Abstract
The Cryptococcus neoformans STE12α gene, a homologue of Saccharomyces cerevisiae STE12, exists only in mating type (MAT)α cells. In S. cerevisiae, STE12 was required for mating and filament formation. In C. neoformans, haploid fruiting on filament agar required STE12α. The ability to form hyphae, however, was not affected by deletion of STE12α when convergently growing MATa strains were present. Furthermore, ste12α disruptants were fertile when mated with MATa strains, albeit with reduced mating frequency. Most importantly, the virulence of a ste12α disruptant of serotype D strain was significantly reduced in a mouse model. When the ste12α locus was reconstituted with the wild-type allele by cotransformation, virulence was restored. Histopathological analysis demonstrated a reduction in capsular size of yeast cells, less severe cystic lesions, and stronger immune responses in meninges of mice infected with ste12α cells than those of mice infected with STE12α cells. Using reporter gene constructs, we found that STE12α controls the expression of several phenotypes known to be involved in virulence, such as capsule and melanin production. These results demonstrate a clear molecular link between mating type and virulence in C. neoformans.
Keywords: haploid fruiting, mating assay, STE12, cotransformation, virulence factor
Introduction
Cryptococcus neoformans is a basidiomycetous yeast pathogen that can cause life-threatening infections in normal as well as immunocompromised individuals 1. In AIDS patients, cryptococcosis is the leading cause of mycotic morbidity and mortality 2. Unlike other well studied heterothallic basidiomycetes, such as Schizophyllum commune and Ustilago maydis, which possess tetrapolar mating systems, C. neoformans possesses a bipolar mating system with two alleles, mating type (MAT)α and MATa 3 4. The sexual state of C. neoformans is characterized by the formation of dikaryotic hyphae, which possess typical basidiomycetous clamp connections and bear terminal basidia. Repeated postmeiotic mitosis and basidiosporogenesis result in the production of four long chains of haploid basidiospores on the apical surface of the basidium. Upon germination, the basidiospores produce yeast cells that multiply by polar budding without hyphal formation. Under specific conditions, such as nitrogen starvation and dehydration, haploid yeast cells can undergo an extensive hyphal differentiation in the absence of the opposite mating type 5. Hyphae produced under these conditions possess unfused clamp connections and develop basidia bearing only MATα-type basidiospores. This phenomenon is called haploid or monokaryotic fruiting, and it occurs only in MATα strains.
The molecular analysis of hyphae production in MATα C. neoformans has resulted in the identification of a gene named STE12α whose sequence displays similarity to the Saccharomyces cerevisiae STE12 gene 6. In S. cerevisiae, STE12 is a transcriptional activator capable of inducing transcription of a number of different genes 7 8 9 10 11 and is well conserved among many fungi, including Aspergillus nidulans, Candida albicans, Kluyveromyces lactis, and S. cerevisiae (EMBL/GenBank/DDBJ accession nos. AF080600, U15152, L21156, and X16112, respectively). In S. cerevisiae, STE12 is the target of a conserved mitogen-activated protein (MAP) kinase signal transduction pathway involved in mating, pseudohyphal development, and haploid invasive growth (for review see references 12–15). The C. neoformans STE12α gene, while having conserved roles in morphogenesis, has one striking difference from the S. cerevisiae STE12 gene: it is found only in MATα cells 6.
It has been observed that clinical as well as environmental isolates of C. neoformans are predominantly MATα 16, and it has been shown that among serotype D strains, MATα cells are more virulent than MATa cells in the murine model 17. Genetic association of virulence and mating type has not been established among serotype A strains due to the absence of MATa strains. The relationship among virulence, mating type, and haploid fruiting at the molecular level is not clear. In this paper, we show that STE12α is important for haploid fruiting but not essential for mating. The importance of STE12α in regulating expression of several virulence-associated genes was also investigated. We show that ste12αΔ serotype D strains are markedly reduced in virulence as compared with the wild-type strain. The decreased virulence of ste12αΔ suggests that STE12α is an important regulator of virulence in serotype D and provides molecular evidence for the important role of mating type–specific gene(s) in the virulence of C. neoformans.
Materials and Methods
Strains, Media, and General Methods.
The strains used in this study were all of serotype D and are listed in Table . B-4500 (MATα) is a wild-type congenic strain of B-4476 (MATa) previously used to investigate genetic association of mating type and virulence 17. All C. neoformans strains used in this study were derived from these two strains. TYCC245F1FO is a ura5 mutant recovered from TYCC245F1 by selection on 5-fluoroorotic acid (5-FOA) medium 18. Yeast extract peptone dextrose (YEPD) contained 1% yeast extract, 2% Bacto-peptone, and 2% glucose. 3,4-dihydroxyphenylalanine (DOPA) medium was prepared as described previously 19. Minimal media contained 6.7 g of yeast nitrogen base without amino acids (Difco Labs., Inc.) with 20 g of glucose, galactose, or raffinose per liter (pH 6.0). RPMI agar 20 and V-8 juice agar 21 were prepared as described previously. Synthetic low ammonia dextrose (SLAD) medium, originally designed to induce pseudohyphal formation in S. cerevisiae 12, was modified by omitting histidine and using unwashed agar. Filament agar, used to induce haploid fruiting in C. neoformans, is similar to SLAD medium, except that filament agar contains 0.5% glucose and 4% agar 5. Culture conditions for RPMI growth were 30°C with 10% CO2 20. All crosses were performed on V-8 juice agar as described previously 21. Phospholipase activity was determined on egg yolk agar as described 22. In brief, the egg yolk agar contained Sabouraud dextrose agar with 1 M sodium chloride, 0.005 M calcium chloride, and 8% sterile egg yolk. Each isolate was tested in triplicate. Cultures were incubated at 30°C for 72 h, and the diameter of the zone of precipitate around the colonies was measured. The ratio of the diameter of the colony to the total diameter of the colony plus precipitation was measured as an index of phospholipase activity. There is an inverse relationship between index and enzyme activity: the smaller the index, the higher the enzyme activity of the strain.
Table 1.
List of Strains Relevant to this Study
| Strain | Genotype/comment | Source |
|---|---|---|
| B4500 | MATα; congenic strain of B-4476 | Reference no. 17 |
| B4476 | MATa; congenic strain of B-4500 | Reference no. 17 |
| B4500FO2 | MATα ura5 | Reference no. 23 |
| LP1 | MATα ura5 ade2 | Reference no. 23 |
| JEC30 | MATa lys1 | Gift of J.C. Edmen |
| JEC32 | MATa lys2 | Gift of J.C. Edmen |
| TYCC259 | MATα ura5 ade2 GAL7(p)::STE12α::ADE2 | This study |
| TYCC245 | MATα ura5 ade2 Δste12a::ADE2; derived from LP1 | This study |
| TYCC245F1 | MATα Δste12::ADE2; F1 of TYCC245 × JEC32 | This study |
| TYCC245F1FO | MATα ura5 Δste12::ADE2; derived from TYCC245F1 | This study |
| TYCC409A | MATα ura5 ade2; STE12α reconstituted | This study |
| TYCC409AF1 | MATα; F1 of TYCC409A × JEC30 | This study |
Plasmids.
Plasmids used in this study are listed in Table . Plasmid pYCC245 was the deletion construct (see Fig. 1) in which the DraIII/BamHI fragment of STE12α in p18-S1 6 was replaced with the 3.0-kb BamHI/EcoRI fragment of the ADE2 gene from pYCC76 23. The URA5 gene was placed at the 5′ flanking region of STE12α in pYCC245. Plasmid pYCC259 was constructed as follows. An NdeI site was created at the first ATG site of the coding region of STE12α by PCR, and the resulting construct was placed under the control of the GAL7 promoter as described 24. Plasmid pYCC328 contained a promoterless β-glucuronidase (GUS) gene and the 1.08-kb stability factor (STAB) sequence 25. STAB confers an autonomous replication sequence–like function, which enhances the episomic maintenance of plasmids in C. neoformans. Incorporation of the STAB sequence in our GUS reporter constructs greatly reduced the variation of the assay results compared with constructs without STAB (our unpublished results). For GUS reporter gene constructs, an NdeI site was created at the first ATG site of the coding region of each gene by PCR. Individual promoters were then cloned into the NdeI site created at the first ATG of the GUS gene. Sequences of all PCR clones were confirmed before use.
Table 2.
List of Plasmids Relevant to This Study
| Plasmid | Gene/description | Source |
|---|---|---|
| pCIP3 | URA5 | Reference no. 26 |
| pNH7 | STE12α URA5 | Reference no. 26 |
| p18-S1 | SphI/EcoRI subclone of STE12α | Reference no. 26 |
| pYCC76 | ADE2 | Reference no. 23 |
| pYCC245 | STE12α deletion construct; see Fig. 1 | This study |
| pYCC259 | GAL7(p)::STE12α in ADE2 plasmid | This study |
| pYCC318 | CAP59(p)::GUS in pYCC328 backbone | This study |
| pYCC319 | CAP64(p)::GUS in pYCC328 backbone | This study |
| pYCC320 | CNLAC1(p)::GUS in pYCC328 backbone | This study |
| pYCC325 | CAP60(p)::GUS in pYCC328 backbone | This study |
| pYCC327 | ADE2(p)::GUS in pYCC328 backbone | This study |
| pYCC328 | 1.1-kb STAB in pCIP3 with promoterless GUS | This study |
| pPM8 | STAB, URA5, telomeres | Gift of P. Mondon |
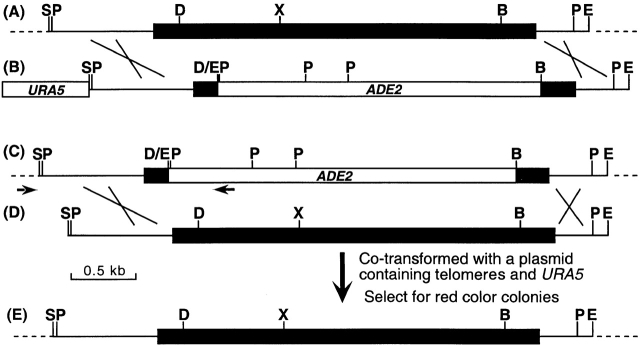
Figure 1.
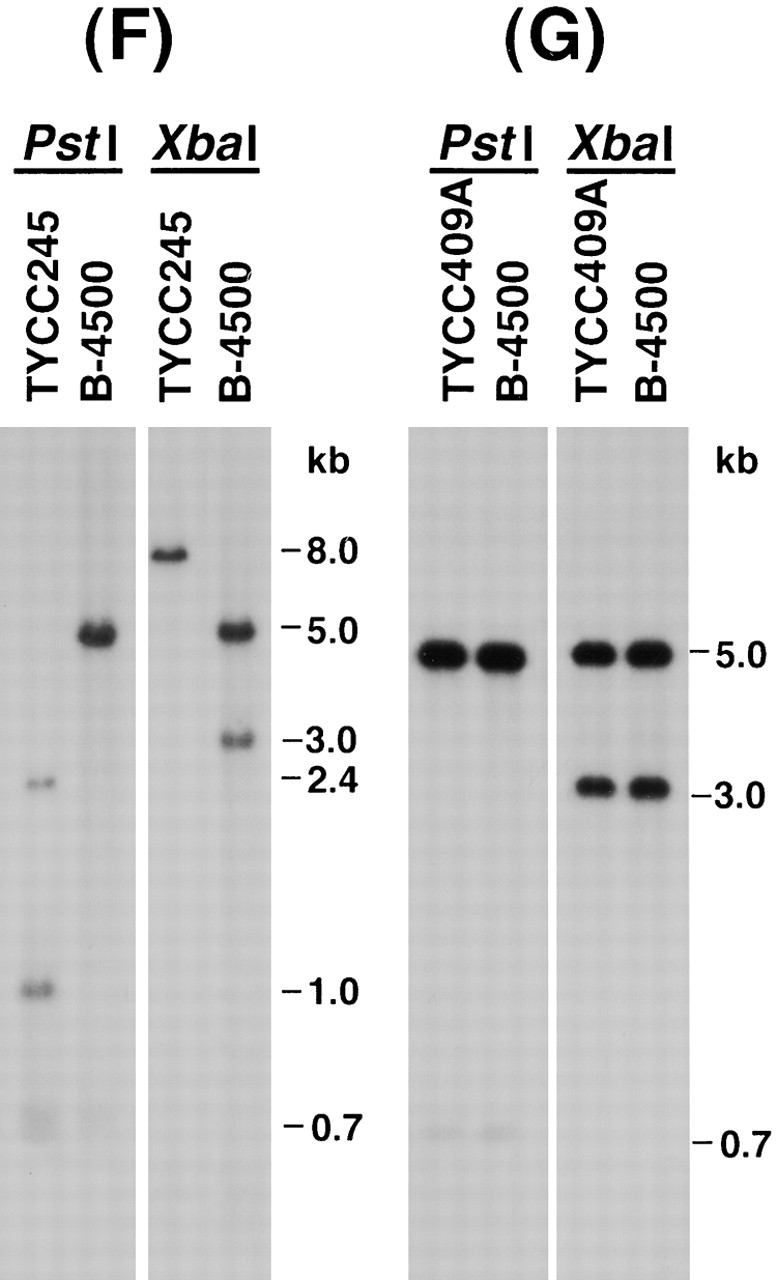
(A) Map of the STE12α gene. (B) Map of STE12α deletion construct. For simplicity, the URA5 gene is not drawn to scale. (C) Map of ste12α in TYCC245. (D) Map of DNA fragment used in cotransformation. (E) Map of reconstituted STE12α. Arrowheads, primer locations. Black boxes, coding region of STE12α. Crosses, crossing over. Dashed lines, chromosomal region flanking STE12α. B = BamHI; D = DraIII; E = EcoRI; P = PstI; S = SphI; X = XbaI. (F) Southern blot of ste12α deletant. (G) Southern blot of STE12α reconstituted strain. B-4500, wild type; TYCC245, ste12α deletant; TYCC409A, STE12α reconstituted strain. DNA was isolated, digested with restriction enzymes, fractionated on 0.8% agarose gel, and analyzed by Southern blot. The blots were hybridized with a probe of the SphI/EcoRI fragment of STE12α.
GUS Activity Assay.
B4500F0, TYCC259, and TYCC245F1FO were transformed with various GUS reporter constructs (Table ). Six independent transformants from each construct were assayed for GUS activity. For overexpression of STE12α, cells were inoculated in 10 ml of minimal medium containing 2% raffinose in a 50-ml Falcon tube and incubated at 30°C with shaking at 200 rpm for 24 h. 1 ml of the culture was inoculated into 9 ml of fresh minimal medium containing either 2% glucose or 2% galactose as a carbon source, and the culture was incubated for an additional 20 h. Cells were harvested, and GUS activity was assayed as previously described 24. Activity was expressed as picomoles of 4-methylumbelliferone produced per minute per 200 μg of protein. To assay GUS activity of cultures in late stationary phase, cells were grown in 10 ml of minimal medium containing 2% raffinose for 24 h, and 1 ml of this culture was inoculated into 9 ml of fresh minimal medium containing 2% glucose. This culture was then grown for an additional 45 h under the same conditions. Cells were harvested, and GUS activity was assayed.
Transformation of C. neoformans and Disruption of STE12α.
The electroporation method described by Edman and Kwon-Chung 26 was used to transform C. neoformans. To disrupt STE12α, the positive–negative selection protocol was used as described previously 20. To identify putative ste12α disruptants, ∼500 transformants that grew on 5-FOA plates after transformation were inoculated into 100 μl of water as pools of 10. An aliquot of each pool was then added to 50 μl of 1× PCR reaction buffer, heated at 95°C for 15 min, and processed for PCR. From pools that gave positive reactions, another round of PCR was performed on individual clones to identify the true positive clones. With this method, we identified two ste12α disruptants that were confirmed by Southern analysis.
Cotransformation Method.
A telomere-based plasmid containing URA5 and STAB (pPM8; a gift from P. Mondon, NIH, Bethesda, MD) was linearized to expose the telomere ends. The linearized DNA was mixed with the 4.5-kb SphI/EcoRI fragment of p18-S1, which contained STE12α. To deliver the DNA into TYCC245, both electroporation and biolistic transformation 27 were tested. After transformation, the cells were plated on minimal medium supplemented with adenine and incubated at 30°C for 1 wk, and colonies showing a red color were isolated. All putative clones were transferred on YEPD three times to cure the telomere-based plasmid. Uracil and adenine auxotrophs were isolated, and DNAs were analyzed by Southern blot. For the electroporation method, two red colonies were isolated from ∼105 transformants, and both clones displayed a Southern pattern of wild-type STE12α. For the biolistic method, five red colonies were isolated from ∼7,000 transformants, and four of them showed Southern patterns identical to that of wild-type STE12α.
Quantitative Assay for Mating Frequency.
The rationale is similar to the quantitative mating assay used in S. cerevisiae 28. All cultures used in the mating assay were <24 h old. About 2 × 106 viable cells from two opposite mating type strains, each carrying different auxotrophic markers, were mixed in 200 μl of 0.9% NaCl in a microfuge tube. The cells were collected on a 0.45-μm filter by suction, and the filters were placed on V-8 agar plates for 7 h at room temperature. Cells were then washed off the filter and spread on minimal media plates, which were incubated for 4 d at 30°C. The number of colonies producing hyphae was then determined. None of the cells from each mating parent would grow on minimal media, because each mating parent carried a different auxotrophic marker. The mating frequency of each strain was determined as the total number of hyphae producing colonies/total number of input cells from both mating type strains. The relative mating frequency was expressed as a percentage of the mating frequency of the wild-type reference strain (LP1). Two different auxotrophic MATa strains (JEC30 and JEC32) were used as tester strains to determine the mating frequency of any given MATα strain. Data were the average of results derived from mating with JEC30 and JEC32. The experiments were repeated at least twice to confirm reproducibility.
Virulence Study.
Female BALB/c mice (6–8 wk old) were injected via the lateral tail vein with 0.2 ml of a suspension of each yeast strain and mortality was monitored. Kaplan-Meier analysis of survival was performed using JMP software for the Macintosh (SAS Institute). To measure the growth rate of each strain in the brain, mice were injected with yeast cells (3 × 105 cells) as above, and then three mice each were killed at several intervals after injection (3 h as the starting point and 3, 6, 9, 12, and 15 d after injection). The brains were homogenized with a mortar and pestle, diluted, and then plated onto YEPD agar plates. Colonies were counted after incubation at 30°C for 2 d.
Histopathology.
Brains were removed and fixed in 10% buffered (neutral) formalin. Paraffin sections of the brains were stained with hematoxylin and eosin and Gomori methenamine silver (performed by American Histolabs).
Results
Disruption of STE12α.
To understand the function of STE12α in serotype D strains, we deleted the STE12α gene from LP1 by a positive–negative selection method 20. With this method, the DraIII/BamHI region, which contains the majority of the STE12α coding region, was replaced by the ADE2 gene in ste12α disruptants by homologous integration (Fig. 1A–C). Because ste12α disruptants would not be expected to have a visible phenotype, we first screened all transformants by PCR using primers designed to detect clones in which STE12α had been deleted (Fig. 1 C, arrowheads). Only clones containing a homologous integration of the deletion construct would yield the predicted PCR product. PCR positive clones were then analyzed by Southern blot to confirm a true gene replacement event. The Southern blot pattern in Fig. 1 F demonstrated that the signals of the wild-type STE12α band in B-4500 were replaced by new signals in one of the putative ste12α disruptants, TYCC245. Therefore, TYCC245 contained a ste12α disruption.
STE12α and Virulence-associated Genes.
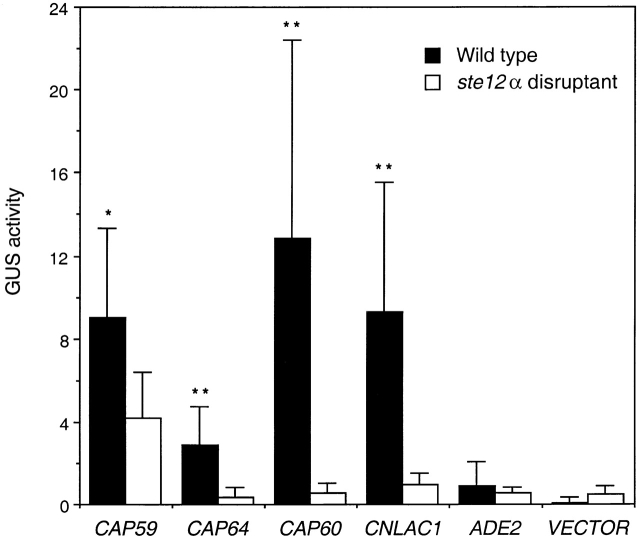
As the STE12α gene is MATα strain specific and MATα is known to be more virulent than MATa strains, it was of interest to determine if deletion of STE12α would change the expression levels of known virulence-associated genes. The laccase gene (CNLAC1) and three capsule-associated genes (CAP59, CAP60, and CAP64) have been identified as virulence factors in C. neoformans 20 23 29 30. The promoters of these genes were individually fused with the GUS gene in a URA5-containing plasmid (Table ). GUS reporter constructs of the four virulence-associated genes were transformed into TYCC245F1FO (ste12α::ADE2, ura5) as well as a wild-type control strain (B-4500FO2), and GUS activity was determined. GUS activity in transformants of B-4500FO2 containing constructs of CAP59, CAP60, CAP64, and CNLAC1 was generally much lower in cells taken from glucose-grown overnight cultures (20-h cultures; Table , column 1) compared with cells from late stationary phase glucose-grown cultures (45-h cultures; Fig. 2, black bars). It has been documented that cells in late stationary phase express higher laccase activity 19 and larger capsules compared with cells in the exponential growth phase 31. Therefore, the observations that cells in the stationary phase displayed higher reporter activity corroborated these reports. When GUS activity of late stationary phase cultures was compared in the ste12α or wild-type background, it was clear that all four constructs produced lower GUS activity in the ste12α background (Fig. 2). In contrast, GUS activity was very low and showed no difference in transformants with constructs containing the ADE2 promoter or in a vector with a promoterless GUS gene (Fig. 2). Reductions in capsule and melanin production, however, were not apparent by visual inspection when the ste12α strain was grown on agar media (YEPD or RPMI for capsule, DOPA for melanin; data not shown).
Table 3.
Effect of Overexpression of STE12α on GUS Activity
| B4500FO2 | TYCC259 | |||
|---|---|---|---|---|
| Glucose | Galactose | Glucose | Galactose | |
| CAP59 | 2.30 ± 1.27 | 3.63 ± 1.11 | 3.00 ± 1.70 | 21.07 ± 4.09 |
| CAP64 | 0.30 ± 0.21 | 2.10 ± 0.30 | 0.53 ± 0.30 | 13.57 ± 2.48 |
| CAP60 | 5.52 ± 2.66 | 6.88 ± 5.05 | 2.73 ± 2.41 | 14.13 ± 4.57 |
| CNLAC1 | 1.92 ± 1.38 | 2.23 ± 1.58 | 1.96 ± 0.62 | 18.97 ± 5.41 |
| ADE2 | 0.20 ± 0.18 | 1.77 ± 0.63 | 0.43 ± 0.39 | 7.92 ± 2.42 |
| Vector | 0.17 ± 0.15 | 1.13 ± 0.45 | 0.40 ± 0.53 | 5.87 ± 0.97 |
Figure 2.
GUS activity assay for stationary phase cells. The GUS reporter constructs were transformed into wild type (B-4500FO; black bars) and ste12α disruptant (TYCC245F1FO; white bars). The cells from stationary phase were used for GUS activity assay. Six independent transformants from each construct were assayed for GUS reporter activity. Error bars represent the sample SD. *P < 0.05; **P < 0.01.
As the ste12α disruptant influenced the expression levels of four virulence-associated genes, we further investigated the effect of overexpression of the STE12α gene. STE12α was placed under control of the C. neoformans GAL7 promoter in an ADE2-containing plasmid. The resulting plasmid was transformed into strain LP1 (MATα, ade2, ura5), and a stable transformant (TYCC259) was obtained (Table ). PCR with construct-specific primers confirmed that the GAL7(p)::STE12α fusion was intact in TYCC259. When TYCC259 was grown on minimal media containing galactose as the sole carbon source, it produced hyphal projections similar to the phenomenon observed by Wickes et al. 6, which was interpreted as being a result of overexpression of STE12α. GUS reporter constructs of four virulence-associated genes were transformed into TYCC259 as well as a wild-type control strain (B-4500FO2), and GUS activity was determined. GUS activity was found to be relatively low in transformants of B-4500FO2 when the cells were grown in either glucose or galactose (Table ). Similarly low levels of GUS activity were detected in transformants of TYCC259 when glucose was used as the sole carbon source. When transformants of TYCC259 were grown in galactose, however, GUS activity increased significantly (Table ). The increase in GUS activity was most dramatic in transformants containing a fusion of CAP64. Although GUS activity also increased in transformants containing constructs with the promoter of ADE2 or in the vector with a promoterless GUS gene, the increase was not as large in the B-4500FO2 background (TYCC259 versus B-4500FO2). Although the reason is not clear, it may be that some sequences in these constructs respond directly or indirectly to the overexpression of STE12α. Therefore, although the influence of overexpression of STE12α on the virulence-associated genes was not as dramatic as that of the ste12α disruptant, it was clear that overexpression of STE12α enhanced the CAP64 reporter activity.
As phospholipase was suggested to be a putative virulence factor for C. neoformans 22, we tested the effect of deletion of ste12α on phospholipase activity. The activity of phospholipase on egg yolk agar was considerably higher in the control strain (B-4500) than that in the ste12α disruptant (TYCC245F1). The index of phospholipase activity of B-4500 was one-half of that produced by TYCC245F1 (0.54 ± 0.04 versus 0.97 ± 0.04). Thus, deletion of ste12α downregulated the activity of extracellular phospholipase.
STE12α and Mating.
In S. cerevisiae, STE12 is required for mating and filament formation, as ste12 mutants are sterile and do not produce pseudohyphae 8 15 32. We first tested if mating ability was altered in C. neoformans when STE12α was deleted. TYCC245 (ste12αΔ) was crossed with a MATa strain (B-4476) on V-8 juice agar. We observed that the mating produced hyphae with fused clamp connections and viable basidiospores, as seen in the wild-type MATα × MATa crosses (control). Thus, surprisingly, TYCC245 was fertile. When the progeny of the TYCC245 × MATa cross were analyzed, a predicted segregation pattern of genetic markers was observed, confirming that ste12α mutants were fertile (data not shown). The number of basidia with healthy spore chains, however, was reduced compared with the control. To assess the extent of reduction in mating, mating frequency was determined quantitatively and found to be reduced almost 93% in ste12α strains (Fig. 3). These results indicated that although the mating frequency of the ste12α disruptant was greatly reduced, deletion of ste12α did not abolish sexual reproduction.
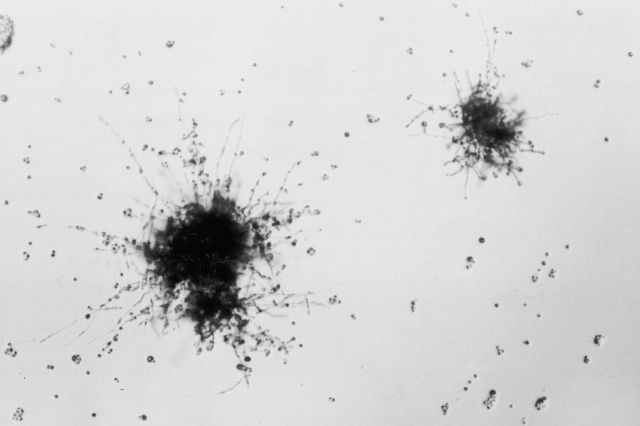
Figure 3.
Quantitative assay for mating frequency. Cells from two opposite mating type strains each carrying different auxotrophic markers were placed on filters and incubated on V-8 agar plates. Cells were washed off the filters and plated on minimal media. Photograph shows colonies producing abundant hyphae as a result of mating. The background cells were the input cells that failed to complement the nutritional deficiency in each mating type.
STE12α and Haploid Fruiting.
In S. cerevisiae, both pseudohyphal formation and haploid invasive growth require the presence of a functional STE12 gene 13 33. Filamentous growth, however, is only partially defective in C. albicans when both copies of CPH1, a STE12 homologue, are disrupted 32. We tested whether MATα strains can undergo haploid fruiting in the ste12α background. We found that the ste12α deletant failed to produce haploid fruiting when it was placed on filament agar. In contrast, the wild-type control strain produced abundant hyphae and possessed unfused clamp connections and spore chains, typical features of haploid fruiting (Fig. 4). These results suggested that while ste12α disruptants were still fertile, they were incapable of undergoing haploid fruiting on filament agar.
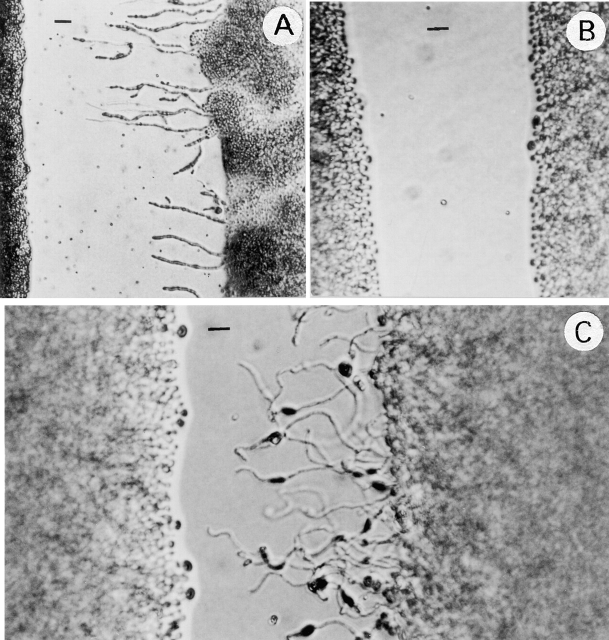
Figure 4.
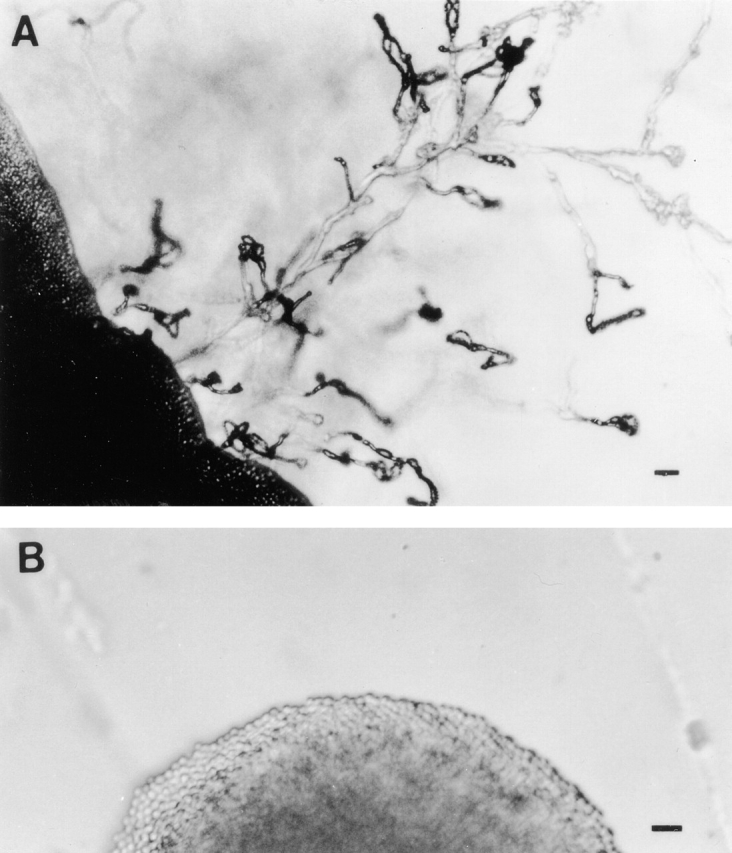
Haploid fruiting. Cells from wild-type B-4500 (A) and TYCC245F1 (B) were inoculated on filament agar and incubated at room temperature for 5 d (bar = 15 μm). Only the wild-type strain produced abundant hyphae and basidia with spore chains.
STE12α and Filament Formation.
We have previously observed that MATα cells produce hyphae within 24 h when streaked in close proximity (<500 μm) with MATa cells on SLAD medium (our unpublished observation). SLAD medium has a composition similar to filament agar and has been shown to induce pseudohyphal formation in S. cerevisiae 12. When MATα cells of C. neoformans were grown alone on SLAD medium, no hyphae were produced. It was of interest to investigate whether ste12α disruptants lost the ability to produce hyphae on SLAD medium in response to the presence of MATa cells. Fig. 5 A shows hyphal production by B-4500 (STE12α, MATα) when streaked in parallel to a streak of B-4476 (MATa) within a 500-μm distance, whereas MATa cells failed to respond in a similar manner (no hyphal filaments were produced). Similarly, no hyphal structures were detected when two streaks of ste12α (TYCC245F1) cells were paired on SLAD agar (Fig. 5 B). TYCC245F1 produced hyphae only when it was streaked in parallel with MATa cells in close proximity (<500 μm distance; Fig. 5 C). The hyphae produced by TYCC245F1 (Fig. 5 C), however, were morphologically different, tending to be more sinuous compared with hyphae produced by B-4500 (Fig. 5 A). When plates containing parallel streaks of opposite mating type strains were incubated for 1 wk, hyphae produced by the MATα strain reached MATa cells, and both basidia and spore chains were produced sporadically at the margin of MATα streaks (data not shown). Similar results were observed on SLAD agar without ammonium sulfate, as well as on minimal medium (SLAD agar with 37.8 mM ammonium sulfate). It is clear, therefore, that there are at least two different signaling pathways leading to hyphal formation in C. neoformans. These observations suggest that although the ste12α disruptant lost its ability to undergo haploid fruiting in response to nitrogen starvation, it retained the ability to form hyphae in response to the presence of MATa cells on SLAD agar.
Figure 5.
Hyphal formation on SLAD medium. (A) B-4500 × B-4476. (B) TYCC245F1 × TYCC245F1. (C) TYCC245F1 × B-4476. Cells of each isolate were streaked in parallel on SLAD medium and incubated at room temperature for 24 h (bar = 15 μm).
Virulence Studies.
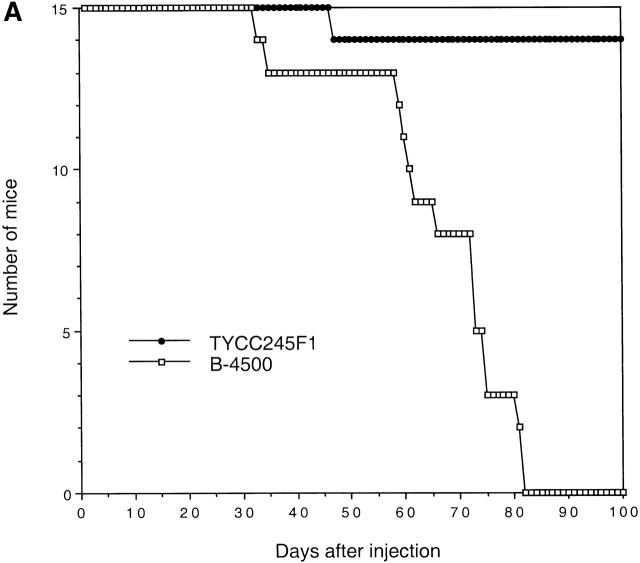
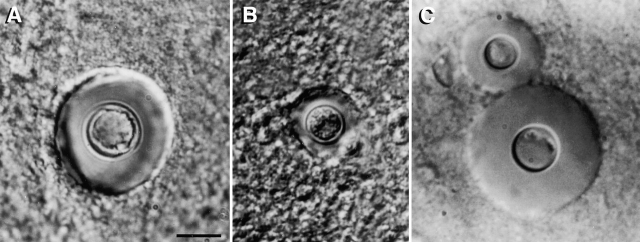
Because the GUS data suggested that STE12α has an effect on the expression of some of the virulence-associated genes under certain growth conditions, it was of interest to test the role of STE12α in virulence using an animal model. Groups of mice were infected with an F1 prototroph of the ste12α disruptant (TYCC245F1) and the wild-type congenic strain (B-4500). The ste12α disruptant produced significantly lower mortality than the wild-type (STE12α) strain, and the average number of days of survival was much longer in mice infected with the ste12α disruptant (log-rank, P < 0.0001; Fig. 6 A). When the growth rate of TYCC245F1 and B-4500 was compared at 37°C in YEPD broth, no obvious difference was observed. To test if the growth rates of these two strains were different in vivo, groups of mice were injected and the fungal burden in the brain was determined at several intervals up to 15 d after infection. Interestingly, similar numbers of CFUs were observed in the brains of mice infected with TYCC245F1 and B-4500, again suggesting no difference in growth rate (data not shown). However, the capsule size of yeast cells in brain smears from mice infected with TYCC245F1 was smaller (ranging from 0.5 to 3 μm) than seen in mice infected with B-4500 (ranging from 1 to 7 μm; Fig. 7). The histopathology of the mouse brains showed that the neuronal parenchyma in both groups of mice was disrupted to a similar degree by large multiloculated lesions that contained both yeast cells and large, foamy macrophages (Fig. 8B and Fig. D). However, mice infected with TYCC245F1 had a greater number of inflammatory cells (lymphocytes, macrophages, and a few neutrophils) surrounding vessels and in meninges (Fig. 8 C) than those infected with the wild-type strain (Fig. 8 A). In addition to being infiltrated by fewer inflammatory cells, the meninges of mice infected with the wild-type strains were more severely expanded by clear cystic spaces and yeast cells (Fig. 8 A) than were those of mice infected with TYCC245F1 (Fig. 8 C). These observations suggested that the ste12α strain provokes a stronger immune response than the STE12α strain. When viewed in light of the GUS experiments, these data implicate the importance of STE12α in regulating the expression of some of the known virulence-associated genes in vivo.
Figure 6.
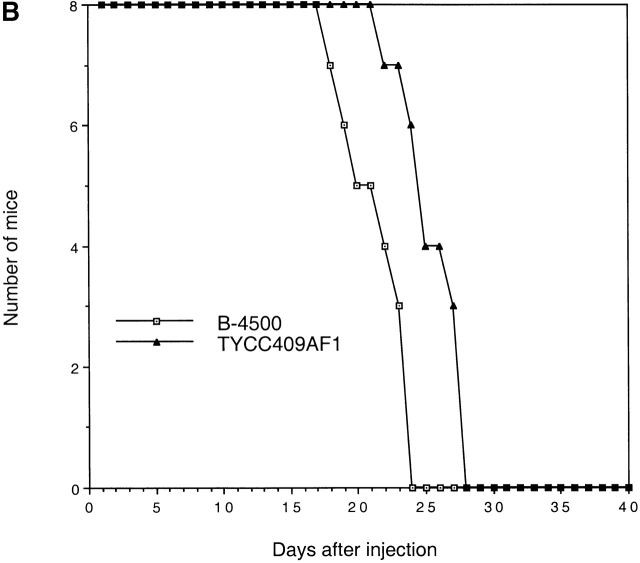
Virulence studies. BALB/c mice were challenged with (A) 5 × 105 yeast cells or (B) 106 yeast cells via the lateral tail vein. The mortality was monitored after the injection. B-4500, wild type; TYCC245F1, ste12α disruptant; TYCC409AF1, STE12α reconstituted strain.
Figure 7.
Brain smear showing cells of B-4500 (A), TYCC245F1 (B), and TYCC409AF1 (C). Brain tissue of mice challenged with different yeast strains was smeared on a microscopic slide and examined under a microscope with a Normalski interference condenser. Yeast cells with the largest capsule observed in the smears were photographed (bar = 10 μm).
Figure 8.
Histopathology of mice infected with wild type (A and B) and ste12α disruptant (C and D) (hematoxylin and eosin staining). (A). The meningeal area showing large cystic space (arrow) containing Cryptococci (light dots) with few immune cells (dark dots) (bar = 100 μm). (B). Neural parenchyma showing extensive formation of locular lesions containing yeast cells (bar = 50 μm). (C) The meningeal area showing smaller cystic space (arrow) and a large number of immune cells (bar = 100 μm). (D). Neural parenchyma with extensive locular lesions containing yeast cells (bar = 50 μm).
Reconstitution of STE12α.
To test if reintroduction of the wild-type STE12α gene into the ste12α disruptant would allow the fungus to regain its virulence, TYCC245F1FO was separately transformed with two URA5-containing plasmids, the vector (pCIP3) and a pCIP3-based plasmid containing wild-type STE12α (pNH7). Stable transformants were selected after repeated transfer on YEPD agar. Because C. neoformans often modifies incoming DNA after electroporation, PCR was used to identify integrative transformants containing an intact STE12α gene. Strains containing an intact STE12α gene were able to produce wild-type hyphae when analyzed by the SLAD assay in the presence of a MATa strain nearby (data not shown). In contrast, transformants that received only the vector (pCIP3) produced sinuous hyphae indistinguishable from hyphae produced by the ste12α disruptant. Considering the fact that transformed DNA tends to integrate randomly at ectopic sites in C. neoformans, three independent stable transformants, each derived from the pCIP3 and pNH7 constructs, were chosen for virulence studies. Two of the three transformants of pNH7 produced mortality earlier, with the total number of mice surviving at the end of the experiment considerably less than that of all the other transformants. One pNH7 transformant, however, produced mortality indiscernible from the transformants of pCIP3. When the results from pNH7 and pCIP3 were pooled and the difference in survival among these two groups was compared, there was a significant difference between mice receiving pNH7 and pCIP3 (log-rank, P = 0.007).
It is possible that an ectopic integration could alter expression, either through positional effects or subtle modifications of the construct, which could lead to unexpected phenotypes. As only two out of the three pNH7 transformants restored virulence, it was necessary to address this variable. This was done by reconstituting the deleted ste12α locus back to the wild type by cotransformation. A DNA fragment containing STE12α and a telomere-based URA5 plasmid were mixed and transformed into TYCC245 (Fig. 1C–E). Because telomere-based plasmids have a very high frequency of transformation and can be easily cured due to their instability 34, one such plasmid was used as a carrier of the URA5 gene in the cotransformation. The resulting transformants were selected on minimal medium supplemented with adenine. If the DNA fragment containing STE12α integrated into the genome homologously and replaced the deleted ste12α gene by double cross-over, the resulting transformants would be auxotrophic for adenine and produce red-colored colonies on minimal medium supplemented with a suboptimal level of adenine. Red-colored transformants were indeed observed. These transformants were isolated and transferred to YEPD medium to cure the cotransformed URA5 plasmid. Fig. 1 G is the Southern blot analysis of one of the transformants (TYCC409A) showing that the hybridization patterns of the wild-type STE12α gene were restored in this strain.
TYCC409AF1, a prototrophic progeny of TYCC409A crossed with JEC30 (MATa, lys1), produced hyphae on filament agar and exhibited a hyphal morphology similar to B-4500 on SLAD medium (data not shown). The mating frequency of TYCC409A was 86.6% of that of LP1. The phospholipase activity of TYY409AF1 was close to the wild-type B-4500 (0.61 ± 0.05 versus 0.50 ± 0.04). Therefore, the in vitro wild-type phenotypes were almost completely restored in the STE12α-reconstituted strains. The virulence of the STE12α-reconstituted strain was compared with that of the wild-type congenic strain, B-4500. Fig. 6 B shows that the virulence was nearly restored when the wild-type STE12α gene was reintroduced into the genome of the ste12α disruptant. Furthermore, the capsule size of yeast cells in brain smears from mice infected with the STE12α-reconstituted strain was the same (1–7 μm) as that of mice infected with B-4500 (Fig. 7 C). Therefore, these observations fulfill the molecular version of Koch's postulates for STE12α as a factor associated with virulence 35.
Discussion
The S. cerevisiae STE12 gene is a key component of two MAP kinase cascades involved in mating and filamentous/invasive growth 12 13 14 15. Although STE12α shows sequence similarity with STE12 of S. cerevisiae, single copy or overexpression of STE12α cDNA in S. cerevisiae failed to complement the ste12 phenotypes, including the ability to mate and form pseudohyphae (data not shown). Recent identification of another C. neoformans MATα-specific gene, STE11α, which belongs to the same cascade, indicates an unusual arrangement of the MAP kinase cascade in C. neoformans 6. One of the intriguing findings in our study is that in contrast to the S. cerevisiae ste12 mutants, the C. neoformans ste12α disruptant was still able to mate, albeit with reduced frequency. The fact that STE12α is dispensable for mating further demonstrates the uniqueness of the mating pathway of C. neoformans.
Mating of C. neoformans is usually performed on V-8 juice agar and requires physical contact of cells from both mating types. In general, it is difficult to measure mating frequency accurately in C. neoformans 36. In our assay, both the ste12α deletant and the reconstituted strain carried auxotrophic markers. The MATa tester strains carried a different nutritional marker to allow the discrimination of mated and unmated cells, as unmated cells could not grow on minimal media. Only cells of the opposite mating type could fuse and produce viable colonies that, in turn, could be detected visually. Thus, our assay measured how frequently cells of opposite mating type fused and formed colonies by compensating for the nutritional deficiency in each individual mating type. This process appeared to require several hours to develop, as no colony grew on minimal medium when the filters containing cells of both mating types were incubated for <2 h (data not shown). We noticed that most of the colonies grew on minimal media as hyphal tufts. This phenotype is typical of heterokaryons formed by fusion of two opposite mating types and has been reported previously 37. This morphology is distinct from the analogous assay in S. cerevisiae in which the diploid grows as yeast. In general, the mating frequency for the wild-type (LP1) ranged from 10−3 to 10−4 of input cells. Deletion of ste12α reduced the mating frequency to 6.7% of LP1, whereas the mating frequency of the reconstituted strain was 86.6% of LP1. Although the 10–15-fold reduction in mating efficiency for ste12α mutants appears substantial, the degree of reduction is negligible compared with S. cerevisiae ste12 mutants 38. It is possible that C. neoformans contains another STE12 homologue that could substitute for the function of STE12α protein during the mating process and would explain the minor effect of ste12α on mating. This possibility has prompted us to further explore the C. neoformans genome. We have tentatively identified a STE12 homologue in MATa strains of C. neoformans (our unpublished results). If this STE12 homologue proves to be functional, C. neoformans would be the first reported species containing two different STE12 homologues in two opposite mating type strains, and this discovery would further portray the uniqueness of the mating pathway in C. neoformans.
One of the intriguing findings in this study was that SLAD medium, independent of the presence of ammonium sulfate, supports hyphal formation in MATα strains if a MATa strain is in close proximity. Induction of hyphal formation occurs on SLAD medium without contact of the two opposite mating types and only occurs in MATα cells. In the SLAD assay, it is reasonable to assume that pheromones or other secreted compounds from MATa strains induce hyphal formation in MATα strains. However, these hyphal strands should not be viewed as typical conjugation tubes produced in response to the opposite mating type. C. neoformans cells do not reach out to cells of the opposite mating type by producing filamentous conjugation tubes 39. C. neoformans cells produce very short conjugation tubes only upon contact with cells of the opposite mating type. This phenomenon is different from many other heterothallic basidiomycetous yeasts, which produce conjugation tubes that resemble germlings in response to the opposite mating type before fusion 40 41. Recent isolation of a gene encoding the pheromone in MATa strains (Wickes, B.L., unpublished results) will allow us to test if hyphal formation can be induced by synthetic a-pheromone in single MATα cultures growing on SLAD medium. It is known that MATa cells have the capability to produce hyphal protrusions when the α-pheromone gene or STE12α is overexpressed in MATa cells 6 42, although these structures are morphologically different from hyphae produced by MATα on filament agar. It is not clear, however, why MATa cells failed to produce hyphal structures when they were streaked closely to MATα cells. The concentration of α-pheromone in such a setting may be inadequate for MATa cells to respond.
In S. cerevisiae, both pseudohyphal formation and haploid invasive growth require the presence of a functional STE12 gene 13 33. In contrast, filamentous growth is only partially defective in C. albicans when both copies of CPH1, a STE12 homologue, are disrupted 32. We found that disruption of STE12α abolishes hyphal formation on filament agar but only slightly affects hyphal morphology on SLAD medium in the presence of the opposite mating type. Thus, as in C. albicans 32, alternative pathways for hyphal formation appear to exist in C. neoformans.
Perhaps the most striking observation of this study was that there is strong evidence that STE12α is required for virulence. Overexpression of STE12α induced the expression of a number of known virulence genes, and disruption of STE12α reduced the expression of these genes. Correspondingly, ste12α mutants were less virulent in mice compared with the wild-type, with the difference in a more vigorous immune response to the mutant strain. In fact, the decrease in ste12α capsule size in vivo was corroborated by the in vitro GUS data on capsular gene expression in these mutants. Conversely, in strains whose ste12α locus was reconstituted, all phenotypes, including virulence (and in vivo capsule size), fertility, phospholipase activity, and haploid fruiting approached wild type. These observations provide an important molecular link between the association of mating type with virulence in C. neoformans and support a previous study showing that MATα cells are more virulent than MATa cells 17.
Since our preliminary study on the effects of the ste12α disruptant of serotype D strain was reported 43, Yue et al. have disrupted the ste12α gene from the strain H99, a serotype A isolate of C. neoformans 44. The ste12α mutant of serotype A strain also showed a lack of haploid fruiting and a reduction in mating frequency. The capsule size of ste12α mutants was smaller than that of the wild type in vivo and in vitro. Unlike in serotype D isolates, however, phospholipase activity and virulence of ste12α mutants were not affected in H99 background. It is most likely that serotype difference was the reason for the discrepancy between our observations and the results of Yue et al. A considerable genetic hiatus between strains of serotype A and D has been observed in sequence differences of rRNA 45 and other genes 46. Our results with serotype D and those of Yue et al. with serotype A suggest possible differences in regulatory pathways. Franzot et al. 46 have even suggested the need for a separate varietal status for C. neoformans serotype A isolates.
The phospholipase activity assay, GUS data and virulence studies strongly suggest that the role of STE12α in virulence involves the regulation of many of the known virulence-associated genes. Thus, STE12α might be a global regulator. However, as deletion of STE12α had no visible effect on capsule size and melanin production in vitro or growth rate at 37°C in vitro or in vivo, it is possible that the effect of deleting STE12α is too subtle to be detected visually when screened on agar. In this case, the number of genes affected, rather than the expression level of each gene, could function cooperatively to reduce virulence. It is also possible that the regulatory effects of STE12α are noticeable only in certain environments, for instance, during stationary phase, reduced O2 tension, increased CO2 concentration, nitrogen starvation, or under certain other stressful conditions. Finally, given that virulence is often multifactorial, STE12α may function in the context of one or more of the above scenarios, which would suggest that fungal genes involved in mating have far more diverse roles than merely for reproduction.
Acknowledgments
We would like to thank Gerry Fink and Hiten Madhani for critical reading of the manuscript.
This work was supported in part by US Public Health Service grant R29AI43522-01 to B.L. Wickes.
Footnotes
Abbreviations used in this paper: GUS, β-glucuronidase; MAP, mitogen-activated protein; SLAD, synthetic low ammonia dextrose; YEPD, yeast extract peptone dextrose.
References
- Kwon-Chung K.J., Bennett J.E. Medical Mycology 1992. Lea & Febiger; Philadelphia: pp. 397–446 [Google Scholar]
- Mitchell T.G., Perfect J.R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans . Clin. Microbiol. Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K.J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans . Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung K.J. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–946. [PubMed] [Google Scholar]
- Wickes B.L., Mayorga M.E., Edman U., Edman J.C. Dimorphism and haploid fruiting in Cryptococcus neoformansassociation with the alpha-mating type. Proc. Natl. Acad. Sci. USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B.L., Edman U., Edman J.C. The Cryptococcus neoformans STE12alpha genea putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol. Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- Kirkman-Correia C., Stroke I.L., Fields S. Functional domains of the yeast STE12 protein, a pheromone-responsive transcriptional activator. Mol. Cell. Biol. 1993;13:3765–3772. doi: 10.1128/mcb.13.6.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell. 1985;42:923–930. doi: 10.1016/0092-8674(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. Regulation by the yeast mating-type locus of STE12, a gene required for cell-type-specific expression. Mol. Cell. Biol. 1987;7:3818–3821. doi: 10.1128/mcb.7.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Dolan J.W., Yuan Y.L., Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- Dolan J.W., Kirkman C., Fields S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA. 1989;86:5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C.J., Ljungdahl P.O., Styles C.A., Fink G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growthregulation by starvation and RAS . Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Fink G.R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell typemating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeastfor mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Fink G.R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K.J., Bennett J.E. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K.J., Edman J.C., Wickes B.L. Genetic association of mating types and virulence in Cryptococcus neoformans . Infect. Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K.J., Varma A., Edman J.C., Bennett J.E. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Mycol. 1992;30:61–69. [PubMed] [Google Scholar]
- Polacheck I., Hearing V.J., Kwon-Chung K.J. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans . J. Bacteriol. 1982;150:1212–1220. doi: 10.1128/jb.150.3.1212-1220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.C., Kwon-Chung K.J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K.J., Bennett J.E., Rhodes J.C. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Van Leeuwenhoek. 1982;48:25–38. doi: 10.1007/BF00399484. [DOI] [PubMed] [Google Scholar]
- Chen S.C., Muller M., Zhou J.Z., Wright L.C., Sorrell T.C. Phospholipase activity in Cryptococcus neoformansa new virulence factor? J. Infect. Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- Chang Y.C., Kwon-Chung K.J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans . Infect. Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B.L., Edman J.C. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol. Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- Varma A., Kwon-Chung K.J. Construction of stable episomes in Cryptococcus neoformans . Curr. Genet. 1998;34:60–66. doi: 10.1007/s002940050366. [DOI] [PubMed] [Google Scholar]
- Edman J.C., Kwon-Chung K.J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti D.L., Rude T.H., Johnston S.A., Durack D.T., Perfect J.R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G.F., Jr. Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- Williamson P.R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformansidentification as a laccase. J. Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.C., Penoyer L.A., Kwon-Chung K.J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger D.L., Perfect J.R., Durack D.T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C.A., Fink G.R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Edman J.C. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch's postulates applied to microbial pathogenicity Rev. Infect. Dis. 10 1988. S274 276(Suppl.) [DOI] [PubMed] [Google Scholar]
- Dong H., Courchesne W. A novel quantitative mating assay for the fungal pathogen Cryptococcus neoformans provides insight into signalling pathways responding to nutrients and temperature. Microbiology. 1998;144:1691–1697. doi: 10.1099/00221287-144-6-1691. [DOI] [PubMed] [Google Scholar]
- Whelan W.L., Kwon-Chung K.J. Genetic complementation in Cryptococcus neoformans . J. Bacteriol. 1986;166:924–929. doi: 10.1128/jb.166.3.924-929.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K., Ganesan K., Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J. Biol. Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- Kwon-Chung K.J. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans . Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- Snetselaar K.M. Microscopic observation of Ustilago maydis mating interaction. Exp. Mycol. 1993;17:345–355. [Google Scholar]
- Bandoni R.J. Conjugation in Tremella mesenterica . Can. J. Bot. 1963;41:468–474. [Google Scholar]
- Moore T.D., Edman J.C. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.C., B. Wickes, and K.J. Kwon-Chung. 1998. The STE12alpha of Cryptococcus neoformans Annu. Meet. Am. Soc. Microbiol., Atlanta, GA. (Abstr. F63).
- Yue C., Cavallo L.M., Alspaugh J.A., Wang P., Cox G.M., Perfect J.R., Heitman J. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans . Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueho E., Improvisi L., Christen R., de Hoog G.S. Phylogenetic relationships of Cryptococcus neoformans and some related basidiomycetous yeasts determined from partial large subunit rRNA sequences. Antonie Van Leeuwenhoek. 1993;63:175–189. doi: 10.1007/BF00872392. [DOI] [PubMed] [Google Scholar]
- Franzot S.P., Salkin I.F., Casadevall A. Cryptococcus neoformans var. grubiiseparate varietal status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]