Abstract
The ability to infect host cells is critical for the survival and replication of intracellular pathogens in humans. We previously found that many genes involved in the ability of Legionella pneumophila to infect macrophages are not expressed efficiently under standard laboratory growth conditions. We have developed an approach using expression of L. pneumophila genes from an exogenous constitutive promoter on a low-copy-number vector that allows identification of genes involved in host cell infection. Through the use of this strategy, we found that expression of a gene, lvhB2, enhances the efficiency of L. pneumophila infection of mammalian cells. The putative protein encoded by lvhB2 has similarity to structural pilin subunits of type IV secretion systems. We confirmed that this gene plays a role in host cell infection by the construction of an in-frame deletion in the L. pneumophila lvhB2 gene and complementation of this mutant with the wild-type gene. The lvhB2 mutant does not display a very obvious defect in interactions with host cells when the bacteria are grown at 37°C, but it has an approximately 100-fold effect on entry and intracellular replication when grown at 30°C. These data suggest that lvhB2 plays an important role in the efficiency of host cell infection by L. pneumophila grown at lower temperatures.
Legionella pneumophila, the causative agent of Legionnaires' disease, replicates primarily intracellularly within monocytes during infections in humans and animals (11, 48). Thus, the ability to productively infect monocytic cells is critical to pathogenesis by L. pneumophila. Entry into monocytes can occur via an unusual mechanism termed “coiling phagocytosis,” where a filipodium wraps asymmetrically around the bacterium (8, 25, 33). Conventional phagocytic events are also observed with L. pneumophila (7, 25, 33); however, coiling phagocytosis correlates with intracellular survival and virulence (7). Although this type of phagocytosis has also been observed in spirochetes, the mechanism by which coiling occurs is not understood (5, 34, 35, 42). In order to better understand the mechanism(s) used by L. pneumophila to productively infect host cells, we set out to identify the bacterial components involved.
Since bacteria are efficient organisms, they express only the genes needed to survive and replicate under specific growth conditions (13, 28, 29). We have recently shown that the frequency of coiling phagocytosis in monocytes increases after intracellular growth in amoebae, suggesting that the genes involved are downregulated on standard laboratory media (7, 8). This information has been used to identify L. pneumophila entry loci through overexpression in wild-type bacteria (10). These studies resulted in the identification of three loci, enh1, enh2, and enh3, that play a role in entry by L. pneumophila into host cells. The rtxA and enhC genes, present in the enh1 and enh2 loci, respectively, have been shown to play a role in entry but when mutated only reduce uptake by approximately 50% (10). Despite this relatively moderate effect in in vitro assays, rtxA plays a critical role in the virulence of L. pneumophila during mouse infections (9). These data suggest that host cell infection by L. pneumophila is a complex process involving a large number of genes and that the genes involved can play an important role in pathogenesis despite having only moderate effects in in vitro virulence models.
In order to obtain a more comprehensive picture of the mechanisms that L. pneumophila uses to infect host cells, we wished to identify more of the genes involved in this process. Since overexpression by gene dosage effects was previously successful for the identification of L. pneumophila entry loci (10), we applied a similar strategy to identify additional genes. A constitutive promoter upstream of random fragments of the L. pneumophila genome was used for construction of a comprehensive expression library. Wild-type L. pneumophila organisms that carry this library can be screened for mutants that display an enhanced ability to infect host cells. We describe the use of this system to identify a locus that, when expressed from a constitutive promoter, enhances host cell infection by L. pneumophila. Mutagenesis and complementation analyses in wild-type bacteria demonstrate that this gene, lvhB2, plays an important role in adherence, entry, and intracellular replication by L. pneumophila when grown at 30°C. This gene is similar to the structural pilin subunit genes of type IV secretion systems and has been previously observed by other investigators (39). These studies support an important role for lvhB2 in the efficiency of L. pneumophila infection of host cells.
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids used in this study are described in Table 1. The streptomycin-resistant variant of L. pneumophila strain AA100 (17), which has been shown to be virulent in both in vitro and in vivo models of infection (30), was used. L. pneumophila strains were passaged no more than twice in the laboratory in order to prevent loss of virulence, and the strains were grown on buffered charcoal yeast extract (BCYE) agar (15) for 3 days at either 30 or 37°C in 5% CO2. Escherichia coli strains were grown in Luria-Bertani medium (Difco, Detroit, Mich.) at 37°C. When appropriate, kanamycin (Sigma) was added at a concentration of 25 μg/ml. The E. coli strain ψec47 has been described previously (10) and was used to propagate plasmids containing an R6K origin of replication. DNA manipulations were done essentially as described previously (37). XL-1 Blue and ψec47 electroporation-competent cells were prepared and frozen as described previously (10, 14).
TABLE 1.
Bacterial strains and plasmids
| Strain of plasmid | Characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1qZΔ M15 Tn10 (Tcr)] | Stratagene |
| ψec47 | XL-1 Blue λpir | 10 |
| L. pneumophila strains | ||
| AA100 | wt, 130b naturally arising Smr strain | 17 |
| ψ1p9 | AA100::L34 | This work |
| ψ1p37 | AA100::pJDC38 | This work |
| ψ1p47 | AA100::pJDC51 | This work |
| ψ1p48 | AA100::pJDC52 | This work |
| ψ1p49 | AA100::pJDC53 | This work |
| ψ1p50 | AA100 ΔlvhB2 | This work |
| ψ1p51 | ψ1p50::pWKS130 | This work |
| ψ1p52 | ψ1p50::pJDC38 | This work |
| ψ1p56 | AA100::pWKS130 | This work |
| Plasmids | ||
| pWKS130 | Kmr Apr pSC101ori Ptac | 43 |
| L34 | Kmr Apr pSC101ori PtaclvhB2 | This work |
| pJDC15 | Kmr R6Kori PL5sacB | 10 |
| pJDC38 | Kmr Apr pSC101ori PtaclvhB2 | This work |
| pJDC51 | Kmr Apr pSC101ori PtaclvhB2 | This work |
| pJDC52 | Kmr Apr pSC101ori Ptac | This work |
| pJDC53 | Kmr Apr pSC101ori Ptac | This work |
Abbreviations: Tc, tetracycline; Sm, streptomycin; Km, kanamycin; Ap, ampicillin; r, resistance; wt, wild-type.
Cell lines and culture conditions.
HEp-2 cells (ATCC CCL23), established from a human epidermoid carcinoma, were grown in RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum (Gibco) and 2 mM l-glutamine at 37°C in 5% CO2. HL-60 (ATCC CCL240) monocyte/macrophage cells were grown at 37°C in 5% CO2 in RPMI 1640 supplemented with 10% fetal bovine serum and 2 mM l-glutamine.
Library construction.
An L. pneumophila total genomic DNA library was constructed in the vector pWKS130 (43). For this library, L. pneumophila genomic DNA was isolated as described previously (10), partially digested with Sau3AI to obtain fragments ranging in length from 0.5 to 2.5 kbp, and ligated into the BamHI site of pWKS130. Plasmid DNA from pools of greater than 5,000 E. coli colonies was then separately transformed into L. pneumophila by electroporation as described previously (10, 14). Six pools of greater than 5,000 L. pneumophila clones (total of 30,000 clones) were enriched for enhanced host cell infection variants using entry assays.
Standard entry assays.
The ability of L. pneumophila to infect host cells was evaluated using standard entry assays, which were carried out as described previously (10). Briefly, 106 HL-60 cells were differentiated in 24-well tissue culture dishes with 100 ng of phorbol 12-myristate 13-acetate (PMA; Sigma)/ml for 48 h prior to use. Nonadherent cells were removed by washing with phosphate-buffered saline (PBS), and 1 ml of culture medium was added. HEp-2 cells were seeded in 24-well dishes at a concentration of 1.5 × 105 cells/ml 18 to 24 h before use. A ratio of 10 bacteria per cell was added to each well and incubated for 30 min with monocytic cells or for 90 min with epithelial cells. All infections were carried out at 37°C. Cells were washed two times with PBS, and fresh medium containing 100 μg of gentamicin/ml was added to kill extracellular bacteria. After 2 h at 37°C, the cells were washed with PBS and lysed with sterile water, and dilutions were plated for CFU determinations. Entry levels were determined by calculating the percentage of the inoculum that became gentamicin resistant over the course of the assay [i.e., percent entry = 100 × (CFU gentamicin resistant/CFU of inoculum)]. To correct for variations in the levels of uptake between experiments, entry is reported relative to that of the wild-type strain (i.e., relative entry = percent entry of test strain/percent entry of wild type).
Enrichment for enhanced host cell infection clones.
Pools of clones from the expression library were enriched for enhanced host cell infection variants in modified entry assays. Entry assays for enrichment of enhanced host cell infection variants were carried out in HEp-2 cells in the same manner as standard entry assays, except that bacteria were coincubated with cells for 5 min prior to the addition of gentamicin. Individual colonies that resulted from the subsequent plating for CFU were then screened individually for an enhanced entry phenotype relative to wild type in standard entry assays.
Adherence assays.
Adherence assays were carried out by two methods, immediate assays (7-9) and formaldehyde assays (9, 16, 21), both of which have been described in detail previously. For immediate assays, HEp-2 and HL-60 cells were seeded and prepared as described for entry assays. After adding the bacteria at a ratio of 10 bacteria per cell, the medium was gently mixed by rocking back and forth, immediately washed five times with PBS to remove nonadherent bacteria, and then lysed by incubation for 10 min in 1 ml of water followed by vigorous pipetting. After lysis, the number of cell-associated bacteria was determined by plating for CFU on BCYE. Adherence assays on formaldehyde-fixed cells were carried out in the same manner except that the cells were fixed in 3.7% formaldehyde for 10 min, washed three times with PBS, and suspended in RPMI prior to addition of the bacteria. The bacteria were coincubated with the cells for 30 min (HL-60) or 90 min (HEp-2). Adherence levels were determined by calculating the percentage of the inoculum that became cell associated over the course of the assay [i.e., percent adherence = 100 × (CFU cell associated/CFU in inoculum)]. In order to correct for variation in levels of uptake between experiments, adherence is reported relative to that of wild-type strain AA100 at 37°C (i.e., relative adherence = percent adherence of test strain/percent adherence of AA100 at 37°C).
Intracellular growth assays.
The growth kinetics of 30°C- and 37°C-grown L. pneumophila in PMA-differentiated HL-60 cells at 37°C was determined as described previously (7, 8). Briefly, bacteria were added to a monolayer of 106 cells/well of HL-60 cells in 24-well tissue culture dishes at a multiplicity of infection of 0.1 and incubated for 30 min at 37°C. Extracellular bacteria were killed by gentamicin treatment for 2 h, the cells were washed twice, and one set of triplicate wells was lysed with water and another set of wells was incubated in fresh medium at 37°C for 72 h before lysis in water. Dilutions of the lysate were plated to determine the CFU present. Survival is expressed as the portion of CFU present at 72 h compared to time zero (T0 = 2.5 h), i.e., mean (CFU T72/ T0) × 100.
Microscopy techniques.
Adherence and entry levels were confirmed through examination of stained coverslips by fluorescence microscopy. We differentially stained intracellular and extracellular bacteria to allow discrimination between adherence and uptake. Intracellular and extracellular bacteria were differentially stained by first prestaining the bacteria prior to infection of cells followed by antibody detection of extracellular bacteria (without permeabilizing the cells) with polyclonal anti-L. pneumophila antibody. The rabbit polyclonal anti-L. pneumophila antibody was prepared and tested as described previously (8). This procedure allows intracellular bacteria to appear green and extracellular bacteria to stain both green and red by fluorescence microscopy. Basically, L. pneumophila cells were prestained with Oregon green (OG) prior to infection in a similar manner to that previously described using fluorescein (18, 47). The bacteria were suspended in PBS plus 5 μg of OG/ml, incubated at 30 or 37°C (depending on the prior growth temperature) for 30 min, and washed five times to remove unbound OG. Stained bacteria were then used to infect cells in the same manner as for standard entry assays, except that no gentamicin incubation was used. After washing, the monolayers were fixed with 2% paraformaldehyde in PBS for 1 h at 4°C followed by staining with primary and secondary antibody as described previously (8), except that the secondary antibody was conjugated with R-phycoerythrin (Becton Dickinson). Dual images of multiple fields were captured using an Optronics charge-coupled device video camera. The OG staining procedure did not affect bacterial viability (data not shown) or the ability of L. pneumophila to adhere to or enter into HL-60 or HEp-2 cells (data not shown). Viability of the labeled bacteria was confirmed using the LIVE-DEAD assay (Molecular Probes, Eugene, Oreg.) and by plating dilutions for CFU on BCYE agar (Difco).
DNA sequence analysis.
DNA sequencing was performed as described previously (10) using a BigDye Terminator (Applied Biosystems) cycle sequencing apparatus and subsequent analysis on an ABI 310 automated sequencing apparatus (Applied Biosystems). Sequence analysis and assembly were carried out using Gene Construction Kit 2 (Textco), searches for homologous sequences were done using BLAST (1), and protein alignments were made using MegAlign (DNASTAR).
Construction of ΔlvhB2 and complementation.
An in-frame deletion in the coding region of lvhB2 was constructed by overlapping PCR as described previously (10). In this construct, the entire LvhB2 coding region was deleted with the exception of the ATG and TGA codons. The resulting PCR product has approximately 1 kbp of DNA flanking the lvhB2 start and stop codons and was cloned into the NotI site of pJDC15 (10). The resulting plasmid was transformed into L. pneumophila, and allelic exchange selected for sucrose and kanamycin as negative and positive selectable markers, respectively, as described previously (10). The presence of the appropriate deletion in the resulting lvhB2 mutant strain (ΔlvhB2) was confirmed by Southern and PCR analyses (data not shown). The complementing plasmid for lvhB2, pJDC38, was constructed using restriction digestion of L34 to delete all potential coding regions other than lvhB2. The resulting plasmid was then transformed into ΔlvhB2 and tested for complementation of the entry phenotype.
Statistical analyses.
All in vitro experiments were carried out in triplicate and repeated at least three times. The significance of the results was analyzed using analysis of variance. P values of <0.05 were considered significant.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of lvhB2 and the surrounding chromosomal region from L. pneumophila strain AA100 is AF410854.
RESULTS
Use of an expression library to identify loci that affect interactions with host cells.
Our previous studies found that a number of proteins are differentially regulated under growth conditions that enhance entry by L. pneumophila (8). We wished to develop a strategy that would allow identification of L. pneumophila genes specifically involved in interactions with host cells rather than other physiological processes that are expressed differently because of growth conditions. Since several genes involved in the efficiency of host cell infection appear to be repressed under standard laboratory conditions, regulatory mutations that increase their expression could result in an enhanced host cell infection phenotype. It is possible to enrich for enhanced host cell infection mutants from a population of bacteria by using entry assays that are based on gentamicin protection, where only those bacteria that enter host cells survive. Our investigators recently used a similar strategy to identify three L. pneumophila loci that are involved in entry (10). In the present study, we constructed a total L. pneumophila genomic DNA library in the low-copy-number vector pWKS130. This vector allows constitutive expression of L. pneumophila genomic DNA fragments from the trp-lac promoter (Ptac). This library was then enriched for L. pneumophila clones that display an enhanced host cell infection phenotype by using entry assays. We optimized these assays for time of coincubation, number of host cells used, host cell type, and multiplicity of infection to result in an uptake of 1 to 10 wild-type bacteria/well (data not shown).
We examined the phenotype of individual clones from the L. pneumophila expression library before and after enrichment. Prior to enrichment, 40 random clones were screened in standard entry assays and compared to wild-type L. pneumophila. None of the preenrichment clones displayed higher levels of entry into host cells than wild type (data not shown). After enrichment, two clones (5%), designated L1 and L34, out of 35 displayed at least twofold-enhanced entry into HEp-2 cells (data not shown) compared to that into the wild type (P < 0.01). In order to characterize the positive clones identified with this approach, plasmids were isolated from them and retransformed into wild-type L. pneumophila cells and the host cell infection phenotype of the resulting transformants was examined. All of the resulting retransformed clones displayed the enhanced host cell infection phenotype of the original clone (data not shown). These data suggest that the plasmids carrying L. pneumophila chromosomal fragments, rather than secondary mutations elsewhere in the chromosome, are responsible for the observed phenotype.
Analysis of clone L34.
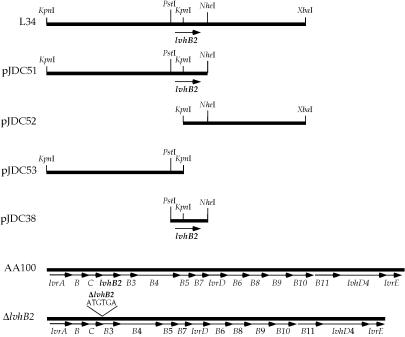
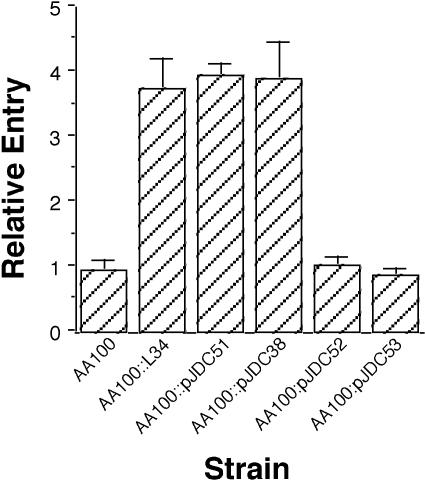
Restriction analysis of the clones that display an enhanced host cell infection phenotype, L1 and L34, with PstI, KpnI, and NheI demonstrated that they have identical physical maps (data not shown). A series of deletions were constructed in L34 to dissect the region responsible for the enhanced host cell infection phenotype (Fig. 1). We found that an internal PstI-NheI fragment is needed for the enhanced host cell infection phenotype conferred by the L34 plasmid (Fig. 2). In addition, neither portion of the L34 plasmid insert to the left (pJDC53) or right (pJDC52) of the KpnI restriction site conferred enhanced host cell infection alone, suggesting that the KpnI restriction site interrupts an open reading frame (ORF) that is necessary for the phenotype conferred by this plasmid. The L. pneumophila chromosomal DNA insert in the L34 plasmid was sequenced and found to contain an ORF encoding a putative protein of 96 amino acids in length that is interrupted by digestion with KpnI. This ORF is in the proper orientation to allow expression from the pWKS130 Ptac promoter. We initially designated this gene enhD to indicate that it has the ability to confer enhanced host cell infection. This gene has recently been sequenced in another strain of L. pneumophila, where it was designated lvhB2 and part of a large putative operon involved in type IV secretion (39). Although the lvh operon appears to play a role in conjugation, its role in virulence has not been investigated.
FIG. 1.
Constructs derived from enhanced entry clone L34 and structures of the lvh region in wild-type (AA100) and the lvhB2 in-frame deletion (ΔlvhB2) mutant L. pneumophila. The chromosomal region surrounding lvhB2 encodes a large putative operon in the wild-type strain. The structure of the in-frame deletion in the lvhB2 coding region is shown above the ΔlvhB2 construct. Arrows indicate putative coding regions and the direction of transcription. All constructs are drawn to scale (L34 is 5.7 kbp) except the wild-type and ΔlvhB2 chromosomal regions, which are scaled relative to each other (the AA100 lvh region is 13.1 kbp).
FIG. 2.
Ability of L34 and the various deletions in this construct to confer enhanced entry into HEp-2 cells by L. pneumophila. Entry of the wild-type strain (AA100) was arbitrarily set to 1. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. Results shown are for a typical experiment.
The protein encoded by lvhB2 is similar to the T-pilin subunit protein encoded by virB2 of Agrobacterium. This gene is part of a growing family of pilin genes that are involved in conjugation and the secretion of virulence proteins (26). Alignment of the similar proteins revealed that they contain several conserved domains (Fig. 3) and a signal peptidase cleavage site (26). Interestingly, LvhB2 has the greatest similarity to the Bordetella pertussis VirB2 homologue (25% identity, 70% similarity over 96 amino acids), which is involved in secretion of virulence proteins (31, 46). The next most similar proteins are from Brucella and Bartonella, both of which have approximately 20% identity with LvhB2. These data suggesting that lvhB2 may play a role in the efficiency of host cell infection are the first to infer a function other than conjugation for lvhB2 and indicate that further investigation of its role in pathogenesis is warranted.
FIG. 3.
Alignment of LvhB2 with other similar bacterial pilin proteins. Conserved amino acid residues are shown in filled boxes. Numbers to the right of the alignment indicate the amino acid position within the protein. The putative signal peptidase cleavage site is indicated by an arrow above the alignment. Abbreviations for aligned proteins and their accession numbers are as follows: LpLvhB2, L. pneumophila LvhB2 (AF410854); BsVirB2, Brucella suis VirB2 (AAD56612); BpPtlA, B. pertussis PtlA (AAB27428); BhVirB2, Bartonella henselae VirB2 (AAD48919); AtVirB2, Agrobacterium tumefaciens VirB2 (AAB28331); pKM101TraM, plasmid pKM101 TraM (I79265); R388TrwM, plasmid R388 TrwM (BAB12656); RP4TrbC, plasmid RP4 TrbC (AAA26429); FTraA, F plasmid TraA (NP_061453); SmVirB2, Sinorhizobium meliloti VirB2 (AAK65376); XfVirB2, Xylella fastidiosa VirB2 (NP_061661).
lvhB2 affects the efficiency of host cell infection.
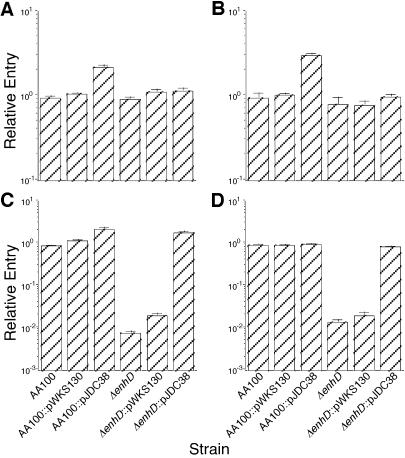
In order to investigate whether lvhB2 plays an important role in the ability of L. pneumophila to infect host cells, we compared the interactions of an lvhB2 mutant and complemented strain with that of the wild type in the human monocyte/macrophage cell line HL-60. Since the lvhB2 gene is part of a potentially large operon, we constructed an in-frame deletion (ΔlvhB2) in this gene to prevent polar effects on downstream genes (Fig. 1). This construct carries a complete in-frame deletion in the LvhB2 coding region, producing an ORF that encodes only the translational start and stop codons for this protein. An L. pneumophila strain carrying this mutation was constructed by allelic exchange using sucrose as a counterselectable marker and confirmed by Southern and PCR analyses (data not shown). We found that the lvhB2 mutant does not display a significant defect in the ability to infect human monocytic or epithelial cell lines when grown at 37°C (Fig. 4A and B). This observation suggests that either this gene plays no role in host cell infection or it is not expressed well at higher temperatures under standard laboratory growth conditions unless it is expressed from an exogenous promoter. This is the case for the L34 construct, where the gene is downstream of the Ptac promoter.
FIG. 4.
Entry of L. pneumophila wild type (AA100) and lvhB2 in-frame deletion (ΔlvhB2) with the vector alone (AA100::pWKS130 ΔlvhB2::pWKS130) or the vector containing lvhB2 (AA100::pJDC38 ΔlvhB2::pJDC38) at 37°C into the epithelial (A and C) or monocytic (B and D) cell line after growth of the bacteria at 37°C (A and B) or 30°C (C and D). Entry of the wild-type strain was arbitrarily set to 1. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. Results shown are for a typical experiment.
T-pilus production and T-DNA transfer in Agrobacterium are optimal at lower temperatures (19, 20). Thus, we examined the effects of bacterial growth temperature on lvhB2 function. When the bacteria were grown at 30°C, the L. pneumophila lvhB2 mutant was ∼100-fold reduced in entry compared to wild type (Fig. 4C and D). This defect in the ability to infect host cells is complemented by the presence of vector containing only the L. pneumophila lvhB2 gene (pJDC38). Since all host cell infections were carried out at 37°C, this difference is likely due to changes in expression of lvhB2 at lower temperatures.
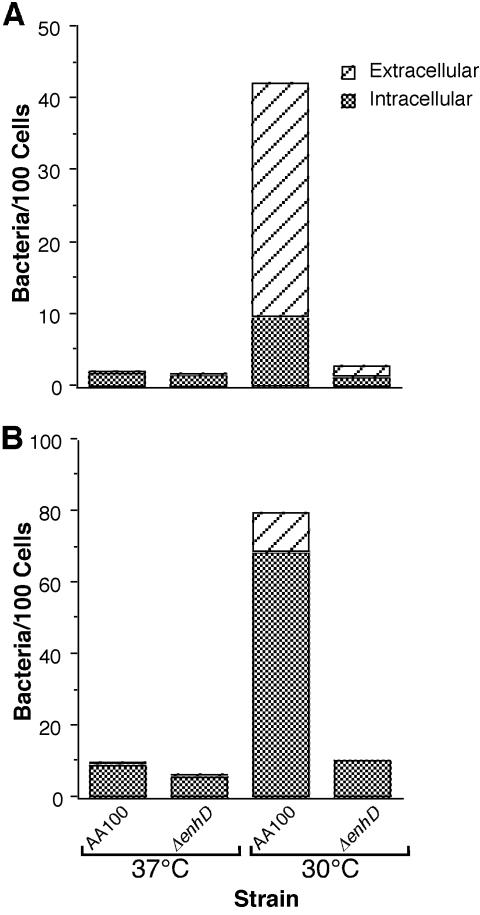
In order to confirm the difference in the ability of the lvhB2 mutant to infect host cells compared to wild-type L. pneumophila, we examined these infections by fluorescence microscopy. We used differential staining with fluorescent markers to determine whether the bacteria were intracellular or extracellular. All bacteria were first prestained with OG, and then only extracellular bacteria were labeled with a secondary antibody conjugated to R-phycoerythrin. This technique allows determination of the total number of cell-associated bacteria and whether they are adherent or intracellular and does not affect bacterial viability or their ability to enter into or adhere to host cells (data not shown). Similar to gentamicin protection assays, there is no apparent difference between the wild type and the ΔlvhB2 L. pneumophila mutant when the bacteria are grown at 37°C, but when grown at 30°C the ΔlvhB2 mutant displays obviously lower (P < 0.01) levels of cell association (Fig. 5). Since the staining technique used for microscopy should not be affected by bacterial viability, as would the CFU-based gentamicin protection assays, rapid killing by host cells following uptake is not solely responsible for the differences observed. The higher percentage of intracellular bacteria observed at 30°C with the monocytic cell line compared to the epithelial cell line is most likely the result of the constitutively phagocytic nature of monocytic cells. Based both on standard gentamicin protection assays for entry and microscopy, we found a significant defect in the ability of the L. pneumophila lvhB2 mutant to infect monocytic and epithelial cell lines.
FIG. 5.
Fluorescence microscopic quantitation of the number of bacteria and the proportion that are extracellular and intracellular per 100 cells for wild-type (AA100) and lvhB2 in-frame deletion (ΔlvhB2) mutant L. pneumophila strains grown at 37 and 30°C and infecting the epithelial (A) or monocytic (B) cell line. Fifty microscopic fields with greater than 10 cells per field were quantitated for each sample (>500 cells). Data points represent the means of assays done in triplicate. Results shown are for a typical experiment.
lvhB2 is involved in adherence.
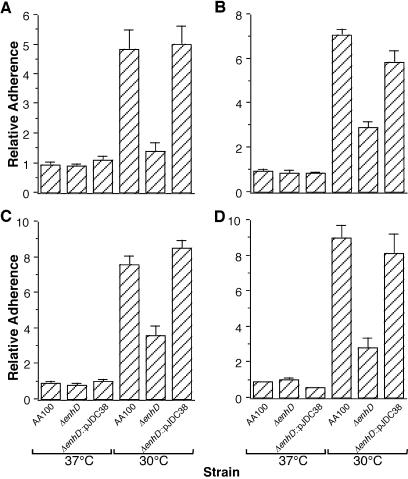
It is possible that the role of lvhB2 in entry is primarily the result of a defect in the ability to adhere to host cells rather than trigger uptake. In order to explore this further, we examined the adherence of ΔlvhB2 and wild-type L. pneumophila to epithelial and monocytic cell lines after growth of the bacteria at both 37 and 30°C (Fig. 6). To completely rule out the possibility that entry and/or intracellular killing were solely responsible for the data obtained, both standard immediate adherence assays and formaldehyde adherence assays, where internalization cannot occur, were used. Both assay methods have been previously shown to allow accurate measurement of adherence to host cells by L. pneumophila (9). Both the wild type and ΔlvhB2 mutant adhere better to epithelial and monocytic cell lines at 30°C than at 37°C (P < 0.01). Furthermore, the ΔlvhB2 mutant displays a defect in adherence at 30°C (P < 0.01) but adheres at the same levels as wild type at 37°C.
FIG. 6.
Adherence of wild-type (AA100), lvhB2 in-frame deletion (ΔlvhB2) mutant, and complemented mutant (ΔlvhB2::pJDC38) L. pneumophila strains grown at 37 and 30°C into the epithelial (A and C) or monocytic (B and D) cell line. Assays were carried out in either normal (A and B) or formaldehyde-fixed (C and D) cells. Adherence of the wild-type strain at 37°C was arbitrarily set to 1. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. Results shown are for a typical experiment.
Intracellular survival and replication.
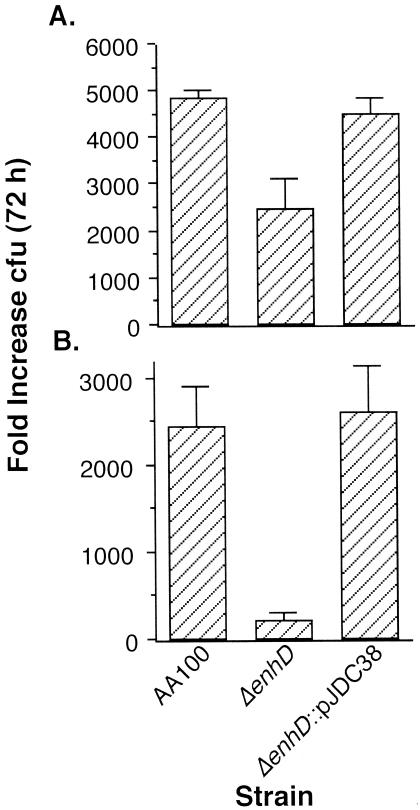
Our laboratory's previous studies have found that the initial interactions with host cells, such as adherence and entry, can correlate with the subsequent ability of L. pneumophila to survive and replicate (7, 9). Since the L. pneumophila lvhB2 gene affects the efficiency of host cell infection, it may also impact the resulting intracellular events. We examined replication of the lvhB2 mutant in HL-60 cells (Fig. 7) and found that although there is a moderate effect on intracellular replication (P < 0.01 after 72 h) when the bacteria are grown at 37°C prior to infection, there is a much greater impact when the bacteria are grown at 30°C (P < 0.001). The generation time in HL-60 cells of wild-type L. pneumophila grown at both 30 and 37°C is between 5 and 6 h, whereas the generation time of the lvhB2 mutant grown at 30°C is between 8 and 9 h and slightly greater than 6 h when grown at 37°C. These observations suggest that lvhB2 is important for intracellular survival and replication when the bacteria are grown at 30°C and less so when they are grown at 37°C.
FIG. 7.
Intracellular survival and replication of 37°C- (A) and 30°C- (B) grown L. pneumophila strain AA100 containing vector alone (AA100::pWKS130), the lvhB2 mutant containing the vector (ΔlvhB2::pWKS130), and the lvhB2 mutant containing the vector carrying lvhB2 (ΔlvhB2::pJDC38) in human HL-60 monocyte-derived macrophages over 72 h. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. Results shown are for a typical experiment.
DISCUSSION
Successful infection of mammalian cells by a bacterial pathogen involves a number of sequential events: the ability to find an appropriate host cell, adherence, entry, and initial intracellular survival. The complexity of this process makes it likely that a large number of bacterial genes are involved. We have recently identified several entry genes in L. pneumophila (10), but it is likely that many more genes involved in the ability to infect host cells remain to be found. Since there is a correlation between the mechanism of entry into host cells by L. pneumophila and virulence (7), loci involved in the ability to infect host cells may play an important role in pathogenesis. This conclusion is supported by the fact that the entry gene rtxA is also important in the ability of L. pneumophila to infect mice (9). In the present study, we have identified an additional gene, lvhB2, that is involved in the ability to efficiently infect and replicate within mammalian epithelial and monocytic cell lines. Since lvhB2 is not expressed optimally under standard laboratory growth conditions, further studies on the effects of growth temperature and other potential regulatory signals on lvhB2 expression and L. pneumophila virulence are necessary to better understand its function.
The lvhB2 gene was identified by enriching for clones that display an enhanced ability to infect host cells from an L. pneumophila expression library. It is not surprising that this type of approach would be effective for the identification of L. pneumophila genes involved in host cell infection, because a similar strategy has been previously used to identify three other loci involved in this process (10). The primary difference between the present strategy and the previous one is that low-copy vectors carrying a constitutive promoter were used instead of a low-copy cosmid vector. Gene dosage effects were most likely responsible for the observed enhanced entry phenotype in our previous study, whereas gene dosage effects and constitutive expression may both contribute to the L34 enhanced host cell infection phenotype.
The putative protein product encoded by the lvhB2 gene is similar to the major pilin subunit for a type IV secretion system. Recently, a type IV secretion system has been implicated in the adherence and entry of Campylobacter jejuni (2), suggesting that this mechanism of host cell infection may be used by other bacterial pathogens. In Campylobacter, another component of the type IV secretion system, a VirB11 homologue, has been shown to be required for both adherence and entry. There are at least four VirB11 homologues in L. pneumophila: LvhB11 in the lvh region, DotB in the dot/icm region, PilB in pilin biosynthesis, and a fourth homologue within the type II secretion system (6, 40). These observations combined with the function of lvhB2 suggest that both the lvh region and dot/icm complex may play a role in the initial interactions of L. pneumophila with host cells. This hypothesis is supported by observations that the effects of the dot/icm complex on intracellular trafficking are seen very early during the interaction of L. pneumophila with monocytes (36, 47), and two of the dot genes, dotH and dotO, can affect the rate of bacterial internalization (44). A role for dot/icm in uptake by macropinocytosis has also been confirmed by two separate groups (24, 45). Since lvhB2 appears to play a role in entry primarily at lower temperatures, possibly the dot/icm complex is an alternative system used at higher growth temperatures. However, detailed studies on the temperature regulation of dot/icm and lvh and the interplay between them are necessary to better understand the roles of these distantly related L. pneumophila type IV secretion systems in host cell infection.
Previous studies in L. pneumophila found that the lvh and dot/icm complex are both involved in conjugation, and some of their components can substitute for each other in this event (39). The lvh region was not found to have a role in the interaction of L. pneumophila with monocytes, in contrast to the critical role of dot/icm in intracellular trafficking and survival. Since these experiments were carried out with bacteria grown at 37°C, they are fairly consistent with our data where a large effect on host cell infection was only observed when L. pneumophila was grown at 30°C. However, we also observed a small, but significant, effect on intracellular growth when the bacteria were grown at 37°C that was not observed in the previous studies. This small discrepancy is most likely due to the different genetic backgrounds of the wild-type strains used in the two studies. Our finding that temperature plays a role in the ability of L. pneumophila to infect host cells is consistent with previous observations that temperature regulates the expression of a type IV pilus in L. pneumophila that also plays a role in adherence (27, 41). Furthermore, temperature has been shown to play an important role in the regulation of flagellum expression (23, 32), and motility has been implicated in the ability of L. pneumophila to infect host cells (12). Three other loci have been identified that play a role in entry by L. pneumophila, but the effects of temperature on their regulation have not been examined (10). Investigation of temperature regulation of these and other L. pneumophila virulence genes should provide useful information regarding the array of genes that are coordinately expressed with lvhB2.
These studies have identified a gene, lvhB2, that affects the efficiency of host cell infection by L. pneumophila when the bacteria are grown at 30°C. This observation might suggest that temperature is important in the ability of L. pneumophila to infect mammalian cells. However, it is equally likely that this gene is primarily involved in the ability to infect organisms living in aquatic environments. A role for environmental temperature in natural L. pneumophila infections has been previously suggested by the fact that they arise after exposure to aerosols from domestic water systems (3, 4, 22), where the optimal growth temperature of the inoculating dose is unclear. Our data combined with the observation that other genes involved in motility and host cell adherence are expressed optimally at lower temperatures (32, 41) suggest that growth of L. pneumophila under these conditions may be relevant to natural infections. Alternatively, these genes may play a role only during infections of environmental hosts, aquatic protozoa. It is likely that genes expressed at temperatures lower than 37°C play an important role in the ability of L. pneumophila to survive and replicate in aquatic environments. However, the theory that these genes are only important in environmental hosts does not fit well with the fact that the lvh region, including lvhB2, is found primarily in Legionella isolates that are the most likely to cause disease in humans (38). Thus, further studies are necessary to evaluate whether temperature is an important regulatory signal for the ability of L. pneumophila to infect humans, environmental protozoa, or both. These studies should provide insight into the complex interplay between Legionella infections in humans and environmental reservoirs for this pathogen.
Acknowledgments
This work was supported by AI40165 from the National Institutes of Health.
Editor: J. D. Clements
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaree, J. M., B. S. Fields, J. C. Feeley, G. W. Gorman, and W. T. Martin. 1986. Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl. Environ. Microbiol. 51:422-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breiman, R. F., B. S. Fields, G. N. Sanden, L. Volmer, A. Meier, and J. S. Spika. 1990. Association of shower use with Legionnaire's disease: possible role of amoebae. JAMA 263:2924-2926. [PubMed] [Google Scholar]
- 5.Cheng, X., J. D. Cirillo, and G. E. Duhamel. 1999. Coiling phagocytosis is the predominant mechanism for uptake of Serpulina pilosicoli by human monocytes. Adv. Exp. Med. Biol. 473:207-214. [DOI] [PubMed] [Google Scholar]
- 6.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo, S. L. G., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirillo, S. L. G., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 11.Davis, G. S., W. C. Winn, Jr., D. W. Gump, and H. N. Beaty. 1983. The kinetics of early inflammatory events during experimental pneumonia due to Legionella pneumophila in guinea pigs. J. Infect. Dis. 148:823-825. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dirita, V. J., and J. J. Mekalanos. 1989. Genetic regulation of bacterial virulence. Annu. Rev. Genet. 23:455-482. [DOI] [PubMed] [Google Scholar]
- 14.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Etr, S. H., L. Yan, and J. D. Cirillo. 2001. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect. Immun. 69:7310-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis, C. L., T. A. Ryan, B. D. Jones, S. J. Smith, and S. Falkow. 1993. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364:639-642. [DOI] [PubMed] [Google Scholar]
- 19.Fullner, K. J., J. C. Lara, and E. W. Nester. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273:1107-1109. [DOI] [PubMed] [Google Scholar]
- 20.Fullner, K. J., and E. W. Nester. 1996. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J. Bacteriol. 178:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannasca, K. T., P. J. Giannasca, and M. R. Neutra. 1996. Adherence of Salmonella typhimurium to Caco-2 cells: identification of a glycoconjugate receptor. Infect. Immun. 64:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henke, M., and K. M. Seidel. 1986. Association between Legionella pneumophila and amoebae in water. Isr. J. Med. Sci. 22:690-695. [PubMed] [Google Scholar]
- 23.Heuner, K., B. C. Brand, and J. Hacker. 1999. The expression of the flagellum of Legionella pneumophila is modulated by different environmental factors. FEMS Microbiol. Lett. 175:69-77. [DOI] [PubMed] [Google Scholar]
- 24.Hilbi, H., G. Segal, and H. A. Shuman. 2001. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603-617. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36:27-33. [DOI] [PubMed] [Google Scholar]
- 26.Lai, E.-M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 27.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mekalanos, J. J. 1992. Environmental signals controlling the expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. F., J. J. Mekalanos, and S. Falkow. 1989. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243:916-922. [DOI] [PubMed] [Google Scholar]
- 30.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 31.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 32.Ott, M., P. Messner, J. Heesemann, R. Marre, and J. Hacker. 1991. Temperature-dependent expression of flagella in Legionella. J. Gen. Microbiol. 137:1955-1961. [DOI] [PubMed] [Google Scholar]
- 33.Rechnitzer, C., and J. Blom. 1989. Engulfment of the Philadelphia strain of Legionella pneumophila within pseudopod coils in human phagocytes. APMIS 97:105-114. [PubMed] [Google Scholar]
- 34.Rittig, M. G., J. C. Jacoda, B. Wilske, R. Murgia, M. Cinco, R. Repp, G. R. Burmester, and A. Krause. 1998. Coiling phagocytosis discriminates between different spirochetes and is enhanced by phorbol myristate acetate and granulocyte-macrophage colony-stimulating factor. Infect. Immun. 66:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rittig, M. G., A. Krause, T. Häupl, U. E. Schaible, M. Modolell, M. D. Kramer, E. Lütjen-Drecoll, M. M. Simon, and G. R. Burmester. 1992. Coiling phagocytosis is the preferential phagocytic mechanism for Borrelia burgdorferi. Infect. Immun. 60:4205-4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Samrakandi, M. M., S. L. Cirillo, D. A. Ridenour, L. E. Bermudez, and J. D. Cirillo. 2002. Genetic and phenotypic differences between Legionella pneumophila strains. J. Clin. Microbiol. 40:1352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 40.Segal, G., and H. A. Shuman. 1999. Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii. Mol. Microbiol. 33:669-670. [DOI] [PubMed] [Google Scholar]
- 41.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szczepanski, A., and H. B. Fleit. 1988. Interaction between Borrelia burgdorferi and polymorphonuclear leukocytes: phagocytosis and the induction of the respiratory burst. Ann. N. Y. Acad. Sci. 539:425-428. [Google Scholar]
- 43.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 44.Watarai, M., H. L. Andrews, and R. R. Isberg. 2001. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 39:313-329. [DOI] [PubMed] [Google Scholar]
- 45.Watarai, M., I. Derre, J. Kirby, J. D. Growney, W. F. Dietrich, and R. R. Isberg. 2001. Legionella pneumophila is internalized by a macropinocytic uptake pathway controlled by the dot/icm system and the mouse Lgn1 locus. J. Exp. Med. 194:1081-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 66:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn, W. C., Jr., and R. L. Myerowitz. 1981. The pathology of Legionella pneumonias. A review of 74 cases and the literature. Hum. Pathol. 12:401-422. [DOI] [PubMed] [Google Scholar]