Abstract
Although the primary function of the DNA mismatch repair (MMR) system is to identify and correct base mismatches that have been erroneously introduced during DNA replication, recent studies have further implicated several MMR components in somatic hypermutation of immunoglobulin (Ig) genes. We studied the immune response in mice deficient in MutS homologue (MSH)3 and MSH6, two mutually exclusive partners of MSH2 that have not been examined previously for their role in Ig hypermutation. In Msh6−/− and Msh3−/−/Msh6−/− mice, base substitutions are preferentially targeted to G and C nucleotides and to an RGYW hot spot, as has been shown previously in Msh2−/− mice. In contrast, Msh3−/− mice show no differences from their littermate controls. These findings indicate that the MSH2–MSH6 heterodimer, but not the MSH2–MSH3 complex, is responsible for modulating Ig hypermutation.
Keywords: DNA mismatch repair, immunoglobulin genes, germinal center, Msh3, Msh6
Introduction
Somatic mutation of Ig V region genes contributes to the generation of antibody diversity and is required for the production of high-affinity neutralizing antibodies 1. In the process of V region hypermutation, single base changes and occasional deletions and insertions are introduced into rearranged V regions and their immediate flanking sequences at extremely high rates of 10−3–10−4/bp/generation 1 2 3. The mutations exhibit a characteristic pattern of hot spot selection and nucleotide substitutions 4 5 6, yet the mechanism by which these base changes are introduced remains a puzzle 7. Considering the high rate and substrate of mutation, it seems likely that normal mechanisms of DNA repair might play a direct role in the mutational process or secondarily affect its outcome 8. Studies of mice and patients who lack components of the nucleotide or base excision repair pathways have not revealed abnormalities in V region mutation (for a review, see reference 9). However, several groups have recently shown that MutS homologue (MSH)2, postmeiotic segregation (PMS)2, and MutL homologue (MLH)1, three components of the DNA mismatch repair (MMR) system, may be involved 8 10 11 12 13 14 15.
The primary function of the MMR system is to identify and correct base mismatches introduced during semiconservative DNA synthesis. Postreplicative MMR is best characterized in bacteria, where MutS first recognizes the mismatched DNA site, then recruits MutL, which in turn binds and activates the MutH endonuclease. A DNA oligonucleotide containing the mismatch is then excised, and a patch roughly 200 nucleotides in length is then resynthesized correctly by DNA polymerase III 16. In yeast and higher eukaryotes, the initiation of MMR is thought to require complexes between subsets of the three MutS homologues, MSH2, MSH3, and MSH6. A complex of MSH2 and MSH6 is required for the repair of single nucleotide mutations, whereas a complex of MSH2 and MSH3 is required for 2–4 bp mutations, although there may be some redundancy in these functions. In a subsequent step, the MLH1–PMS2 heterodimer is recruited, and ultimately the mismatch-containing DNA is excised and resynthesized correctly 17.
Previous studies of MSH2-deficient mice revealed that V region mutations were preferentially targeted to G and C nucleotides and to hot spot motifs 8 11 12. Although earlier studies did not show selective targeting to G and C in PMS2- or MLH1-deficient mice 8 13 18, a more recent study, in which a passenger L chain transgene contained an artificial target within its V region, did reveal that G and C were also preferentially targeted in PMS2- and MLH1-deficient mice 14. The overall frequency of mutations during the primary response was similar in the Msh2−/ − and wild-type mice. There was a decrease in the accumulation of base changes in chronically immunized Peyer's patch B cells 11 12.
We present here our studies on V region hypermutation in mice genetically deficient in the MSH3 and MSH6 components of the MMR system, two proteins that have not been examined previously for their role in the immune response. Our studies reveal that Msh6−/− and Msh3−/−/Msh6−/− mice resemble the Msh2−/− mice, in that V region mutations are primarily targeted to G and C nucleotides and are focused in RGYW hot spots. Furthermore, although the mutation frequency is unchanged in the splenic primary response, fewer base changes accumulate in Ig genes of chronically stimulated Peyer's patch germinal center (GC) cells. In contrast, the frequency and characteristics of V region mutation in Msh3−/− mice are the same as in their wild-type or heterozygous littermates. Taken together with the earlier studies on Msh2−/− mice, these results indicate that in B cells, the MSH2–MSH6 complex and additional downstream factors are collectively responsible for the balance of mutations between G/Cs and A/Ts. These studies thus confirm that MMR plays a role in the outcome of V region hypermutation, and extend our knowledge of the individual factors that are involved.
Materials and Methods
MMR-deficient Mice.
Mice deficient in MSH6 and MSH3 were generated previously 19 20. Single-knockout Msh6 and Msh3 mice (129/Sv × C57BL/6 × SJL) were backcrossed to C57BL/6 for four to six generations before use. Mice deficient in both MSH3 and MSH6 were obtained by breeding Msh3−/− mice to Msh6−/− mice that had been backcrossed twice to C57BL/6. Mice were prescreened for IgHb allotype before use. In all cases, MMR-deficient animals were analyzed in parallel with wild-type or heterozygous sex-matched littermate controls, so that any residual mouse strain background or environmental differences would be controlled for in each analysis. All mice were maintained in specific pathogen-free housing in microisolator cages.
Mouse Immunization.
4-hydroxy-3-nitrophenyl (NP) conjugated to chicken γ-globulin (CGG) at a ratio of 17:1 was purchased from Biosearch Technologies. Mice 8–12 wk old that were free of obvious disease were given a primary immunization with 50 μg of alum-precipitated NP17-CGG via the intraperitoneal route 21. Mice were tail-bled at weekly intervals, and then boosted intraperitoneally with 100–200 μg NP17-CGG in PBS. Spleens were obtained 10 d after the primary immunization or 4 d after the boost.
Isolation of B Cells.
Single cell suspensions of either spleen or Peyer's patch lymphocytes were sequentially incubated with anti–mouse CD45R (B220), MACS MicroBeads (Miltenyi Biotec), phycoerythrin-labeled anti–mouse CD45R (B220) antibody (GIBCO BRL), and fluorescein-labeled peanut agglutinin (Vector Labs). After binding to a MACS VS+ column, the B220+ fraction was separated by FACS® (FACStarPLUS™; Becton Dickinson), yielding 90–95% pure B220+ peanut agglutinin–high cells. Genomic DNA was processed as described 22.
PCR Amplification of Ig Sequences.
Anti-NP V regions (V186.2 and V3 gene families) rearranged to JH1 and JH2 segments were amplified by nested PCR using published primers as the outer pair 21. The nested forward primer had the sequence 5′-CAG GTC CAA CTG CAG CAG C-3′, with reverse primer sequence 5′-TGA GGA GAC GAT GAC CGT GG-3′. Cycling parameters were 30 cycles of 45 s at 95°C, 45 s at 61°C, and 3 min at 72°C for the first round. In the second round, 1/50 of the PCR product was amplified for another 30 cycles of 45 s at 95°C, 45 s at 55°C, and 2 min at 72°C (Perkin-Elmer Corp.). The fidelity of Pfu DNA polymerase was verified by sequencing V regions from splenic peanut agglutinin–low B220+ cells, yielding a mutation frequency of 1.81 × 10−4/bp (5 mutations in 27,636 bp). JH4 flanking regions were amplified according to the methods of Frey et al. 11, except that restriction site sequences were omitted from the primers. Only JH4 segments from genes with unique rearrangements were analyzed. PCR products were cloned into the Bluescript vector (Stratagene) and sequenced (ABI 377; Applied Biosystems) at the Albert Einstein College of Medicine Sequencing Facility.
Serological Assays.
Sera were screened by ELISA for NP reactivity using plates coated with 1 μg/ml NP13-BSA to assay the total response, or 1 μg/ml NP2.5-BSA to detect high affinity antibodies (Biosearch Technologies). Anti-NP antibodies were detected with goat anti–mouse IgM or IgG1 (Southern Biotechnology Associates). Sera were plated at an initial dilution of 1:100, then serially diluted to 1:102,400. The titer was assigned at one half the maximum absorbance, measured at 405 nm.
Statistical Analysis.
To assess the statistical significance of hot spot targeting in mutated V regions, the frequency of clones containing a mutation in the G92 hot spot was compared with the frequency of mutated clones lacking this RGYW hot spot mutation, using the χ2 and Fisher's exact tests. The frequency of mutated clones was also compared with the frequency of unmutated V regions recovered, using the same statistical tests. For the statistical analysis of anti-NP IgG1 titers, the two-sample t test and the Wilcoxon rank sum test were both carried out, using the titer measured in each mouse as the input. Both tests yielded comparable results. All statistical analyses were performed with StatXact v4.0 (Cytel Software Corp.).
Results
Mutation Frequency in Ig Genes of MMR-deficient Mice.
Mice lacking MSH3 or MSH6 were challenged with the hapten NP, and GC B cells from the spleen were analyzed 10 d after primary immunization. The proportion of mutated V regions and the frequency of base changes in Msh3−/−, Msh6−/−, and Msh3−/−/Msh6−/− mice were not significantly different from each other (Table ). In addition, mutation frequencies in each of these gene-targeted mice were indistinguishable from those observed in their respective littermate controls during the early primary response. Our results are consistent with those reported by others for MSH2- and PMS2-deficient mice 8 11 12 18. Collectively, the data suggest that these defects in MMR do not greatly affect the frequency of somatic mutation when B cells have undergone relatively few rounds of replication and mutation. Msh6−/− mice were also examined 4 d after secondary immunization (Table ). In the Msh6−/− mice, a significantly lower proportion of the V regions contained mutations (p < 0.015), yielding fivefold fewer mutations overall. However, the frequency of base changes in those few V regions that did mutate was not statistically different between Msh6−/− and Msh6+/+ mice.
Table 1.
Mutation Frequency in the Anti-NP Response
| V sequences analyzed | Mutation frequency (×10−3) | |||||
|---|---|---|---|---|---|---|
| Total No. of sequences | Percent mutated sequences | Total No. of mutations | All sequences | Mutated sequences | ||
| Primary response | ||||||
| Msh3−/− | 35 | 71 | 91 | 9.3 | 13.0 | |
| Msh3+/− | 5 | 80 | 16 | 11.4 | 14.3 | |
| Msh6−/− | 16 | 69 | 36 | 8.0 | 12.0 | |
| Msh6+/− | 27 | 59 | 36 | 4.8 | 8.0 | |
| Msh3−/−/Msh6−/− | 30 | 50 | 30 | 3.6 | 7.1 | |
| Msh3+/−/Msh6+/− | 11 | 29 | 20 | 6.5 | 17.9 | |
| Secondary response | ||||||
| Msh6−/− | 22 | 5 | 1 | 0.2 | 3.6 | |
| Msh6+/− | 14 | 43 | 12 | 3.1 | 7.1 | |
Canonical V186.2 mutated regions and V186.2 analogues have been included in this analysis.
Since B cells in the intestinal Peyer's patches are constantly responding to a wide variety of gut antigens rather than to one specific immunogen, many different V regions are being mutated. Mutation in these different V regions can be analyzed by examining the noncoding intronic sequence flanking a rearranged JH4 segment, using the PCR strategy developed by Jolly et al. 23. Using this approach in slightly modified form, Frey et al. 11 observed threefold and fivefold decreases in mutation frequency in the Peyer's patch GC B cells of Pms2−/− mice and Msh2−/− mice, respectively. Rada et al. 12 independently observed a fivefold reduction in mutation frequency in Msh2−/− mice compared with their littermate controls.
As shown in Table , JH4 flanking regions from the Peyer's patches of Msh6−/− mice exhibit a decrease in mutation frequency, which, although not statistically significant, is similar to the findings of other researchers for Pms2−/− mice 10 11. For the Msh3−/− mice, we had a larger number of sequences to analyze than in Table , and the frequency of mutation was statistically indistinguishable from their Msh3+/+ littermates, as assessed with the two-tailed Fisher's exact test. This indicates that even after many rounds of cell replication, the loss of MSH3 does not affect the frequency of V region mutation. In contrast, mice deficient in both MSH3 and MSH6 have fewer mutated V regions (p < 0.037), and a lower frequency of mutations in those few V regions that are mutated, than their heterozygous littermates. In these Msh3−/−/Msh6−/− mice, the proportion of mutated JH4 regions and the mutation frequency are also lower than in the Msh3−/− and Msh6−/− single-knockout mice (Table ).
Table 2.
Mutation Frequency in JH4 Flanking Regions from Peyer's Patch GC B Cells
| V sequences analyzed | Mutation frequency (×10−3) | |||||
|---|---|---|---|---|---|---|
| Total No. of sequences | Percent mutated sequences | Total No. of mutations | All sequences | Mutated sequences | ||
| Msh3−/− | 11 | 36 | 17 | 5.9 | 16.3 | |
| Msh3+/− | 10 | 30 | 13 | 5.0 | 16.6 | |
| Msh6−/− | 18 | 22 | 7 | 1.5 | 6.7 | |
| Msh6+/+ | 8 | 14 | 4 | 1.9 | 15.4 | |
| Msh3−/−/Msh6−/− | 14 | 14 | 2 | 0.5 | 3.8 | |
| Msh3+/−/Msh6+/− | 6 | 66 | 19 | 12.2 | 18.3 | |
The Anti-NP Humoral Response in MMR-deficient Mice.
To evaluate whether these small differences in the frequency of mutation were associated with detectable changes in the immune response, we measured anti-NP Ig serum levels and examined the B and T cell compartments in MMR-deficient mice. Mice lacking MSH3 alone responded to NP immunization in a manner statistically indistinguishable from that of their littermates. In contrast, in Msh6−/− mice, IgG1 anti-NP titers were at one half the level measured in wild-type littermates in both the primary (p < 0.008) and secondary (p < 0.032) response (data not shown). Mice lacking both MSH3 and MSH6 also had significantly lower titers of anti-NP IgG1 at day 21 of the primary response (p < 0.015), with titers at one quarter the levels observed in littermate controls. The average affinity, as measured by the titer of IgG1 binding low-substituted NP-BSA, was also depressed. Similar decreases in hapten-specific IgG1 antibodies have been reported in MSH2-, PMS2-, and MLH1-deficient mice 11 12 24 25 26. Since hapten-specific IgM levels were normal in all of the mice tested, the lower level of IgG1 may be due to a switching defect 25 26 or genetic instability caused by a deficiency in MMR in memory B or T cells that have undergone many rounds of replication 11 24. We detected no gross defects in the number or maturation of peripheral T cell subsets in spleen or Peyer's patches in any of the MMR-deficient mice (data not shown). There was a moderate decrease in the absolute and relative number of CD45R+ B cells in the spleens of Msh3−/−/Msh6−/− mice, whereas Msh3 or Msh6 single-mutant mice showed no difference compared with wild-type controls (data not shown). In addition, we observed fewer mature B cells in both the bone marrow and spleens of Msh3−/−/Msh6−/− mice (data not shown), as has been reported in MSH2-deficient mice 24.
Targeting to G and C Bases and to Hot Spots in MMR-deficient Mice.
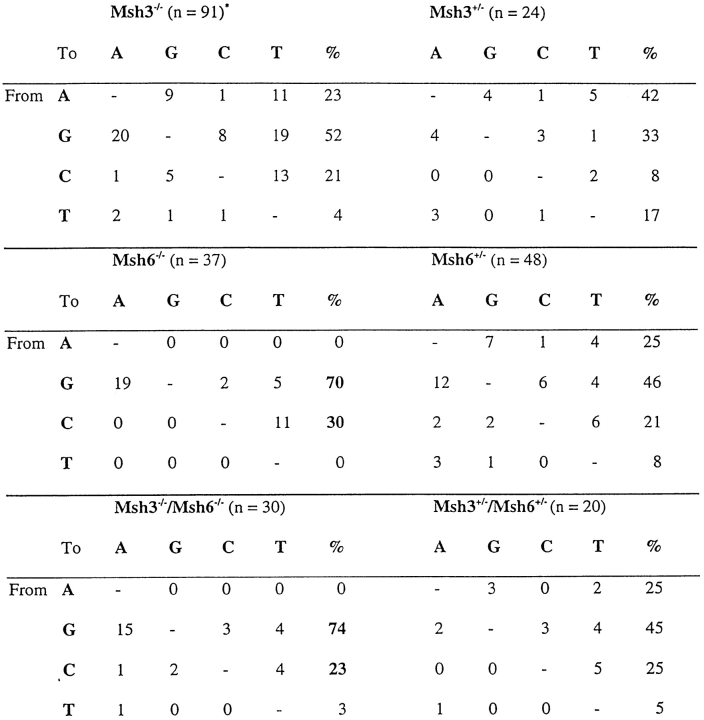
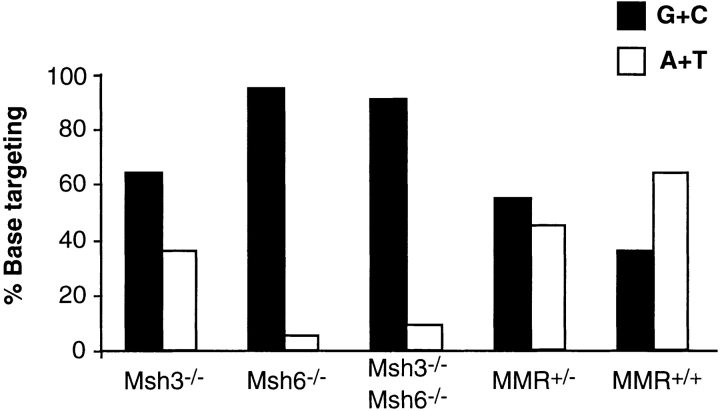
In MSH3-deficient mice, the spectrum of mutations was very similar and not significantly different from their heterozygous littermates (Table ). In contrast, Msh6−/− mice resembled Msh2−/− mice, in that 100% of mutations in the V regions from splenic B GC cells occurred in G or C, compared with 67% for the Msh6+/− littermates (Table ). The difference in the targeting of A and T nucleotides was significant at P < 0.0004. For Msh3−/−/Msh6−/− mice, 95% of mutations were in G and C, compared with 67% in the double-heterozygote littermates, and the differences in the proportion of mutations in A and T were significant at P < 0.004. When we included the base substitutions recovered from JH4 regions (Peyer's patches), there were a few additional mutations in A and T, but the vast majority of the mutations were still in G and C bases (Fig. 1). As shown in Table , there is a strong bias for transitions over transversions, a hallmark of somatic hypermutation. Thus, mice lacking MSH2, MSH6, or both MSH3 and MSH6 show a skewed targeting for G and C, whereas Msh3−/− mice exhibit a normal pattern of base substitutions. The results reported here complement and extend the observations published by others on the MSH2, PMS2, and MLH1 proteins 8 10 11 12 13 14 18.
Table 3.
Base Substitutions in Mutated V Regions from Splenic GC B Cells
Primary and secondary anti-NP responses in Msh3−/− and Msh6−/− mice are pooled in this analysis.
Figure 1.
G and C nucleotides are preferentially mutated in Msh6−/− and Msh3−/−/Msh6−/− mice, but not in Msh3−/− or in heterozygous mice. The proportions of mutations in G and C bases, relative to those targeting A and T bases, are expressed as a percentage of the total mutations and are depicted as black and white bars, respectively. As discussed in the text, mutations in A and T are significantly fewer in Msh6−/− and Msh3−/−/Msh6−/− mice, compared with Msh3−/− or wild-type control mice. Base substitutions collected from both spleen and Peyer's patch GC B cells are combined in this figure. Results from heterozygous littermates of all three genotypes are pooled in this figure; details of each MMR littermate control can be found in Table Table Table . The wild-type sample represents data from Msh6+/+ control mice. These sequence data are available from GenBank under accession nos. AF183260–AF183322.
Rada et al. 12 have also shown that in MSH2-deficient mice, the base substitutions are clustered in a relatively small number of hot spots. In agreement with the findings of others who have studied the NP response 21 27 28, we found that the RGYW hot spot at base 92 and codon 31 exhibited a high frequency of mutations. This hot spot accounts for 17 and 24% of the mutations in Msh6−/− and Msh3−/−/Msh6−/− mice, and preferential mutation at this position is significantly different between the null mice and their littermates (p < 0.02 and P < 0.003, respectively). In contrast, mutations at the G92 hot spot accounted for only 4% of the mutations in Msh3−/− mice, and were not significantly different from the wild-type control sample (7%).
Discussion
Since MSH2 dimerizes with MSH3 or MSH6, we were interested in whether the lack of either one of these molecules would differentially affect V region hypermutation. The phenotypes of the MSH3 and MSH6 single-mutant mice are quite different 19 20. Msh6−/− mice have a reduced life span and an increased susceptibility to cancer, whereas Msh3−/− mice have neither a decrease in survival nor a significant increase in cancer risk. Nuclear extracts derived from the Msh6−/− mice are deficient in the repair of single base mismatches, but are proficient in the repair of single base insertions and dinucleotide insertions and deletions. In contrast, Msh3−/− extracts are proficient in the repair of single base mismatches and certain types of single base insertions, but are deficient in the repair of dinucleotide insertions and deletions. In the studies reported here, we have shown that the loss of MSH3 does not have a detectable effect on the characteristics or frequency of V region mutation. In contrast, the absence of MSH6 results in the restriction of mutations to G and C in some hot spots, as has been reported for the Msh2−/− mice 8 12 29. In addition, the absence of both MSH3 and MSH6 has a similar effect as the loss of either MSH6 or MSH2. These results suggest that the mismatches that are created during the primary process of V region mutation in B cells are recognized by the MSH2/MSH6 dimer, rather than by the MSH2/MSH3 complex. This is consistent with the fact that most of the somatic mutations that have been observed in normal mice are single base changes. The increase in tumor load, the earlier onset of tumors, and the apparently greater decrease in the frequency of mutation in the Peyer's patch GC cells of the Msh3−/−/Msh6−/− mice reported here collectively suggest that there may be a role for MSH3, at least in the absence of MSH6, and that it is expressed in GC B cells.
Somatic hypermutation was studied in NP-immunized mice, as this hapten elicits a well-studied immune response in C57BL/6 mice, where the V186.2 gene and close relatives thereof, termed analogues, play a predominant role 27 28. The NP response of mice lacking both MSH3 and MSH6 is atypical in that we recovered very few canonical V186.2 regions (7% of the mutated V regions). This was not observed in the Msh3−/− mice, where the canonical V186.2 region, along with the affinity-enhancing W33L mutation, was recovered from 74% of the mutated VDJ rearrangements. In Msh6−/− mice, 55% of the mutated anti-NP rearrangements contained the V186.2 gene. One explanation for the low representation of V186.2 in the double-mutant mice is that they were incompletely backcrossed to C57BL/6, unlike the Msh3 and Msh6 single mutants.
There is no simple explanation for why there should be a selective decrease in A and T mutations in MSH2- and MSH6-deficient mice. One possibility is that V region hypermutation occurs as a series of separable events, and that the processes that cause mutation in G and C are different from those that target A and T. One example of such a model is that MMR is not involved in the primary mutational event that affects primarily G and C, but MSH2 and its associated proteins do recognize the mismatches created by that initial mutagenic event, and in the course of repairing them, mutations are introduced in A and T bases 12. It is intriguing that mutational targeting to G and C bases is an intrinsic feature of mutation in lower vertebrates and in hypermutating cell lines 30 31 32. Although the consensus is that MMR is not involved in the primary mutational event, Cascalho et al. 10, Wilson et al. 3, and Kong and Maizels 15 have suggested that MMR may play a primary role in the mutation process.
In conclusion, the studies reported here confirm a role for MMR in the outcome of the process of V region hypermutation. They show that in addition to MSH2, MLH1, and PMS2, MSH6 plays a role in this event. Although they do not eliminate the possibility that one or more of the MMR proteins play a direct role in the mutational process, they are consistent with the proposal that the initial target of the mutational process is single base changes in G or C, and that the mutations in A and T are the result of a subsequent error-prone repair process 8 12. Although we do not know what proteins are responsible for the primary mutational event, these studies indicate that MSH6 is among the transacting proteins that are required for the mutational process.
Acknowledgments
We wish to thank Dave Gebhard for the FACStarPLUS™ cell isolation and for his assistance in FACScan™ analysis (Albert Einstein College of Medicine FACS® Facility), and Elena Avdievich for expert technical assistance in the maintenance of the mouse colony. We also wish to thank Dr. Betty Diamond for her critical reading of the manuscript. Statistical analyses were performed by Drs. C.J. Chang and Yun Gu, Department of Epidemiology and Social Medicine, Albert Einstein College of Medicine.
This work was supported by National Institutes of Health grants 5R37 CA39838, R01 CA72649, R01 CA76329, 5P30 CA1330, and 5T32 CA09173. M.D. Scharff is also supported by the Harry Eagle Chair in Cancer Research, provided by the National Women's Division of the Albert Einstein College of Medicine.
References
- Kocks C., Rajewsky K. Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proc. Natl. Acad. Sci. USA. 1988;85:8206–8210 . doi: 10.1073/pnas.85.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens T., Klein U., Kuppers R. Frequent occurrence of deletions and duplications during somatic hypermutationimplications for oncogene translocations and heavy chain disease. Proc. Natl. Acad. Sci. USA. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P., Liu Y.J., Banchereau J., Capra J.D., Pascual V. Amino acid insertions and deletions contribute to diversify the human Ig repertoire. Immunol. Rev. 1998;162:143–151. doi: 10.1111/j.1600-065x.1998.tb01437.x. [DOI] [PubMed] [Google Scholar]
- Dorner T., Foster S.J., Brezinschek H.-P., Lipsky P.E. Analysis of the targeting of the hypermutational machinery and the impact of subsequent selection on the distribution of nucleotide changes in human VHDJH rearrangements. Immunol. Rev. 1998;162:161–171. doi: 10.1111/j.1600-065x.1998.tb01439.x. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Kolchanov N.A. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- Milstein C., Neuberger M.S., Staden R. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Kong Q., Maizels N. Somatic hypermutation and the three R'srepair, replication and recombination. Mutat. Res. 1999;436:157–178. doi: 10.1016/s1383-5742(99)00003-4. [DOI] [PubMed] [Google Scholar]
- Phung Q.H., Winter D.B., Cranston A., Tarone R.E., Bohr V.A., Fishel R., Gearhart P.J. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Storb U. The role of DNA repair in somatic hypermutation of immunoglobulin genes. J. Exp. Med. 1998;187:1729–1733. doi: 10.1084/jem.187.11.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascalho M., Wong J., Steinberg C., Wabl M. Mismatch repair co-opted by hypermutation. Science. 1998;279:1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- Frey S., Bertocci B., Delbos F., Quint L., Weill J.C., Reynaud C.A. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- Phung Q.H., Winter D.B., Alrefai R., Gearhart P.J. Hypermutation in Ig V genes from mice deficient in the MLH1 mismatch repair protein. J. Immunol. 1999;162:3121–3124. [PubMed] [Google Scholar]
- Kim N., Bozek G., Lo J.C., Storb U. Different mismatch repair deficiencies all have the same effects on somatic hypermutationintact primary mechanism accompanied by secondary modifications. J. Exp. Med. 1999;190:21–30. doi: 10.1084/jem.190.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Maizels N. PMS2-deficiency diminishes hypermutation of a lambda1 transgene in young but not older mice. Mol. Immunol. 1999;36:83–91. doi: 10.1016/s0161-5890(99)00027-9. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- Winter D.B., Phung Q.H., Umar A., Baker S.M., Tarone R.E., Tanaka K., Liskay R.M., Kunkel T.A., Bohr V.A., Gearhart P.J. Altered spectra of hypermutation in antibodies from mice deficient for the mismatch repair protein PMS2. Proc. Natl. Acad. Sci. USA. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W., Yang K., Umar A., Heyer J., Lau K., Fan K., Liedtke W., Cohen P.E., Kane M.F., Lipford J.R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- Edelmann W., Umar A., Yang K., Heyer J., Kucherlapati M., Lia M., Kneitz B., Avdievich E., Fan K., Wong E. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 2000;In press [PubMed] [Google Scholar]
- Weiss U., Rajewsky K. The repertoire of somatic antibody mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J. Exp. Med. 1990;172:1681–1689. doi: 10.1084/jem.172.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Przylepa J., Miller C., Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med. 1993;178:1293–1307. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C.J., Klix N., Neuberger M.S. Rapid methods for the analysis of immunoglobulin gene hypermutationapplication to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora K.A., Tumas-Brundage K.M., Lentz V.M., Cranston A., Fishel R., Manser T. Severe attenuation of the B cell immune response in Msh2-deficient mice. J. Exp. Med. 1999;189:471–481. doi: 10.1084/jem.189.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Edelmann W., Kucherlapati R., Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein M.R., Neuberger M.S. Deficiency in msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombinationparallels with somatic hypermutation. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.G., Light A., Nossal G.J., Tarlinton D.M. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Dutta P.R., Cerasoli D.M., Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J. Exp. Med. 1998;187:885–895. doi: 10.1084/jem.187.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H., Fukita Y., van der Horst G.T., de Boer J., Weeda G., Essers J., de Wind N., Engelward B.P., Samson L., Verbeek S. Hypermutation of immunoglobulin genes in memory B cells of DNA repair–deficient mice. J. Exp. Med. 1998;187:1735–1743. doi: 10.1084/jem.187.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M., Flajnik M.F. Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol. Rev. 1998;162:13–24. doi: 10.1111/j.1600-065x.1998.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Bachl J., Wabl M. An immunoglobulin mutator that targets G.C base pairs. Proc. Natl. Acad. Sci. USA. 1996;93:851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E., Neuberger M.S. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]