Abstract
After the productive rearrangement of immunoglobulin (Ig) heavy chain genes, precursor (pre-)B lymphocytes undergo a limited number of cell divisions in response to interleukin (IL)-7. Here, we present evidence that this phase of IL-7–dependent expansion is constrained by an inhibitory signal initiated by antigen receptor assembly. A line of pre-B cells from normal murine bone marrow that expresses a μ heavy chain with a D-proximal VH7183.2 region divides continuously in IL-7. IL-7 responsiveness ceases upon differentiation to the μ1, κ1 stage, despite continuing expression of the IL-7 receptor (IL-7R), suggesting that antigen receptor assembly inhibits IL-7 responsiveness. This is confirmed by introduction of a rearranged λ light chain gene, which inhibits proliferative signaling through the IL-7R. Inhibition is specific to the IL-7R, because it is overcome by replacement of the IL-7R cytoplasmic domain with corresponding sequences from the closely related IL-2Rβ chain. Alteration of a single tyrosine residue, Tyr410, in the IL-7R cytoplasmic domain to phenylalanine also prevents the inhibition of proliferation after antigen receptor assembly. Thus, the loss of IL-7 responsiveness after antigen receptor assembly may be mediated through the recruitment of an inhibitory molecule to this residue. Our findings identify a novel mechanism that limits cytokine-dependent proliferation during B lymphopoiesis. This mechanism may be essential for the proper regulation of peripheral B lymphocyte numbers.
Keywords: interleukin 7 receptor, B lymphocyte differentiation, signal transduction, antigen receptor, immunoglobulin
Introduction
A potent stimulus for the proliferation of immature B lymphocytes is provided by IL-7 1, a cytokine secreted by stromal cells in the bone marrow. IL-7 triggers cell division by engagement of a heterodimeric receptor (IL-7R) that contains a ligand-specific α chain 2, and a common γ chain shared with the receptors for IL-2, IL-4, IL-9, IL-13, and IL-15 3 4 5 6 7. We have shown elsewhere that proliferative signaling through the IL-7R is critically dependent on the integrity of a single tyrosine residue in the cytoplasmic domain of the α chain 8. This tyrosine residue is essential for the ligand-induced recruitment of the effector enzyme phosphatidylinositol 3-kinase (PI 3-k), through the Src homology 2 (SH2) domains in its regulatory subunit, p85 9.
In mice, the ability of immature B cells to divide in response to IL-7 changes with their developmental progression. Progenitor (pro-)B cells, which have yet to complete Ig heavy chain (IgH) gene rearrangement, require stromal cell contact as well as IL-7 to divide 10 11. Productive IgH rearrangement triggers progression to the precursor (pre-)B cell stage, at which IL-7 alone is sufficient to provoke proliferation 1 12 13. Pre-B cells undergo only a limited number of cell divisions before they differentiate into IL-7–unresponsive, mature B lymphocytes, which express a functional antigen receptor 14 15. It remains unclear how IL-7–dependent proliferation can be limited during these developmental transitions to regulate the number of mature cells emerging to the periphery. Here, we present evidence that the IL-7–dependent proliferation of pre-B cells is constrained by an inhibitory signal initiated through antigen receptor assembly.
Materials and Methods
Cells.
IL-7–dependent cells derived from normal bone marrow 16 were maintained in growth medium with 10 ng/ml of recombinant IL-7 (Genzyme Diagnostics). The retroviral packaging cell lines CRE 17, AM12 GP + env 18, and Φnx 19 were used, and were maintained in DMEM with 10% FCS.
Immunofluorescence Staining and Flow Cytometric Analysis.
Cells were washed in ice-cold PBS. Staining for CD19 and CD43 was with FITC-conjugated rat anti–mouse CD19 and with a biotinylated rat anti–mouse CD43, followed by streptavidin-R-PE. Staining for CD2 was with PE-conjugated rat anti–mouse CD2, and for CD25, with an FITC-conjugated rat anti–mouse CD25. All of these reagents were from PharMingen. Staining for μ was with FITC-conjugated goat anti–mouse μ (μ chain specific; Southern Biotechnology Associates), whereas staining for κ light chain was with PE-conjugated goat anti–mouse κ (κ chain specific; Southern Biotechnology Associates). Staining for λ1 light chain was with FITC-conjugated goat anti–mouse λ (λ chain specific; Southern Biotechnology Associates). Receptors for human IL-4R were detected with biotinylated human IL-4 (R&D Systems), followed by streptavidin-PE (PharMingen). Staining for the murine IL-7R was with rat anti–mouse IL-7R (A7R34; gift of S. Nishikawa, Kyoto University, Kyoto, Japan), followed by a biotinylated anti–rat antibody (PharMingen) and streptavidin-FITC (Jackson ImmunoResearch Labs). Negative control staining was with the matched isotype controls (PharMingen). All samples were analyzed on a FACScalibur™ flow cytometer (Becton Dickinson) by standard methods using CELLQuest™ analysis software.
Sequencing of the Productive IgH Rearrangement.
A PCR-based assay for VH rearrangements was first carried out. High-molecular-weight genomic DNA was extracted by the Proteinase K method, and subjected to two rounds of PCR. Nested 3′ primers from the intron downstream of the JH4 and nested 5′ primers corresponding to VH7183 and VHJ558 V region families have already been described 20. Southern blotting of PCR products using a VH7183 family–specific probe 20 confirmed that the rearrangement utilized a member of this family. The PCR product was then gel purified and sequenced using a nested primer corresponding to the 5′ end of VH7183 20. Sequence comparisons were carried out using the BLAST search algorithms (available at http://www.ncbi.nlm.nih.gov/BLAST).
Pre-B Cell Cloning Assay.
The pre-B cell line was cloned by limiting dilution in the presence of 10 ng/ml recombinant IL-7 (Genzyme Diagnostics). The pre-B cells were plated at 0.3 cells per well in 96-well plates and then cultured in recombinant IL-7 containing medium for 2 wk. The colonies were then removed, and 100 were analyzed for the expression of κ light chain, as described above.
Retroviral Constructs and Their Transfection into the Ecotropic Packaging Cell Line.
The human (hu)IL-4R/IL-7R and the huIL-4R/IL-2Rβ are the same constructs that were used in Corcoran et al. 8. The huIL-4R/IL-7R Tyr410Phe and the huIL-4R/IL-7R Tyr456Phe were generated with a PCR-based strategy using oligonucleotides encoding the desired point mutations. The Tyr456Phe mutation is at the extreme 3′ end of the coding sequence, and so the huIL-4R/IL-7RY456 mutant was constructed simply by PCR amplification using the wild-type huIL-4R/IL-7R receptor 8 as a template (forward, 5′-AGCTGAACGCGTCCATGGGGTGGCTTTGCTCTGGGCTC-3′; reverse, 5′-AACTCTGAGTCTCACTGGTTTTGGAAGCTGGAC-3′). For the Tyr410Phe mutation, the oligonucleotide encoding the mutation (5′-CTAGACTGCAGGGAGAGTGGCAAGAATGGGCCTCAT-3′) and a reverse primer from the 3′ end of the coding sequence (5′-AGCTTGTCTTCACTGGTTTTGGTAGAA-3′) were used in PCR to generate a cytoplasmic domain fragment encoding the desired alteration. After digestion with PstI and BglII, the fragment was cloned into the corresponding sites in the cytoplasmic domain of the huIL-4R/IL-7R wild-type receptor. The huIL-4R/IL-7R Tyr410Phe and the huIL-4R/IL-7R Tyr456Phe were then digested with MluI and BglII and ligated into the MluI and BamHI sites of a vector based on the Moloney leukemia provirus 8 rendered replication defective by deletion of the pol and env genes.
The bicistronic expression vectors containing the rearranged λ1 light chain gene cDNA and the huIL-4R/IL-7R, huIL-4R/IL-2Rβ, and huIL-4R/IL-7R Tyr410Phe chimeric receptors were also constructed using a PCR-based approach. The λ1 light chain cDNA was used as a template for two oligonucleotides (forward, 5′-TCTACAACGCGTCCATGGCCTGGATTTCACTTATA-3′; reverse, 5′-TCTAGAGGATCCGCGGCCGCTAGGAACAGTCAGCAC-3′), which contained MluI and BamHI sites, respectively. The λ1 light chain fragment was linked through an internal ribosome entry site (IRES) from the EMC virus to a cDNA encoding the huIL-4R/IL-7R, huIL-4R/IL-2Rβ, or huIL-4R/IL-7R Tyr410Phe chimeric receptors. A three-way ligation was performed using: λ1 fragment digested MluI, BamHI; IRES fragment digested BamHI, NcoI; and the chimeric receptor cDNAs digested NcoI, NotI. These chimeric receptor sequences were generated with the correct template using oligonucleotides containing NcoI and NotI sites at the 5′ and 3′ ends of the fragment, respectively. The three-way digestion products were then inserted into the MluI and NotI sites of the retroviral vector. The integrity of all constructs was confirmed by nucleotide sequencing.
Constructs were cotransfected by the calcium phosphate method into an ecotropic packaging cell line 17 with a plasmid encoding NeoR. Stable transfectants were isolated by selection in 1 mg/ml of G418 sulfate (GIBCO BRL).
Retrovirus-mediated Gene Transfer in Cultures of the Pre-B Cell Line.
Pre-B cells were incubated at 106 cells/ml, on a 20% confluent monolayer of 2,000-rad γ-irradiated retrovirus-secreting packaging cells, in RPMI 1640 medium containing 10% FCS, 5 × 10−5 β-mercaptoethanol (growth medium), and 10 ng/ml recombinant IL-7 (Genzyme Diagnostics) at 37°C in a 5% CO2 atmosphere. After 2 d, cultures were washed and transferred to growth medium containing 10 ng/ml IL-4 (Genzyme Diagnostics) and 5 × 10−5 β-mercaptoethanol (growth medium) and then fed every 3–4 d by removal of spent medium and replacement with new.
Proliferation Assays.
Cells (5 × 105) were washed and resuspended in 3 ml of growth medium. The cells were then incubated for 4 d at 37°C in 96-well microtiter plates in 100 μl of growth medium containing the indicated quantities of recombinant human IL-4 or recombinant mouse IL-7 (Genzyme Diagnostics). Proliferation was quantified using the MTT assay 21.
Results and Discussion
A Line of Pre-B I Cells from Normal Murine Bone Marrow Divides Continuously in IL-7.
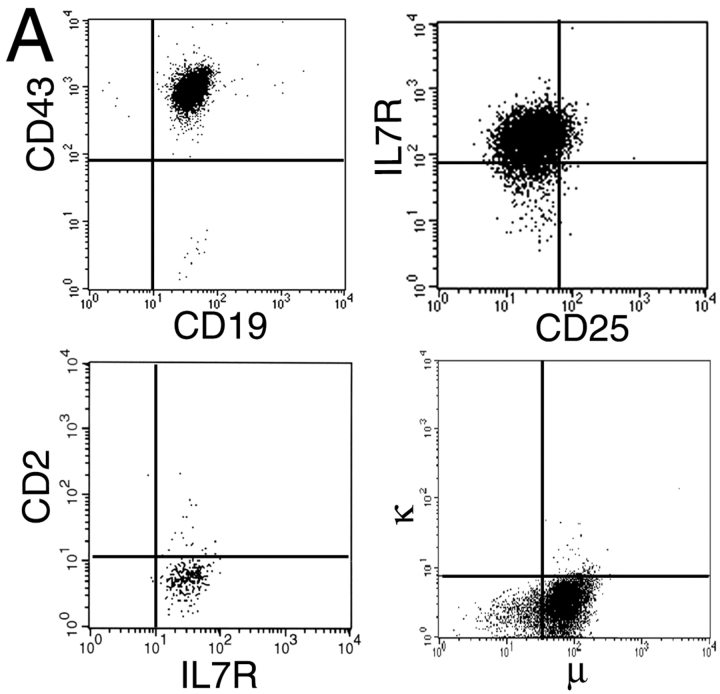
We used a line of pre-B cells isolated from normal murine bone marrow 16 to study the mechanisms that limit IL-7 responsiveness during B lymphopoiesis. These cells express markers characteristic of the pre-B I stage in development (Fig. 1 A). Thus, in addition to the CD19 marker typical of the B lineage 22, the cells display the marker CD43, whose expression is lost upon differentiation to the pre-B II stage 10. They also express BP-1 16, whose expression commences at Hardy stage C 10. Finally, the cells fail to display CD25, a marker that commences its expression at the pre-B II stage of B cell development 23 24.
Figure 1.
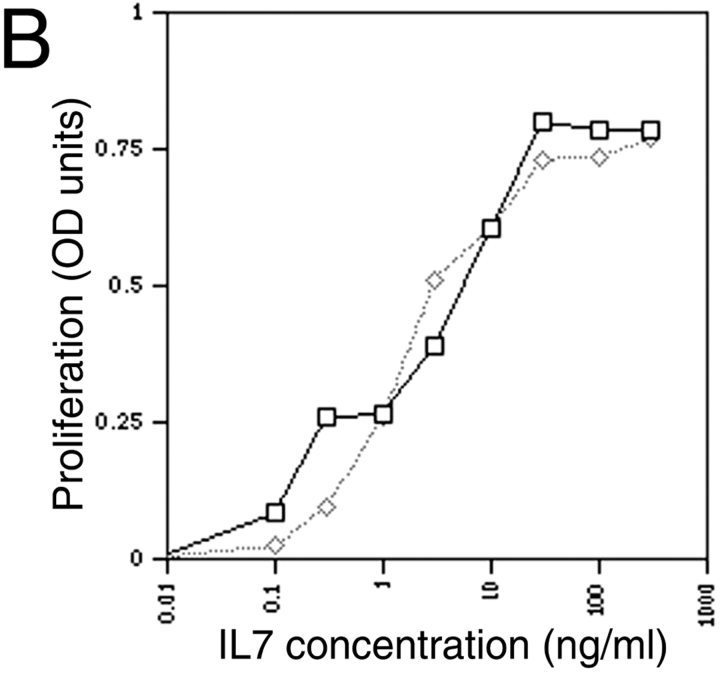
(A) Shows that markers characteristic of the pre-B I stage in differentiation are expressed by the pre-B cell line used in these studies. The panels show the results of flow cytometric analyses of 10,000 cells after staining with antibodies against the indicated markers. Forward and side light scatter profiles were used to exclude dead cells. (B) Demonstrates the dose-dependent proliferation of the pre-B cells in response to IL-7. Proliferation quantified by the MTT assay is plotted on the y-axis against cytokine dose on the x-axis. The solid and dotted lines compare the proliferation of a pre-B cell culture tested at an interval of >1 yr, showing that IL-7–dependent proliferation is not lost after prolonged passage in culture. Results are typical of at least three independent experiments.
This pre-B I cell line divides continuously in the presence of IL-7 (Fig. 1 B). We have found no alteration in IL-7 responsiveness after multiple rounds of passage over a period of 3 yr (data not shown).
In these cells, a productive IgH gene rearrangement results in the synthesis of membrane-inserted μ heavy chains (Fig. 1 A). We have sequenced this productive rearrangement (Fig. 2 A). It involves a VH gene segment from the VH7183 family located in the D-proximal region of the murine IgH locus. Comparison of the sequence with known members of the family reveals a precise match with VH7183.2 (Fig. 2 B; reference 25). In contrast, the IgL genes remain in the germline configuration 16.
Figure 2.

(A) The sequence of the productive IgH rearrangement from the pre-B cell line. (B) The V region sequence, shaded grey in A, is aligned with the sequence of VH7183.2. The D segment (shaded black) and the J region (unshaded) are also marked in A.
Differentiation to the μ 1, κ1 Stage Results in the Loss of IL-7 Responsiveness.
A small fraction of cells in this pre-B I cell line spontaneously undergoes differentiation to the μ1, κ1 stage during propagation in culture (Fig. 1 A). Although ≥95% of the cells lack κ expression, between 0.5 and 5% are consistently κ1 at any given time. Interestingly, the fraction of κ1 cells fails to increase even after prolonged periods in culture (data not shown), indicating that they do not grow out in an IL-7–dependent manner. This suggested to us that differentiation to the μ1, κ1 stage might be accompanied by the loss of IL-7 responsiveness.
To confirm that this was indeed the case, the pre-B I cells were cloned at limiting dilution in the presence of IL-7, and the resulting clones were analyzed for expression of a rearranged κ chain gene. As shown in Table , this procedure gave rise exclusively to cell clones that were ≥95% κ2, just as are the parental cells. Thus, cells that have differentiated to the μ1, κ1 stage cannot be cloned out in IL-7, indicating that IL-7 responsiveness is indeed lost.
Table 1.
κ+ Cells Responsive to IL-7 Cannot Be Cloned Out
| Cell dilution | No. of clones analyzed | No. of κ− clones (≤5% κ+) | No. of κ+ clones |
|---|---|---|---|
| 0.3 cells/well | 100 | 100 | 0 |
Pre-B cells were cloned at limiting dilution (0.3 cells/well) in 96-well plates using culture medium containing 10 ng/ml of recombinant IL-7. Cell surface expression of μ and κ in individual clones was determined by flow cytometry as described above.
Loss of IL-7 Responsiveness Occurs Despite the Continuing Expression of IL-7R.
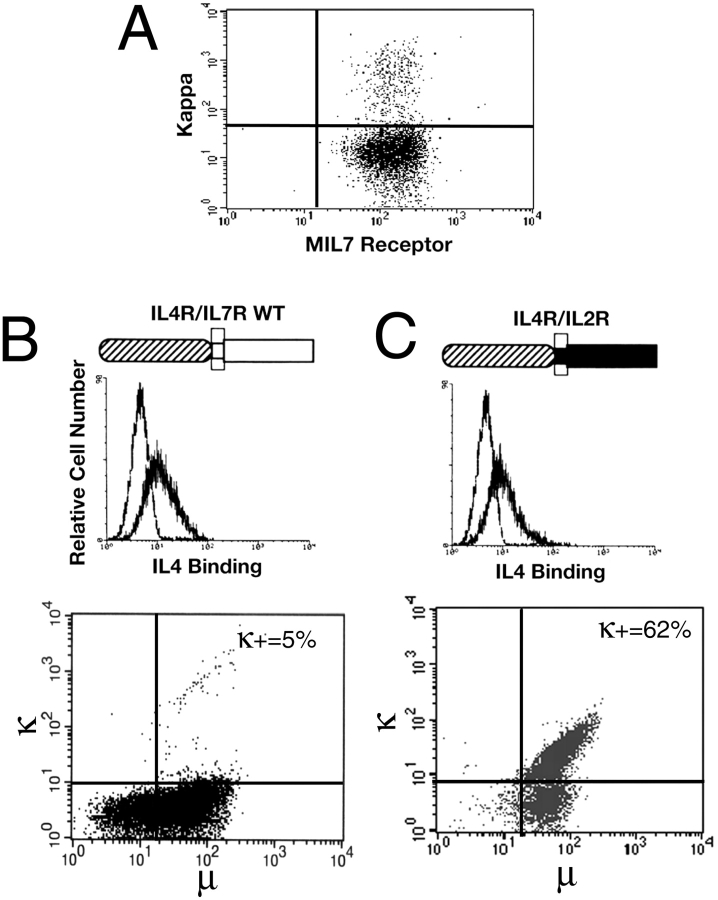
We reasoned that the loss of IL-7 responsiveness upon differentiation might result from the downregulation of IL-7R expression. To test this possibility, surface expression of the IL-7R was compared in κ1 and κ2 cells using an mAb specific to the α chain of the mouse IL-7R. Fig. 3 A demonstrates that IL-7R expression is detected to a similar level in both the κ1 and the κ2 fractions. These results suggest that, contrary to expectation, the loss of IL-7 responsiveness in μ1, κ1 cells cannot simply be ascribed to the lack of IL-7R expression.
Figure 3.
(A) Shows that κ+ and κ− pre-B cells stain with equal frequency and intensity for the murine IL-7 (MIL7) receptor. (B and C) The results of experiments with pre-B cells transduced with retrovirally encoded huIL-4R/IL-7R wild-type (WT) or huIL-4R/IL-2Rβ chimeric receptors. The top panel in each case shows that expression of the transduced receptors is equivalent, whereas the bottom panels demonstrate the marked difference in the capacity of the two chimeric receptors to support the outgrowth of κ+ cells. Results are typical of at least three independent experiments.
Retroviral Transfer of a Chimeric HuIL-4R/IL-7R Receptor Fails to Rescue μ1, κ1 Cell Outgrowth.
To subject the validity of these observations to further test, we introduced a cDNA encoding a huIL-4R/IL-7R chimeric receptor into the pre-B I cells by retroviral gene transfer. The chimeric molecule (Fig. 3 B) contains the extracellular sequences of the human IL-4R, conferring strict, species-specific responsiveness to human IL-4 26. However, its transmembrane and cytoplasmic domains are derived from the IL-7R. Expression of the chimeric receptor is promoted by the long terminal repeat sequence of the Moloney murine leukemia virus, active in a wide range of cell types 17. Therefore, it is not subject to the same transcriptional constraints as is the endogenous IL-7R.
Transduced pre-B I cells were selected in human IL-4 for expression of the huIL-4R/IL-7R before analysis. As shown in Fig. 3 B, the huIL-4R/IL-7R supports the proliferation of the κ2 pre-B I cell population. However, the chimeric receptor does not rescue the outgrowth of μ1, κ1 cells, further substantiating that the loss of IL-7 responsiveness at this developmental stage is not simply due to the lack of IL-7R expression. Rather, our results suggest that differentiation to the μ1, κ1 stage is accompanied by the inhibition of proliferative signaling through the IL-7R.
Inhibition of Proliferative Signaling at the μ1, κ1 Stage Is Specific to the IL-7R Cytoplasmic Domain.
Accordingly, we tested if replacement of the IL-7R cytoplasmic domain with sequences from the related IL-2R β chain 27 could overcome this inhibition. Pre-B I cells transduced with a huIL-4R/IL-2Rβ chimeric receptor were selected in human IL-4 before analysis for κ expression. As shown in Fig. 3 C, the huIL-4R/IL-2Rβ supports the outgrowth not only of κ2 pre-B I cells but also of cells that have differentiated to the μ1, κ1 stage. This is in marked contrast to the huIL-4R/IL-7R (Fig. 3 B). Thus, the loss of proliferative signaling at this developmental stage is specific to the IL-7R cytoplasmic domain. Taken together with the results in Table , this finding confirms that the differentiation of pre-B I cells to the μ1, κ1 stage triggers the inhibition of proliferative signaling through the IL-7R, despite continuing expression of the receptor.
Antigen Receptor Assembly Initiates the Inhibition of Proliferative Signaling through the IL-7R.
Our results suggest that the assembly of a functional antigen receptor initiates the inhibition of proliferative signaling through the IL-7R. To test this hypothesis directly, we created bicistronic gene constructs in which a rearranged λ1 light chain gene was linked through an IRES to a cDNA encoding either the huIL-4R/IL-7R or huIL-4R/IL-2Rβ chimeric receptors (Table ). These constructs were introduced into pre-B I cells by retroviral transduction, and λ1-expressing cells were selected in human IL-4.
Table 2.
Ability of Chimeric Receptors to Support the Outgrowth of Cells Expressing a μ, λ1 Antigen Receptor
| Construct | n | Outgrowth of μ+, λ+ cells |
|---|---|---|
| λ1–IRES–huIL-4R/IL-7R | 5 | − |
| λ1–IRES–huIL-4R/IL-2Rβ | 3 | + |
| λ1–IRES–huIL-4R/IL-7RY410F | 3 | + |
Pre-B cells were transduced with bicistronic retroviruses encoding a λ1 light chain cDNA linked through an IRES to chimeric cytokine receptors. Transduced cells were selected in human IL-4 (10 ng/ml) for 7 d, and the surface expression of μ and λ was determined by flow cytometry (see Materials and Methods). n, the number of independent experiments.
As shown in Table , λ1-expressing cells were readily detected in cultures transduced with the λ1–IRES–huIL-4R/IL-2Rβ construct. These λ1 cells could be maintained in IL-4–dependent culture for >1 mo without loss of λ expression (data not shown). In contrast, λ1 cells could not be isolated from cultures transduced with the λ1–IRES–huIL-4R/IL-7R construct in multiple independent experiments, suggesting that their outgrowth was not supported by signaling through the huIL-4R/IL-7R. Taken together, our results indicate that assembly of a μ, λ1 antigen receptor results in the loss of responsiveness to proliferative signaling through the huIL-4R/IL-7R, and that this inhibition is specific to the cytoplasmic sequences of the IL-7R but not the IL-2Rβ.
Inhibition of Proliferative Signaling Is Dependent on Tyr410.
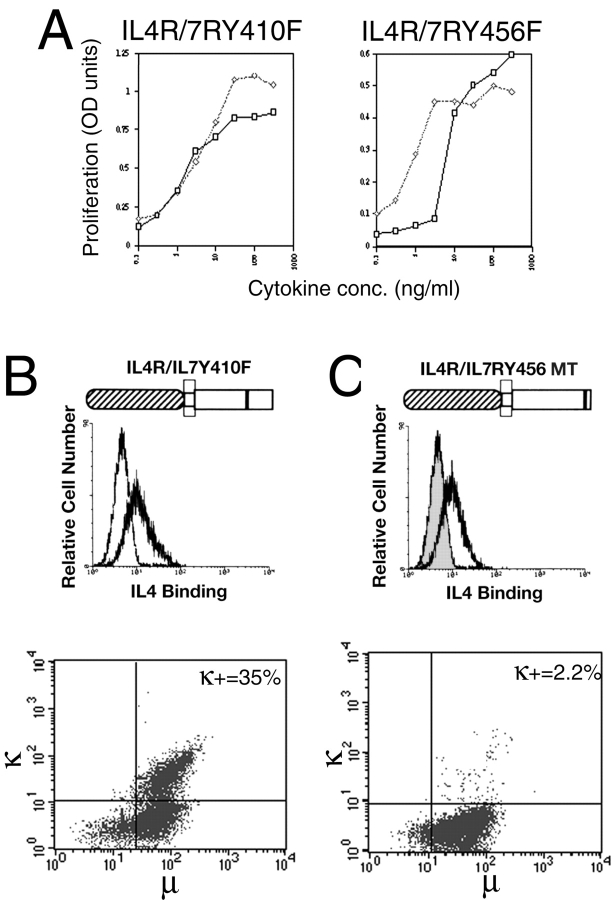
To investigate the mechanism by which this inhibitory signal might act, we constructed a series of mutant huIL-4R/IL-7R constructs in which cytosolic Tyr residues were altered to Phe. The cytoplasmic domain of the IL-7R contains three Tyr residues (Tyr410, Tyr449, and Tyr456) conserved in evolution between the murine and human receptors, suggesting an important function. The Tyr410Phe and Tyr456Phe mutants were tested in these experiments because we have already shown that Tyr449, which recruits the effector enzyme PI 3-k, is essential for proliferative signaling through the IL-7R 8 9.
Fig. 4 A shows that both the huIL-4R/IL-7R Tyr 410Phe and huIL-4R/IL-7R Tyr456Phe mutants are equally proficient at supporting the proliferation of pre-B I cells. Thus, neither residue is essential for proliferative signaling. However, in contrast to both the wild-type receptor (Fig. 3 B) and the Tyr456Phe mutant (Fig. 4 C), the huIL-4R/IL-7R Tyr410Phe mutant alone could support the outgrowth of μ1, κ1 cells (Fig. 4 B). This implies that mutation of a single Tyr residue can overcome the inhibition of proliferative signaling through the IL-7R.
Figure 4.
(A) Shows that pre-B cells transduced with huIL-4R/IL-7RY410F and huIL-4R/IL-7RY456F receptors proliferate as well in response to stimulation of the mutant receptors with IL-4 (dotted line) as they do in response to stimulation of their endogenous IL-7R by IL-7 (solid line). Proliferation quantified by the MTT assay is plotted on the y-axis against cytokine dose on the x-axis. (B and C) Compare the phenotype of pre-B cells transduced with retrovirally encoded huIL-4R/IL-7RY410F or huIL-4R/IL-7RY456F mutant chimeric receptors. The top panel in each case shows that expression of the transduced receptors is equivalent, whereas the bottom panels demonstrate the marked difference in the capacity of the two mutant chimeric receptors to support the outgrowth of κ+ cells. Results are typical of at least three independent experiments.
Accordingly, we performed a further experiment in which a rearranged λ1 light chain gene was linked in a bicistronic expression vector to the huIL-4R/IL-7R Tyr 410Phe mutant receptor (Table ). In contrast to the huIL-4R/IL-7R, but like the huIL-4R/IL-2Rβ, the Tyr410Phe mutant was competent to support the outgrowth of λ1-expressing cells. Taken together, these results confirm that the inhibitory signal initiated by antigen receptor assembly is critically dependent on the integrity of Tyr410 in the cytoplasmic domain of the IL-7R α chain. Therefore, we propose that the inhibitory signal may work through the recruitment of an inhibitory molecule to this residue.
In summary then, the results we have presented in this paper identify a novel mechanism that limits the expansion of pre-B lymphocytes in the murine bone marrow. By suppressing proliferative signaling through the IL-7R cytoplasmic domain upon antigen receptor assembly, this mechanism provides a feedback loop that may regulate the throughput of cells reaching the mature B lymphocyte stage. The assembly of a functional pre-B cell receptor may itself conceivably be sufficient to trigger this growth inhibitory process 28 29, restricting the number of divisions pre-B cells can undergo before commencing IgL rearrangement. Our findings demonstrate for the first time a homeostatic mechanism that limits cytokine-dependent cell expansion during hematopoiesis.
IL-7R Signaling Inhibited by Antigen Receptor Assembly
Acknowledgments
We thank Dr. J. Sims (Immunex Corp., Seattle, WA), Prof. S.I. Nishikawa (Kyoto University, Kyoto, Japan), Dr. A. Corcoran (The Cambridge Institute for Medical Research), and Dr. G. Nolan (Stanford University, Stanford, CA) for the generous gift of reagents used in this work, Dr. A. Corcoran and Dr. K.J. Patel for much helpful discussion, and Andrew Riddell for assistance with flow cytometry.
F.M. Smart received a Ph.D. Studentship from the Medical Research Council, UK. A.R. Venkitaraman holds a Professorship generously endowed by the late Dr. F.A. Zoellner. Work in A.R. Venkitaraman's laboratory is supported by the Medical Research Council, UK.
References
- Namen A.E., Schmierer A.E., March C.J., Overell R.W., Park L.S., Urdal D.L., Mochizuki D.Y. B cell precursor growth-promoting activity. Purification and characterization of a growth factor active on lymphocyte precursors. J. Exp. Med. 1988;167:988–1002. doi: 10.1084/jem.167.3.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin R.G., Friend D., Ziegler S.F., Jerzy R., Falk B.A., Gimpel S., Cosman D., Dower S.K., March C.J., Namen A.E. Cloning of the human and murine interleukin-7 receptorsdemonstration of a soluble form and homology to a new receptor superfamily. Cell. 1990;60:941–951. doi: 10.1016/0092-8674(90)90342-c. [DOI] [PubMed] [Google Scholar]
- Takeshita T., Asao H., Ohtani K., Ishii N., Kumaki S., Tanaka N., Munakata H., Nakamura M., Sugamura K. Cloning of the gamma chain of the human IL-2 receptor. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- Noguchi M., Nakamura Y., Russell S.M., Ziegler S.F., Tsang M., Cao X., Leonard W.J. Interleukin-2 receptor gamma chaina functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- Russell S.M., Keegan A.D., Harada N., Nakamura Y., Noguchi M., Leland P., Friedmann M.C., Miyajima A., Puri R.K., Paul W.E. Interleukin-2 receptor gamma chaina functional component of the interleukin-4 receptor. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- Giri J.G., Ahdieh M., Eisenman J., Shanebeck K., Grabstein K., Kumaki S., Namen A., Park L.S., Cosman D., Anderson D. Utilization of the beta and gamma chains of the IL-2 receptor by the novel cytokine IL-15. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Takeshita T., Higuchi M., Nakamura M., Sudo T., Nishikawa S., Sugamura K. Functional participation of the IL-2 receptor gamma chain in IL-7 receptor complexes. Science. 1994;263:1453–1454. doi: 10.1126/science.8128231. [DOI] [PubMed] [Google Scholar]
- Corcoran A.E., Smart F.M., Cowling R.J., Crompton T., Owen M.J., Venkitaraman A.R. The interleukin-7 receptor α chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:1924–1932. [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman A.R., Cowling R.J. Interleukin-7 induces the association of phosphatidylinositol 3-kinase with the alpha chain of the interleukin-7 receptor. Eur. J. Immunol. 1994;24:2168–2174. doi: 10.1002/eji.1830240935. [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Carmack C.E., Shinton S.A., Kemp J.D., Hayakawa K. Resolution and characterization of pro-B and pre–pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Kunisada T., Ogawa M., Sudo T., Kodama H., Suda T., Nishikawa S., Nishikawa S. Stepwise progression of B lineage differentiation supported by interleukin 7 and other stromal cell molecules. J. Exp. Med. 1990;171:1683–1695. doi: 10.1084/jem.171.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Namen A.E., Gillis S., Ellingsworth L.R., Kincade P.W. Normal B cell precursors responsive to recombinant murine IL-7 and inhibition of IL-7 activity by transforming growth factor β. J. Immunol. 1989;142:3875–3883. [PubMed] [Google Scholar]
- Sudo T., Ito M., Ogawa Y., Iizuka M., Kodama H., Kunisada T., Hayashi S., Ogawa M., Sakai K., Nishikawa S. Interleukin 7 production and function in stromal cell–dependent B cell development. J. Exp. Med. 1989;170:333–338. doi: 10.1084/jem.170.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker D.J., Boyle N.E., Koziol J.A., Klinman N.R. The expression of the Ig H chain repertoire in developing bone marrow B lineage cells. J. Immunol. 1991;146:350–361. [PubMed] [Google Scholar]
- Suda T., Okada S., Suda J., Miura Y., Ito M., Sudo T., Hayashi S., Nishikawa S., Nakauchi H. A stimulatory effect of recombinant murine interleukin-7 (IL-7) on B-cell colony formation and an inhibitory effect of IL-1 alpha. Blood. 1989;74:1936–1941. [PubMed] [Google Scholar]
- Park L.S., Friend D.J., Schmierer A.E., Dower S.K., Namen A.E. Murine interleukin 7 (IL-7) receptor. Characterization on an IL-7–dependent cell line. J. Exp. Med. 1990;171:1073–1089. doi: 10.1084/jem.171.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos O., Mulligan R.C. Safe and efficient generation of recombinant retroviruses with amphotropic and ecotropic host ranges. Proc. Natl. Acad. Sci. USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D., Goff S., Bank A. Construction and use of a safe and efficient amphotropic packaging cell line. Virology. 1988;167:400–406. [PubMed] [Google Scholar]
- Kinsella T.M., Nolan G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;1:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Corcoran A.E., Riddell A., Krooshoop D., Venkitaraman A.R. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survivalapplication to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;66:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Zhou L.J., Ord D.C., Hughes A.L., Tedder T.F. Structure and domain organization of the CD19 antigen of human, mouse, and guinea pig B lymphocytes. Conservation of the extensive cytoplasmic domain. J. Immunol. 1991;147:1424–1432. [PubMed] [Google Scholar]
- Rolink A., Grawunder U., Winkler T.H., Karasuyama H., Melchers F. IL-2 receptor alpha chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int. Immunol. 1994;8:1257–1264. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- Chen J., Ma A., Young F., Alt F.W. IL-2 receptor α chain expression during early B lymphocyte differentiation. Int. Immunol. 1994;6:1265–1268. doi: 10.1093/intimm/6.8.1265. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D., Desiderio S.V., Paskind M., Kearney J.F., Baltimore D., Alt F.W. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- Idzerda R.L., March C.J., Mosley B., Lyman S.D., Vanden Bos T., Gimpel S.D., Din W.S., Grabstein K.H., Widmer M.B., Park L.S. Human interleukin 4 receptor confers biological responsiveness and defines a novel receptor superfamily. J. Exp. Med. 1990;171:861–873. doi: 10.1084/jem.171.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M., Mori H., Doi T., Taniguchi T. A restricted cytoplasmic region of IL-2 receptor beta chain is essential for growth signal transduction but not for ligand binding and internalization. Cell. 1989;59:837–845. doi: 10.1016/0092-8674(89)90607-7. [DOI] [PubMed] [Google Scholar]
- ten Boekel E., Melchers F., Rolink A.G. Changes in the VH repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity. 1997;7:357–368. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- Wasserman R., Li Y.S., Shinton S.A., Carmack C.E., Manser T., Wiest D.L., Hayakawa K., Hardy R.R. A novel mechanism for B cell repertoire maturation based on response by B cell precursors to pre-B receptor assembly. J. Exp. Med. 1998;187:259–264. doi: 10.1084/jem.187.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]