Abstract
The iron-binding protein lactoferrin is a ubiquitous and abundant constituent of human exocrine secretions. Lactoferrin inhibits bacterial growth by sequestering essential iron and also exhibits non-iron-dependent antibacterial, antifungal, antiviral, antitumor, anti-inflammatory, and immunoregulatory activities. All of these non-iron-dependent activities are mediated by the highly charged N terminus of lactoferrin. In this study we characterized a Lys/Arg polymorphism at position 29 in the N-terminal region of human lactoferrin that results from a single nucleotide polymorphism in exon 1 of the human lactoferrin gene. We expressed cDNAs encoding both lactoferrin variants in insect cells and purified the two proteins by ion exchange chromatography. The two lactoferrin variants exhibited nearly identical iron-binding and iron-releasing activities and equivalent bactericidal activities against a strain of the gram-negative bacterium Actinobacillus actinomycetemcomitans. When tested against the gram-positive species Streptococcus mutans and Streptococcus mitis, however, lactoferrin containing Lys at position 29 exhibited significantly greater bactericidal activity than did lactoferrin containing Arg. In addition, the Lys-containing lactoferrin stimulated bovine tracheal epithelial cells to synthesize much higher levels of tracheal antimicrobial peptide mRNA than did the Arg-containing variant. A genotyping assay that distinguished between the two alleles based on a polymorphic EarI restriction site showed that the Lys and Arg alleles had frequencies of 24% and 76%, respectively, among 17 healthy human subjects, and 72% and 28%, respectively, among nine patients with localized juvenile periodontitis. Our findings suggest that these two lactoferrin variants are functionally different and that these differences may contribute to the pathogenesis of localized juvenile periodontitis.
Lactoferrin is an 80-kDa iron-binding glycoprotein found in the saliva, milk, tears, and other exocrine secretions of mammals (30). Lactoferrin is also released from the specific, secondary granules of neutrophils during inflammatory responses (31). Lactoferrin exhibits bacteriostatic activity against a wide range of gram-negative and gram-positive bacteria due to its ability to chelate iron, which is essential for microbial growth (2, 3). In addition, lactoferrin exhibits non-iron-dependent antibacterial, antifungal, antiviral, antitumor, anti-inflammatory, and immunoregulatory activities (4, 7, 12, 44). Lactoferrin constitutes an important component of the innate immune system (34, 41) and may represent a novel therapeutic with broad-spectrum potential (45).
Human lactoferrin is a single polypeptide chain containing 692 amino acids that is folded into two symmetric lobes connected by a hinge region (1). Each lobe is capable of binding one Fe3+ ion together with one CO32− ion in a deep intralobe cleft that contains a nonheme iron-binding site (1). The non-iron-dependent activities of human lactoferrin have been shown to be mediated by a 47-residue peptide (corresponding to residues 1 to 47 of the mature human lactoferrin protein) that is released upon cleavage of human lactoferrin with pepsin (5, 6, 8, 27, 28, 35, 46). These residues form a β-α-β unit located on the exposed surface of the human lactoferrin protein that is distinct from the iron-binding region. This exposed region contains nine basic amino acid side chains projecting from its surface, most of which are located in a highly charged N-terminal tail (residues 1 to 5; GRRRR) and an amphipathic region near the C terminus of helix A (residues 28 to 31; RKVR) that are in close proximity in the folded human lactoferrin protein (1, 6). These basic residues have been shown to participate directly in bacterial killing, probably by binding to lipopolysaccharide (13), porins (38), or other bacterial surface molecules (20), and disrupting the cell membrane (14). This N-terminal region has also been shown to bind to specific DNA sequences located upstream from various genes (21), resulting in transcriptional activation (43).
In this study we characterized a Lys/Arg polymorphism that occurs at position 29 in the N-terminal alpha-helical region of human lactoferrin. This polymorphism results from an A↔G transition in exon 1 of the human lactoferrin gene. Using proteins purified from insect cells infected with recombinant baculoviruses, we show that both lactoferrin variants bind and release iron with equal efficiency and display equivalent bactericidal activities against gram-negative bacteria. Lactoferrin containing Lys at position 29, however, exhibited significantly greater bactericidal activity against gram-positive bacteria and significantly greater transcriptional activation activity when tested against primary bovine tracheal epithelial cells. With a PCR-based genotyping assay, we also show that the allele encoding the Lys variant of human lactoferrin occurs more frequently in localized juvenile periodontitis patients than in healthy subjects.
MATERIALS AND METHODS
Cloning and sequencing of human lactoferrin cDNA.
Total RNA was purified from blood collected from a healthy African-American female subject with the QIAamp RNA blood mini kit (Qiagen). RNA was reverse transcribed into cDNA with SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primer. The cDNA was amplified by PCR with primers 5′-CAAGTCGCCTCCAGACC-3′ and 5′-CTGGGCCATCTTCTTCGG-3′, which hybridize to sequences upstream and downstream from the human lactoferrin coding region (corresponding to coordinates 272 to 288 and 2433 to 2450, respectively, in GenBank accession no. NM_002343). The reaction mixture contained 50 pmol of each primer, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 2.5 U of Taq DNA polymerase (PE Biosystems), and 40 ng of cDNA as a target in a 100-μl reaction overlaid with 100 μl of mineral oil. Thirty cycles of PCR were performed under the following conditions: denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The resulting PCR product was ligated into plasmid LITMUS28 (New England Biolabs) and subjected to DNA sequence analysis as previously described (25).
Site-directed mutagenesis.
A human lactoferrin cDNA that contained an AGA (Arg) codon at position 29 was obtained from Anthony Schryvers (University of Calgary). This cDNA also contained a BamHI restriction site located 3 bp upstream from the human lactoferrin start codon and a HindIII site located 3 bp downstream from the human lactoferrin stop codon. This cDNA was digested with BamHI and HindIII and ligated into the BamHI/HindIII sites of LITMUS28, resulting in the plasmid LITMUS-hLf(R). The mutagenic oligonucleotide 5′-GCAAAGGAATATGAGAAAAGTGCGTGGCCCTCCTG-3′ (corresponding to coordinates 126 to 160 in GenBank no. AY137470) and its complement were used to mutate codon 29 in LITMUS-hLf(R) from AGA to AAA (italic) with a primer-mediated mutagenesis procedure (23). The PCR product was cloned into LITMUS28, and the A→G nucleotide substitution in the resulting plasmid, designated LITMUS-hLf(K), was confirmed by DNA sequence analysis.
Insect cell culture and production of recombinant human lactoferrin.
Stock cultures of Spodoptera frugiperda Sf9 cells were grown at 28°C as monolayers in Sf 900 II SFM medium (Life Technologies) containing 10 μg of gentamicin per ml. Confluent monolayers were subcultured by removing the cells from the flask by gentle pipetting and diluting the cells to 2 × 106 cells/ml in fresh medium. To construct recombinant baculoviruses, the BamHI/HindIII DNA inserts from LITMUS-hLf(K) and LITMUS-hLf(R) were ligated into the BamHI and HindIII sites of plasmid pFastBac1 (Life Technologies), which placed each cDNA under control of the baculovirus polyhedrin promoter. The resulting plasmids, pFB-hLf(K) and pFB-hLf(R), respectively, were used to generate recombinant baculoviruses by site-specific transposition into a baculovirus shuttle vector as described in the Bac-to-Bac baculovirus expression system manual (Invitrogen). Recombinant baculoviruses (5 μg each) were transfected into Sf9 cells (2 × 106 cells in a Corning 25-cm2 tissue culture flask) with Cellfectin reagent (Life Technologies) according to the instructions supplied by the manufacturer. Cells were incubated for 7 h at 28°C, the transfection medium was removed, 10 ml of fresh 900 II SFM medium was added, and the cells were incubated for an additional 5 days. The supernatant was harvested by centrifugation at 5,000 × g for 15 min, and several recombinant viral plaques were amplified. For large-scale production of recombinant human lactoferrin, 100 ml of Sf9 cells (2 × 106 cells/ml) in a 500-ml Erlenmeyer flask was infected with recombinant viral stocks (at a multiplicity of infection of 1:10), and the flasks were incubated at 28°C with gentle shaking for 4 days.
Protein purification.
All of the following procedures were carried out at room temperature. The supernatant from 1 liter of culture was collected by centrifugation at 5,000 × g for 15 min and concentrated to 100 ml with a 30-kDa cutoff ultrafiltration unit (model RA2000; Amicon). The concentrated supernatant was loaded directly onto a 15-ml bed volume SP Sepharose Fast Flow column (Pharmacia) that had been washed with 100 ml of water and equilibrated with 75 ml of 0.2 M sodium acetate (pH 5.6). The column was washed sequentially with 50 ml of 0.2, 0.4, and 0.7 M NaCl (in 0.2 M sodium acetate, pH 5.6). The recombinant human lactoferrin protein was eluted with a linear NaCl gradient (0.7 to 0.8 M in 0.2 M sodium acetate, pH 5.6). The presence of recombinant human lactoferrin in the eluted fractions was assessed by measuring the optical density of each fraction (at 280 nm) and by Western blot analysis with rabbit anti-human lactoferrin IgG as described below. Positive fractions were concentrated with a Centriprep-30 concentrator (Amicon) and loaded onto a 10-ml bed volume Sephadex G-25 column (Pharmacia) equilibrated with phosphate-buffered saline in order to remove excess salts. The recombinant human lactoferrin protein was eluted from the column with phosphate-buffered saline, and fractions were assayed for the presence of protein by spectrophotometry at 280 nm and Western blotting as described below. Pooled fractions were concentrated with a Centricon-30 concentrator. Purified proteins were stored at −20°C until further use.
Mass spectrometric analysis of 1 μl (100 pmol) of purified human lactoferrin was carried out on a Perspective Biosystems DE Pro matrix-assisted laser desorption ionization (MALDI) mass spectrometer as previously described (39). N-terminal amino acid sequence analysis of 10 μg of purified protein was performed with the Edman degradation procedure on an Applied BioSystems 477 protein sequencer.
Polyacrylamide gel electrophoresis and Western blotting.
Proteins were separated on precast 10% polyacrylamide gels with 5% stacking gels (Bio-Rad) with the buffer system of Laemmli (40). Proteins were either stained with a silver staining kit (Pharmacia) or transferred to Immobilon-P membranes (Millipore) and incubated with a 1:1,000 dilution of rabbit anti-human lactoferrin IgG (Sigma). Binding of antibody was detected with alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma) and a p-nitrophenylphosphate colorimetric detection reagent.
Iron binding assay.
Iron binding assays were carried out in 1.5-ml microcentrifuge tubes containing 30 μg of recombinant human lactoferrin protein per ml and various concentrations of 59Fe (as FeCl2, 500 MBq/ml) in 50 μl of phosphate-buffered saline. Tubes were incubated for 30 min at room temperature. Aliquots were removed and electrophoresed through polyacrylamide gels as described above. 59Fe-labeled recombinant human lactoferrin proteins were visualized by autoradiography, cut from the gel with a razor blade, and placed in liquid scintillation vials containing 5 ml of scintillation fluid. Radioactivity was measured in a TriCarb-1500 liquid scintillation counter (Packard).
Iron release assay.
Recombinant human lactoferrin proteins were incubated in 59Fe (110 MBq/ml) for 30 min as described above. Aliquots of the iron-saturated recombinant human lactoferrin solutions were dialyzed for 24 h at room temperature against four separate buffers at four different pH values: 0.2 M NaCl-0.05 M Tris-HCl (pH 6.0), 0.05 M MES (pH 5.0), 0.05 M sodium acetate (pH 3.5) and 0.05 M glycine-HCl (pH 2.0). The iron content of recombinant human lactoferrin proteins at different pHs was measured in a liquid scintillation counter as described above.
Bactericidal assays.
The bacterial strains used in this study were Actinobacillus actinomycetemcomitans CU1060 (17), Escherichia coli NCTC 8007 (obtained from the National Collection of Type Cultures, Colindale, United Kingdom), Streptococcus mutans ATCC 25175 (obtained from the American Type Culture Collection, Manassas, Va.), and Streptococcus mitis NJ9705 (24). A. actinomycetemcomitans and streptococcal strains were grown in Trypticase soy broth (BD Biosystems) supplemented with 6 g of yeast extract and 8 g of glucose per liter at 37°C in 10% CO2. E. coli was grown in Luria-Bertani broth at 37°C in air. The lactoferrin killing assay was carried out in 1.5-ml microcentrifuge tubes containing 1 ml of bacterial cell suspension (107 to 109 CFU/ml) and 500 μg of recombinant human lactoferrin per ml as previously described (15). Briefly, cell suspensions were incubated with gentle agitation at 37°C. At various times, 100-μl aliquots were removed from each tube, serially diluted, and plated on agar plates with a spiral plater. Plates were incubated for 3 days at 37°C in 10% CO2, and colonies were enumerated.
Measuring tracheal antimicrobial peptide mRNA levels.
Bovine tracheal epithelial cells were obtained and cultured as previously described (47, 48). The cultures used for experiments were from the first passage and were approximately 80 to 100% confluent. Tracheal epithelial cells were stimulated with 100 ng of Pseudomonas aeruginosa lipopolysaccharide (Sigma Chemical Co.) or with 5 to 50 μg of different recombinant human lactoferrin proteins per ml for 18 h. The recombinant human lactoferrin samples contained less than 12 pg of endotoxin per ml as determined by a Pyrotell Limulus amebocyte lysate assay (Associates of Cape Cod, Falmouth, Mass.). Following the incubation, mRNA was extracted with the RNeasy mini kit (Qiagen).
cDNA was synthesized from 1 mg of mRNA with 200 units of Superscript II reverse transcriptase (Invitrogen) according to the protocol supplied by the manufacturer. PCR was performed with 12.5 units of Taq DNA polymerase (Invitrogen), with a standard protocol. The reaction was carried out with an initial denaturing step at 95°C for 3 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. An additional extension step of 72°C for 15 min followed the 30 cycles. The sequences of the bovine β-defensin oligonucleotide primers were 5′-GCCAGCATGAGGCTCCAT-3′ (sense) and 5′-AACAGGTGCCAATCTGT-3′ (antisense). The sequences of the bovine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) oligonucleotide primers were 5′-TGGCAAAGTGGACATCGTCG-3′ (sense) and 5′-TGGCGTGGACAGTGGTCATAAGTC-3′ (antisense). The PCR product was electrophoresed through a 2% agarose gel and stained with ethidium bromide. The amplified tracheal antimicrobial peptide product was visualized by Southern blot hybridization with 32P-labeled oligonucleotides as previously described (40). The bands were visualized by PhosphorImage analysis (Molecular Dynamics, Sunnyvale, Calif.).
PCR genotyping.
Epithelial cells were collected from healthy volunteers and localized juvenile periodontitis patients by scraping the inside of a cheek with a tongue depressor. Epithelial cell genomic DNA was isolated with a DNeasy tissue kit (Qiagen). A total of 100 ng of genomic DNA was amplified by PCR with 50 pmol each of primers 5′-CTCGTCCTGCTGTTCCTC-3′ and 5′-AGCATCGGCCCTGTTTTCC-3′ (corresponding to coordinates 16 to 33 and 210 to 228 in GenBank accession no. AY137470, respectively) in a 100-μl reaction as described above. Ten cycles of PCR at 94°C, 55°C, and 72°C (1 min, 1 min, and 20 s, respectively) were performed. The PCR product (213 bp) was purified with a QIAquick PCR purification kit (Qiagen) and eluted in 50 μl of Tris-EDTA buffer. Then 2 μl of the purified PCR product was reamplified under the same conditions for 20 cycles with PCR primers 5′-GTTCCTCGGGGCCCTCGG-3′ and 5′-TGTTTTCCGCAATGGCCTG-3′ (corresponding to coordinates 27 to 44 and 199 to 217 in GenBank accession no. AY137470, respectively). The PCR product (190 bp) was purified with a QIAquick PCR purification kit and eluted in 30 μl of Tris-EDTA buffer; 1 μl of the purified PCR product was digested with EarI and the digestion products were electrophoresed through a 5% acrylamide-0.17% bisacrylamide gel in 1× Tris-borate-EDTA buffer and visualized by staining with ethidium bromide.
Nucleotide sequence accession numbers.
The human lactoferrin cDNA sequence determined in this study was submitted to GenBank under accession no. AY137470.
RESULTS
Identification of a polymorphism in the antibacterial region of human lactoferrin.
We collected blood from a healthy African-American female subject, purified total RNA from the blood, reverse transcribed the RNA into cDNA, and amplified a full-length human lactoferrin cDNA by PCR with primers that hybridize upstream and downstream from the human lactoferrin coding region. The DNA sequence of this human lactoferrin cDNA (GenBank accession no. AY137470) contained an AGA (Arg) codon at position 29 of the mature human lactoferrin protein. This sequence differed from that of the reference human lactoferrin cDNA sequence (accession no. X52941), which contains AAA (Lys) at codon position 29. A search of the National Center for Biotechnology single-nucleotide polymorphism database (www.ncbi.nlm.nih.gov/SNP) revealed that this A/G nucleotide polymorphism corresponded to a previously identified but nonvalidated human single-nucleotide polymorphism (rs no. 1126478). The AGA (Arg) codon was present in 2 out of 10 human lactoferrin sequences in the GenBank database (accession nos. M93150 and AF332168).
Cloning, expression, and purification of recombinant human lactoferrin in insect cells.
We cloned human lactoferrin cDNAs that contained AGA (Arg) and AAA (Lys) codons at position 29 into a baculovirus expression plasmid. These plasmids were used to construct two recombinant baculoviruses that expressed the Lys-29 and Arg-29 human lactoferrin variants (designated rhLf-K and rhLf-R, respectively) under control of the baculovirus polyhedrin promoter. Ion exchange chromatography was used to purify the recombinant human lactoferrin proteins from the conditioned medium of S. frugiperda cell cultures infected with these two recombinant baculoviruses.
Both recombinant human lactoferrin proteins eluted as a sharp peak at 0.75 M NaCl. Positive fractions were pooled and concentrated. Salt was removed from the purified recombinant proteins by chromatography on a Sephadex G-25 column. When analyzed by mass spectrometry, both proteins migrated as a peak at 77.9 kDa (data not shown), which corresponded to the predicted molecular mass of the recombinant proteins. N-terminal sequence analysis of both purified proteins yielded the amino acid sequence GRRRRSVQWCAV, which corresponded to the N-terminal sequence of lactoferrin isolated from human milk (36) and indicated that the signal sequences of the two proteins were correctly processed in insect cells.
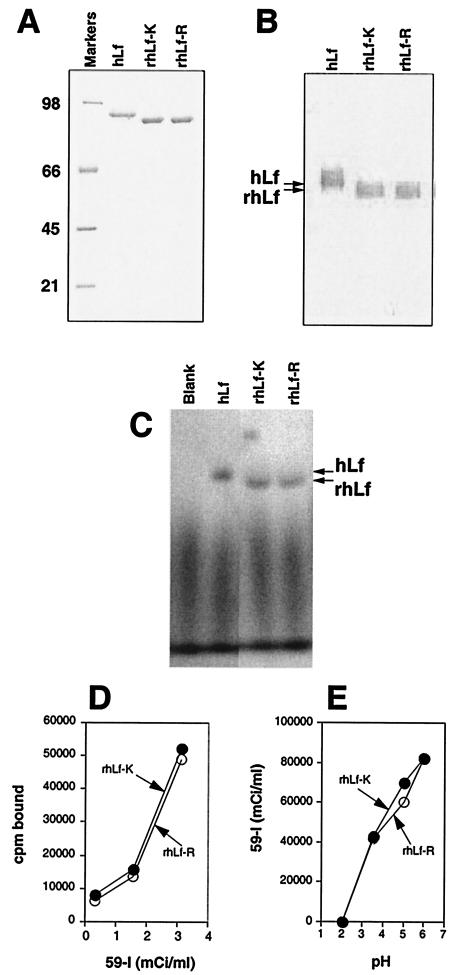
Figure 1A shows 1 μg of each recombinant human lactoferrin protein along with 1 μg of purified milk human lactoferrin, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by silver staining. The electrophoretic mobilities of the recombinant proteins were slightly faster than that of human lactoferrin from milk (80 kDa), probably due to differences in the glycosylation patterns of the proteins (39). Figure 1B shows milk human lactoferrin, rhLf-K, and rhLf-R analyzed by Western blotting with rabbit anti-human lactoferrin antiserum as a probe. All three lactoferrin proteins reacted equally with the anti-human lactoferrin antiserum.
FIG. 1.
Characterization of recombinant human lactoferrin proteins purified from insect cells. (A) SDS-PAGE analysis of 1 μg of human milk lactoferrin (hLf) and 1 μg each of rhLf-K and rhLf-R. The proteins were detected by silver staining. The sizes (in kilodaltons) of markers are indicated on the left. (B) Western blot analysis of milk human lactoferrin, rhLf-K, and rhLf-R with rabbit anti-human lactoferrin antiserum. The electrophoretic mobilities of human lactoferrin and recombinant human lactoferrin proteins are indicated on the left. (C) SDS-PAGE analysis of human lactoferrin and recombinant human lactoferrin proteins saturated with 59Fe. The proteins were visualized by autoradiography. The electrophoretic mobilities of human lactoferrin and recombinant human lactoferrin proteins are indicated on the right. (D) Iron-binding activities of recombinant human lactoferrin proteins in the presence of various concentrations of 59Fe. (E) Iron-releasing activities of recombinant human lactoferrin proteins saturated with 59Fe.
Iron-binding and iron-releasing activities of recombinant human lactoferrin proteins.
The ability of rhLf-K and rhLf-R to bind and release iron was measured (Fig. 1C to E). The proteins exhibited equivalent iron-binding properties as determined by autoradiography (Fig. 1C) and direct radiometric measurement (Fig. 1D). The ability of rhLf-K and rhLf-R to release iron was measured by dialyzing the 59Fe-saturated recombinant proteins against buffers of different pHs (39). Both proteins exhibited equivalent iron-releasing activities (Fig. 1E).
Antibacterial activities of recombinant human lactoferrin proteins.
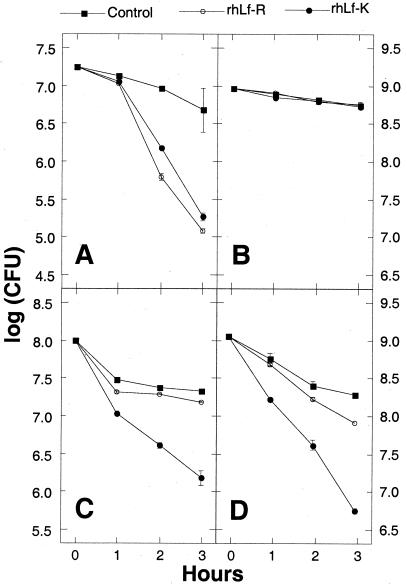
The ability of rhLf-K and rhLf-R to kill four species of bacteria was measured by incubating cells with the recombinant proteins for various lengths of time and measuring the CFU remaining in the culture (Fig. 2). Both recombinant human lactoferrin proteins exhibited equivalent bactericidal activities against a strain of the gram-negative bacterium Actinobacillus actinomycetemcomitans (Fig. 2A), and neither protein killed a lactoferrin-resistant strain of E. coli (Fig. 2B). When tested against the gram-positive species Streptococcus mutans (Fig. 2C) and S. mitis (Fig. 2D), rhLf-K exhibited significantly greater bactericidal activity than did rhLf-R at all three time points examined (P < 0.01 as determined by a Fisher's protected least-significant-difference test).
FIG. 2.
Antibacterial activities of human lactoferrin proteins. The bacterial species tested were A. actinomycetemcomitans (A), E. coli (B), S. mutans (C), and S. mitis (D). Bacterial cells were incubated in the absence of lactoferrin (control) or in the presence of 1 mg of each recombinant human lactoferrin protein per ml for the indicated times, and the numbers of CFU were measured.
Transcriptional activation activities of recombinant proteins.
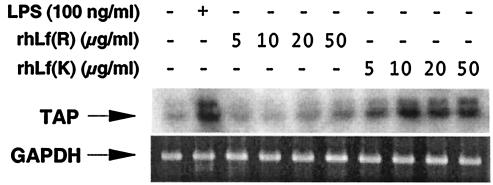
We tested the ability of rhLf-K and rhLf-R to stimulate transcription of tracheal antimicrobial peptide, a member of the β-defensin family of antimicrobial peptides (12). Previous studies have shown that transcription of tracheal antimicrobial peptide is activated in response to lipopolysaccharide and inflammatory cytokines such as tumor necrosis factor alpha and interleukin-1β (9, 10). Bovine tracheal epithelial cells were stimulated for 18 h with various amounts of rhLf-K and rhLf-R, and the amount of tracheal antimicrobial peptide mRNA present in the cells was determined by semiquantitative RT-PCR (Fig. 3). As expected, control tracheal epithelial cells stimulated with 10 ng of lipopolysaccharide per ml exhibited a significant increase in the amount of tracheal antimicrobial peptide mRNA. Tracheal epithelial cells stimulated with recombinant human lactoferrin proteins exhibited a dose-dependent stimulation of tracheal antimicrobial peptide mRNA levels. Tracheal epithelial cells stimulated with rhLf-K contained much higher levels of tracheal antimicrobial peptide mRNA than did cells stimulated with rhLf-R.
FIG. 3.
Induction of tracheal antimicrobial peptide mRNA in bovine tracheal epithelial cells stimulated for 18 h with lipopolysaccharide (LPS) or with various concentrations of rhLf-K or rhLf-R as determined by RT-PCR. PCR products amplified with tracheal antimicrobial peptide (TAP)-specific primers were visualized by autoradiography. PCR products amplified with glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primers were visualized by staining with ethidium bromide.
PCR genotyping.
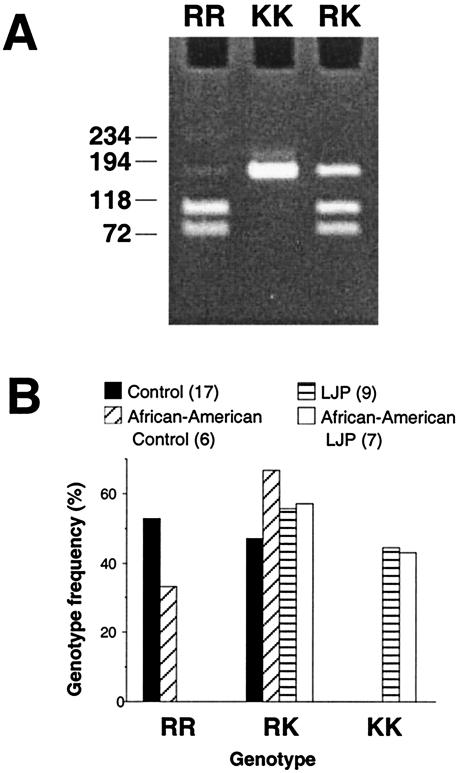
The A/G polymorphism in codon 29 of human lactoferrin results in the presence of an EarI restriction site (5′-GAAGAG-3′) in sequences that contain the G nucleotide (italic) which is absent in sequences that contain the A nucleotide. We developed a genotyping assay that utilized nested PCR followed by digestion with EarI (Fig. 4A) to detect the presence of this A/G polymorphism in a sample population of 17 healthy subjects and nine localized juvenile periodontitis patients. The frequencies of the KK, KR, and RR genotypes in these two groups were significantly different (P < 0.02 as determined by a Fisher's exact test) (Fig. 4B). No healthy subject harbored the KK genotype, and no subject from the localized juvenile periodontitis group harbored the RR genotype. The frequencies of the different genotypes in African-American subjects, who exhibit an increased incidence of localized juvenile periodontitis compared to other ethnic groups (29), followed a similar pattern.
FIG. 4.
Lactoferrin genotyping assay. (A) Examples of the genotyping assay carried out on subjects with the RR, KK, and RK genotypes are shown. The sizes (in base pairs) of molecular size markers run in an adjacent lane are shown on the left. (B) Lactoferrin genotype frequencies among 17 healthy subjects and nine localized juvenile periodontitis (LJP) patients. Also shown are the genotype frequencies among the subset of African-American subjects. The numbers in parentheses indicate the number of subjects in each group.
DISCUSSION
The results of the present study validate the existence of a Lys/Arg polymorphism at position 29 in the highly charged N-terminal region of human lactoferrin. Although the chemical nature of the polymorphic amino acid is conserved, our findings indicate that the Lys-containing variant (hLf-K) exhibits significantly greater non-iron-dependent antibacterial activity against gram-positive bacteria than does the Arg-containing variant (hLf-R). These data suggest that human lactoferrin may bind to different or slightly different receptors on gram-positive and gram-negative bacteria. Our findings also indicate that hLf-K exhibits significantly greater transcriptional activation activity than hLf-R. These data are consistent with the proposed interaction between the highly charged N-terminal region of human lactoferrin and eukaryotic cells (8, 27, 28, 46).
Localized juvenile periodontitis is a rapid and aggressive form of periodontitis that primarily affects African-American adolescents (51). Previous studies have reported increased lactoferrin levels (18, 19) and decreased lactoferrin iron saturation levels (16) in localized juvenile periodontitis patients, suggesting that lactoferrin may play a role in pathogenesis. In the present study, genotypic analysis of a small population of localized juvenile periodontitis patients and healthy subjects revealed a dramatic difference in the frequencies of the hLf-K and hLf-R alleles in these two groups (Fig. 4). The hLf-K allele occurred much more frequently in localized juvenile periodontitis patients than in healthy subjects (72% versus 24%). Significantly, no healthy subject was homozygous for the hLf-K allele, whereas no localized juvenile periodontitis patient was homozygous for the hLf-R allele. These data suggest that the polymorphism at position 29 of human lactoferrin may be a marker for susceptibility to localized juvenile periodontitis. These findings are consistent with previous studies which indicated that genetic variation plays an important role in periodontal disease (29, 32). A study testing the association between human lactoferrin allele frequencies and localized juvenile periodontitis in a larger population is in progress.
There are several possible mechanisms by which the observed functional differences between hLf-K and hLf-R could contribute to the pathogenesis of localized juvenile periodontitis. Localized juvenile periodontitis is associated with high levels of the gram-negative bacterium A. actinomycetemcomitans in the periodontal pocket (51). Localized juvenile periodontitis patients also exhibit significantly less proximal caries than control patients (42). These carious lesions result from colonization of interproximal tooth surfaces by the gram-positive bacterium S. mutans. It is possible that the oral microbiota of subjects harboring one or two hLf-K alleles contains lower levels of S. mutans due to the increased antibacterial activity of hLf-K against gram-positive bacteria (Fig. 2C and D). This alteration in the oral flora could account for the decreased incidence of proximal caries in localized juvenile periodontitis patients. It is also possible that the increased transcriptional activation activity of hLf-K contributes to an altered microenvironment which favors A. actinomycetemcomitans colonization. β-Defensins such as tracheal antimicrobial peptide and its human homologue hBD2 are broad-spectrum antimicrobial peptides that are active against various periodontal bacteria (33). Modulations in the expression of genes encoding these peptides could therefore result in changes in the host defensive environment against A. actinomycetemcomitans. Also, β-defensins are chemotactic for a variety of inflammatory cells (49, 50). A chronic elevation in β-defensin gene expression in the gingival epithelium, as may be seen with KK individuals, could result in an increased inflammatory response, such as is seen in localized juvenile periodontitis.
Rose et al. (37) recently showed that human lactoferrin cleaves an adhesin located on the surface of A. actinomycetemcomitans and that this cleavage results in decreased binding of the bacteria to epithelial cells. This cleavage may result from a serine protease activity exhibited by human lactoferrin (22). These data are consistent with previous studies which showed that treatment of A. actinomycetemcomitans with human lactoferrin results in decreased binding to epithelial cells (15). These findings raise the possibility that differences in the serine protease activities of hLf-K and hLf-R may influence the ability of A. actinomycetemcomitans to colonize the periodontal pocket.
Acknowledgments
We thank Tony Schryvers for providing a human lactoferrin cDNA and Scott Diehl for helpful discussions.
This work was supported in part by Public Health Service grant DE14897 (to G.D.).
Editor: F. C. Fang
REFERENCES
- 1.Anderson, B. F., H. M. Baker, G. E. Norris, D. W. Rice, and E. N. Baker. 1989. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 A resolution. J. Mol. Biol. 209:711-734. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, R. R., M. Brewer, and J. J. Gauthier. 1980. Bactericidal activity of human lactoferrin: sensitivity of a variety of organisms. Infect. Immun. 28:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, R. R., M. F. Cole, and J. R. McGhee. 1977. A bactericidal effect for human lactoferrin. Science 197:263-265. [DOI] [PubMed] [Google Scholar]
- 4.Baveye, S., E. Elass, J. Mazurier, G. Spik, and D. Legrand. 1999. Lactoferrin: a multifunctional glycoprotein involved in the modulation of the inflammatory process. Clin. Chem. Lab. Med. 37:281-286. [DOI] [PubMed] [Google Scholar]
- 5.Bellamy, W., M. Takase, K. Yamauchi, H. Wakabayashi, K. Kawase, and M. Tomita. 1992. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta 1121:130-136. [DOI] [PubMed] [Google Scholar]
- 6.Chapple, D. S., D. J. Mason, C. L. Joannou, E. W. Odell, V. Gant, and R. W. Evans. 1998. Structure-function relationship of antibacterial synthetic peptides homologous to a helical surface region on human lactoferrin against Escherichia coli serotype O111. Infect. Immun. 66:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conneely, O. M. 2001. Antiinflammatory activities of lactoferrin. J. Am. Coll. Nutr. 20:389S-395S. [DOI] [PubMed] [Google Scholar]
- 8.Di Biase, A. M., A. Pietrantoni, A. Tinari, R. Siciliano, P. Valenti, G. Antonini, L. Seganti, and F. Superti. 2003. Heparin-interacting sites of bovine lactoferrin are involved in anti-adenovirus activity. J. Med. Virol. 69:495-502. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, G., V. Kaiser, J. Rhodes, J. P. Russell, and C. L. Bevins. 2000. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect. Immun. 68:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond, G., J. P. Russell, and C. L. Bevins. 1996. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc. Natl. Acad. Sci. 93:5156-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond, G., M. Zasloff, H. Eck, M. Brasseur, W. L. Maloy, and C. L. Bevins. 1991. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc. Natl. Acad. Sci. 88:3952-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elass, E., M. Masson, J. Mazurier, and D. Legrand. 2002. Lactoferrin inhibits the lipopolysaccharide-induced expression and proteoglycan-binding ability of interleukin-8 in human endothelial cells. Infect. Immun. 70:1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elass-Rouchard, E., A. Roseanu, D. Legrand, M. Trif, V. Salmon, C. Motas, J. Montreuil, and G. Spik. 1995. Lactoferrin-lipopolysaccharide interaction: involvement of the 28-34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli O55B5 lipopolysaccharide. Biochem. J. 312:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellison, R. T., T. J. Giehl, and F. M. LaForce. 1988. Damage of the outer membrane of enteric gram-negative bacteria by lactoferrin and transferrin. Infect. Immun. 56:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fine, D. H., and D. Furgang. 2002. Lactoferrin iron levels affect attachment of Actinobacillus actinomycetemcomitans to buccal epithelial cells. J. Periodontol. 73:616-623. [DOI] [PubMed] [Google Scholar]
- 16.Fine, D. H., D. Furgang, and F. Beydouin. 2002. Lactoferrin iron levels are reduced in saliva of patients with localized juvenile periodontitis. J. Periodontol. 73:624-630. [DOI] [PubMed] [Google Scholar]
- 17.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, S., I. Mandel, and M. Herrera. 1983. Lysozyme and lactoferrin quantitation in the crevicular fluid. J. Periodontol. 54:347-350. [DOI] [PubMed] [Google Scholar]
- 19.Groenink, J., E. Walgreen-Weterings, K. Nazmi, J. G. M. Bolscher, E. C. I. Veerman, A. J. van Winkelhoff, and A. V. Nieuw Amerongen. 1999. Salivary lactoferrin and low-Mr mucin MG2 in Actinobacillus actinomycetemcomitans-associated periodontitis. J. Clin. Periodontol. 26:269-275. [DOI] [PubMed] [Google Scholar]
- 20.Håkansson, A., H. Roche, S. Mirza, L. S. McDaniel, A. Brooks-Walter, and D. E. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, J., and P. Furmanski. 1995. Sequence specificity and transcriptional activation in the binding of lactoferrin to DNA. Nature 373:721-724. [DOI] [PubMed] [Google Scholar]
- 22.Hendrixson, D. R., J. Qiu, S. C. Shewry, D. L. Fink, S. Petty, E. N. Baker, A. G. Plaut, and J. W. St. Geme III. 2003. Human milk lactoferrin is a serine protease that cleaves Haemophilus surface proteins at arginine-rich sites. Mol. Microbiol. 47:607-617. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan, J. B., and D. H. Fine. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl. Environ. Microbiol. 68:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 69:5375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Löe, H., and L. J. Brown. 1991. Early onset periodontitis in the United States of America. J. Periodontol. 62:608-616. [DOI] [PubMed] [Google Scholar]
- 27.Lupetti, A., A. Paulusma-Annema, S. Senesi, M. Campa, J. T. van Dissel, and P. H. Nibbering. 2002. Internal thiols and reactive oxygen species in candidacial activity exerted by an N-terminal peptide of human lactoferrin. Antimicrob. Agents Chemother. 46:1634-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann, D. M., E. Romm, and M. Migliorini. 1994. Delineation of the glycosaminoglycan-binding site in the human inflammatory response protein lactoferrin. J. Biol. Chem. 269:23661-23667. [PubMed] [Google Scholar]
- 29.Marazita, M. L., J. A. Burmeister, J. C. Gunsolley, T. E. Koertge, K. Lake, and H. A. Schenkein. 1994. Evidence for autosomal dominant inheritance and race-specific heterogeneity in early-onset periodontitis. J. Periodontol. 65:623-630. [DOI] [PubMed] [Google Scholar]
- 30.Masson, P. L., J. F. Heremans, and C. Dive. 1966. An iron-binding protein common to many external secretions. Clin. Chim. Acta 14:735-739. [Google Scholar]
- 31.Masson, P. L., J. F. Heremans, and E. Schonne. 1969. Lactoferrin, an iron-binding protein in neutrophilic leukocytes. J. Exp. Med. 130:643-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalowitz, B. S., S. R. Diehl, J. C. Gunsolley, B. S. Sparks, C. N. Brooks, T. E. Koertge, J. V. Califano, J. A. Burmeister, and H. A. Schenkein. 2000. Evidence of a substantial genetic basis for risk of adult periodontitis. J. Periodontol. 71:1699-1707. [DOI] [PubMed] [Google Scholar]
- 33.Mineshiba, F., S. Takashiba, J. Mineshiba, K. Matsuura, S. Kokeguchi, and Y. Murayama. 2003. Antibacterial activity of synthetic human β-defensin-2 against periodontal bacteria. J. Int. Acad. Periodontol. 5:35-40. [PubMed] [Google Scholar]
- 34.Nuijens, J. H., P. H. C. van Berkel, and F. L. Schanbacher. 1996. Structure and biological actions of lactoferrin. J. Mammary Gland Biol. Neoplasia 1:285-295. [DOI] [PubMed] [Google Scholar]
- 35.Odell, E. W., R. Sarra, M. Foxworthy, D. S. Chapple, and R. W. Evans. 1996. Antibacterial activity of peptides homologous to a loop region in human lactoferrin. FEBS Lett. 382:175-178. [DOI] [PubMed] [Google Scholar]
- 36.Powell, M. J., and J. E. Ogden. 1990. Nucleotide sequence of human lactoferrin cDNA. Nucleic Acids Res. 18:4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose, J. E., D. H. Meyer, and P. M. Fives-Taylor. 2003. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 71:2384-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallmann, F. R., S. Baveye-Descamps, F. Pattus, V. Salmon, N. Branza, G. Spik, and D. Legrand. 1999. Porins OmpC and PhoE of Escherichia coli as specific cell-surface targets of human lactoferrin. J. Biol. Chem. 274:16107-16114. [DOI] [PubMed] [Google Scholar]
- 39.Salmon, V., D. Legrand, B. Georges, M.-C. Slomianny, B. Coddeville, and G. Spik. 1997. Characterization of human lactoferrin produced in the baculovirus expression system. Protein Expr. Purif. 9:203-210. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sanchez, L., M. Calvo, and J. H. Brock. 1992. Biological role of lactoferrin. Arch. Dis. Child. 67:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sioson, P. B., D. Furgang, L. M. Steinberg, and D. H. Fine. 2000. Proximal caries in juvenile periodontitis patients. J. Periodontol. 71:710-716. [DOI] [PubMed] [Google Scholar]
- 43.Son, K.-N., J. Park, C.-K. Chung, D. K. Chung, D.-Y. Yu, K.-K. Lee, and J. Kim. 2002. Human lactoferrin activates transcription of IL-1β gene in mammalian cells. Biochem. Biophys. Res. Commun. 290:236-241. [DOI] [PubMed] [Google Scholar]
- 44.Vorland, L. H. 1999. Lactoferrin: a multifunctional glycoprotein. APMIS 107:971-981. [DOI] [PubMed] [Google Scholar]
- 45.Weinberg, E. D. 2001. Human lactoferrin: a novel therapeutic with broad spectrum potential. J. Pharm. Pharmacol. 53:1303-1310. [DOI] [PubMed] [Google Scholar]
- 46.Wu, H. M., and F. C. Church. 2003. Arginine 25 and arginine 28 of lactoferrin are critical for effective heparin neutralization in blood. Arch. Biochem. Biophys. 412:121-125. [DOI] [PubMed] [Google Scholar]
- 47.Wu, R., E. Nolan, and C. Turner. 1985. Expression of tracheal differentiated functions in serum-free hormone-supplemented medium. J. Cell. Physiol. 125:167-181. [DOI] [PubMed] [Google Scholar]
- 48.Wu, R., J. Yankaskas, E. Cheng, M. R. Knowles, and R. Boucher. 1985. Growth and differentiation of human nasal epithelial cells in culture: serum-free, hormone-supplemented medium and proteoglycan synthesis. Am. Rev. Respir. Dis. 132:311-320. [DOI] [PubMed] [Google Scholar]
- 49.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder. J. M. Wang, and O. M. Howard. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 50.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. Participation of mammalian defensins and cathelicidins in anti-microbial immunity: receptors and activitites of human defensins and cathelicidin (LL-37). J. Leukocyte Biol. 69:691-697. [PubMed] [Google Scholar]
- 51.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]