R., Sher A., Varmus H.E., Lenardo M.J., Schwartzberg P.L. Requirement for Tec kinases Rlk and Itk in T cell receptor signaling and immunity. Science. 1999;284:638–641. doi: 10.1126/science.284.5414.638. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sloan-Lancaster J., Kitchen J., Trible R.P., Samelson L.E. LATthe ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- Chan A.C., Iwashima M., Turck C.W., Weiss A. ZAP-70a 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell. 1992;71:649–662. doi: 10.1016/0092-8674(92)90598-7. [DOI] [PubMed] [Google Scholar]
- Spits H., Blom B., Jaleco A.C., Weijer K., Verschuren M.C., van Dongen J.J., Heemskerk M.H., Res P.C. Early stages in the development of human T, natural killer and thymic dendritic cells. Immunol. Rev. 1998;165:75–86. doi: 10.1111/j.1600-065x.1998.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Milia E., Di Somma M.M., Baldoni F., Chiari R., Lanfrancone L., Pelicci P.G., Telford J.L., Baldari C.T. The aminoterminal phosphotyrosine binding domain of Shc associates with ZAP-70 and mediates TCR dependent gene activation. Oncogene. 1996;13:767–775. [PubMed] [Google Scholar]
- Murphy M.A., Schnall R.G., Venter D.J., Barnett L., Bertoncello I., Thien C.B., Langdon W.Y., Bowtell D.D. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell. Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramura M., Kole H.K., Hu R.J., Gu H. Altered thymic positive selection and intracellular signals in cbl-deficient mice. Proc. Natl. Acad. Sci. USA. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- August A., Sadra A., Dupont B., Hanafusa H. Src-induced activation of inducible T cell kinase (ITK) requires phosphatidylinositol 3-kinase activity and the Pleckstrin homology domain of inducible T cell kinase. Proc. Natl. Acad. Sci. USA. 1997;94:11227–11232. doi: 10.1073/pnas.94.21.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S., August A., Kawakami Y., Kawakami T., Dupont B., Mills G.B. The EMT/ITK/TSK (EMT) tyrosine kinase is activated during TCR signalingLCK is required for optimal activation of EMT. J. Immunol. 1996;156:2716–2722. [PubMed] [Google Scholar]
- Heyeck S.D., Wilcox H.M., Bunnell S.C., Berg L.J. Lck phosphorylates the activation loop tyrosine of the Itk kinase domain and activates Itk kinase activity. J. Biol. Chem. 1997;272:25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- August A., Gibson S., Kawakami Y., Kawakami T., Mills G.B., Dupont B. CD28 is associated with and induces the immediate tyrosine phosphorylation and activation of the Tec family kinase ITK/EMT in the human Jurkat leukemic T-cell line. Proc. Natl. Acad. Sci. USA. 1994;91:9347–9351. doi: 10.1073/pnas.91.20.9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant S., Pfeiffer C., Woodard A., Pasqualini T., Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J. Exp. Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]