Abstract
Aurora/Ipl1-related kinases are a conserved family of enzymes that have multiple functions during mitotic progression. Although it has been possible to use conventional genetic analysis to dissect the function of aurora, the founding family member in Drosophila (Glover, D.M., M.H. Leibowitz, D.A. McLean, and H. Parry. 1995. Cell. 81:95–105), the lack of mutations in a second aurora-like kinase gene, aurora B, precluded this approach. We now show that depleting Aurora B kinase using double-stranded RNA interference in cultured Drosophila cells results in polyploidy. aurora B encodes a passenger protein that associates first with condensing chromatin, concentrates at centromeres, and then relocates onto the central spindle at anaphase. Cells depleted of the Aurora B kinase show only partial chromosome condensation at mitosis. This is associated with a reduction in levels of the serine 10 phosphorylated form of histone H3 and a failure to recruit the Barren condensin protein onto chromosomes. These defects are associated with abnormal segregation resulting from lagging chromatids and extensive chromatin bridging at anaphase, similar to the phenotype of barren mutants (Bhat, M.A., A.V. Philp, D.M. Glover, and H.J. Bellen. 1996. Cell. 87:1103–1114.). The majority of treated cells also fail to undertake cytokinesis and show a reduced density of microtubules in the central region of the spindle. This is accompanied by a failure to correctly localize the Pavarotti kinesin-like protein, essential for this process. We discuss these conserved functions of Aurora B kinase in chromosome transmission and cytokinesis.
Keywords: Aurora B kinase, chromosome condensation, phospho-histone H3, barren, cytokinesis
Introduction
The segregation of chromosomes with high fidelity requires exquisite coordination of cellular processes. The mechanisms that coordinate the cycle of chromosome condensation and decondensation with the assembly, function, and subsequent disassembly of the mitotic spindle are poorly understood. Highly conserved genes essential for chromosome condensation have been found through genetic screens in yeasts and Drosophila (Bhat et al. 1996; Sutani et al. 1999). This screen has identified five members of a protein complex known as condensin that is functionally and structurally conserved. Mutants exhibit incomplete chromosome condensation associated with failure of segregation and the stretching of chromatin upon the spindle. Biochemical approaches also identified the protein complex in Xenopus (Hirano et al. 1997) and showed that it could promote chromatin condensation by directing the supercoiling of the DNA in an ATP-dependent manner (Kimura and Hirano 1997; Kimura et al. 1999). Chromosome condensation is also accompanied by phosphorylation of histones H1 and H3 (Bradbury 1992; Hendzel et al. 1997; de la Barre et al. 2000). Indeed, mutation of the mitotic phosphorylation site of histone H3 of Tetrahymena leads to both chromosome condensation and segregation defects (Wei et al. 1999). A direct link between histone H3 phosphorylation and condensin recruitment onto chromosomes has recently been suggested by the colocalization of members of the condensin complex with phosphorylated histone H3 during the early stages of mitotic chromosome condensation (Schmiesing et al. 2000). However, the generality of the requirement for the phosphorylation of histone H3 for chromosome condensation and segregation must be questioned by the finding that budding yeast cells in which serine 10 of histone H3 is replaced with alanine show no apparent defects in cell cycle progression or chromosome transmission. Nevertheless, maximal chromosome condensation in meiosis does correlate with maximal levels of phospho-histone H3 in wild-type cells. The enzyme required for histone H3 phosphorylation in Saccharomyces cerevisiae is the aurora-related protein kinase Ipl1p (Hsu et al. 2000). Moreover, one of its two counterparts from Caenorhabditis elegans, the air-2 protein kinase, was shown to have the same function.
The Aurora- and Ipl1-like protein kinases form a conserved family of enzymes, the founding members of which are encoded by the S. cerevisiae and Drosophila genes, IPL1 and aurora, respectively (Francisco and Chan 1994; Glover et al. 1995; for review see Giet and Prigent 1999). While the yeast genome encodes only one such kinase required for accurate chromosome segregation, metazoan genomes have at least two subfamilies of aurora-like kinases. One is associated with centrosomes and is activated in early mitosis, and a second is associated with chromosomes and the spindle midbody and is activated later. We will refer to these families as Aurora-like kinases A and B, respectively, as recently suggested by Nigg 2001. The precise effects of loss of function of either of these enzymes varies a little between different organisms and cell types. Broadly speaking, however, the A-type enzymes are required to maintain the separation of centrosomes to give normal bipolar spindle structure. This is shown, for example, in Drosophila from the phenotype of aurora mutants (which we now propose to call aurora A; Glover et al. 1995); or in Xenopus, where the corresponding pEg2 kinase can be eliminated using antibodies or inactive mutants (Roghi et al. 1998; Giet and Prigent 2000). The B-type Aurora-like kinases, on the other hand, appear to be required for cytokinesis, as shown, for example, by transfection of an inactive kinase mutant into cultured mammalian cells (Tatsuka et al. 1998; Terada et al. 1998). An affect on cytokinesis has also been reported in mutants of the gene for the C. elegans B-type enzyme, Air-2, or after RNA interference (RNAi)1 (Schumacher et al. 1998; Kaitna et al. 2000; Severson et al. 2000). The air-2 encoded kinase is required for the positioning of Zen-4, a kinesin-like protein required at the midzone of the late central spindle for cytokinesis. Abnormal chromosome segregation is also observed after reduction of air-2 function.
The dynamics of the localization of the Aurora B class of enzymes can be partially explained by recent findings showing they exist in a complex with an inner centromere protein (INCENP) (Adams et al. 2000; Kaitna et al. 2000). INCENPs are one example of so-called “passenger proteins” that localize to the centromeric regions of chromosomes at metaphase and are then redistributed to the central spindle during cytokinesis. Defects in INCENP function lead to failure of chromosome congression and cytokinesis defects (Mackay et al. 1998). These findings, and the fact that B-type Aurora kinase becomes incorrectly localized in human cells expressing mutant INCENPs that fail to localize, has led to the idea that INCENP functions to target the B-type kinases, first to chromosomes and then to the spindle midzone (Adams et al. 2000). A physical interaction is also seen between the Air-2 kinase and the counterpart of INCENP in C. elegans, ICP-1. Moreover, the disruption of icp-1 function by RNAi leads to the same phenotype as air-2 RNAi (Kaitna et al. 2000). This direct functional interaction between the Aurora-like kinases and INCENP occurs not only in metazoan cells, but also in budding yeast where the counterpart of INCENP, Sli15p, was identified through a screen for genes that interact with Ipl1 (Kim et al. 1999).
Although a B-type Aurora kinase gene has been identified in Drosophila, the lack of mutants at this locus has prevented any analysis of its potential mitotic function (Reich et al. 1999). In this paper we report that levels of the Aurora B kinase can be reduced by RNAi in cultured Drosophila S2 cells. This leads to cytokinesis failure, together with chromosome condensation and segregation defects strikingly similar to those we have described previously for mutations in the condensin gene barren (Bhat et al. 1996). The segregation defects are accompanied by aberrant chromatin condensation, a reduction in the phosphorylated form of histone H3, and a failure to recruit the Barren protein onto condensed chromosomes.
Materials and Methods
Cell Culture
S2 cells were grown in Schneider's Drosophila medium (GIBCO BRL) supplemented with 10% fetal calf serum (GIBCO BRL) and 50 μg/ml streptomycin and penicillin. For FACS® analysis, the cells were recovered by trypsin treatment. They were then washed with PBS and fixed in 90% ice-cold ethanol. The cells were incubated at 37°C for 30 min in PBS containing 40 μg/ml boiled RNase A and 1 μg/ml propidium iodide before analysis. For protein analysis, an aliquot of the cells was resuspended and boiled in Laemmli buffer.
Double-stranded RNA Synthesis
The aurora B cDNA was amplified by PCR from a testis cDNA library (Hazelrigg and Tu 1994) using the primers 5′-CAGAATTCCGCCATGACGCTTTCCCGCGCG-3′ containing the EcoRI site, and 5′-CAAAAGCTTCCTGGCCGTGTTCTCCTTGCC-3′ containing the HindIII site. The PCR amplification product was cloned into pGEMt easy vector (Promega). The clones containing the opposite orientations were selected, mixed in equal proportions, and digested by SpeI. The digested plasmids were used to produce double-stranded RNA (dsRNA) using Megascript T7 transcription kit (Ambion). The RNA was purified according to the manufacturer's instructions, denaturated for 5 min at 94°C, and annealed overnight by a slow cooling. dsRNA was analyzed by 1% electrophoresis in agarose gel to ensure that the RNA migrated as a single band (∼1 kb).
Condition for RNAi in S2 Cultured Cells
RNAi was carried out using a modification of the method of Clemens (2000). Cells were inoculated the day before transfection into 6-well plates. For RNAi, the cells were washed three times with medium without serum and antibiotics and transfected with Transfast lipid reagent (Promega) according to the manufacturer's instructions. 3.3 μg was used to transfect each well plate. Cells were incubated for 3 d to allow the turnover of the aurora B protein kinase. Using these conditions, effects on cell cycle progression could first be seen 2 d after transfection. Samples were taken at various times after transfection and analyzed by FACS®, Western blot, or immunofluorescence.
Antibodies and Western Blotting
The rat antitubulin antibody (clone YL1/2) was obtained from Sigma-Aldrich and the antiphospho-histone H3 from Upstate Biotechnology. Rabbit anticyclin B (Rb271), antibarren, and antipavarotti (Rb3301) have been described previously (Whitfield et al. 1990; Bhat et al. 1996; Adams et al. 1998). Anti-Prod antibody (Torok et al. 1997) was given to us by Peter Bryant (University of California at Irvine, Irvine, CA). FITC- or Texas red–conjugated goat anti–rat, or anti–rabbit, secondary antibodies used in immunofluorescence were obtained from Jackson ImmunoResearch Laboratories. Alkaline phosphatase–conjugated goat anti–rabbit or anti–rat antibodies used in Western blotting were from Sigma-Aldrich. To produce antibodies against Aurora B, the 15 COOH-terminal amino acids of the aurora B protein kinase (QLKQKRDRGKENTARN) were coupled to keyhole limpit hemocyanin and injected into rabbits. After three boosts, the serum was purified by affinity chromatography using a recombinant histidine-tagged protein blotted on a nitrocellulose membrane as described previously (Giet et al. 1999). Recombinant AurB-his6 was produced by first cloning PCR product into pGEM, and then subcloning it into pET23b. Protein was isolated from inclusion bodies of bacteria expressing the kinase from pET23b by resuspension in 8 M urea, followed by dialysis in PBS. The antibody was used at a concentration of 100 ng/ml. In peptide competition experiments, the peptide concentration was 10 μg/ml.
Immunofluorescence Analysis
S2 cells grown on glass coverslips were fixed in PHEM buffer (60 mM Pipes, 25 mM Hepes, pH 6.8, 10 mM EGTA, 4 mM MgCl2). The cells were permeabilized using PBST (PBS containing 0.1% Triton X-100). All incubations were performed in PBST containing 1% BSA. DNA was stained by TOTO3-iodide (Molecular Probes) or propidium iodide. Vectashield mounting medium H-1000 was purchased from Vector Laboratories. Images were acquired using a confocal scanning head (model 1024; Bio-Rad Laboratories) mounted on an Optiphot microscope (Nikon) and prepared for publication using Adobe Photoshop®.
Results
Aurora B Is a Chromosome Passenger Protein
To study the localization of the Aurora B protein kinase in Drosophila cells, we raised an antipeptide antibody against its specific 15 COOH-terminal amino acids. This antibody does not recognize recombinant Aurora A protein (data not shown) or endogenous Aurora A protein of 50 kD, but does stain a single band of ∼40 kD in Western blots of extracts of S2 cells that is greatly reduced when cells are treated with the aurora B dsRNA (see Fig. 2 C, middle). This staining, together with the immunostaining of mitotic cells (see below), can be competed out by the peptide used to raise the antibody (Fig. 1G and Fig. H). Aurora B cannot be detected by this antibody in interphase cells (Fig. 1 A), but it is readily apparent with a punctate distribution throughout all regions of condensing chromosomes in prophase cells (Fig. 1 B). By metaphase, the concentration of the protein has strongly increased in the centromeric regions (Fig. 1 C). Some of this centromeric staining persists in early anaphase, at which time the enzyme appears to become relocated on the central region of the spindle (Fig. 1 D). During anaphase B, when the poles start to move apart, labeling of the central spindle has become predominant (Fig. 1 E) and the enzyme is essentially confined to the midbody during cytokinesis (Fig. 1 F). Proteins showing this dynamic pattern of localization during mitosis have been termed “passengers”; they ride upon the chromosomes until metaphase, whereupon they alight to the platform of the central spindle.
Figure 2.
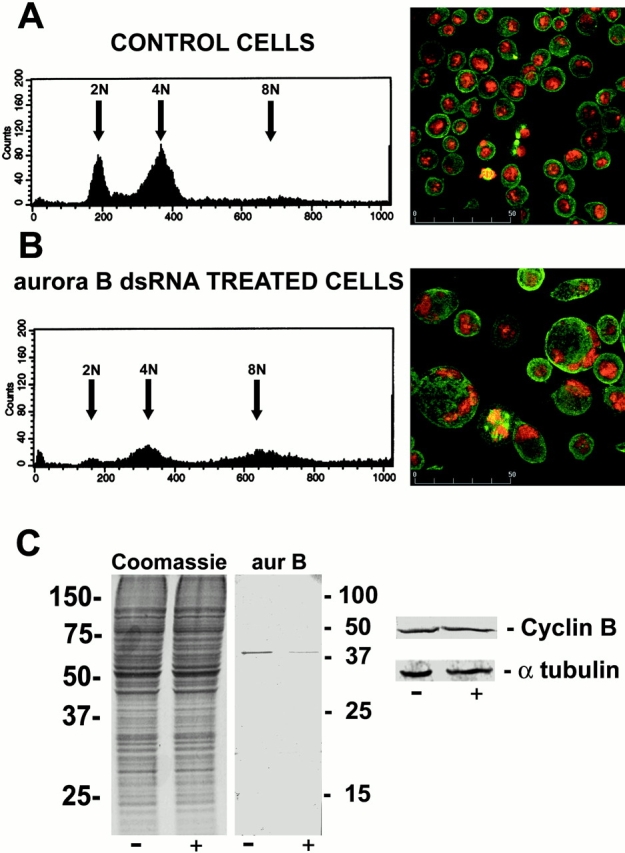
Polyploidy in Drosophila S2 cells treated with aurB dsRNA. FACS® profiles (left) and immunostaining (right) of control (A) or aurB RNAi (B) in S2 cells 3 d after addition of dsRNA to the experimental culture. Cells were stained to reveal DNA (red) and microtubules (green). (C) Protein profiles of similar preparations of S2 cells. Total Coomassie-stained proteins separated by SDS-PAGE are shown on the left, together with Western blots revealing Aurora B, cyclin B, and α-tubulin as indicated.
Figure 1.
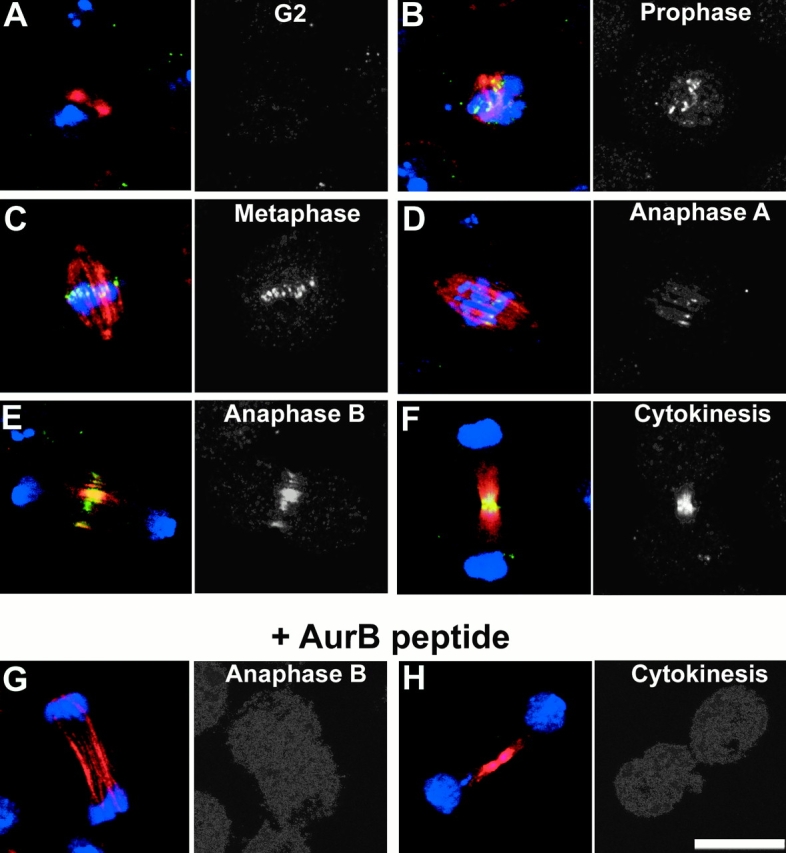
Aurora B is a chromosome passenger protein. Cultured Drosophila S2 cells were grown on glass coverslips, fixed, and stained to reveal DNA (blue), microtubules (red), and Aurora B kinase (green) (see Materials and Methods). Images are shown in pairs with the merged image on the left and the channel showing anti–Aurora B staining alone in monochrome on the right. The indicated successive stages of mitotic progression are shown in A–F. A detailed description of the Aurora B localization pattern is given in the text. In brief, it is first found throughout the condensing chromatin, concentrates at centromeres, and is then redistributed to the central spindle and midbody in late mitosis. G and H show cells stained in the presence of the peptide used to raise the anti–Aurora B antibody. Bar, 10 μm.
aurora B dsRNAi Leads to Polyploidy in Drosophila S2 Cells
To determine whether decrease in expression levels of the aurora B gene would affect the progression of Drosophila Schneider S2 cells through their division cycle, we treated such cells with dsRNA synthesized from the aurora B cDNA (see Materials and Methods). FACS® analysis of control cells showed two predominant peaks of G1 (2N) and G2/M (4N) cells (Fig. 2 A). 3 d after transfection of aurB dsRNA, the number of G1 cells with a 2N complement of DNA is strongly reduced and the profile shows prominent peaks at 4N and 8N (Fig. 2 B). This indicates a doubling in ploidy of a substantial proportion of the population of cells, such that G1 cells now fall within the 4N peak and G2/M cells in the 8N peak. This was confirmed by staining the cells to reveal DNA, whereupon it could be seen that most of the transfected interphase cells were significantly larger than the controls and that they contained two or more nuclei or a single large nucleus (Fig. 2A and Fig. B, right). Although the level of Aurora B protein within the population of cells is reduced by ∼90% after aurB RNAi, amounts of total cellular protein appear unchanged, as do levels of α-tubulin or the mitotically labile protein cyclin B (Fig. 2 C). This suggests that the cells are capable of progressing through S phase, but that defects occur in either or both chromosome segregation at mitosis or during cytokinesis.
Defects in Chromosome Segregation and Cytokinesis Occur in Multiple Cycles after aurora B RNAi
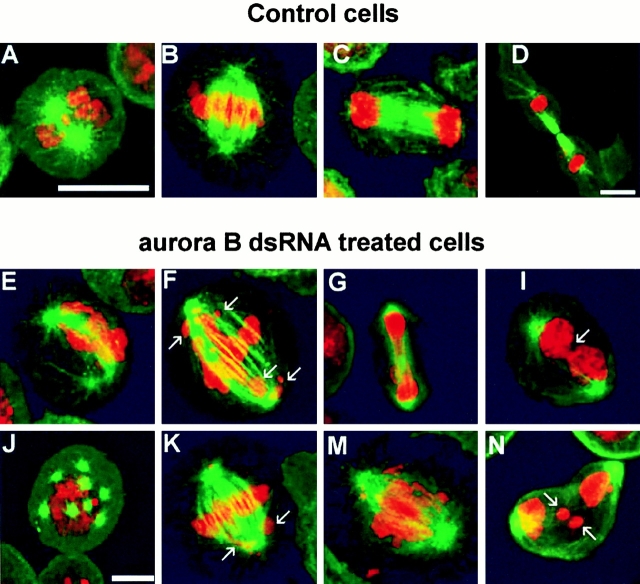
To assess the nature of the defects in cell cycle progression, we stained cells to reveal DNA and microtubules and quantified defects in the mitotic cells within the asynchronous population 3 d after treatment with aurB dsRNA. Whereas >90% of control interphase cells appear to have normal DNA content, as judged by the size of their nuclei, 70% of cells become polyploid after aurB RNAi, in agreement with the results reported above. Of these, 19% had a single abnormally large nucleus and 52% were multinucleate. The mitotic index of the population of aurB dsRNA–treated cells (5%) was not significantly different from control cells, indicating that in spite of the mitotic defects, cell cycle progression was not affected. Within the population of aurB RNAi cells a proportion of cells showed mitotic figures comparable to control cells (Fig. 3, A–D). However, the proportion of these cells undergoing apparently normal mitosis was greatly reduced (Table ). These apparently normal cells may not have taken up the dsRNA and their proportion relative to abnormal mitoses is comparable to the reduction in level of Aurora B kinase detected by Western blotting. Examples of the mitotic defects seen in the aurB RNAi cells are shown in Fig. 3E–N. The most striking feature is that incomplete chromosome condensation is seen in essentially all of these mitotic cells, albeit to varying extents. No obvious defects were detected at the spindle poles in prophase (Fig. 3E and Fig. J) and microtubules are well nucleated by the centrosomes at this and other mitotic stages. The prophase cell in Fig. 3 J has eight centrosomes, suggesting this is a 16N cell that has failed the two previous rounds of cytokinesis. There were also failures in the alignment of chromosomes on the metaphase plate. This feature is very clear in the cell in Fig. 3 F (arrows), but is also apparent in the cell in Fig. 3 K in which the majority of chromatin is on the metaphase plate. The extent to which chromatin can segregate to the spindle poles at anaphase varies dramatically, from situations in which there appear to be lagging chromatids, to cases in which massive chromatin bridges are formed (Fig. 3G and Fig. M). Such bridges fail to resolve at telophase (Fig. 3 I) and are presumably one means by which cells can arise that have a single polyploid nucleus. In other cases, lagging chromatids fail to resolve and will eventually form micronuclei, and cytokinesis appears to be blocked (Fig. 3 N). The proportion of binucleate cells at these late mitotic stages is elevated ∼15-fold over control cells. The density of microtubules in the central region of the mitotic spindle in these cells appears much lower than in control cells able to undergo cytokinesis (see below). Thus, defects in chromosome condensation after aurB RNAi are accompanied with abnormalities in chromosome segregation and failure of cytokinesis. Interestingly, the cells are not subject to checkpoint arrest and are able to undertake at least two to three rounds of polyploidization within the time frame of the experiment.
Figure 3.
Mitotic abnormalities in S2 cells depleted of Aurora B. (A–D) Control cells at prophase, metaphase, anaphase, and cytokinesis, respectively. Microtubules are stained green and DNA red. (E–N) Abnormal mitoses in aurora B–depleted cells in states resembling prophase (E and J), metaphase (F and K), anaphase (G and M), and telophase (I and N). The arrows in F and K indicate chromosomes that have not aligned on the metaphase plate. The arrows in I and N indicate a conspicuous chromatin bridge or lagging chromatids at telophase, respectively. Bars, 10 μm.
Table 1.
Quantitation of Mitotic and Interphase Defects after aurB RNAi
| Mitosis | Interphase | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mitotic index | Prophase | Metaphase | Anaphase | Cytokinesis | Polyploid | Abnormal | Normal cells | Cells with a single large nucleus | Cells with two or more nuclei | |
| Control cells | 6.5 ± 2.5 | 19.5 ± 2.0 | 22.3 ± 0.7 | 7.0 ± 0.2 | 44.3 ± 2.6 | 7.0 ± 2.5 | 0 | 93.5 ± 3.2 | 3.0 ± 1.5 | 3.5 ± 1.7 |
| aurB RNAi | 5.2 ± 1.1 | 15.1 ± 1.4 | 2.6 ± 2.1 | 1.2 ± 1.0 | 3.5 ± 2.0 | 70.1 ± 7.0 | 7.5 ± 0.8 | 28.2 ± 6.3 | 19.2 ± 5.0 | 52.6 ± 2.8 |
Failure in the Resolution of Centromeres in aurora B–depleted Cells
The defects in chromosome segregation after aurB RNAi suggested that the centromeric regions of chromosomes may not form correct attachments to be able to move along the spindle microtubules. As centromeric heterochromatin is difficult to identify within these poorly condensed chromosomes, we chose to immunolabel the region using antibodies to Prod, the product of the gene proliferation disrupter (Torok et al. 1997). In control cells, Prod localizes to centromeric regions of congressed metaphase chromosomes, and in late anaphase this centromeric marker is clearly seen near the poles of the spindle (Fig. 4A and Fig. B). In aurB RNAi cells, Prod shows only a slight tendency to be associated with the spindle poles at anaphase and there is conspicuous punctate staining given by anti-Prod throughout the poorly condensed chromatin mass (Fig. 4, C–F). Fig. 4 F displays a presumptive 16N cell in which there appears to be little segregation of the centromeric regions on the octapolar spindle. These observations indicate that in addition to being required for cytokinesis, Aurora B also functions to regulate chromatin dynamics during mitosis, in particular in directing the organization and function of centromeric regions.
Figure 4.
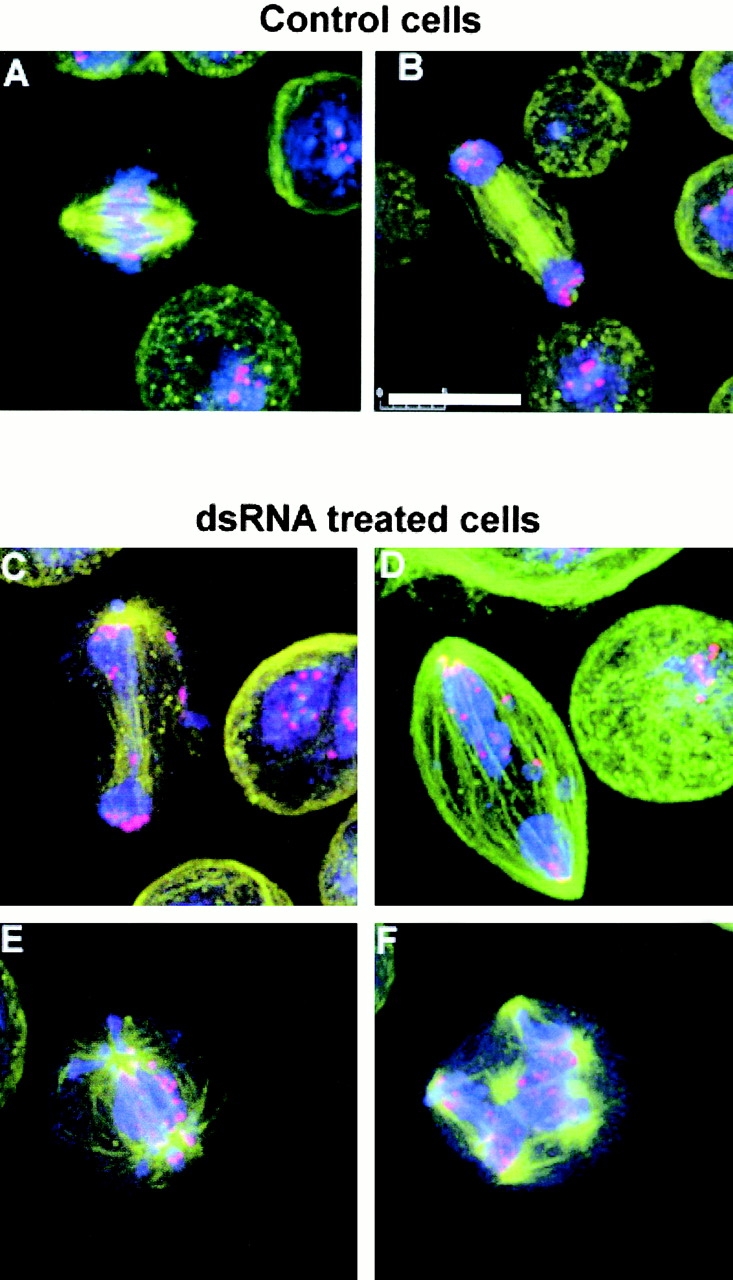
Centromere segregation is defective in Aurora B–depleted cells. (A and B) Control cells at metaphase and anaphase, respectively. (C–F) Abnormal mitoses in aurora B–depleted cells in an anaphase-like state showing centromeric regions (stained red with anti-Prod antibody) that fail to reach the spindle poles. DNA is stained blue and microtubules green. Bar, 10 μm.
Phosphorylation of Histone H3 Is Reduced and the Condensin Protein Barren Fails to Associate with Mitotic Chromosomes
It has recently been shown that Aurora-related kinases direct the phosphorylation of histone H3 in meiosis and mitosis (Hsu et al. 2000). As the Ipl1 aurora-like kinase is required for accurate chromosome transmission in budding yeast, and as phospho-histone H3 is found on mitotic chromosomes, we chose to examine the phosphorylation state of this histone in aurB RNAi cells that show defects in chromosome condensation and segregation. To this end, we carried out immunostaining and Western blotting using an antibody specific to this phosphoepitope (Fig. 5). The immunostaining of control cells indicates that histone H3 begins to become phosphorylated at the onset of prophase (Fig. 5 A) and increases to give intense signals at metaphase (Fig. 5 B) and anaphase (Fig. 5 C). Staining levels decrease during telophase (Fig. 5 D) and then disappear during cytokinesis (Fig. 5 E). Western blotting (Fig. 5 F) of asynchronous cultures of control or RNAi-treated cells indicated a pronounced decrease in levels of phosphorylated histone H3 in parallel with reductions in the level of Aurora B kinase. Immunostaining indicates that phosphorylation of histone H3 in mitotic cells is dramatically reduced after aurB RNAi but not completely extinguished (Fig. 5, G–J). It remains present in foci distributed throughout the poorly condensed chromatin mass, rather than being uniformly associated with the chromosomes.
Figure 5.
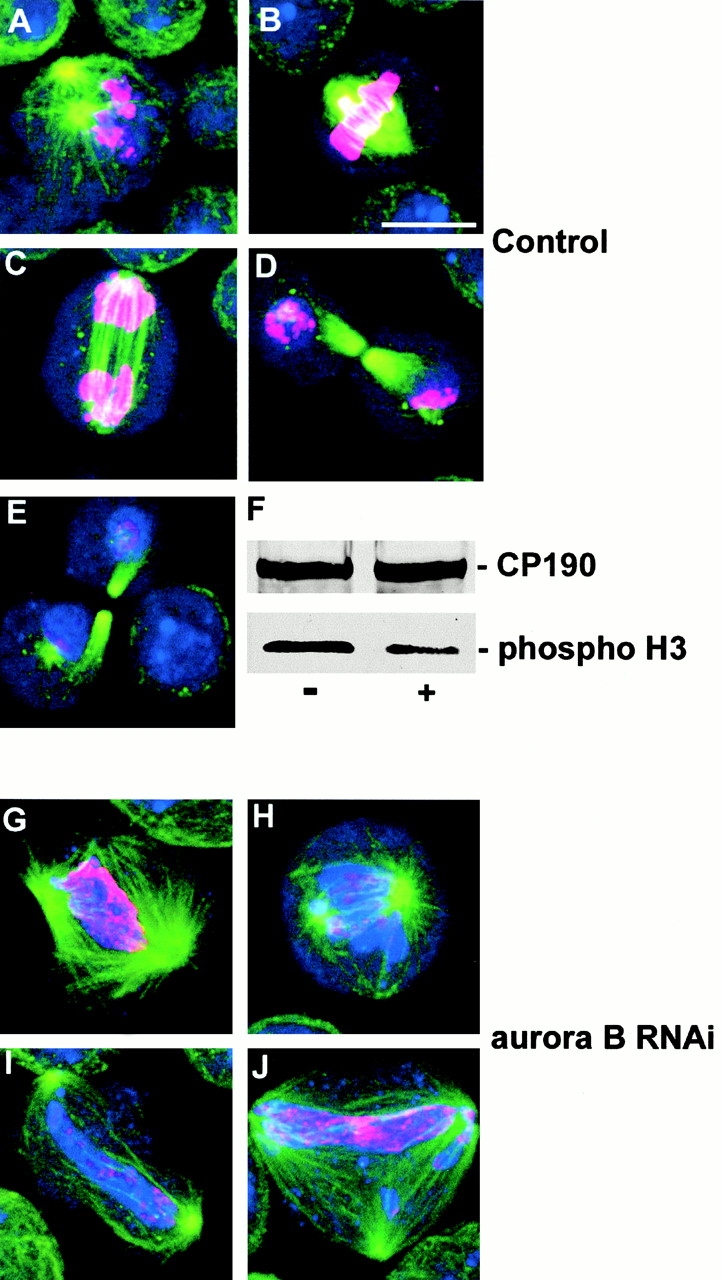
Histone H3 phosphorylation in untreated and aurB RNAi S2 cells. (A–E) The cycle of histone H3 phosphorylation revealed by antiphospho-histone H3 antibody (red) in control S2 cells. DNA is stained blue and microtubules green. Cells are shown at prophase (A), metaphase (B), anaphase (C), and telophase/cytokinesis (D and E). Immunostaining of the phosphoepitope builds to a maximum during metaphase and early anaphase (see text). (F) A Western blot that shows reduction in phospho-histone H3 staining (bottom) in dsRNA-treated cells (+) compared with control cells (−), whereas CP190 levels (top) remain constant. (G–J) Abnormal mitotic figures in aurB RNAi cells showing reduced levels of phospho-histone H3 staining compared with control mitoses. Bar, 10 μm.
It has been proposed that the modification of histone H3 by phosphorylation on serine 10 could lead to the recruitment of condensation factors on the DNA (Wei et al. 1999). To test the feasibility of this model, we decided to follow the dynamics by which condensin would associate with mitotic chromosomes in Drosophila cells using Barren protein as a marker. The Drosophila Barren protein is a component of the condensin complex, homologous to the XCAPH protein of Xenopus (Hirano et al. 1997). Mutations in barren are characterized by an absence of chromosome condensation and the formation of chromatin bridges during anaphase that are very similar to defects seen in aurB RNAi cells (Bhat et al. 1996). We found that Barren protein appears on the chromosomes with the same timing and dynamics as phosphorylation of histone H3 (Fig. 6, A–E). It is first detected on condensing chromosomes at prophase as multiple foci (Fig. 6 A); it is maximal during metaphase (Fig. 6 B) and anaphase A (Fig. 6 C); and it disappears from chromosomes as they decondense at late anaphase/telophase (Fig. 6D and Fig. E). In aurB RNAi cells, we found a dramatic decrease of Barren protein associated with chromosomes (Fig. 6, G–J), although Western blotting indicated that there was no diminution in levels of the protein (Fig. 6 F). Thus, Aurora B activity is essential to recruit Barren protein to chromosomes during mitosis and the dynamics of the process are consistent with a model in which the phosphorylation of histone H3 is required for this process.
Figure 6.
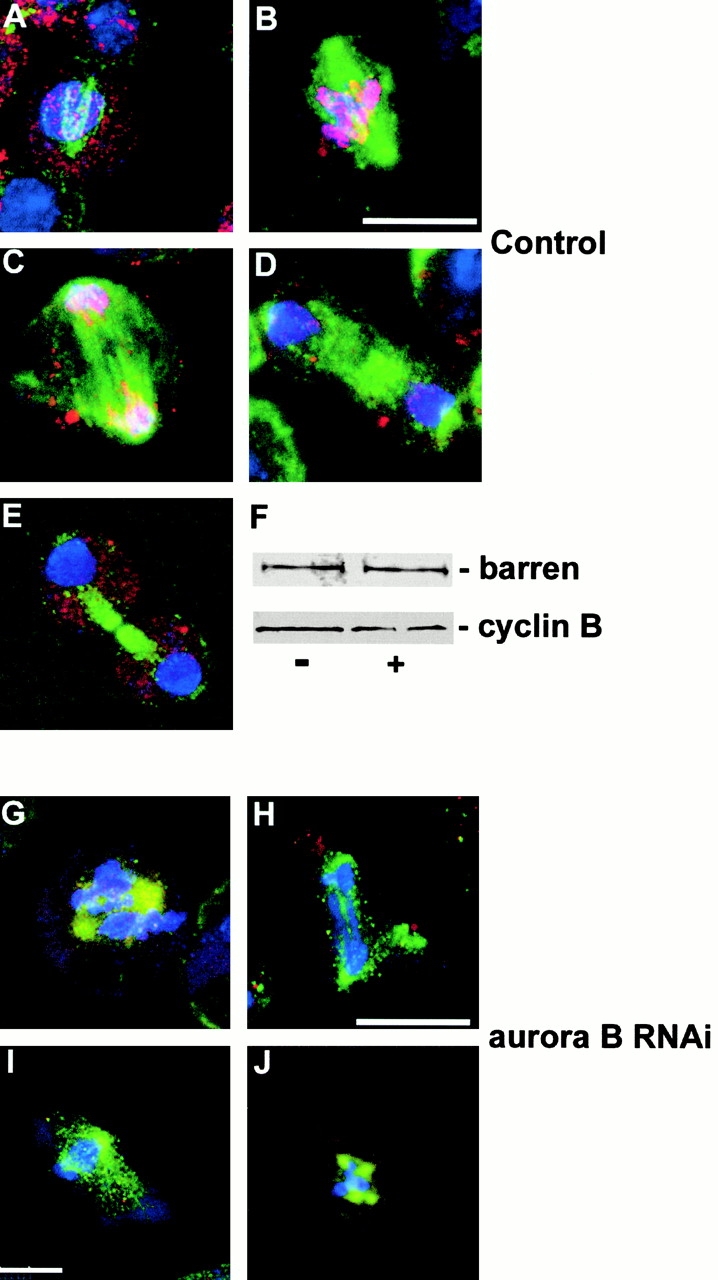
Barren fails to localize to mitotic chromosomes after depletion of Aurora B. (A–E) The cycle of association of Barren protein (red) onto mitotic chromosomes in control S2 cells. DNA is stained blue and microtubules green. Cells are shown at prophase (A), metaphase (B), anaphase (C), telophase (D), and cytokinesis (E). Association of Barren with chromosomes follows the same dynamics as histone H3 phosphorylation (see text). (F) Western blot showing that levels of Barren protein (top) are unchanged from control cells (−) after aurB RNAi (+) as are levels of cyclin B (bottom). (G–J) Abnormal mitotic figures in aurB RNAi cells in which barren does not localize to chromosomes. Bars, 10 μm.
Abnormalities of the Central Spindle and Failure to Recruit Pav-KLP after aurB RNAi
Examination of spindle microtubules in aurB RNAi cells indicated abnormalities in the organization of the central spindle as indicated above. We have shown previously that a kinesin-like protein encoded by the gene pavarotti (Pav-KLP) is required for this aspect of spindle organization before cytokinesis (Adams et al. 1998). As the C. elegans orthologue of Pav-KLP, Zen4-klp, is not recruited to the spindle in conditional mutants for the Aurora B–like kinase Air-2, we wished to determine whether the localization of Pav-KLP would be affected in aurB RNAi cells. As described previously, Pav-KLP normally localizes to the central spindle during anaphase (Fig. 7 A) and to the midbody during telophase at the onset of cytokinesis (Fig. 7 B). However, when S2 cells were subjected to aurB RNAi we observed a marked decrease of Pav-KLP immunostaining from the diminished central spindle (Fig. 7D and Fig. E) and in many cells localized Pav-KLP could not be detected (Fig. 7 C). Total Pav-KLP levels are not affected by aurB RNAi (Fig. 7 F). This strongly suggests that in normal mitosis the presence of Aurora B kinase on the central spindle is essential for its correct organization and the recruitment of Pav-KLP in order for cytokinesis to take place.
Figure 7.
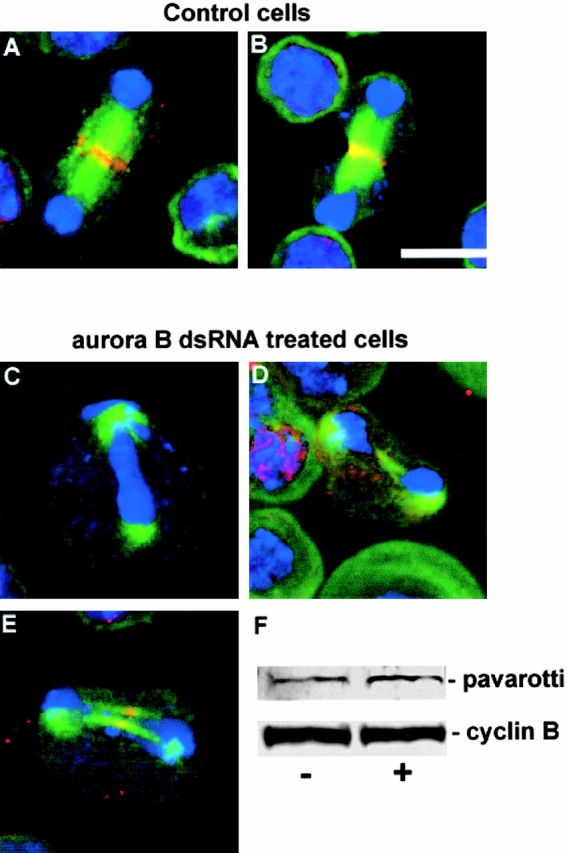
Failure of Pav-KLP to fully localize to the central spindle in Aurora B–depleted cells. (A and B) Pav-KLP (red) localizes to the central spindle/midbody in control S2 cells at late anaphase (A) and early cytokinesis (B). DNA is blue and microtubules are green. (C–E) Abnormal late mitotic figures in Aurora B–depleted cells in which the density of microtubules in the central spindle region is greatly reduced and in which the Pav-KLP either fails to localize (C) or localizes very poorly (D and E). (F) Western blot showing that levels of Pav-KLP (top) are unchanged from control cells (−) after treatment with aurB dsRNA (+), as are levels of cyclin B (bottom). Bar, 10 μm.
Discussion
At the outset of this work, the existence of a second aurora-like gene in Drosophila, aurora B, was already known (previously named ial; Reich et al. 1999), but there were no known mutations at the locus and the enzyme had not been localized within cells. The proximity of other genes made the prospect of generating such mutations by classical genetic means somewhat daunting and although a model system for the targeted disruption of genes has recently been developed (Rong and Golic 2000), this is also a time-consuming and not yet widely used procedure. In several organisms dsRNAi has offered opportunities for studying the genetic basis of many aspects of cell biology in the absence of mutations in the genes directing these activities. This has been perhaps best demonstrated in C. elegans, where the technique is most widely used (Hunter 1999, Hunter 2000). A simple system for dsRNAi in cultured Drosophila cells not only offers the means of eliminating specific gene expression (Caplen et al. 2000; Clemens et al. 2000), but also of studying the dynamics of such processes in relation to cell cycle progression (Hammond et al. 2000). Using this approach we have shown that the Aurora B kinase is required for mitotic chromosome condensation and segregation, and subsequently for cytokinesis. We have also carried out RNAi in S2 cells on aurA (data not shown). This results in a mitotic phenotype similar to the aurA mutants (Glover et al. 1995) and a characteristic FACS® profile in which the proportion of G2/M cells increases at the expense of G1 cells. The protein levels of Aurora A are unaffected by aurB RNAi and, conversely, Aurora B is unaffected by aurA RNAi. This points to the specificity of RNAi and the observed phenotypes reflect the distinct mitotic functions of the two kinases.
The Aurora B enzyme becomes perfectly positioned to execute these processes as mitosis proceeds. It is distributed throughout the chromatin as it condenses at prophase, then becomes concentrated around the centromeric regions of the condensed chromosomes at metaphase, and finally leaves for the company of the central spindle region during anaphase. As such it behaves as a so-called passenger protein. It appears from recent studies to be in an intimate relationship with a travelling companion INCENP (Adams et al. 2000; Kaitna et al. 2000). The interaction of INCENP, or its yeast counterpart Sli5p, with Aurora-like kinases in yeast, C. elegans, and Xenopus suggests that this interaction is universal. The dynamic association of INCENP with chromosome arms at prometaphase, the centromeric region at metaphase, and then the spindle midzone at anaphase makes it an attractive candidate for targeting the Aurora B kinase to these regions. Indeed, dominant mutants of INCENP in human cells disrupt the localization of the Aurora B–like kinase AIL2 (Adams et al. 2000). The finding of abnormal chromosome segregation and cytokinesis after depletion of either the C. elegans INCENP, Icp-1, or its Aurora B–like kinase, Air-2, suggests the two passengers are about similar business (Schumacher et al. 1998; Woolard and Hodgekin 1999; Kaitna et al. 2000).
One striking effect of aurB RNAi is to permit progression through mitosis with improperly condensed chromosomes. We are able to account for these condensation defects by a diminution of the phosphorylation of serine 10 of histone H3 and a failure to localize condensin on the chromosomes. The former finding is consistent with several studies that now implicate a requirement for the phosphorylation of the NH2-terminal region of histone H3 at this residue for chromosome condensation. Not only does the formation of mitotic chromosomes in a Xenopus cell-free extract by a nucleosome-associated kinase correlate with histone H3 phosphorylation (de la Barre et al. 2000), but when the serine 10 residue is mutated to alanine it results in abnormal segregation and chromosome loss during mitosis and meiosis in Tetrahymena (Wei et al. 1999). One enzyme credited with the ability to phosphorylate histone H3 at mitosis is the NIMA kinase of Aspergillus (De Souza et al. 2000). However, our finding that levels of histone H3 phosphorylation are reduced after aurB RNAi in Drosophila cells is more in keeping with the report that the Aurora-like kinase homologues, Ipl1 of yeast and Air-2 (but not Air-1) of C. elegans, are required for histone H3 phosphorylation in these organisms (Hsu et al. 2000). The finding of some residual histone H3 phosphorylation either could reflect the incomplete elimination of Aurora B by RNAi, or could indicate that an alternative kinase has this capability, offering an explanation of the partial chromosome condensation seen in the RNAi-treated cells. Our current data are important in emphasizing the importance of histone H3 phosphorylation for chromosome transmission and as such are in line with the findings in Tetrahymena. This differs from the effects seen in budding yeast cells that continue through division cycles in the absence of histone H3 phosphorylation without showing defects in chromosome transmission. As an explanation, it has been suggested that other histones could be phosphorylated in addition to the histone H3 in the yeast cell and that such phosphorylation events could be sufficient to ensure normal chromosome dynamics. A major role of the yeast enzyme Ipl1p is to regulate the function of the kinetochore-associated protein Ndc10p through its phosphorylation (Biggins et al. 1999; Sassoon et al. 1999). Therefore, the increase in ploidy reported in ipl1 mutant cells has been attributed more to inappropriate kinetochore function, and consequently the effects of Air-2 depletion upon chromosome condensation in C. elegans have been a little overshadowed. It seems likely that the abnormal chromosome segregation in Drosophila cells after aurB RNAi is due to incomplete condensation, since a similar phenotype is seen in mutants of the condensin subunit Barren (Bhat et al. 1996). Of course, this does not exclude the possibility that defects in the organization of the centromeric regions and kinetochores arise directly as a result of aurB RNAi or as either a direct or indirect consequence of condensation defects. The increase in ploidy seen after aurora B RNAi is reminiscent of the Ipl1 phenotype in budding yeast, but differs in that it arises from both chromosome segregation and cytokinesis defects.
The resemblance of the mitotic phenotype of cells after RNAi with aurB to that previously reported for Drosophila barren mutants (Bhat et al. 1996) can be further explained by the failure of Barren protein to be recruited to the mitotic chromosomes after aurB RNAi. Originally recognized through this mutant defect, it was later realized that Barren is the fly homologue of a member of the pentameric complex, condensin, first shown to be required for mitotic chromosome condensation in Xenopus (Hirano et al. 1997). It is possible that Barren or other members of the condensin complex could themselves be directly phosphorylated by Aurora B during chromosome condensation. However, the process seems likely to involve a plethora of phosphorylation events: the nuclear A-kinase anchoring protein (AKAP95) appears to target the human hCAP-D2 condensin to chromosomes (Collas et al. 1999; Steen et al. 2000) and phosphorylation of condensin subunits by cdk1 has been associated both with their nuclear accumulation and activation (Kimura et al. 1998; Sutani et al. 1999). It has been proposed that phosphorylation of the NH2 terminus of histone H3 leads to the recruitment or the activation of the condensin complex to the chromosome, where it can modify DNA topology. Our data indicate that phosphorylation of histone H3 by the Aurora B kinase and the localization of Barren onto chromosomes are associated events in mitosis. They support and extend a recent observation that human condensin proteins hCAP-E, hCAP-C, and hCAP-D2 colocalize with phosphorylated histone H3 in clusters in partially condensed regions of chromosomes in early prophase (Schmiesing et al. 2000). The similarity of the effects seen on chromosome condensation resulting from loss of either aurora B or barren function is striking and points to the value of studying these processes in a single model organism amenable to both genetic manipulation and RNAi. It is perhaps surprising that in both cases partial chromosome condensation is achieved and that there can be some degree of segregation of chromatin to the poles.
The second major mitotic abnormality that we observe after aurB RNAi in Drosophila cells is a failure of cytokinesis. Thus, like its mammalian and nematode counterparts AIM-1 and AIR-2, the enzyme encoded by aurora B appears essential for this process. Two proteins that play a role in cytokinesis have recently been shown to associate with the Aurora B–like kinases: INCENP, as discussed above (Adams et al. 2000; Kaitna et al. 2000), and the Zen-4 kinesin-like protein of C. elegans (Kaitna et al. 2000; Severson et al. 2000). The localization of the latter is disrupted after disruption of air-2 function using RNAi or conditional mutant alleles. Zen-4 is the C. elegans homologue of the Pavarotti KLP of Drosophila, which we now show likewise to be mislocalized on the central spindle from anaphase onwards after aurB RNAi. Pav-KLP also cooperates with Polo kinase to achieve its localization and function in Drosophila (Adams et al. 1998), suggesting that multiple mitotic kinases may be required to coordinate central spindle formation before cytokinesis, just as several kinases appear to be required for centrosome maturation and separation and chromosome condensation.
It is striking that aurB RNAi cells are not arrested by a mitotic checkpoint, given the abnormalities that they show in chromosome alignment at metaphase and the subsequent disorganization of the later mitotic spindle. However, the treated cells do undergo multiple cell cycles, as is clearly demonstrated in this cell culture system in which one can monitor the shift in ploidy by FACS® analysis and the increase in chromosome and centrosome complements by immunocytology. It is possible that these abnormalities arise too late in the mitotic cycle to trigger checkpoint arrest, although this seems unlikely for the chromosome segregation defect. Although it is possible that Aurora B is itself required for checkpoint functions, it could also be that the kinetochore regions of chromosomes are insufficiently well organized after aurB RNAi to promote the checkpoint activity of the complex of Bub/Mad proteins that associate with unaligned centromeres. It is noteworthy that the C. elegans baculovirus inhibitor of apoptosis (IAP)-related repeat protein Bir-1 appears to be required for the localization of Air-2. Bir-1 localizes to chromosomes and then the spindle midzone and Air-2 fails to localize to these same sites in the absence of Bir-1 (Speliotes et al. 2000). These IAP proteins, also known as survivin, are caspase inhibitors and as such counteract apoptosis (Li et al. 1998). Is it possible that B-type Aurora kinases might play a role alongside survivin in an apoptotic checkpoint to promote mitosis?
It is of considerable interest to know the multiple substrates of Aurora B kinase and to understand its mode of regulation in mitotic progression. It seems that subcellular localization of the enzyme could be one critical means of controlling access to its substrates. The enzyme localizes throughout condensing chromosomes when histone H3 is required to be phosphorylated. Its subsequent concentration at centromeres could direct enzyme activity towards specific chromosomal proteins at these sites, but may be instrumental in its movement onto the central spindle at anaphase, thereby providing an effective way of removing the enzyme from the chromatin to facilitate chromosome decondensation at telophase. Understanding the intricacies of these processes will be a future challenge.
Acknowledgments
We would like to thank Mark Jackman and Jonathan Pines for their great help in FACS® analysis with S2 cells and Hugo Bellen and Karen Schultze for continued interest and for kindly sending the antibarren antibody.
The work was supported by a Marie-Curie Research Fellowship from the European Commission to R. Giet and by a program grant from the Cancer Research Campaign.
Footnotes
1Abbreviations used in this paper: dsRNA, double-stranded RNA; INCENP, inner centromere protein; RNAi, RNA interference.
References
- Adams R.R., Tavers A.A., Salzberg A., Bellen H.J., Glover D.M. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.R., Wheatleya S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Bhat M.A., Philp A.V., Glover D.M., Bellen H.J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E.M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- Caplen N.J., Fleenor J., Fire A., Morgan R.A. dsRNA-mediated gene silencing in cultured Drosophila cellsa tissue culture model for the analysis of RNA interference. Gene. 2000;252:95–105. doi: 10.1016/s0378-1119(00)00224-9. [DOI] [PubMed] [Google Scholar]
- Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P., Le Guellec K., Tasken K. The A-kinase–anchoring protein AKAP95 is a multivalent protein with a key role in chromatin condensation at mitosis. J. Cell Biol. 1999;147:1167–1180. doi: 10.1083/jcb.147.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Barre A.E., Gerson V., Gout S., Creaven M., Allis C.D., Dimitrov S. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:379–391. doi: 10.1093/emboj/19.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C.P., Osmani A.H., Wu L.P., Spotts J.L., Osmani S.A. Mitotic histone H3 phosphorylation by the NIMA kinase in Asperigillus nidulans . Cell. 2000;102:293–302. doi: 10.1016/s0092-8674(00)00035-0. [DOI] [PubMed] [Google Scholar]
- Francisco L., Chan C.S. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell. Mol. Biol. Res. 1994;40:207–213. [PubMed] [Google Scholar]
- Giet R., Prigent C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 1999;112:3591–3601. doi: 10.1242/jcs.112.21.3591. [DOI] [PubMed] [Google Scholar]
- Giet R., Prigent C. The Xenopus laevis aurora/Ip11p-related kinase pEg2 participates in the stability of the bipolar mitotic spindle. Exp. Cell Res. 2000;258:145–151. doi: 10.1006/excr.2000.4903. [DOI] [PubMed] [Google Scholar]
- Giet R., Uzbekov R., Cubizolles F., Le Guellec K., Prigent C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 1999;274:15005–15013. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- Glover D.M., Leibowitz M.H., Mclean D.A., Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T., Tu C. Sex specific processing of the Drosophila exuperentia transcript is regulated in male germline cells by the tra-2 gene. Proc. Natl. Acad. Sci. USA. 1994;91:10752–10756. doi: 10.1073/pnas.91.22.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi R., Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Hunter C.P. Geneticsa touch of elegance with RNAi. Curr. Biol. 1999;9:R440–R442. doi: 10.1016/s0960-9822(99)80276-0. [DOI] [PubMed] [Google Scholar]
- Hunter C.P. Gene silencingshrinking the black box of RNAi. Curr. Biol. 2000;10:R137–R140. doi: 10.1016/s0960-9822(00)00325-0. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Mendoza M., Jantsch-Plunger V., Glotzer M. Incenp and an Aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kang J.S., Chan C.S. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae . J. Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensina biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Kimura K., Hirano M., Kobayashi R., Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K., Rybenkov V.V., Crisona N.J., Hirano T., Cozzarelli N.R. 13S condensin actively reconfigures DNA by introducing global positive writheimplications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Li F.Z., Ambrosini G., Chu E.Y., Plescia J., Tognin S., Marchisio P.C., Altieri D.C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Mackay A.M., Ainsztein A.M., Eckley D.M., Earnshaw W.C. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 1998;140:991–1002. doi: 10.1083/jcb.140.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nature Reviews Molecular Cell Biology. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Reich A., Yanai A., Mesilaty-Gross S., Chen-Moses A., Wides R., Motro B. Cloning, mapping, and expression of ial, a novel Drosophila member of the Ipl1/aurora mitotic control kinase family. DNA Cell Biol. 1999;18:593–603. doi: 10.1089/104454999315141. [DOI] [PubMed] [Google Scholar]
- Roghi C., Giet R., Uzbekov R., Morin N., Chartrain I., Le Guellec R., Couturier A., Doree M., Philippe M., Prigent C. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 1998;111:557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- Rong Y.S., Golic K.G. Gene targeting by homologous recombination in Drosophila . Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Sassoon I., Severin F.F., Andrews P.D., Taba M.R., Kaplan K.B., Ashford A.J., Stark M.J., Sorger P.K., Hyman A.A. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing J.A., Gregson H.C., Zhou S., Yokomori K. A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell. Biol. 2000;20:6996–7006. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J.M., Golden A., Donovan P.J. AIR-2an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A.F., Hamill D.R., Carter J.C., Schumacher J., Bowerman B. The Aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Speliotes E.K., Uren A., Vaux D., Horvitz H.R. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Steen R.L., Cubizolles F., Le Guellec K., Collas P. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J. Cell Biol. 2000;149:531–536. doi: 10.1083/jcb.149.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T., Yuasa T., Tomonaga T., Dohmae N., Takio K., Yanagida M. Fission yeast condensin complexessential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuka M., Katayama H., Ota T., Tanaka T., Odashima S., Suzuki F., Terada Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58:4811–4816. [PubMed] [Google Scholar]
- Terada Y., Tatsuka M., Suzuki F., Yasuda Y., Fujita S., Otsu M. AIM-1a mammalian midbody-associated protein required for cytokinesis. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok T., Harvie P.D., Buratovich M., Bryant P.J. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila . Genes Dev. 1997;11:213–225. doi: 10.1101/gad.11.2.213. [DOI] [PubMed] [Google Scholar]
- Wei Y., Yu L., Bowen J., Gorovsky M.A., Allis C.D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Whitfield W.G., Gonzalez C., Maldonado-Codina G., Glover D.M. The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:2563–2572. doi: 10.1002/j.1460-2075.1990.tb07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolard A., Hodgekin J. Stu-7/air-2 is a C. elegans aurora homologue essential for chromosome segregation during embryonic and postembryonic development. Mech. Dev. 1999;82:95–108. doi: 10.1016/s0925-4773(99)00020-9. [DOI] [PubMed] [Google Scholar]