Abstract
We have investigated in vivo roles of CCAAT/enhancer binding protein γ (C/EBPγ) by gene targeting. C/EBPγ-deficient (C/EBPγ2/−) mice showed a high mortality rate within 48 h after birth. To analyze the roles of C/EBPγ in lymphoid lineage cells, bone marrow chimeras were established. C/EBPγ2/− chimeras showed normal T and B cell development. However, cytolytic functions of their splenic natural killer (NK) cells after stimulation with cytokines such as interleukin (IL)-12, IL-18, and IL-2 were significantly reduced as compared with those of control chimera NK cells. In addition, the ability of C/EBPγ−/− chimera splenocytes to produce interferon (IFN)-γ in response to IL-12 and/or IL-18 was markedly impaired. NK cells could be generated in vitro with normal surface marker expression in the presence of IL-15 from C/EBPγ2/− newborn spleen cells. However, they also showed lower cytotoxic activity and IFN-γ production when stimulated with IL-12 plus IL-18 than control NK cells, as observed in C/EBPγ2/− chimera splenocytes. In conclusion, our study reveals that C/EBPγ is a critical transcription factor involved in the functional maturation of NK cells.
Keywords: gene targeting, natural killer cells, C/EBPγ, interleukin 15, interferon γ
The CCAAT/enhancer binding proteins (C/EBPs) belong to a family of basic leucine zipper transcription factors 1 2. So far six members are isolated and share well conserved COOH-terminal regions, which contain a basic region for DNA binding and a leucine zipper motif for dimerization. Gene targeting revealed that each member plays a critical role in cellular proliferation and differentiation in various tissues. For example, nuclear factor (NF)-IL6, also designated C/EBPβ, is essential for macrophage function 3 4 and involved in adipocyte differentiation, working coordinately with C/EBPδ 5. C/EBPα is essential for hepatocyte function and adipocyte differentiation 6 7. Furthermore, C/EBP∈, expressed specifically in myeloid lineage cells, plays a critical role in granulopoiesis 8.
C/EBPγ is a member of the C/EBP family. C/EBPγ can bind to the μEBP-E binding sites (E sites) in both the IgH enhancer and the Vh1 promotor and was originally identified as an Ig enhancer binding protein, Ig/EBP-1 9 10. C/EBPγ can also bind to the regulatory elements for other genes such as G-CSF and IL-4 11 12. Although other members are expressed in a relatively tissue-specific or inducible manner, expression of C/EBPγ is ubiquitous and constitutive 9. In addition, C/EBPγ does not contain transcription activating domains. It can nevertheless interact with other transcription factors through the leucine zipper domain and function as a dominant negative form 10. In some cases, C/EBPγ can augment the DNA binding ability of other transcription factors 12. Thus, C/EBPγ is considered to be a regulator for transcription factors with the leucine zipper domain. However, the biological roles of C/EBPγ remain unclear.
To clarify in vivo functions of C/EBPγ, we have generated C/EBPγ-deficient (C/EBPγ2/−) mice by gene targeting. C/EBPγ2/− mice showed a high mortality rate within 48 h after birth, indicating that neonatal survival requires C/EBPγ. By establishing bone marrow chimeras, we found that C/EBPγ is critical for NK cell functions. NK cells are large granular lymphocytes derived from the bone marrow and essential for innate immunity 13 14. NK cells can mediate cytotoxicity against tumor cells and virus-infected cells and produce immunomodulatory cytokines such as IFN-γ. These activities were impaired in C/EBPγ2/− NK cells, establishing that C/EBPγ is a novel regulator for functional NK cell maturation.
Materials and Methods
Generation of C/EBPγ2/− Mice.
A BALB/c-derived genomic fragment including the C/EBPγ gene was provided by Dr. S. Nagata (Osaka University, Osaka, Japan). As the long arm of the homology region, an 8.8-kb genomic fragment including the first exon, the first intron, and part of the second exon was used. The neomycin resistance gene derived from pMC1Neo-poly(A) 15 was inserted into the targeting vector to disrupt the basic DNA binding and leucine zipper regions. A short arm of the homology region was amplified by PCR. The MC1 herpes simplex virus thymidine kinase 16 was inserted 5′ upstream of the homologous region.
E14-1 embryonic stem cells, which were derived from 129/SvJ (129) mice, were transfected with a linearized targeting vector by electroporation. Homologous recombinants were identified among double-resistant clones against G418 and gancyclovir by PCR and Southern blot analysis. Generation of chimeras and mutant mice was essentially as described 17.
Reverse Transcriptase–PCR for C/EBPγ.
Splenic B and T cells were purified from B6 spleen cells using Magnetic Cell Sorter (MACS®; Miltenyi Biotec) with B220 and CD3 microbeads, respectively. For purification of NK cells, DX5+ cells were first prepared from B6 spleen cells using MACS® with biotinylated DX5 and streptavidin microbeads. Then the cells were stained with anti-CD3 and anti-DX5. CD3−DX5+ cells were sorted with EPICS Elite (Coulter Immunology) and used as NK cells for reverse transcriptase (RT)-PCR. Total RNAs were extracted from tissues or cells using Sepazol-RNA I (Nacalai Tesque). Oligo-dT–primed reverse transcription was performed with Superscript-RT (GIBCO BRL). Then cDNAs were subjected to PCR analysis. Primers for C/EBPγ were as follows: sense primer, 5′-GGCCGCTCGGAGTGGAGGCCGTCTGGG-3′; antisense primer, 5′-ACGTTGTCTGCGAGGCTGTGCGCATGC-3′. The length of the amplified product was 547 bp. These primers cannot amplify the genomic DNA because sense and antisense primers are located in the first and second exons, respectively. Cycling conditions were 35 cycles of 94°C for 30 s, 65°C for 30 s, and 75°C for 1 min. Primers for β-actin were as follows: sense primer, 5′-CCCACACTGTGCCCATCTAC-3′; antisense primer, 5′-TACGGATGTCAACGTCACAC-3′. Cycling conditions were 24 cycles of 94°C for 15 s and 60°C for 30 s.
Generation of Radiation Chimeras.
The C/EBPγ genotype of neonates obtained by intercrossing heterozygous mutant mice was determined by PCR. The splenocytes were taken from the neonates within 12–18 h after birth and used as sources for hematopoietic stem cells. They were injected intravenously into recombination activating gene (RAG)2−/− B6 mice 18 that had received 12 Gy from an X-ray irradiation system, MBR-1520R (Hitachi Medical Corp.), before transfer. The recipient mice were given 1 mg/ml neomycin sulfate and 1,000 U/ml polymixin B in their drinking water after irradiation and analyzed 6–10 wk after reconstitution. To evaluate the chimerism in NK cells, we used surface marker Ly9.1, the allele of which is carried by 129 but not B6 mice, as used by Ogasawara et al. 19. The genetic background of the donors in our experiments was a mixture of 129 and B6. Therefore, chimeras, of which the donor was Ly9.1+, were used for our experiments. In all reconstituted mice used, >99% of CD3−DX5+ cells were positive for Ly9.1 expression (data not shown).
Flow Cytometric Analysis.
Single-cell suspensions from thymus, spleen, or cultured cells were incubated with anti-CD16/32 (PharMingen) to minimize nonspecific staining. They were then stained with cocktails of mAbs conjugated to FITC, PE, biotin, or Cy-Chrome for 20 min at 4°C. The biotinylated Abs were developed with streptavidin conjugated to PE or Cy-Chrome (PharMingen). All mAbs, with the exception of PE-labeled anti-IgD (Southern Biotechnology Associates), were purchased from PharMingen. Flow cytometric analysis was performed using a FACSCalibur™ with CELLQuest™ software (Becton Dickinson).
Proliferation Assay.
Fresh splenocytes (105 cells per well) were cultured in complete RPMI 1640 (RPMI 1640 medium supplemented with 10% FCS, 2-ME, penicillin, and streptomycin) with 10 μg/ml anti-IgM (Zymed Labs.) plus 0.5 μg/ml anti-CD40 (PharMingen), 10 μg/ml LPS (055:B5; Sigma Chemical Co.), 20 ng/ml IL-2 (Genzyme Corp.), 0.1 μg/ml anti-CD3 (PharMingen) plus 0.1 μg/ml anti-CD28 (PharMingen), and 2.5 μg/ml Con A (Sigma Chemical. Co.) in 96-well plates. 48 h later, they were pulsed with 0.2 μCi of [3H]thymidine (NEN Research Products) and cultured for a further 15 h.
Analysis of NK Cell Activity.
Splenocytes from bone marrow chimeras were incubated with 2 ng/ml IL-12, 20 ng/ml IL-18, and 2 ng/ml IL-12 plus 20 ng/ml IL-18 or 500 U/ml IL-2 for 24 h, and their cytotoxic activities against YAC-1 cells were measured as described previously 20 21. Spontaneous cytotoxic activity was measured by incubating splenocytes with 51Cr-labeled YAC-1 cells in the absence of cytokines for 4 h.
ELISA and RT-PCR for IFN-γ.
Splenic cells at 105 cells per well in 96-well plates were cultured in complete RPMI 1640 in the presence or absence of 2 ng/ml IL-12 and/or 20 ng/ml IL-18 for 24 h. Amounts of IFN-γ in harvested supernatants were measured by ELISA using Duoset (Genzyme Corp.) according to the manufacturer's instructions. The lowest detection limit of ELISA is 10 pg/ml. Semiquantitative RT-PCR for IFN-γ was performed as described previously 22.
In Vitro Culture of Newborn Spleen Cells.
Newborn spleen cells were cultured in complete RPMI 1640 with 300 ng/ml IL-15 (Genzyme Corp.) at 106 cells per well in 24-well plates. 10 d later, cells were harvested, washed four times, and used for further analysis. For cytotoxic activities, harvested cells were incubated with labeled YAC-1 cells in the absence of cytokines for 4 h. Their cytotoxic activities against YAC-1 cells were then measured as described previously 20. For IFN-γ measurement, harvested cells were cultured at 105 cells per well in 96-well plates with 2 ng/ml IL-12 and 20 ng/ml IL-18 for a further 24 h. Then, amounts of IFN-γ in the culture supernatants were measured with ELISA.
RT-PCR Analysis of Cultured Spleen Cells.
Total RNAs were purified from 10-d–cultured newborn spleen cells with IL-15, reverse transcribed, and amplified. Primers and amplifying conditions for IL-12Rβ1, IL-12Rβ2, and IL-18R were described previously 21.
Western Blot Analysis of Cultured Spleen Cells.
Newborn spleen cells cultured with IL-15 for 10 d were stimulated with 2 ng/ml IL-12 or 20 ng/ml IL-18 for 20 min and lysed in buffer containing 20 mM Tris/HCl, pH 8.0, 137 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM PMSF, 20 mg/ml aprotinin, 20 mg/ml leupeptin, 1 mM Na3VO4, 1 mM EGTA, 10 mM NaF, 1 mM Na4P2O7, and 10 mM β-glycerophosphate. For signal transducer and activator of transcription (STAT)4 activation, the cell lysates were immunoprecipitated with anti-STAT4 Ab (Santa Cruz Biotechnology) and Protein A–Sepharose (Amersham Pharmacia Biotech). The immunoprecipitates were separated on SDS-PAGE, transferred onto a nitrocellulose membrane, and blotted with antiphosphotyrosine 4G10 (Upstate Biotechnology Inc.) or anti-STAT4 Ab. For c-Jun NH2-terminal kinase (JNK) activation, the cell lysates were separated on SDS-PAGE, followed by blotting with antiphospho-JNK Ab (Promega Corp.) or anti-JNK1 Ab (Santa Cruz Biotechnology). Proteins bound to the Abs were visualized using the enhanced chemiluminescence system (Dupont).
Results and Discussion
Generation of C/EBPγ2/− Mice.
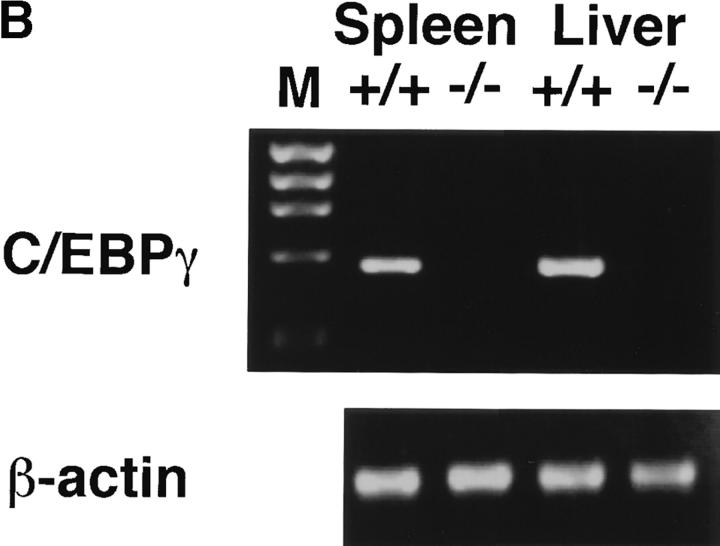
The C/EBPγ genomic locus was disrupted by inserting the neomycin resistance gene into the second exon (Fig. 1 A). This insertion resulted in disruption of the basic and leucine zipper domains, which are essential for DNA binding and dimer formation. Homologous recombinants were obtained through a double selection with G418 and gancyclovir. Targeted clones were injected into B6 blastocysts to generate chimeric mice, which were bred to achieve germline transmission. By mating heterozygous mutants, homozygous mutants were obtained at a frequency of the expected Mendelian ratio (22.6% of littermates). C/EBPγ expression was detected in wild-type newborn spleens and livers, but not in those of homozygous mutants as expected (Fig. 1 B). General appearance at birth was indistinguishable among C/EBPγ1/+, C/EBPγ1/−, and C/EBPγ2/− mice. However, only 11% of homozygous mutants could survive >60 h, although they were healthy until 12 h after they were born (Table ). These results indicate that C/EBPγ is involved in early neonatal survival but not embryonic survival. By histological examination, C/EBPγ2/− mice at 24 h showed emphysematous changes in their lungs (data not shown). However, it is not clear at the time of this writing whether the lung lesions alone can account for the high mortality of homozygous mutants at the early neonatal stage.
Figure 1.
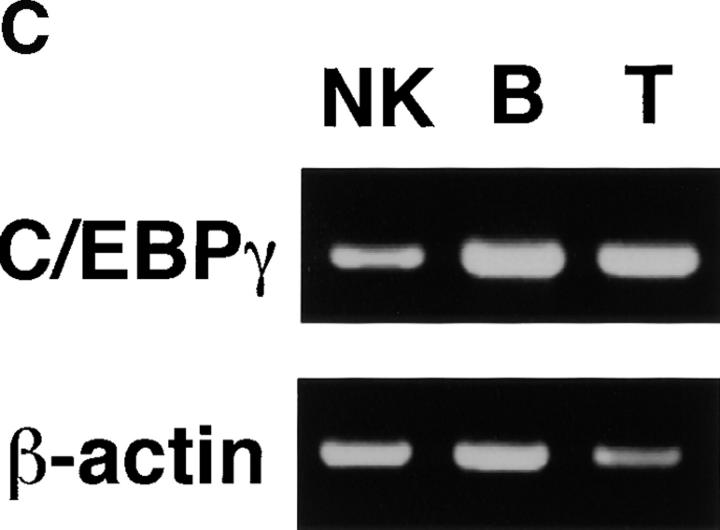
Generation of C/EBPγ2/− mice and expression of C/EBPγ mRNA. (A) Schematic representation of the C/EBPγ targeting vector, the C/EBPγ genomic locus, and the targeted C/EBPγ allele. The targeting vector contains the herpes simplex virus thymidine kinase (HSV-TK) gene 5′ upstream of the long arm homology region and the neomycin resistance gene (NEO) in the middle of the second exon. (B) RT-PCR analysis of newborn spleen and liver RNAs from C/EBPγ1/+ and C/EBPγ2/− mice. Total RNAs were isolated and analyzed for C/EBPγ and β-actin expression by RT-PCR. M, ΦX174/HaeIII digest marker. (C) RT-PCR analysis for C/EBPγ in lymphoid lineage cells. Total RNAs were isolated from splenic CD3−DX5+ (NK), B220+ (B), and CD3+ (T) cells and analyzed for C/EBPγ and β-actin expression by RT-PCR.
Table 1.
Postnatal Survival of C/EBPγ2/− Mice
| Time | Alive (%) | No. dead |
|---|---|---|
| h | ||
| At birth | 18 (100) | 0 |
| 0–12 | 18 (100) | 0 |
| 12–24 | 12 (66.7) | 6 |
| 24–36 | 9 (50.0) | 3 |
| 36–48 | 3 (16.7) | 6 |
| 48–60 | 2 (11.1) | 1 |
| >60 | 2 (11.1) | 0 |
C/EBPγ Deficiency Does Not Affect T or B Cell Development.
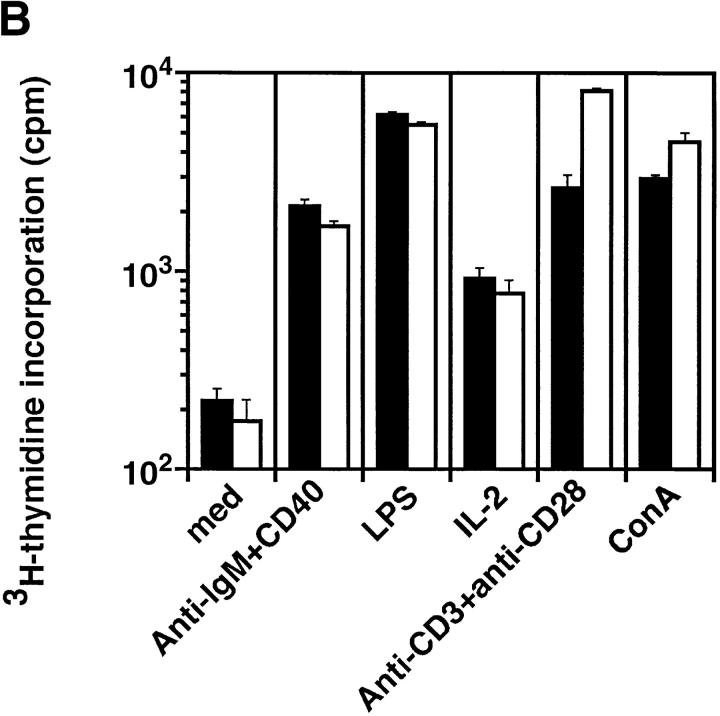
C/EBPγ is expressed in T and B cells 9. Furthermore, NK cells, which belong to a distinct lymphoid lineage from T and B cells 13 14, also expressed C/EBPγ mRNA (Fig. 1 C). To investigate in vivo roles of C/EBPγ in these lymphoid lineage cells, we generated bone marrow chimeric mice by transferring newborn spleen cells into lethally irradiated, RAG2−/− B6 mice. Thymocytes from C/EBPγ2/− chimeras showed normal T cell development (Fig. 2 A). Splenocytes from C/EBPγ2/− chimeras also showed normal population of CD4+8− and CD4−8+ T cells (Fig. 2 A). Furthermore, analysis of surface markers such as B220, IgM, and IgD revealed that B cell maturation was not disturbed in C/EBPγ2/− chimeras (Fig. 2 A). In addition to surface phenotype, proliferative responses to mitogens or stimulating Abs were not significantly different between C/EBPγ1/+ and C/EBPγ2/− chimeras (Fig. 2 B). Taken together, these results suggest that C/EBPγ is not essential for functional T and B cell development.
Figure 2.
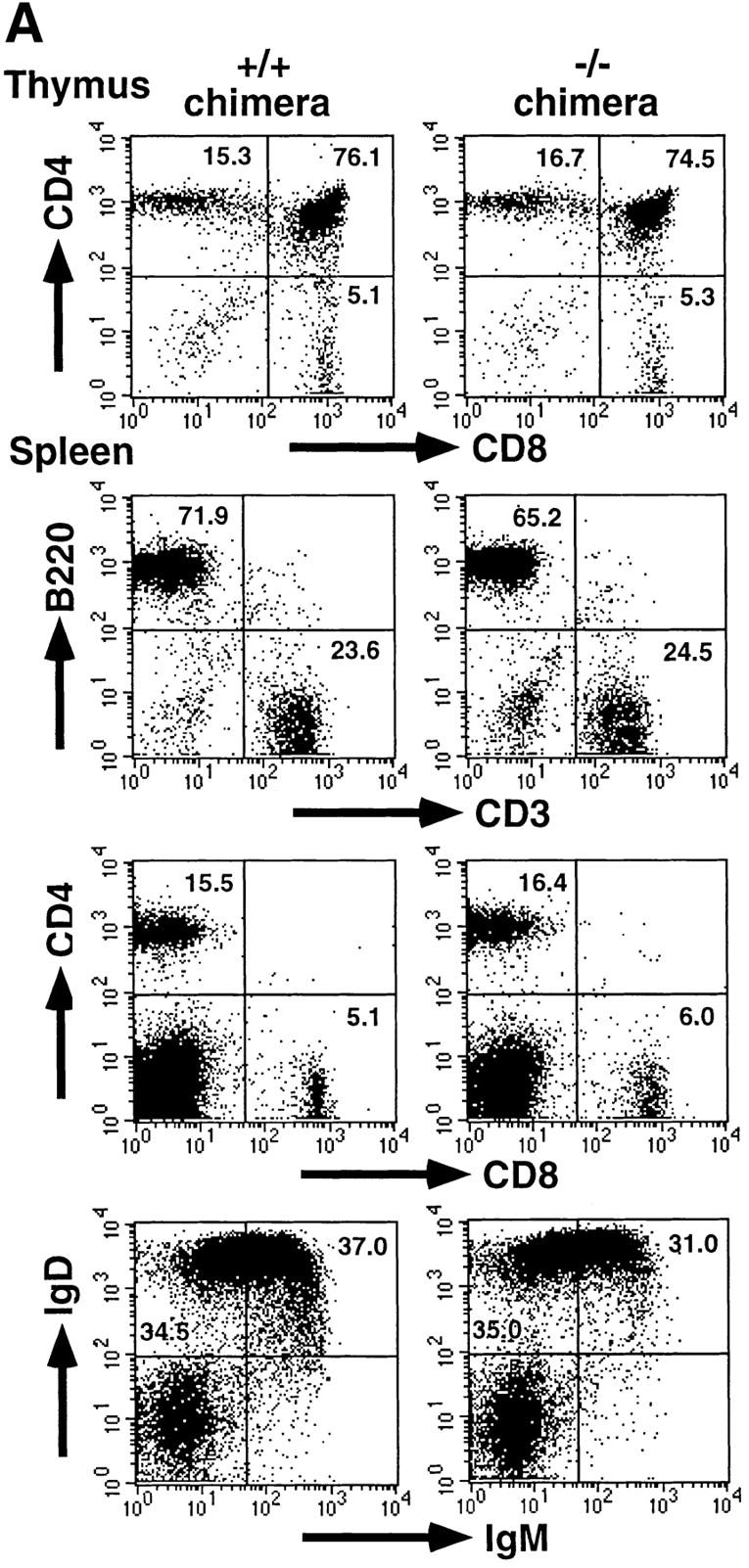
Development and function of T and B cells in C/EBPγ1/+ and C/EBPγ2/− chimeras. (A) Flow cytometric analysis of thymi and spleens from C/EBPγ1/+ and C/EBPγ2/− chimeras. Single-cell suspensions from the thymus and spleen were stained with indicated Abs and analyzed using FACSCalibur™ with CELLQuest™ software. The percentages of the quadrants are shown. (B) C/EBPγ1/+ (filled columns) and C/EBPγ2/− (open columns) chimera splenocytes were cultured with indicated mitogens or Abs and analyzed for their proliferation by measuring [3H]thymidine incorporation. Five independent experiments were performed with similar results. The data indicate mean ± SD of triplicate samples of one representative experiment.
Impaired Splenic NK Cell Activity in C/EBPγ2/− Chimeras.
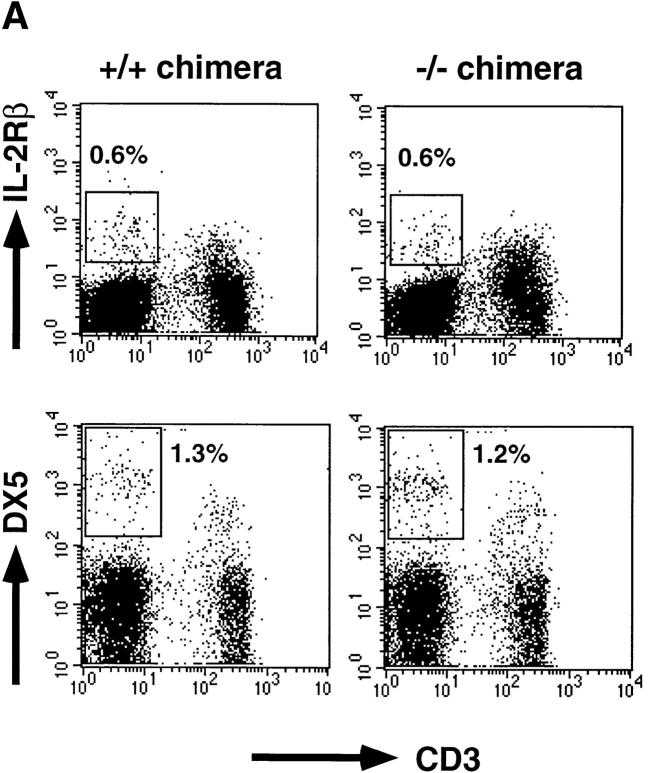
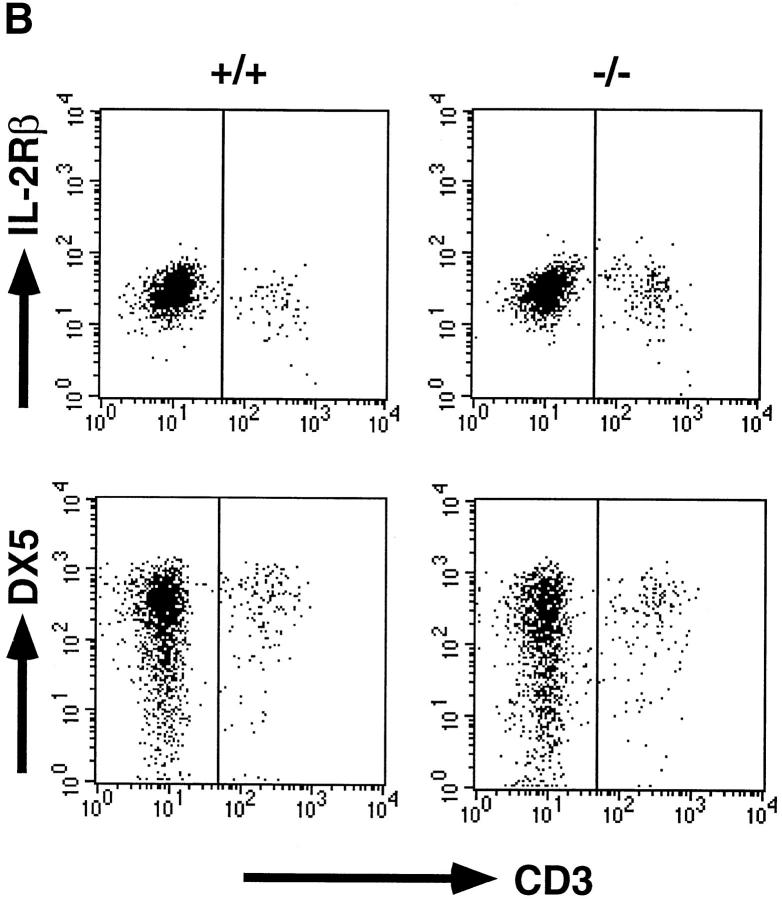
NK cells can be identified as CD3−IL-2Rβ1, CD3−NK1.1+, or CD3−DX5+ cells by flow cytometry. However, the NK1.1 analysis in the chimeras is limited, because only some mice in a mixed genetic background of 129 and B6 carried the NK1.1 allele, of which expression is detected on B6 but not 129 NK cells. Therefore, the NK cell population was analyzed for IL-2Rβ and DX5 expression. Splenic CD3−IL-2Rβ1 cells in C/EBPγ2/− chimeras were detected at levels equivalent to those in C/EBPγ1/+ chimeras (Fig. 3 A). Furthermore, the frequency of CD3−DX5+ cells was also comparable between C/EBPγ1/+ and C/EBPγ2/− chimeras (Fig. 3 A). Percentages of CD3−DX5+ cells were larger than those of CD3−IL-2Rβ1 cells in both control and C/EBPγ2/− chimera splenocytes, as described previously 23.
Figure 3.
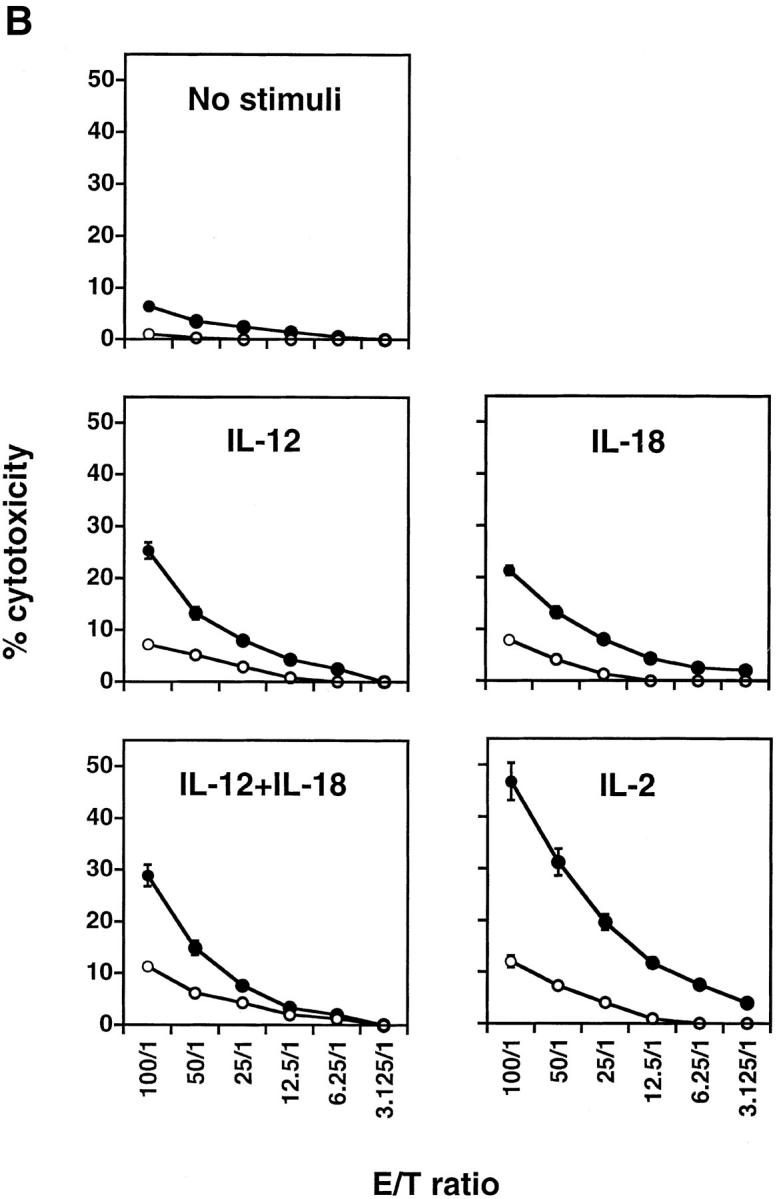
NK cell population and cytolytic activity of C/EBPγ1/+ and C/EBPγ2/− chimeras. (A) Single-cell suspensions from the chimera splenocytes were stained with FITC–anti-CD3 and PE–anti-IL-2Rβ Ab (top panels) or with FITC–anti-CD3 and biotin–DX5 followed by PE–streptavidin (bottom panels). The percentages of NK cell population are shown. (B) YAC-1 killing analysis of spleen cells. C/EBPγ1/+ (•) and −/− (○) chimera spleen cells were incubated without any cytokines for 4 h or with 2 ng/ml IL-12, 20 ng/ml IL-18, 2 ng/ml IL-12 plus 20 ng/ml IL-18, or 500 U/ml IL-2 for 24 h, and their cytotoxic activities against YAC-1 cells were determined. E/T ratios are shown on the x-axis. Six independent experiments were performed with similar results. The data indicate mean ± SD of triplicate samples of one representative experiment.
Next, NK cytotoxicity was measured by YAC-1 cell–killing activity in the absence or presence of various cytokines (Fig. 3 B). Spontaneous cytotoxicity of C/EBPγ2/− chimera splenocytes (1.0% at 100:1) was impaired as compared with that of control chimera splenocytes (6.4% at 100:1; Fig. 3 B). IL-12 and/or IL-18 act on NK cells and can enhance their cytotoxic activity 21. When stimulated with these cytokines, C/EBPγ2/− chimera splenocytes also showed impaired killing activity as compared with control splenocytes (Fig. 3 B). IL-2 is another stimulatory cytokine for NK cell activity 24. Decreased killing activity of C/EBPγ2/− chimeras was also observed in the presence of IL-2 (Fig. 3 B). Poly (I:C)-stimulated C/EBPγ2/− chimera splenocytes also showed lower cytotoxic activity than control splenocytes (data not shown).
Reduced Ability of C/EBPγ2/− Chimera Splenocytes to Produce IFN-γ in Response to IL-12 and/or IL-18.
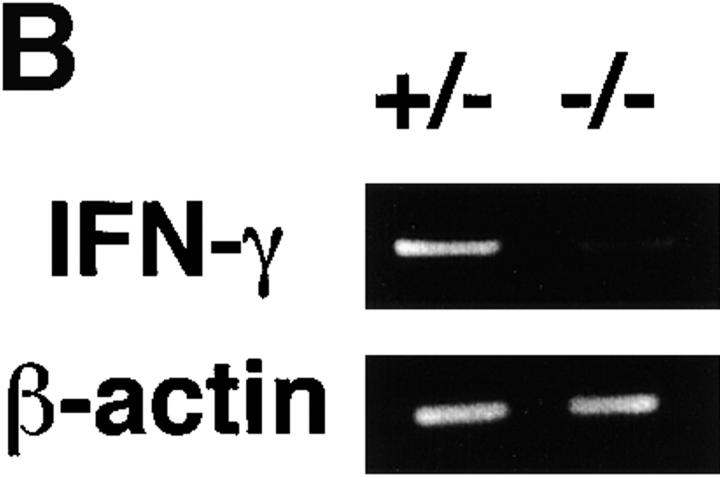
NK cells constitutively express both functional IL-12R and IL-18R, whereas naive T cells do not 21 25 26. Therefore, splenic IFN-γ production by stimulation with IL-12 and IL-18 is dependent on NK but not T cells. To evaluate the ability of NK cells from chimeric mice to produce IFN-γ, splenocytes were cultured with or without IL-12 and/or IL-18. 24 h later, cell-free supernatants were harvested and assayed for IFN-γ production with ELISA. Under this condition, C/EBPγ2/− chimera splenocytes produced much lower amounts of IFN-γ than C/EBPγ1/+ chimera splenocytes (Fig. 4 A). IL-12 or IL-18 can induce IFN-γ production at the transcriptional level in NK cells 27. Consistent with reduced IFN-γ production, induction of IFN-γ mRNA was markedly decreased in C/EBPγ2/− chimera splenocytes (Fig. 4 B). Taken together, these results suggest that C/EBPγ is required for induction of IFN-γ by IL-12 and/or IL-18 in NK cells.
Figure 4.
IFN-γ production by chimera splenocytes in response to IL-12 and/or IL-18. (A) Spleen cells from C/EBPγ1/+ (filled columns) and C/EBPγ −/− (open column) chimera splenocytes were cultured in the absence (med) or presence of 2 ng/ml IL-12 and/or 20 ng/ml IL-18 for 24 h. Amounts of IFN-γ in the culture supernatants were measured by ELISA. Experiments were independently performed six times with similar results. The data indicate mean ± SD of triplicate samples of one representative experiment. N.D., not detected. (B) RT-PCR analysis of C/EBPγ1/− and C/EBPγ2/− chimera splenocytes cultured with 2 ng/ml IL-12 and 20 ng/ml IL-18 for 24 h. Total RNAs were isolated and analyzed for IFN-γ and β-actin expression by RT-PCR. Experiments were independently performed three times with similar results.
IL-15–induced NK Cells from C/EBPγ2/− Newborn Spleens Showed Impairment of Both Cytotoxic Activity and IFN-γ Production.
NK cell population was analyzed in newborn splenocytes. Both CD3−IL-2Rβ1 and CD3−DX5+ cells were equivalently detected in C/EBPγ1/+ and C/EBPγ −/− newborn splenocytes (Fig. 5 A). IL-15 can stimulate NK cell activity and proliferation and is essential for NK cell development 28 29. Adult bone marrow cells can generate NK cells when cultured with IL-15 28 30. Wild-type newborn spleen cells could also give rise to CD3−IL-2Rβ1 DX5+ cells in the presence of IL-15 (Fig. 5 B). In this culture condition, harvested cell numbers from control (C/EBPγ1/+, n = 5, and C/EBPγ1/−, n = 3) and C/EBPγ2/− (n = 7) mouse spleen cells were 5.7 ± 4.1 × 105 and 4.9 ± 6.7 × 105 per well, respectively. Surface phenotype of cultured C/EBPγ2/− cells was identical to that of wild-type cells (Fig. 5 B). However, NK cells generated from C/EBPγ2/− spleens showed impaired cytotoxic activity against YAC-1 cells as compared with those from C/EBPγ1/+ spleens (Fig. 5 C). Furthermore, C/EBPγ2/− NK cells produced lower amounts of IFN-γ in response to IL-12 plus IL-18 than C/EBPγ1/+ and C/EBPγ1/− NK cells (Fig. 5 D). Taken together, two major NK cell activities, cytotoxic activity and IFN-γ production, were impaired in both C/EBPγ2/− chimera splenocytes and C/EBPγ2/− NK cells generated in the presence of IL-15 in vitro.
Figure 5.
Characterization of IL-15–induced NK cells from newborn spleen cells. (A) Flow cytometric analysis of newborn spleen cells. Splenocytes were taken within 12–18 h after birth and analyzed for IL-2Rβ, DX5, and CD3 expressions as described in the legend to Fig. 3 A. The percentages of NK cell population are shown. (B) Flow cytometric analysis of in vitro NK cells. Newborn spleen cells cultured with IL-15 for 10 d were analyzed using FACSCalibur™ with CELLQuest™ software. (C) YAC-1 killing analysis of in vitro NK cells. C/EBPγ1/+ (•) and C/EBPγ2/− (○) NK cells were analyzed for their cytotoxic activities. The data indicate mean ± SD of triplicate samples of one representative experiment. (D) IFN-γ production by C/EBPγ1/+, C/EBPγ1/−, and C/EBPγ2/− NK cells. IL-15–induced NK cells were harvested and cultured at 105 cells/well in 96-well plates for 24 h in the presence of 2 ng/ml IL-12 and 20 ng/ml IL-18. Amounts of IFN-γ in the culture supernatants were measured by ELISA. The representative data from the same littermates are shown as mean ± SD of triplicate samples. We analyzed four C/EBPγ1/+, eight C/EBPγ1/−, and five C/EBPγ2/− mice for A, five C/EBPγ1/+, three C/EBPγ1/−, and seven C/EBPγ2/− mice for B and D, and three C/EBPγ1/+, one C/EBPγ1/−, and four C/EBPγ2/− mice for C and obtained similar results.
IL-12 and IL-18 Signaling Analysis of C/EBPγ1/+ and C/EBPγ2/− NK Cells.
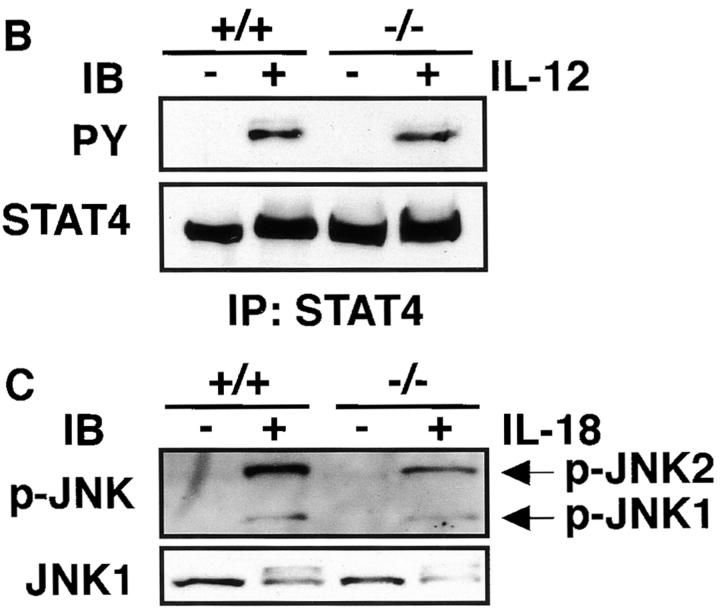
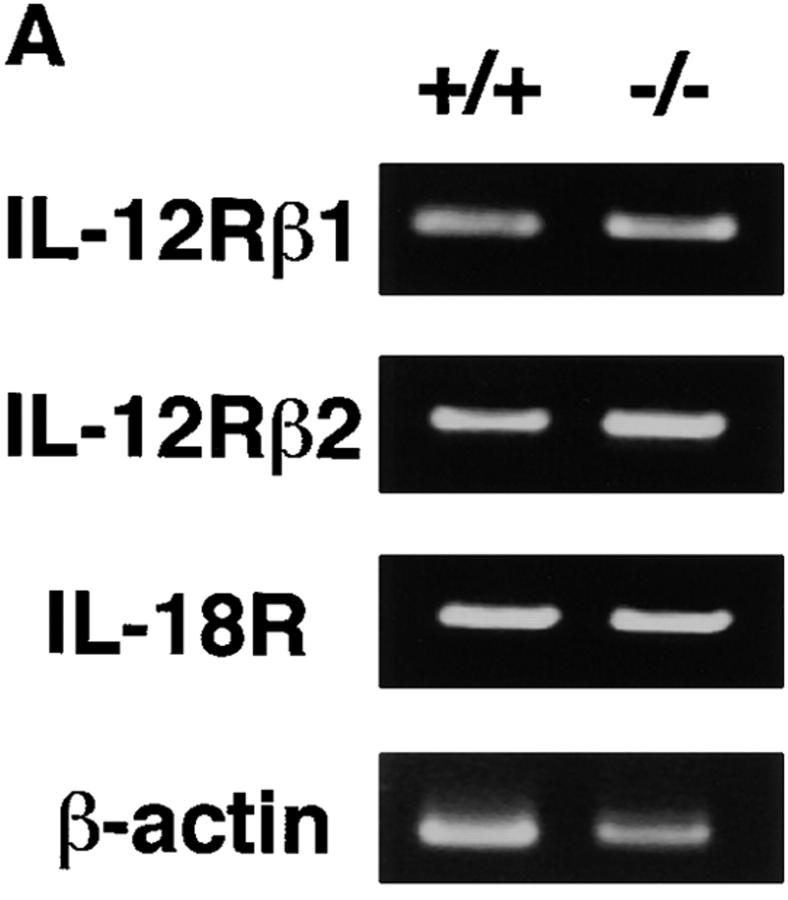
To determine if impaired induction of IFN-γ by IL-12 and IL-18 is caused by decreased expression of these receptors, we examined their mRNA expression in C/EBPγ1/+ and C/EBPγ2/− NK cells. IL-12Rβ1, IL-12Rβ2, and IL-18R expression was equivalent in C/EBPγ1/+ and C/EBPγ2/− NK cells (Fig. 6 A). IL-12 can induce tyrosine phosphorylation and activation of STAT4 31 32. This pathway was not impaired in C/EBPγ2/− NK cells (Fig. 6 B). IL-18 can cause phosphorylation and activation of JNK 33. Equivalent JNK phosphorylation was observed in C/EBPγ1/+ and C/EBPγ2/− NK cells when stimulated by IL-18 (Fig. 6 C). Taken together, these results suggest that signaling pathways proximate to these cytokine receptors are intact in C/EBPγ2/− NK cells.
Figure 6.
IL-12 and IL-18 signaling of IL-15–induced NK cells. (A) Total RNAs were purified from 10-d IL-15–cultured newborn spleen cells and analyzed for IL-12Rβ1, IL-12Rβ2, IL-18R, and β-actin expression by RT-PCR. (B) Cells were stimulated with or without IL-12 for 20 min, and the cell lysates were immunoprecipitated with anti-STAT4 Ab. The immunoprecipitates were separated on SDS-PAGE and blotted with antiphosphotyrosine Ab (top) or anti-STAT4 Ab (bottom). (C) Cells were or were not stimulated with IL-18 for 20 min. The cell lysates were blotted with antiphospho-JNK Ab (top) or anti-JNK1 Ab (bottom). Experiments were independently performed three times with similar results.
In this study, we demonstrate that C/EBPγ2/− NK cells have defects in IFN-γ production and cytotoxicity. It has been shown that several regulatory elements such as the activator protein (AP)-1 or NF-κB sites are essential for IFN-γ gene expression by IL-12 and/or IL-18 34 35. Although NF-IL6 is a candidate for heterodimerizing with C/EBPγ 36, it seems unlikely that the heterodimer plays an essential role. First, no C/EBP sites have been shown to be important for IFN-γ induction. Second, splenic IFN-γ production in response to IL-12 and/or IL-18 is not impaired in NF-IL6−/− mice (our unpublished data). It is possible that C/EBPγ plays a critical role in IFN-γ gene regulation by dimerizing with AP-1 components. AP-1 components are activated by IL-12 or IL-18 and essential for both IL-12– and IL-18–induced IFN-γ promotor activation 34. Fos or Jun is shown to require C/EBPγ in order to efficiently bind to the regulatory element in the IL-4 promotor 12. Although further studies are necessary, our results suggest that C/EBPγ regulates IFN-γ gene expression. At present, the possibility that C/EBPγ is necessary for expression of other gene(s) critical for IFN-γ gene induction cannot formally be excluded.
IFN-γ does not seem to be involved in cytotoxic activity of NK cells, because NK activity is not remarkably impaired in IFN-γ2/− mice 21. Therefore, impaired IFN-γ production cannot account for decreased cytotoxic activity of C/EBPγ2/− NK cells. Although CD18 is important for NK cells to recognize target cells 37, surface expression of CD18 was not decreased in C/EBPγ2/− chimera splenocytes (data not shown). Furthermore, IL-12 and IL-18 could induce expression of a critical cytolytic mediator, perforin, in C/EBPγ2/− chimera splenocytes (data not shown). These results indicate the presence of other target(s) of C/EBPγ that are involved in the cytotoxicity of NK cells.
It is noteworthy that in spite of NK cell dysfunction, NK cell generation is intact in the absence of C/EBPγ. This means that C/EBPγ should play critical roles in a late maturation step rather than in an early developmental step. In contrast, mutant mice established so far manifested impaired NK cell generation. The IL-15 receptor consists of IL-15Rα, IL-2Rβ, and the common γ chain 38 39 40 41 42. Deficiency of one of these components caused reduction of NK cell numbers in vivo and impaired NK cell expansion in vitro 43 44 45. Janus kinase (JAK)-3 and STAT5 are essential for IL-15 signaling components. JAK-3−/− or STAT5−/− mice also showed impairment of NK cell generation 23 46 47 48 49. Furthermore, IFN regulatory factor (IRF)-1 was found to be critical for expansion of NK cells 19. This can be explained by impaired IL-15 production in the bone marrow microenvironment 19. In these mutants, deficient NK cell generation is caused by disturbance of IL-15 signaling or decreased IL-15 production. Membrane lymphotoxin (LT)α also plays essential roles for NK cell development by acting independently of IL-15/IRF-1 or upstream of the IL-15/IRF-1 pathway 50. NK cell generation was severely impaired in LTα2/− mice. Furthermore, NK cells, which were generated in vitro with IL-15 from LTα2/− bone marrow cells, showed intact cytotoxic activity. These characteristics in LTα2/− mice are distinct from those in C/EBPγ2/− mice. In addition, there are two more mutants with impaired NK cell development, the mechanism of which is not yet clear. One is deficiency of a winged helix-turn-helix transcriptional factor, Ets-1 51. The other is deficiency of Id2, an inhibitor for transcription factors with helix-loop-helix domains 52.
At present, little is known about the molecular mechanisms that regulate NK cell functions or development. Our study clearly reveals that C/EBPγ is critically involved in functional NK cell maturation. Identification of the target genes regulated by C/EBPγ will clarify the molecular mechanism of NK cell functions.
Acknowledgments
We thank Ms. Akiko Maekawa, Ms. Nana Iwami, and Ms. Shizue Futatsugi for excellent technical assistance and Drs. Mitsuru Matsumoto, Yoshiko Murakami, Teruhito Yasui, and Hitoshi Kikutani for useful advice about transfer experiments. We are also grateful to Drs. Frederick W. Alt, Shigekazu Nagata, and Makoto Matsumoto for providing RAG2−/− mice, the plasmid for C/EBPγ genomic fragments, and PCR primers for β-actin, respectively.
This work was supported in part by grants from the Ministry of Education, Science, and Culture of Japan, Uehara Memorial Foundation, Novartis Foundation for the Promotion of Science, Kato Memorial Bioscience Foundation, and CREST (Core Research for Evolutional Science and Technology) of Japan Science and Technology Corporation.
Footnotes
T. Kaisho's current address is Dept. of Host Defense, Research Institute for Microbial Diseases, Osaka University, Osaka 565-0871, Japan.
Abbreviations used in this paper: AP, activator protein; C/EBPs, CCAAT/enhancer binding proteins; IRF, IFN regulatory factor; LT, lymphotoxin; NF, nuclear factor; RAG, recombination activating gene; RT, reverse transcriptase; STAT, signal transducer and activator of transcription.
References
- Lekstrom-Himes J., Xanthopoulos K.G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- Screpanti I., Romani L., Musiani P., Modesti A., Fattori E., Lazzaro D., Sellitto C., Scarpa S., Bellavia D., Lattanzio G. Lymphoproliferative disorder and imbalanced T-helper response in C/EBPβ-deficient mice. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Akira S., Yoshida K., Umemoto M., Yoneda Y., Shirafuji N., Fujiwara H., Suematsu S., Yoshida N., Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Yoshida N., Kishimoto T., Akira S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.D., Finegold M.J., Bradley A., Ou C.N., Abdelsayed S.V., Wilde M.D., Taylor L.R., Wilson D.R., Darlington G.J. Impaired energy homeostasis in C/EBPα knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Flodby P., Barlow C., Kylefjord H., Ahrlund-Richter L., Xanthopoulos K.G. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J. Biol. Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- Yamanaka R., Barlow C., Lekstrom-Himes J., Castilla L.H., Liu P.P., Eckhaus M., Decker T., Wynshaw-Boris A., Xanthopoulos K.G. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein ∈-deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman C., Platero J.S., Shuman J., Calame K. Ig/EBP-1a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes Dev. 1990;4:1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- Cooper C., Henderson A., Artandi S., Avitahl N., Calame K. Ig/EBP (C/EBPγ) is a transdominant negative inhibitor of C/EBP family transcriptional activators. Nucleic Acids Res. 1995;23:4371–4377. doi: 10.1093/nar/23.21.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Wakabayashi-Ito N., Nagata S. Molecular cloning of cDNA and a chromosomal gene encoding GPE1-BP, a nuclear protein which binds to granulocyte colony-stimulating factor promoter element 1. FEBS Lett. 1991;282:95–97. doi: 10.1016/0014-5793(91)80452-9. [DOI] [PubMed] [Google Scholar]
- Davydov I.V., Bohmann D., Krammer P.H., Li-Weber M. Cloning of the cDNA encoding human C/EBPγ, a protein binding to the PRE-I enhancer element of the human interleukin-4 promoter. Gene. 1995;161:271–275. doi: 10.1016/0378-1119(95)00271-7. [DOI] [PubMed] [Google Scholar]
- Moore T.A., Bennett M., Kumar V. Murine natural killer cell differentiationpast, present, and future. Immunol. Res. 1996;15:151–162. doi: 10.1007/BF02918504. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Biology of natural killer cells. Adv. Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K.R., Capecchi M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Mansour S.L., Thomas K.R., Capecchi M.R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cellsa general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Kawabe T., Naka T., Yoshida K., Tanaka T., Fujiwara H., Suematsu S., Yoshida N., Kishimoto T., Kikutani H. The immune responses in CD40-deficient miceimpaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Ogasawara K., Hida S., Azimi N., Tagaya Y., Sato T., Yokochi-Fukuda T., Waldmann T.A., Taniguchi T., Taki S. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature. 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Nakanishi K., Matsui K., Higashino K., Okamura H., Miyazawa Y., Kaneda K. IFN-γ-inducing factor up-regulates Fas ligand-mediated cytotoxic activity of murine natural killer cell clones. J. Immunol. 1996;157:3967–3973. [PubMed] [Google Scholar]
- Hyodo Y., Matsui K., Hayashi N., Tsutsui H., Kashiwamura S., Yamauchi H., Hiroishi K., Takeda K., Tagawa Y., Iwakura Y. IL-18 up-regulates perforin-mediated NK activity without increasing perforin messenger RNA expression by binding to constitutively expressed IL-18 receptor. J. Immunol. 1999;162:1662–1668. [PubMed] [Google Scholar]
- Svetic A., Finkelman F.D., Jian Y.C., Dieffenbach C.W., Scott D.E., McCarthy K.F., Steinberg A.D., Gause W.C. Cytokine gene expression after in vivo primary immunization with goat antibody to mouse IgD antibody. J. Immunol. 1991;147:2391–2397. [PubMed] [Google Scholar]
- Imada K., Bloom E.T., Nakajima H., Horvath-Arcidiacono J.A., Udy G.B., Davey H.W., Leonard W.J. Stat5b is essential for natural killer cell–mediated proliferation and cytolytic activity. J. Exp. Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesano A., Visonneau S., Clark S.C., Santoli D. Cellular and molecular mechanisms of activation of MHC nonrestricted cytotoxic cells by IL-12. J. Immunol. 1993;151:2943–2957. [PubMed] [Google Scholar]
- Robinson D., Shibuya K., Mui A., Zonin F., Murphy E., Sana T., Hartley S.B., Menon S., Kastelein R., Bazan F. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-γ production and activates IRAK and NFκB. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T., Takeda K., Tanaka T., Ohkusu K., Kashiwamura S., Okamura H., Akira S., Nakanishi K. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cellssynergism with IL-18 for IFN-γ production. J. Immunol. 1998;161:3400–3407. [PubMed] [Google Scholar]
- Hunter C.A., Timans J., Pisacane P., Menon S., Cai G., Walker W., Aste-Amezaga M., Chizzonite R., Bazan J.F., Kastelein R.A. Comparison of the effects of interleukin-1 α, interleukin-1 β and interferon-γ-inducing factor on the production of interferon-γ by natural killer. Eur. J. Immunol. 1997;27:2787–2792. doi: 10.1002/eji.1830271107. [DOI] [PubMed] [Google Scholar]
- Mrozek E., Anderson P., Caligiuri M.A. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- Williams N.S., Klem J., Puzanov I.J., Sivakumar P.V., Schatzle J.D., Bennett M., Kumar V. Natural killer cell differentiationinsights from knockout and transgenic mouse models and in vitro systems. Immunol. Rev. 1998;165:47–61. doi: 10.1111/j.1600-065x.1998.tb01229.x. [DOI] [PubMed] [Google Scholar]
- Puzanov I.J., Bennett M., Kumar V. IL-15 can substitute for the marrow microenvironment in the differentiation of natural killer cells. J. Immunol. 1996;157:4282–4285. [PubMed] [Google Scholar]
- Bacon C.M., Petricoin E.F.R., Ortaldo J.R., Rees R.C., Larner A.C., Johnston J.A., O'Shea J.J. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc. Natl. Acad. Sci. USA. 1995;92:7307–7311. doi: 10.1073/pnas.92.16.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson N.G., Szabo S.J., Weber-Nordt R.M., Zhong Z., Schreiber R.D., Darnell J.E., Jr., Murphy K.M. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Barbulescu K., Becker C., Schlaak J.F., Schmitt E., Meyer zum Buschenfelde K.H., Neurath M.F. IL-12 and IL-18 differentially regulate the transcriptional activity of the human IFN-γ promoter in primary CD4+ T lymphocytes. J. Immunol. 1998;160:3642–3647. [PubMed] [Google Scholar]
- Kojima H., Aizawa Y., Yanai Y., Nagaoka K., Takeuchi M., Ohta T., Ikegami H., Ikeda M., Kurimoto M. An essential role for NF-κB in IL-18-induced IFN-γ expression in KG-1 cells. J. Immunol. 1999;162:5063–5069. [PubMed] [Google Scholar]
- Cooper C.L., Berrier A.L., Roman C., Calame K.L. Limited expression of C/EBP family proteins during B lymphocyte development. Negative regulator Ig/EBP predominates early and activator NF-IL-6 is induced later. J. Immunol. 1994;153:5049–5058. [PubMed] [Google Scholar]
- Helander T.S., Carpen O., Turunen O., Kovanen P.E., Vaheri A., Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- Grabstein K.H., Eisenman J., Shanebeck K., Rauch C., Srinivasan S., Fung V., Beers C., Richardson J., Schoenborn M.A., Ahdieh M. Cloning of a T cell growth factor that interacts with the β chain of the interleukin-2 receptor. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- Giri J.G., Kumaki S., Ahdieh M., Friend D.J., Loomis A., Shanebeck K., DuBose R., Cosman D., Park L.S., Anderson D.M. Identification and cloning of a novel IL-15 binding protein that is structurally related to the a chain of the IL-2 receptor. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:3654–3663. doi: 10.1002/j.1460-2075.1995.tb00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M., Nakamura Y., Russell S.M., Ziegler S.F., Tsang M., Cao X., Leonard W.J. Interleukin-2 receptor γ chaina functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- Tagaya Y., Bamford R.N., DeFilippis A.P., Waldmann T.A. IL-15a pleiotropic cytokine with diverse receptor/signaling pathways whose expression is controlled at multiple levels. Immunity. 1996;4:329–336. doi: 10.1016/s1074-7613(00)80246-0. [DOI] [PubMed] [Google Scholar]
- Carson W.E., Giri J.G., Lindemann M.J., Linett M.L., Ahdieh M., Paxton R., Anderson D., Eisenmann J., Grabstein K., Caligiuri M.A. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J. Exp. Med. 1994;180:1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodolce J.P., Boone D.L., Chai S., Swain R.E., Dassopoulos T., Trettin S., Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Duncan G.S., Takimoto H., Mak T.W. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor β chain. J. Exp. Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Santo J.P., Colucci F., Guy-Grand D. Natural killer and T cells of innate and adaptive immunitylymphoid compartments with different requirements for common γ chain-dependent cytokines. Immunol. Rev. 1998;165:29–38. doi: 10.1111/j.1600-065x.1998.tb01227.x. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Saijo K., Takahashi T., Osawa M., Arase H., Hirayama N., Miyake K., Nakauchi H., Shirasawa T., Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- Nosaka T., van Deursen J.M., Tripp R.A., Thierfelder W.E., Witthuhn B.A., McMickle A.P., Doherty P.C., Grosveld G.C., Ihle J.N. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- Thomis D.C., Gurniak C.B., Tivol E., Sharpe A.H., Berg L.J. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- Moriggl R., Topham D.J., Teglund S., Sexl V., McKay C., Wang D., Hoffmeyer A., van Deursen J., Sangster M.Y., Bunting K.D. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- Iizuka K., Chaplin D.D., Wang Y., Wu Q., Pegg L.E., Yokoyama W.M., Fu Y.-X. Requirement for membrane lymphotoxin in natural killer cell development. Proc. Natl. Acad. Sci. USA. 1999;96:6336–6340. doi: 10.1073/pnas.96.11.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K., Muthusamy N., Fischer C., Ting C.N., Walunas T.L., Lanier L.L., Leiden J.M. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]