Abstract
Interferon (IFN)-γ–induced cells express the proteasome subunits low molecular weight protein (LMP)2, LMP7, and MECL-1 (multicatalytic endopeptidase complex–like 1), leading to the formation of immunoproteasomes. Although these subunits are thought to optimize MHC class I antigen processing, the extent of their role and the mechanistic aspects involved remain unclear. Herein, we study the proteolytic generation of an human histocompatibility leukocyte antigen (HLA)-Aw68–restricted hepatitis B virus core antigen (HBcAg) cytotoxic T lymphocyte (CTL) epitope that is recognized by peripheral blood lymphocytes from patients with acute self-limited but not chronic hepatitis B virus (HBV). Immunological data suggest that IFN-γ–induced rather than uninduced HeLa cells process and present the HBV CTL epitope upon infection with HBcAg-expressing vaccinia viruses. Analyses of 20S proteasome digests of synthetic polypeptides covering the antigenic HBcAg peptide demonstrate that only immunoproteasomes efficiently perform the cleavages needed for the liberation of this HBV CTL epitope. Although the concerted presence of the three immunosubunits appears essential, we find that both catalytically active LMP7 and inactive LMP7 T1A support CTL epitope generation. We conclude that LMP7 influences the structural features of 20S proteasomes, thereby enhancing the activity of the LMP2 and MECL-1 catalytic sites, which provide cleavage specificity. Thus, LMP7 incorporation is of greater functional importance for the generation of an HBV CTL epitope than cleavage specificity.
Keywords: antigen processing, LMP7 T1A, hepatitis B virus, immunoproteasome, MHC class I
Introduction
The MHC class I Ag processing pathway allows CTLs to survey the inner cellular protein contents. Cytosolic endogenous and foreign proteins are degraded into peptides that bind the TAP (transporter associated with Ag processing) and are translocated into the endoplasmic reticulum. Peptides containing an appropriate motif are bound by newly synthesized MHC class I molecules, which then travel to the cell surface (for review see reference 1).
Proteasomes, the major cytosolic proteinase complexes, are essential for the generation of most MHC class I–presented peptides. Indeed, treatment of cells with membrane-permeable proteasome inhibitors inhibits cytosolic Ag degradation, production of antigenic peptides, and, due to a shortage of peptides in the endoplasmic reticulum, markedly impairs MHC class I assembly and cell surface transport 2 3 4 5 6. Proteasomes consist of a 20S catalytic core that is built from 28 (14 different) subunits, arranged as four heptameric rings (for review see reference 7). The two outer rings contain the structural alpha subunits (α1–α7); the inner rings contain β subunits (β1–β7), of which three (β1, β2, and β5) exert catalytic activity. Binding of 20S proteasomes to 19S regulatory complexes (PA700) confers protein degrading capacity and substrate specificity, including that for ubiquitin-conjugated proteins.
Stimulation of cells with IFN-γ induces the expression of the proteasome activator PA28αβ and three additional catalytic proteasome subunits, e.g., the subunits low molecular weight protein (LMP)2 and LMP7, encoded in the MHC class II region, and MECL-1 (multicatalytic endopeptidase complex–like 1; for review see references 8 and 9). These subunits replace the constitutive subunits delta (β1), MB1 (β5), and MC14 (mouse) or Z (human) (β2) in newly formed so-called immunoproteasomes. Although the association with PA28 has been shown to promote the production of MHC class I binding peptides, most likely by increasing the rate of dual cleavages by 20S proteasomes 10 11, the role of the IFN-γ–induced catalytic subunits is still unclear. In vitro studies using fluorogenic tri- and tetrapeptides as substrates have shown that the incorporation of immunosubunits alters the catalytic activity of 20S proteasomes 12 13 14 15 16. However, the physiological relevance of the effected changes is unclear.
Surprisingly, accurate results were obtained with polypeptides as substrates to document proteasomal cleavage site preferences. Several studies have shown that 20S proteasomes liberate MHC class I ligands out of larger polypeptides or denatured proteins in vitro 13 17 with rates that correlate well with those of production of the antigenic peptide in intact cells 18 19 20 21. This became especially clear in a study examining the generation of an immunodominant CTL epitope from the murine (m)CMV pp89 protein 18. Flanking of the pp89 antigenic peptide with alanine residues resulted in enhanced cleavage out of a pp89 polypeptide, both if synthetic polypeptide was degraded by purified proteasomes and if polypeptide-encoding sequences were expressed from a minigene in intact cells. Furthermore, the presence of preferred proteasomal cleavage sites was found to prevent efficient MHC class I presentation of OVA and of a murine leukemia virus CTL epitope 19 20, whereas a point mutation destroying a specific cleavage site in the p53 protein inhibited CTL epitope generation 21. Remarkably, whereas digestion with purified proteasomes in either substrate studied generated the exact COOH terminus of MHC class I binding peptides, the NH2-terminal cleavage was not always as precise. Thus, as also indicated by other studies 22, additional nonproteasomal proteases may trim the NH2 terminus of antigenic peptides in intact cells.
The influence of proteasome composition on antigenic peptide generation was studied for the pp89 substrate 14 23. It was found that the association of the LMP subunits with the proteasome changed cleavage specificity, resulting in the generation of a qualitatively and quantitatively different set of peptides. However, an obvious effect on the generation of the pp89 antigenic peptide was not observed 8 10 14. In other studies using cell lines lacking the LMPs, the production of certain viral CTL epitopes appeared diminished, whereas the presentation of other epitopes was not influenced 24 25 26 27. Furthermore, LMP2 and LMP7 knockout mice showed partial deficits in MHC class I Ag presentation 28 29. Thus, these data indicate that immunoproteasomes contribute to MHC class I Ag processing, but the extent of their role is presently unclear.
The question arises of how the IFN-γ–inducible subunits affect MHC class I Ag processing. The simultaneous expression of both constitutive and inducible subunits in IFN-γ–stimulated cells could, by enhancing the choices for active site subunits, simply function to increase the heterogeneity of the cytosolic proteasome population 30. Assuming that each subunit adds its own unique cleavage specificity to the proteasome, this would enhance the likelihood that antigenic peptides would be generated. Alternatively, the association of the IFN-γ–inducible subunits may in a more specific way alter the cleavage properties of proteasomes, such as to enhance the generation of ligands suitable for binding to MHC class I molecules.
To test above hypotheses, we digested a synthetic 32-mer polypeptide from the hepatitis B virus (HBV) core Ag (HBcAg) with purified proteasomes and analyzed the proteolytic generation of a contained CTL epitope, HBcAg141-151, first described by Missale et al. 31. This peptide binds the MHC class I HLA-A31 and HLA-Aw68 molecules and is recognized by CD8+ CTLs from patients with acute self-limited or convalescent HBV (reference 31 and our results). Our studies show that only immunoproteasomes liberate the antigenic peptide efficiently. Although the association of the immunosubunits with the proteasome is essential, we find that antigenic peptide production also occurs in the presence of a mutated, catalytically inactive form of one of the LMPs and thus does not require the full enzymatic activity of proteasomes. We conclude that structural changes induced by the incorporation of, in particular, LMP7 influence the proteasome-mediated cleavages, resulting in an enhanced production of MHC class I–presentable peptides.
Materials and Methods
Cell Lines.
C4 (BALB/c) mouse fibroblast cells 14 were cultured in Iscove's MEM (Biochrom), and the human cervical carcinoma cell line HeLa, the human lymphoblastoid cell line T2 32, and T2 transfectant cell lines were cultured in RPMI (Biochrom). All media were supplemented with 10% FCS, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Stimulation with 20 U/ml of murine (C4 cells) or human (HeLa cells) recombinant (r)IFN-γ (Boehringer Mannheim) was performed for 72 h before harvesting. T2 transfectant lines expressing the mouse proteasome subunits LMP2, LMP7, mutant LMP2 T1A, or mutant LMP7 T1A and combinations thereof have been described (23, 33, 34, and Schmidt, M., and P.-M. Kloetzel, manuscript in preparation).
Recovery of HBVcAg141-151–specific PBLs and CTL Assays.
PBMCs of a HLA-Aw68+ patient who had recovered from acute HBV (hepatitis B core [HBc] antibody–positive, hepatitis B surface [HBs] antibody–positive, hepatitis B envelope [HBe] antibody–positive, HBeAg-negative, HBsAg-negative) were resuspended in RPMI 1640 supplemented with l-glutamine, penicillin, streptomycin, and 10% heat-inactivated human AB serum and stimulated in 96-well round-bottomed plates at 0.2 × 106 cells per well with 10 μg/ml synthetic peptide, 10 ng/ml of rIL-7, and 10 pg/ml of rIL-12 (PeproTech Inc.). 20 U/ml of rIL-2 (Chiron Corp.) was added every 3–4 d. The cultures were restimulated with 10 μg/ml of peptide, 20 U/ml of rIL-2, and 105 irradiated (3,000 rads) autologous PBMCs in the presence of rIL-7 on day 7 and in the absence of IL-7 on day 14. A standard CTL assay with peptide-pulsed autologous PHA blasts was performed on days 20–24 as described previously 35. Alternatively, HeLa cells cultured for 2 d in the absence or presence of 20 U/ml of IFN-γ were infected with recombinant vaccinia viruses expressing HBcAg (C-Vac; provided by Dr. B. Moss, National Institute of Allergy and Infectious Diseases, National Institutes of Health) and HBxAg, respectively (X-Vac; provided by Dr. H. Schaller, ZMBH, Heidelberg, Germany) at a multiplicity of infection of 10 and used as target cells in the CTL assay the next day.
Proteasome Isolations.
20S proteasomes were purified as described 14 33. The purity of the preparates was checked by Coomassie-stained SDS-PAGE and was >90%. Aliquots were frozen at −80°C until use.
Two-dimensional Gel Analysis.
Purified proteasomes were analyzed by nonequilibrium pH gradient gel electrophoresis (NEPHGE)–SDS-PAGE and Coomassie staining as described 14.
Western Blot Analysis.
Purified proteasomes were quantified according to Bradford et al. 36, and samples of 150 ng (for the detection of LMP7) and 500 ng (for the detection of LMP2) were separated on 15% SDS–polyacrylamide gels. Immunoblots were probed with mouse LMP2– and mouse LMP7–specific polyclonal rabbit antisera as described 37.
Peptides.
The synthetic 32-mer polypeptide AYRPPNAPILSTLPETTVVRRRGRSPRRRTPS derived from HBV (subtype ayw) cAg, amino acids 131–162, and the 11-mer peptide STLPETTVVRR (HBcAg141-151) were provided by Dr. P. Henklein at our institute in Berlin.
Peptide Digestion Assays.
20 μg of HBV 32-mer polypeptide and 3 μg of purified proteasomes was incubated in 300 μl of assay buffer (20 mM Hepes/KOH, pH 7.8, 2 mM MgAc2, 1 mM dithiothreitol) at 37°C for the time periods specified in the figure legends and then frozen at −20°C. 20 μl of digests was separated by reverse-phase (RP)-HPLC (SMART system equipped with a μRPC C2/C18 SC 2.1/10 column, Pharmacia; eluent A, 0.05% TFA; eluent B, 70% acetonitrile containing 0.045% TFA; gradient: 5–63% eluent B in 30 min, 63–95% eluent B in 4 min; flow rate: 50 μl/min). Analysis was performed on line by a tandem quadrupol mass spectrometer equipped with an electrospray ion source (TSQ 7000; Finnigan MAT). Each scan was acquired over the range m/z = 300–1,300 in 3 s. The peptides were identified by their molecular masses calculated from the m/z peaks of the single or multiple charged ions.
Results
IFN-γ Treatment Induces Production of HBcAg141-151 CTL Epitopes in Infected Cells.
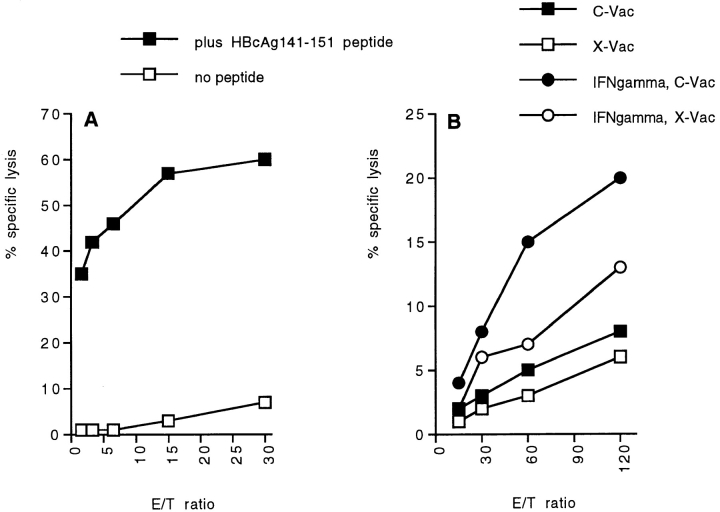
We previously demonstrated that LMP2 and LMP7 alter the proteasomal cleavage site preference in a 25-mer peptide from the mCMV pp89 protein 14 23. To further analyze the impact of the IFN-γ–inducible β subunits on Ag cleavage and antigenic peptide production, we selected a CTL epitope from the HBV core Ag, HBcAg141-151, originally identified by Missale et al. 31. Using peptide variants covering the HBcAg140-155 region, this group showed that the HBcAg141-151 11-mer represents the sequence optimally recognized by HLA-A31– and HLA-Aw68–restricted CTLs from a patient with acute self-limited hepatitis B 31. We now demonstrate that this epitope is also recognized by an HLA-Aw68+ patient who has clinically and serologically recovered from HBV infection (Fig. 1 A). HBcAg141-151–specific CTLs, expanded from PBMCs during 2 wk of in vitro culture, efficiently lysed autologous HBcAg141-151–coated PHA blasts with high specific cytotoxicity even at a low E/T ratio (Fig. 1 A). Thus, HBcAg141-151 is an HBV epitope that induces MHC class I–restricted CTLs during acute HBV infection that then persist in the peripheral blood even after recovery from hepatitis B.
Figure 1.
HBcAg141-151 is processed naturally in IFN-γ–induced HeLa cells. PBMCs of an HLA-Aw68+ patient who had recovered from acute HBV infection were stimulated with 10 μg/ml of synthetic HBcAg141-151 as described in Materials and Methods. Cytotoxicity of 3-wk cultures was tested in a 4-h 51Cr-release assay against autologous PHA blasts loaded overnight with synthetic HBcAg141-151 or left unloaded (A) or against uninduced or IFN-γ–induced HeLa cells infected with C-Vac or X-Vac recombinant vaccinia virus as described in Materials and Methods (B). Mean percentages of specific lysis of triplicate wells (y-axis) for different E/T ratios (x-axis) are plotted.
To analyze the effect of immunosubunit expression on HBcAg141-151 generation, untreated HeLa cells lacking the immunosubunits and IFN-γ–treated HeLa cells were infected with recombinant vaccinia virus encoding HBcAg (C-Vac) or with X-Vac virus as a control and then tested for recognition by HBcAg141-151–specific CTLs (Fig. 1 B). Whereas untreated C-Vac–infected cells were not recognized, the lysis percentages obtained with IFN-γ–treated, C-Vac–infected HeLa cells exceeded the background lysis levels of X-Vac–infected induced cells. Thus, only IFN-γ–treated HeLa cells process HBcAg efficiently into HBcAg141-151 CTL epitopes.
Generation of HBcAg141-151 Requires the Presence of Immunoproteasomes.
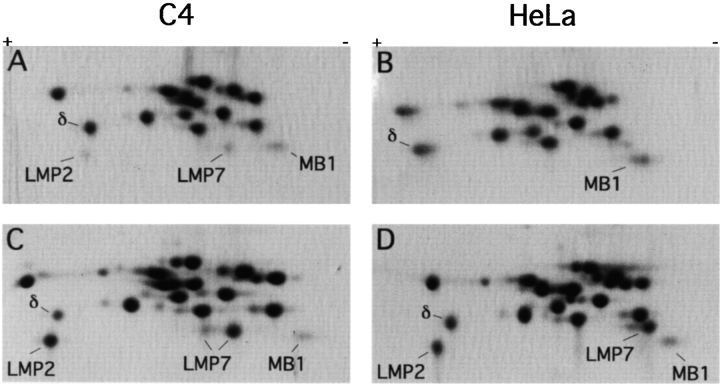
To study the proteolytic generation of HBcAg141-151, proteasomes were purified from mouse C4 fibroblast cells and human HeLa cells, in which the expression levels of immunosubunits are low under normal conditions but highly inducible by IFN-γ 13 38. A synthetic 32-mer polypeptide corresponding to HBcAg131-162 (Table ) was incubated with purified proteasomes from untreated and IFN-γ–treated C4 and HeLa cells (Fig. 2) and with proteasomes from T2/mouse LMP2+7 transfectant cells (see below). The digestion products were separated by RP-HPLC and on-line analyzed with a mass spectrometer. In each sample, >25 peptide products were detected, from which 13 were generated by two cleavages (Fig. 3). The digestion products containing either the correct COOH terminus or the correct NH2 terminus of the antigenic HBV peptide are depicted in Table . No detectable amounts of the HBV 32-mer were found in any of the digests, indicating that substrate turnover was complete. As judged from the HPLC 214-nm UV profiles (not shown), peptide Ala131–Leu140 was the predominant product independent of the type of proteasome used, identifying the Leu140-Ser141 bond, immediately NH2-terminal of the CTL epitope, as a preferred cleavage site. Strikingly, despite effective and correct usage of the NH2-terminal cleavage site of the epitope in the 32-mer polypeptide substrate, the correct COOH-terminal cleavage resulting in the generation of HBcAg141-151 only took place in the presence of proteasomes from IFN-γ–treated cells and from LMP2+7 T2 transfectant cells (Table ). In these digests, double cleavage products (DCPs) included the peptides Ser141–Val149, Ser141–Arg150, Ser141–Arg151, and Ser141–Arg152 (Table ). In contrast, constitutive proteasomes from untreated cells produced only the DCPs Ser141–Val149 and Ser141–Arg152 in detectable amounts and thus failed to generate the HBV CTL epitope. Our analyses did not reveal the presence of peptide products containing the correct epitope COOH terminus but elongated at the NH2 terminus, representing potential epitope precursors (Fig. 3).
Table 1.
Relative Quantities of Peptide Fragments Generated by Digestion of HBcAg131-162 with Purified 20S Proteasomes
| Peptide fragment | Analysis 1 | Analysis 2 | ||||
|---|---|---|---|---|---|---|
| C4 | C4+IFN-γ | HeLa | T2 LMP2+7 | HeLa+IFN-γ | T2 LMP2+7 | |
| Ala131–Ser162 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ala131–Leu140 | 1,201 | 1,629 | 578 | 1,299 | 2,865 | 3,851 |
| Ser141–Ser162 | 14 | 0 | 0 | 0 | 12 | 13 |
| Ser141–Val149 | 151 | 243 | 111 | 194 | 584 | 705 |
| Ser141–Arg150 | 26 | 214 | 17 | 140 | 444 | 460 |
| Ser141–Arg151 | 35 | 201 | 0 | 149 | 478 | 436 |
| Ser141–Arg152 | 190 | 260 | 59 | 152 | 794 | 413 |
Figure 2.
Subunit composition of 20S proteasomes purified from uninduced (A and B) and IFN-γ–induced (C and D) mouse C4 and human HeLa cells. Cells were cultured in the absence or presence of 20 U/ml of mouse or human rIFN-γ for 72 h and then harvested. 20S proteasomes were purified as described in Materials and Methods, and 50-μg aliquots were subjected to two-dimensional NEPHGE/PAGE. Separated proteins were stained with Coomassie blue. Proteasome subunits were assigned on the basis of their migrational behavior (reference 54). The identity of the human LMP7 subunit, which runs differently from its mouse equivalent, was verified by microsequencing (our unpublished observations).
Figure 3.
Peptide fragments detected after digestion of HBcAg131-162 with purified 20S proteasomes. The amino acid sequence of HBcAg131-162 containing the antigenic epitope HBcAg141-151 is shown in single-letter code. This 32-mer polypeptide was incubated with 20S proteasome for 24 h at 37°C. Cleavage products were identified by mass spectrometry after HPLC separation as described in Materials and Methods and are indicated by bars.
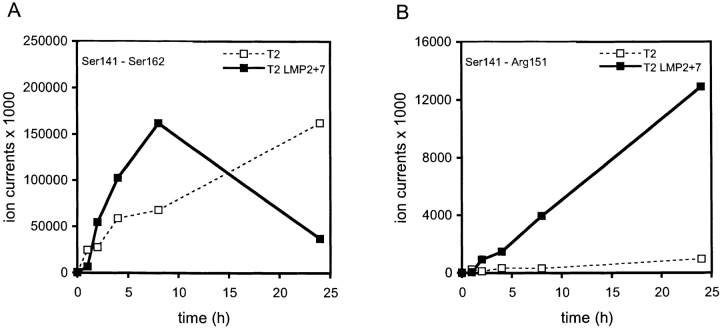
To examine whether the absence of Ser141–Arg151 in digests by constitutive proteasomes is caused by lack of generation or by further degradation, we compared the kinetics of product generation upon digestion of the 32-mer with constitutive proteasomes of T2 cells (see below) and immunoproteasomes of T2 LMP2+7 cells (Fig. 4). Although both types of proteasomes preferentially cleaved the Leu140–Ser141 bond (not shown), immunoproteasomes used this cleavage site most effectively, resulting in a rapid accumulation of Ser141–Ser162 single cleavage products (SCPs; Fig. 4 A), which at later time points of digestion (8–24 h) were further degraded into DCPs, including the Ser141–Arg151 CTL epitope (Fig. 4 B). Importantly, the early accumulation of Ser141–Arg151, as quickly as within the first hours of digestion (Fig. 4 B), suggests that this DCP is also liberated by two simultaneous cuts and is thus not dependent on the Ser141–Ser162 processing intermediate for its generation. In contrast, T2 proteasomes cleaved the Leu140–Ser141 bond at a relatively slow but constant rate over the whole course of digestion, as evident from a continuous accumulation of the Ala131–Leu140 product (not shown). Remarkably, the amounts of Ser141–Ser162 SCPs also continued to increase over the 24 h of digestion (Fig. 4 A), indicating a poor further degradation into DCPs. Only barely detectable amounts of Ser141–Arg151 were generated (Fig. 4 B), even at later time points of digestion, dismissing Arg151 as an important cleavage site for constitutive proteasomes. Taken together, we conclude from this data that the association of LMP2, LMP7, and MECL-1 with proteasomes changes the cleavage site specificity and enhances the frequency of cleavages, leading to efficient production of HBcAg141-151 CTL epitopes.
Figure 4.
Time-dependent generation of a potential precursor peptide HBcAg141-162 and of HBcAg141-151. HBcAg131-162 was digested for 0.5, 1, 2, 4, 8, and 24 h with 20S proteasome isolated from T2 cells and T2 LMP2+7 transfectant cells. Cleavage products were separated by HPLC as described in Materials and Methods but analyzed on-line by an ion trap mass spectrometer (ThermoQuest) equipped with an electrospray ion source, which results in increased sensitivity. Time course of the potential precursor peptide HBcAg141-162, which is generated by a single cleavage after Leu140 (A), and of the antigenic HBcAg141-151 (B) is depicted. Plotted values were confirmed by repeated analysis of the same digests.
Interestingly, the quality and relative quantities of peptides yielded by digestion with proteasomes from untreated mouse C4 and human HeLa cells or with proteasomes from IFN-γ–treated mouse C4 and human HeLa and human T2 cells expressing the mouse LMP2 and LMP7 proteins were markedly similar (Table and data not shown). Thus, our results do not support the idea of species-specific proteasomal cleavage properties.
Establishment of a Panel of Proteasomes with Varying Subunit Composition.
To study the contribution of individual catalytic sites to antigenic peptide generation, we took advantage of a panel of T2 transfectant cells that had been generated in our laboratory. T2 cells lack the endogenous expression of LMP2 and LMP7, whereas MECL-1 is expressed but not incorporated into the 20S proteasome due to the absence of LMP2 37. Proteasomes were isolated from parental T2 cells and from T2 cells transfected with mouse-derived cDNAs encoding LMP2, a mutated, catalytically inactive form of LMP2 (LMP2 T1A), mouse LMP7, LMP2+7, or LMP2 plus catalytically inactive LMP7 T1A. Incorporation of the introduced LMPs was verified by immunoblot analyses using specific rabbit antisera (Fig. 5). As shown in Fig. 5 A, LMP2 was detected as a 21-kD product in proteasome preparations from LMP2, LMP2+7, and LMP2+7 T1A transfectant cells and as a product with slower migration properties in proteasomes from LMP2 T1A transfectant cells. The lower electrophoretic mobility of the T1A mutant forms of LMP2 and LMP7 is due to incomplete processing of the NH2-terminal prosequence, which is an autocatalytic process 33. Likewise, LMP7 (Fig. 5 B) was detected as a 23-kD protein in purified proteasomes from LMP7 and LMP2+7 transfectant cells and as a 26-kD product in proteasomes from LMP2+7 T1A transfectant cells. The exact ratios between constitutive and introduced β subunits in the used T2 proteasome populations have been published recently 34 or have been submitted for publication (Schmidt, M., and P.-M. Kloetzel, manuscript in preparation) and are of minor importance for the interpretation of our results. In summary, all transfected subunits replaced their homologous constitutive subunits with >50% efficiency, with the exception of LMP7 T1A. The incorporation of this subunit could not be quantified due to its unknown migrational behavior in two-dimensional gels but was less efficient than that of LMP7 as judged from immunoblots (Fig. 5 and data not shown). Consistent with our earlier observations 34 39, we found that the association of both LMP2 and LMP2 T1A with the proteasome complex induced the simultaneous incorporation of MECL-1 with equal efficiency. Thus, taken together, the isolated set of proteasomes contained the immunosubunits LMP2 or LMP2 T1A, each in combination with MECL-1 and with or without LMP7 or LMP7 T1A.
Figure 5.
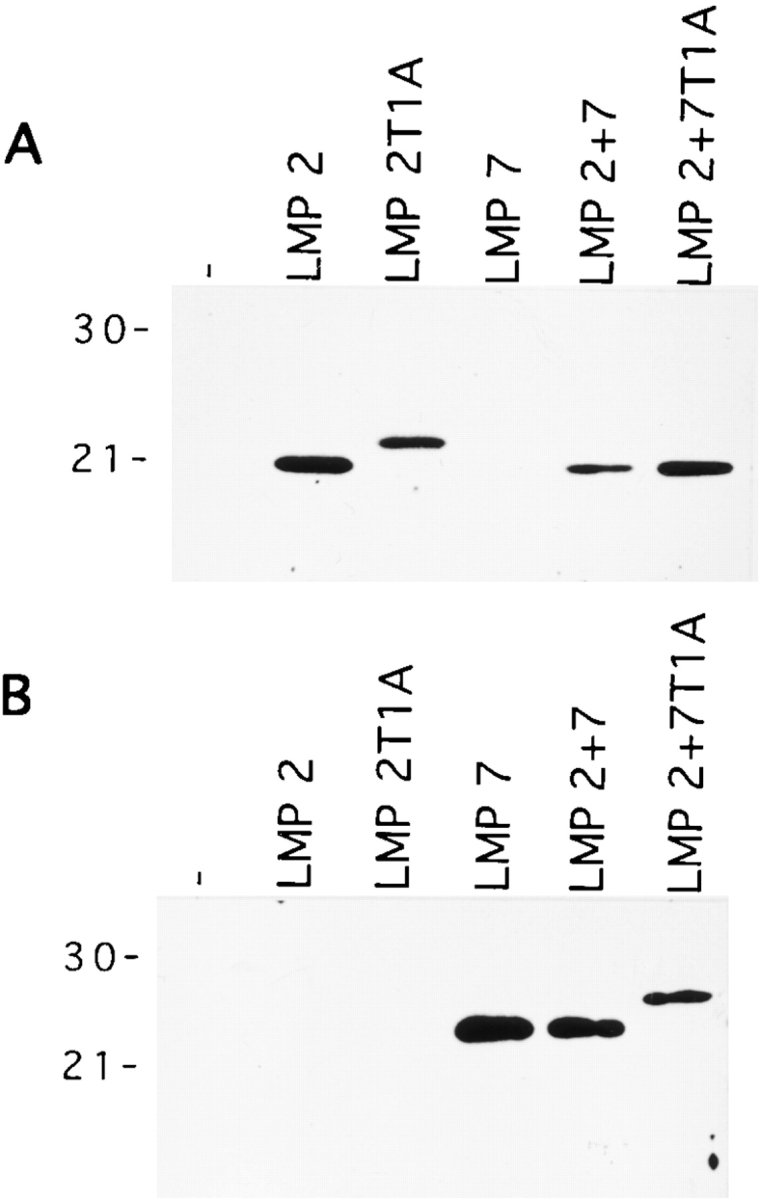
Incorporation of the LMP2 and LMP7 subunits in 20S proteasomes of T2 transfectant lines. 500 ng (A) or 150 ng (B) of purified 20S proteasomes was separated by SDS-PAGE and probed with mouse LMP2–specific (A) or mouse LMP7–specific (B) polyclonal rabbit antisera. Molecular mass markers (kD) are shown at left.
Efficient HBcAg141-151 Generation Requires the Structural Information of LMP7.
We next examined how the individual catalytic sites contribute to the efficient generation of HBcAg141-151 by immunoproteasomes. The HBV 32-mer was digested with proteasomes from the different T2 transfectant cells, and the cleavage products were analyzed (Table ). In agreement with the data shown in Table and Fig. 4, we found that in the absence of IFN-γ–inducible subunits, as in proteasomes from T2 cells, HBcAg141-151 was not generated. Remarkably, already the presence of either LMP2 in combination with MECL-1 or LMP7 alone was sufficient to induce the production of small amounts of antigenic peptide. In the case of LMP2-containing proteasomes, MECL-1 was most likely not responsible for the observed effect. No antigenic peptide was generated by digestion with proteasomes containing catalytically inactive LMP2 T1A and MECL-1. Remarkably, the combined presence of LMP2, LMP7, and MECL-1 resulted in a significantly more efficient generation of HBcAg141-151 than observed with either of the subunits present alone. These findings were highly consistent among different experiments (Table and Table , Fig. 4, Fig. 6) and reproducible upon repeated mass spectrometric analysis of the same digests or of new digests generated with the same or newly prepared proteasomes of other T2 clones obtained from the same transfections (not shown).
Table 2.
Impact of IFN-γ–inducible Proteasome Subunits on the Generation of HBcAg141-151
| Peptide fragment | T2 | T2 LMP2 | T2 LMP2 T1A | T2 LMP7 | T2 LMP2+7 | T2 LMP2+7 T1A | Mock |
|---|---|---|---|---|---|---|---|
| Ala131–Ser162 | 0 | 27 | 499 | 0 | 0 | 0 | 1,629 |
| Ala131–Leu140 | 885 | 1,299 | 287 | 1,540 | 2,330 | 2,667 | 0 |
| Ser141–Ser162 | 22 | 27 | 12 | 49 | 6 | 43 | 0 |
| Ser141–Val149 | 66 | 169 | 0 | 42 | 270 | 157 | 0 |
| Ser141–Arg150 | 0 | 0 | 0 | 67 | 259 | 239 | 0 |
| Ser141–Arg151 | 0 | 24 | 0 | 68 | 183 | 245 | 0 |
| Ser141–Arg152 | 69 | 62 | 0 | 180 | 305 | 227 | 0 |
| Thr142–Arg151 | 60 | 117 | 0 | 0 | 172 | 185 | 0 |
| Pro138–Val149 | 497 | 503 | 593 | 329 | 265 | 504 | 0 |
Figure 6.
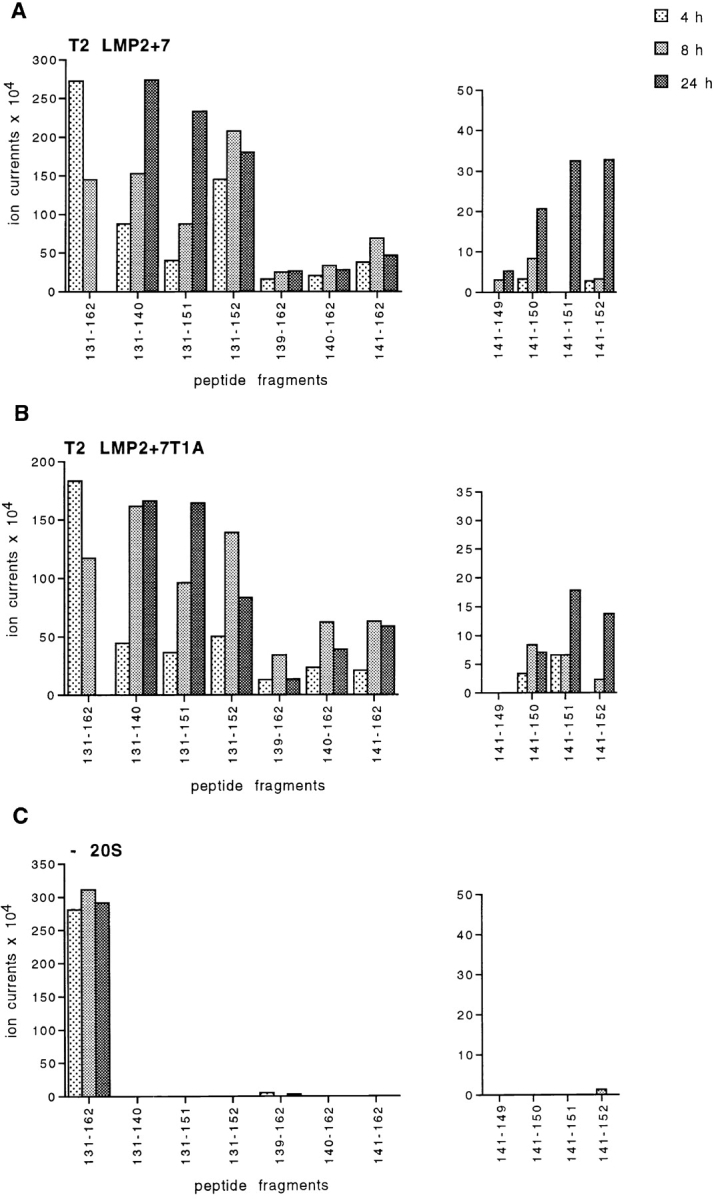
Kinetic analysis of the production of HBcAg141-151 and of related peptide fragments. HBcAg131-162 was digested for 4, 8, or 24 h with 20S proteasomes isolated from T2 LMP2+7 (A) and T2 LMP2+7 T1A transfectant cells (B) or mock-digested (C). Cleavage products were analyzed as described in Materials and Methods. Relative abundances of 11 selected peptides are plotted. Shown data were reproduced in a second, independent experiment.
Our results argue that the concerted function of immunosubunits is essential for the generation of the HBcAg141-151 peptide. Strikingly, proteasomes containing a mutated inactive LMP7 (LMP7 T1A) in addition to wild-type LMP2 and MECL-1 still generated the HBV CTL epitope with similar efficiency as in the presence of wild-type LMP7 (Table ). We therefore conclude that the physical presence of the LMP7 protein in combination with the other two immunosubunits already contributes to an efficient antigenic peptide production and that the activity of the LMP7 hydrolytic site is of minor importance.
Kinetic Analysis of Proteasome-mediated Turnover of HBV Polypeptide.
To analyze the importance of LMP7 for kinetics and relative abundance of peptide fragments generated, we performed time course experiments using proteasomes from T2 LMP2+7 and T2 LMP2+7 T1A transfectants. As shown in Fig. 6, the relative abundance of any of the peptide fragments analyzed was largely similar in the two proteasomal digests. Whereas the 32-mer substrate disappeared within the 24 h of digestion, the SCP Ala131–Leu140 accumulated with reciprocal kinetics. The amount of SCP Ser141–Ser162 increased at early time points but then stabilized or slightly diminished, indicating its further cleavage into the smaller peptides Ser141–Val149, Ser141–Arg150, the CTL epitope Ser141–Arg151, and Ser141–Arg152. Peptide fragment Ala131–Arg151, also a potential precursor of the HBV CTL epitope Ser141–Arg151, continued to accumulate over the whole period of digestion, indicating that this SCP was rather resistant to further cleavage behind Leu140 and was not a major source of antigenic peptide. Thus, whereas LMP2 and MECL-1 provide the required cleavage site specificity (e.g., ability to cleave after Arg 151), the incorporation of LMP7 in LMP2/MECL-1–containing proteasomes changes the frequencies of cleavage site usage such to support the production of antigenic peptides out of the HBV polypeptide. As the activity of the LMP7 catalytic site is not required for the observed effect, we assume that the altered cleavage properties depend on LMP7-induced structural changes in the 20S proteasome complex.
Discussion
The efficiency and specificity of proteasome-mediated cytosolic Ag degradation largely shape the pool of peptides available for binding to MHC class I molecules. Here, we show that only by incorporating the IFN-γ–inducible catalytic subunits proteasomes gain the ability to liberate an HBcAg CTL epitope from a 32-mer polypeptide.
We used a panel of T2 LMP2/7 transfectant cells to analyze the impact of individual proteasomal catalytic sites on peptide cleavage under conditions that bypass other effects of IFN-γ. Whereas the isolated incorporation of LMP2/MECL-1 and LMP7 had only minor effects on the generation of the antigenic peptide, the concerted presence of LMP2, MECL-1, and LMP7 dramatically enhanced the production of the relevant peptides. This change in the quality of peptides generated is not simply explained by an additive effect of cleavages mediated by the catalytic sites of these subunits; instead, our results suggest that the immunosubunits must cooperate in the generation of the HBV CTL epitope.
Remarkably, although the presence of the LMP7 subunit is essential, the LMP7-restricted proteolytic activity is apparently not required for effective epitope generation. Even in the absence of a catalytically active LMP7 subunit (LMP7 T1A), the generation of HBcAg141-151 is enhanced (Table ). It should be noted that the efficiency of replacement of especially the constitutive subunit MB1 for LMP7 or LMP7 T1A differed among our proteasome preparates (Fig. 5 and data not shown). However, as constitutive or LMP2+MECL-1–containing proteasomes generate none or only low quantities of HBcAg141-151, the proteasome populations containing either LMP7 or LMP7 T1A must be responsible for the efficient production of the antigenic peptide. As the LMP7 capacity to cleave is not essential for the generation of the HBV CTL epitope, we infer that structural changes imposed by the incorporation of this immunosubunit determine the altered cleavage pattern of immunoproteasomes. Efficient generation of HBcAg141-151 further requires the cleavage specificities of the LMP2 and MECL-1 active site subunits, as the presence of the constitutive subunits in these positions results in a relatively poor antigenic peptide liberation (compare T2 LMP7 and T2 LMP2[/MECL-1]+7 in Table ).
This unexpected finding that structural features besides functional specificities determine the cleavage properties of proteasomes finds support from other studies. For example, we recently reported that the replacement of subunit delta in constitutive proteasomes by either LMP2 or LMP2 T1A likewise influences the catalytic activity of the two other pairs of catalytic sites 39. The groups of Heinemeyer et al. 40 and Dick et al. 41 recently demonstrated specific activities for the individual catalytic subunits using yeast mutant proteasomes. Interestingly, whereas β1 was found responsible for cleavage after acidic residues and after branched chain amino acids and β2 for cleavage after basic residues, cleavages after hydrophobic residues appeared to be exerted by each of the catalytic subunits (β1, β2, and β5). These cleavage site preferences, initially documented for short peptide substrates, also applied for a protein substrate 42. Likewise, we were able to assign the proteasomal peptidyl glutamyl hydrolyzing activity (cleavage after acidic residues) to δ (β1) and trypsin-like activity (cleavage after basic residues) to MC14 (β2) in mammalian proteasomes 33 43. However, our preliminary data also show that the generation, for example, of HBcAg fragments containing arginine at the COOH terminus occurs less efficiently but is not abrogated in the absence of the MC14/MECL-1 catalytic site after the incorporation of MECL-1 T1A. Therefore, we infer that the individual catalytic sites of mammalian proteasomes have overlapping specificities and that so far unknown structural features appear to tune their activity to an important extent.
Remarkably, the replacement of MB1 by the transfection of LMP7 T1A into T2 LMP2 cells resulted in a growth disadvantage (Schmidt, M., and P.-M. Kloetzel, manuscript in preparation). Thus, although LMP7 T1A– and LMP7-containing proteasomes similarly cleave the HBV polypeptide studied here, the inactivity of the catalytic site or the presence of an only partially cleaved prosequence interferes with the function of proteasomes in intact cells.
Cellular substrates are specifically targeted for proteolysis by 26S proteasomes, for example by linkage to ubiquitin. Captured substrates are unfolded by the 19S component and subsequently degraded by the 20S proteasome. The process of proteolysis may be modulated by PA28 bound to the opposite site of 20S 44. To avoid the complexity of 26S proteasome isolations and in vitro substrate targeting, we and other groups classically use purified 20S proteasomes and polypeptides as model substrates to study proteolytic peptide generation. This experimental approach is believed to accurately document cleavage specificities of cellular proteasomes, as demonstrated by the comparison of peptide products generated in vitro and in intact cells 18 19 20 21. Moreover, in vitro degradation of denatured OVA protein 17 and of OVA polypeptides 19 by 20S proteasomes as well as the degradation of antizyme-targeted ornithine decarboxylase–OVA hybrid protein by purified 26S proteasomes 45 resulted in the generation of the immunodominant OVA257-264 peptide, indicating that the steps upstream of 20S-mediated proteolysis and substrate length are unlikely to influence specificity and frequency of cleavage site usage.
Proteasomes of mouse and of human origin cleave the HBV model substrate identically (Table ), which, considering >90% homology between the subunits from the two species, is not too surprising. These results are in line with a recent study by Niedermann et al. 46 demonstrating that the processing of polypeptides by proteasomes is conserved in evolution. In contrast, our data seem inconsistent with the recent work by Braud et al. 47, who showed that an MHC class I HLA-A3–presented nonamer peptide derived from the influenza nucleoprotein (NP) was generated in human but not in mouse cells. However, the proteolytic processes involved in the generation of the NP peptide were not studied in depth, which will be needed to determine whether and how proteasome subunits contribute to the processing defect in mouse cells. Still, although fundamental differences in cleavage specificities between proteasomes of human and mouse origin seem unlikely, we can not exclude the possibility that certain differences do exist and are detected only when suitable specific substrates are analyzed.
How does the synthesis of three additional catalytic proteasome subunits contribute to the formation of MHC class I ligands in IFN-γ–induced cells? As discussed by Nandi et al. 30, the expression of six different catalytic subunits creates the theoretical possibility for 36 different proteasome subpopulations. As these subpopulations are likely to exhibit distinct cleavage properties, their presence could lead to a diversification of cleavage site usage in Ag degradation and thus broaden the repertoire of antigenic peptides. This model, however, is opposed by recent findings that LMP2 and MECL-1 and also, to a lesser extent, LMP7 incorporate interdependently into proteasomes 34 37 48. Thus, less variety and the formation of homogenous immunoproteasomes is favored, suggesting that the immunosubunits fulfill a specific function. Based on our data, we propose that both cleavage specificities and structural changes imposed by immunosubunit incorporation lead to an altered cleavage site usage by immunoproteasomes, broadening the spectrum of peptides that is generated and available for association with MHC class I molecules. Such a model is supported by the findings of Cardozo and Kohaski 49, who showed that immunosubunit incorporation was associated with an acceleration of proteasomal cleavages after branched chain residues (Leu, Ile, and Val) in lysozyme substrate, leading to an enhanced generation of potential MHC class I ligands. Recent results in our laboratory further demonstrate that purified immunoproteasomes degrade the pp89 polypeptide faster than constitutive proteasomes, with more dual cleavage products generated at early time points of digestion (Kuckelkorn, U., T. Ruppert, and P.-M. Kloetzel, unpublished observations), like observed with the HBV 32-mer (Fig. 4). Thus, the immunosubunits and the proteasome activator PA28 could mediate similar effects, i.e., modification of the cleavage mechanism of the 20S proteasomes to enhance the formation of peptides via coordinated double cleavage events (reference 11 and our unpublished observations). Likely, both of these mechanisms act together to enhance the antigenic peptide production in IFN-γ–stimulated cells.
LMP2, LMP7, and MECL-1 are constitutively highly expressed in many cells of lymphoid origin, probably including APCs. Other cells do not express these subunits unless stimulated by IFN-γ. Thus, the set of antigenic peptides generated by professional APCs to prime CTLs may differ from the set of peptides produced by and presented on cells of nonlymphoid origin. This may have important biological consequences. The work of Restifo et al. 50 and Seliger et al. 51 indicated that the expression of the IFN-γ–inducible proteasome subunits influenced the antigenicity of tumor cells. They found that the downregulation of LMP2 and LMP7 in tumor cells was associated with enhanced immune escape. Differences in induced and constitutive proteasome composition may affect the CTL-mediated immune defence against intracellular pathogens. For example, while HBcAg141-151 is processed by immunoproteasomes only, hepatocytes mostly contain constitutive proteasomes (reference 52 and Seifert, U., A.J.A.M. Sijts, and P.-M. Kloetzel, unpublished observations). Thus, HBcAg141-151 can be produced in HBV-infected hepatocytes during the acute phase of infection only in the presence of lymphokines that induce the synthesis of LMP2, LMP7, and MECL-1. The peptide may not be generated during chronic stages of infection, when lymphokine levels have declined. Thus, the failure of hepatocytes to generate certain viral peptides could potentially contribute to the success of HBV to persist in chronically infected patients. Indeed, an HBV-specific CTL response is weak and only rarely detectable in the blood of patients with chronic HBV infection, whereas it is strong and multispecific in patients with acute hepatitis B 35. Another example is provided by the finding that in absence of immunocompetence, the lack of IFN-γ precludes the efficient presentation of antigens and CTL control in vivo 53. Further studies addressing the tissue distribution of distinct proteasome populations and their cleavage properties under in vivo conditions will contribute to our understanding of CTL-mediated immunity and can perhaps shed light on the failure of immune surveillance during chronic viral disease. These studies may therefore provide a basis for the rational design of immunotherapeutic strategies.
Acknowledgments
We thank Dr. U. Kuckelkorn for sharing her proteasome preparate of T2 LMP2+7 transfectant cells, Mrs. S. Standera and D. Zantopf for wonderful technical assistance, and Dr. P. Henklein for peptide synthesis.
This work was supported by Deutsche Forschungsgemeinschaft grants SFB 469 to T. Ruppert and U. Koszinowski and SFB421 to P.-M. Kloetzel and by European Community contract B104-CT97-0505 to P.-M. Kloetzel and U. Koszinowski.
Footnotes
Abbreviations used in this paper: DCPs, double cleavage products; HBcAg, hepatitis B virus core antigen; HBV, hepatitis B virus; LMP, low molecular weight protein; MECL, multicatalytic endopeptidase complex–like; r, recombinant; SCPs, single cleavage products.
References
- Pamer E., Cresswell P. Mechanisms of MHC class-I restricted antigen processing. Annu. Rev. Immunol. 1998;16:323–358 . doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Sijts A.J., Villanueva M.S., Pamer E.G. CTL epitope generation is tightly linked to cellular proteolysis of a Listeria monocytogenes antigen. J. Immunol. 1996;156:1497–1503. [PubMed] [Google Scholar]
- Cerundolo V., Benham A., Braud V., Mukherjee S., Gould K., Macino B., Neefjes J., Townsend A. The proteasome-specific inhibitor lactacystin blocks presentation of cytotoxic T lymphocyte epitopes in human and murine cells. Eur. J. Immunol. 1997;27:336–341. doi: 10.1002/eji.1830270148. [DOI] [PubMed] [Google Scholar]
- Craiu A., Gaczynska M., Akopian T., Gramm C.F., Fenteany G., Goldberg A.L., Rock K.L. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- Vinitsky A., Anton L.C., Snyder H.L., Orlowski M., Bennink J.R., Yewdell J.W. The generation of MHC class I-associated peptides is only partially inhibited by proteasome inhibitors. Involvement of nonproteasomal cytosolic proteases in antigen processing? J. Immunol. 1997;159:554–564. [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Soza A., Kuckelkorn U., Kloetzel P.-M. Peptide antigen production by the proteasomecomplexity provides efficiency. Immunol. Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Tanahashi N., Tsurumi C., Yokota K.-Y., Shimbara N. Proteasomes and antigen processing. Adv. Immunol. 1997;64:1–38. doi: 10.1016/s0065-2776(08)60885-8. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Soza A., Eggers M., Kuehn L., Dick T.P., Schild H., Rammensee H.-G., Koszinowski U.H., Kloetzel P.-M. A role for the proteasome regulator PA28α in antigen presentation. Nature. 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- Dick T.P., Ruppert T., Groettrup M., Kloetzel P.M., Kuehn L., Koszinowski U.H., Stevanovic S., Schild H., Rammensee H.G. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- Gaczynska M., Rock K.L., Goldberg A.L. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- Boes B., Hengel H., Ruppert T., Multhaup G., Koszinowski U.H., Kloetzel P.M. Interferon γ stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J. Exp. Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M., Ruppert T., Kuehn L., Seeger M., Standera S., Koszinowski U., Kloetzel P.M. The interferon-γ-inducible 11S regulator (PA28) and the LMP2/LMP7 subunits govern the peptide production by the 20S proteasome in vitro. J. Biol. Chem. 1995;270:23808–23815. doi: 10.1074/jbc.270.40.23808. [DOI] [PubMed] [Google Scholar]
- Ustrell V., Pratt G., Rechsteiner M. Effects of interferon γ and major histocompatibility complex-encoded subunits on peptidase activities of human multicatalytic proteases. Proc. Natl. Acad. Sci. USA. 1995;92:584–588. doi: 10.1073/pnas.92.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleuteri A.M., Kohanski R.A., Cardozo C., Orlowski M. Bovine spleen multicatalytic proteinase complex (proteasome). Replacement of X, Y, and Z subunits by LMP7, LMP2, and MECL1 and changes in properties and specificities. J. Biol. Chem. 1997;272:11824–11831. doi: 10.1074/jbc.272.18.11824. [DOI] [PubMed] [Google Scholar]
- Dick L.R., Aldrich C., Jameson S.C., Moomaw C.R., Pramanik B.C., Doyle C.K., DeMartino G., Bevan M.J., Forman J.M., Slaughter C.A. Proteolytic processing of ovalbumin and β-galactosidase by the proteasome to yield antigenic peptides. J. Immunol. 1994;152:3884–3894. [PMC free article] [PubMed] [Google Scholar]
- Eggers M., Boes-Fabian B., Ruppert T., Kloetzel P.-M., Koszinowski U.H. The cleavage preference of the proteasome governs the yield of antigenic peptides. J. Exp. Med. 1995;182:1865–1870. doi: 10.1084/jem.182.6.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermann G., Butz S., Ihlenfeldt H.G., Grimm R., Lucchiari M., Hoschützky H., Jung G., Maier B., Eichmann K. Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity. 1995;2:289–299. doi: 10.1016/1074-7613(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Ossendorp F., Eggers M., Neisig A., Ruppert T., Groettrup M., Sijts A., Mengedé E., Kloetzel P.M., Neefjes J., Koszinowski U. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- Theobald M., Ruppert T., Kuckelkorn U., Hernandez J., Haussler A., Ferreira E.A., Liewer U., Biggs J., Levine A.J., Huber C. The sequence alteration associated with a mutational hotspot in p53 protects cells from lysis by cytotoxic T lymphocytes specific for a flanking peptide epitope. J. Exp. Med. 1998;188:1017–1028. doi: 10.1084/jem.188.6.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craiu A., Akopian T., Goldberg A., Rock K.L. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl. Acad. Sci. USA. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckelkorn U., Frentzel S., Kraft R., Kostka S., Groettrup M., Kloetzel P.-M. Incorporation of major histocompatibility complex-encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-γ. Eur. J. Immunol. 1995;25:2605–2611. doi: 10.1002/eji.1830250930. [DOI] [PubMed] [Google Scholar]
- Arnold D., Driscoll J., Androlewicz M., Hughes E., Cresswell P., Spies T. Proteasome subunits encoded in the MHC are not generally required for the processing of peptides bound by MHC class I molecules. Nature. 1993;360:171–174. doi: 10.1038/360171a0. [DOI] [PubMed] [Google Scholar]
- Momburg F., Ortiz-Navarrete V., Neefjes J., Goulmy E., van-de-Wal Y., Spits H., Powis S.J., Butcher G.W., Howard J.C., Walden P. Proteasome subunits encoded by the major histocompatibility complex are not essential for antigen presentation. Nature. 1992;360:174–177. doi: 10.1038/360174a0. [DOI] [PubMed] [Google Scholar]
- Yewdell J., Lapham C., Bacik I., Spies T., Bennink J. MHC-encoded proteasome subunits LMP2 and LMP7 are not required for efficient antigen presentation. J. Immunol. 1994;152:1163–1170. [PubMed] [Google Scholar]
- Cerundolo V., Kelly A., Elliot T., Trowsdale J., Townsend A. Genes encoded in the major histocompatibility complex affecting the generation of peptides for TAP transport. Eur. J. Immunol. 1995;25:554–562. doi: 10.1002/eji.1830250238. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Swat W., Laplace C., Kuehn R., Rajewsky K., Mueller U., von Boehmer H. MHC class I expression in mice lacking proteasome subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- Van Kaer L., Ashton-Rickardt P.G., Eichelberger M., Gaczynska M., Nagashima K., Rock K.L., Goldberg A.L., Doherty P.C., Tonegawa S. Altered peptidase and viral-specific T cell response in LMP 2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- Nandi D., Jiang H., Monaco J.J. Identification of MECL-1 (LMP-10) as the third IFN-gamma-inducible proteasome subunit. J. Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- Missale G., Redeker A., Person J., Fowler P., Guilhot S., Schlicht H.J., Ferrari C., Chisari F.V. HLA-A31– and HLA-Aw68–restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J. Exp. Med. 1993;177:751–761. doi: 10.1084/jem.177.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter R.D., Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO (Eur. Mol. Biol. Organ.) J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke G., Kraft R., Kostka S., Henklein P., Frömmel C., Löwe J., Huber R., Kloetzel P.-M., Schmidt M. Analysis of mammalian 20S proteasome biogenesisthe maturation of β-subunits is an ordered two-step mechanism involving autocatalysis. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:6887–6898. [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Zantopf D., Kraft R., Kostka S., Preissner R., Kloetzel P.-M. Sequence information within proteasomal prosequences mediates efficient integration of β-subunits into the 20S proteasome complex. J. Mol. Biol. 1999;288:117–128. doi: 10.1006/jmbi.1999.2660. [DOI] [PubMed] [Google Scholar]
- Rehermann B., Fowler P., Sidney J., Person J., Redeker A., Brown M., Moss B., Sette A., Chisari F.V. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J. Exp. Med. 1995;181:1047–1058. doi: 10.1084/jem.181.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Standera S., Stohwasser R., Kloetzel P.M. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc. Natl. Acad. Sci. USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh K., Gossen M., Wang K., Bujard H., Peterson P.A., Yang Y. Displacement of housekeeping proteasome subunits by MHC-encoded LMPsa newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:3236–3244. doi: 10.1002/j.1460-2075.1994.tb06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke G., Eggers M., Ruppert T., Groettrup M., Koszinowski U.H., Kloetzel P.-M. Inactivation of a defined active site in the mouse 20S proteasome complex enhances major histocompatibility complex class I antigen presentation of a murine cytomegalovirus protein. J. Exp. Med. 1998;187:1641–1646. doi: 10.1084/jem.187.10.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D.H. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Dick T.P., Nussbaum A.K., Deeg M., Heinemeyer W., Groll M., Schirle M., Keilholz W., Stevanovic S., Wolf D.H., Huber R. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J. Biol. Chem. 1998;273:25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- Nussbaum A.K., Dick T.P., Keilholz W., Schirle M., Stevanovic S., Dietz K., Heinemeyer W., Groll M., Wolf D.H., Huber R. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc. Natl. Acad. Sci. USA. 1998;95:12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann U., Kral S., Braun B., Standera S., Schmidt M., Kloetzel P.-M., Sijts A. Mutational analysis of subunit iβ2 (MECL-1) demonstrates conservation of cleavage specificity between yeast and mammalian proteasomes. FEBS Lett. 1999;454:11–15. doi: 10.1016/s0014-5793(99)00768-1. [DOI] [PubMed] [Google Scholar]
- Hendil K.B., Khan S., Tanaka K. Simultaneous binding of PA28 and PA700 activators to 20 S proteasomes. Biochem. J. 1998;15:749–754. doi: 10.1042/bj3320749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar S., Komlosh A., Nadav E., Shaked I., Ziv T., Admon A., DeMartino G.N., Reiss Y. 26S proteasome-mediated production of an authentic major histocompatibility class I-restricted epitope from an intact protein substrate. J. Biol. Chem. 1999;274:21963–21972. doi: 10.1074/jbc.274.31.21963. [DOI] [PubMed] [Google Scholar]
- Niedermann G., Grimm R., Geier E., Maurer M., Realini C., Gartmann C., Soll J., Omura S., Rechsteiner M.C., Baumeister W. Potential immunocompetence of proteolytic fragments produced by proteasomes before evolution of the vertebrate immune system. J. Exp. Med. 1997;186:209–220. doi: 10.1084/jem.186.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud V.M., McMichael A.J., Cerundolo V. Differential processing of influenza nucleoprotein in human and mouse cells. Eur. J. Immunol. 1998;28:625–635. doi: 10.1002/(SICI)1521-4141(199802)28:02<625::AID-IMMU625>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Griffin T.A., Nandi D., Cruz M., Fehling H.J., Kaer L.V., Monaco J.J., Colbert R.A. Immunoproteasome assemblycooperative incorporation of interferon γ (IFN-γ)-inducible subunits. J. Exp. Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo C., Kohanski R.A. Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the “immunoproteasome.”. J. Biol. Chem. 1998;273:16764–16770. doi: 10.1074/jbc.273.27.16764. [DOI] [PubMed] [Google Scholar]
- Restifo N.P., Esquivel F., Kawakami Y., Yewdell J.W., Mule J.J., Rosenberg S.A., Bennink J.R. Identification of human cancers deficient in antigen processing. J. Exp. Med. 1993;177:265–272. doi: 10.1084/jem.177.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger B., Höhne A., Knuth A., Bernard H., Meyer T., Tampe R., Momburg F., Huber C. Analysis of the major histocompatibility complex class I antigen presentation machinery in normal and malignant renal cellsevidence for deficiencies associated with transformation and progression. Cancer Res. 1995;56:1756–1760. [PubMed] [Google Scholar]
- Brown M.G., Driscoll J., Monaco J.J. MHC-linked low-molecular mass polypeptide subunits define distinct subsets of proteasomesimplications for divergent function among distinct proteasome subsets. J. Immunol. 1993;151:1193–1204. [PubMed] [Google Scholar]
- Geginat G., Ruppert T., Hengel H., Holtappels R., Koszinowski U.H. IFNγ is a prerequisite for optimal antigen processing of viral peptides in vivo. J. Immunol. 1997;158:3303–3310. [PubMed] [Google Scholar]
- Groettrup M., Kraft R., Kostka S., Standera S., Stohwasser R., Kloetzel P.-M. A third interferon-γ-induced subunit exchange in the 20S proteasome. Eur. J. Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]