Abstract
By analyzing T cell responses against foreign major histocompatibility complex (MHC) molecules loaded with peptide libraries and defined self- and viral peptides, we demonstrate a profound influence of self-MHC molecules on the repertoire of alloreactive T cells: the closer the foreign MHC molecule is related to the T cell's MHC, the higher is the proportion of peptide-specific, alloreactive (“allorestricted”) T cells versus T cells recognizing the foreign MHC molecule without regard to the peptide in the groove. Thus, the peptide repertoire of alloreactive T cells must be influenced by self-MHC molecules during positive or negative thymic selection or peripheral survival, much like the repertoire of the self-restricted T cells. In consequence, allorestricted, peptide-specific T cells (that are of interest for clinical applications) are easier to obtain if T cells and target cells express related MHC molecules.
Keywords: peptide library, T cell repertoire, molecular basis of alloreactivity, limiting dilution, positive selection
Introduction
Alloreactive T lymphocytes respond to allelic MHC variants in the apparent absence of nominal antigen and are the cause for important clinical problems such as graft rejection and graft versus host disease. In vitro, alloreactive T cells are activated easily by foreign MHC molecules without previous in vivo priming and are detectable in amazingly high frequencies 1. The molecular basis for these phenomena has not been elucidated, despite extensive characterization of peptide requirements in alloreactivity and the solution of several molecular structures of the 2C and AHIII12.2 TCR allo- and xenomodels 2 3 4 5.
Several hypotheses have been proposed to explain alloreactivity 2. Matzinger and Bevan 6 suggested that alloreactive T cells recognize many different cellular antigens, e.g., peptides, together with the foreign MHC. This model explains the strong alloresponse by multiple binary interactions between allorestricted T cell clones and new peptides not presented by self-MHC. Later, Kaye and Janeway 7 and Bevan 8 proposed that alloreactive T cells see all cell surface MHC molecules regardless of peptides as antigens, leading to a high determinant density per target cell. In the meantime, evidence for peptide-dependent and peptide-independent allorecognition has accumulated. On the one hand, for several alloreactive T cell clones the exact peptides recognized, which can be present at both high and low copy numbers, have been identified 9 10 11 12 13. On the other hand, recognition of MHC molecules in the absence of peptides has been clearly demonstrated 14 15. There are also cases where the peptide changes structural characteristics of the MHC molecule and leads to recognition by certain T cell clones in a peptide-dependent rather than peptide-specific manner 16 17 18. Today, the two models of allorecognition may be seen as two extremes on a scale where MHC and peptide contribute in different degrees to the overall binding energy between the TCR and its ligand. Accordingly, Daniel et al. explained the decreased peptide specificity of the alloreactive T cell clone 2.102 with certain allelic α-helical residues of the foreign MHC, which supposedly supply more binding energy to the TCR than the respective self-MHC molecule 19.
The involvement of peptides in alloreactivity that are present on stimulator cells in low copy numbers 3 13 raised the possibility of generating allorestricted CTLs against tumor-associated self-peptides (self as seen from the tumor's host) presented by MHC molecules (nonself for the T cells) for adoptive immunotherapy 20 21 22. For this purpose, referring to allogeneic repertoires is necessary because T cell tolerance is MHC restricted 20 23 24 25. Stauss and colleagues and our group demonstrated high avidity allorestricted CTLs against tumor-associated peptides and libraries from allogeneic mouse and human repertoires 21 26 27 28. However, a T cell repertoire, which contains T cells recognizing peptides bound to nonself-MHC molecules with high avidity contradicts the well-established bias of T cells to react against antigens preferentially in the context of self-MHC 29.
To analyze the peptide repertoire of alloreactive T cells and the potential influence of self-MHC molecules, we stimulated naive splenic T cells of MHC class I mutant or H2 recombinant mice against RMA-S cells (H2b) loaded with defined Kb- or Db-specific peptide libraries (KbL, DbL) under limiting dilution conditions. The relative frequencies of peptide-specific versus peptide-nonspecific T cells were determined, and several T cells of interesting peptide specificity were expanded and further analyzed. The results allow an unexpected insight into the relations between self-MHC and the repertoire of alloreactive T cells.
Materials and Methods
Mice, Cell Lines, and Virus.
C57BL/6 (abbreviated B6) and BALB/c mice were purchased from Charles River. B10.A(5R) (5R), B10.HTG (HTG), and B6.C-H2bm1 (bm1) animals were obtained from The Jackson Laboratory and maintained in our animal facility. B6.C-H2bm13 (bm13) and B6.C-H2bm14 (bm14) mice were received from Drs. R. Brandt and C.J.M. Melief (Leiden University Medical Center, The Netherlands). FVB/N transporter associated with antigen processing 1−/− (TAP)-1−/− 30) mice were obtained from Drs. H. ter Rile and A. Berns (Cancer Institute, Amsterdam, The Netherlands) and also bred in our animal facility. EL4, RMA (H2b, TAP+), and RMA.S (H2b, TAP−) cell lines were maintained in RPMI 1640 (Sigma Chemical Co.) supplemented with 2 mM l-glutamine (BioWhittaker), 2 μM 2-ME, and 10% FCS (Sigma Chemical Co.). For generating blasts, 0.5–2 × 107 splenocytes (in 5–10 ml RPMI) were stimulated for 2 d with 2.5–5 μg/ml Con A (Boehringer Mannheim), replated in CTL medium (see below), and used on days 3–5 of culture. Vesicular stomatitis virus (VSV) was obtained from Dr. R. Zawatzky (German Cancer Research Center, Heidelberg, Germany).
Peptides.
Peptides and libraries were synthesized and analyzed as described 27. The KbL has been described previously 27, and the DbL consisted of the amino acids indicated in Table (anchor positions in bold). Both libraries bind to the respective MHC allele as strongly as control peptides from OVA and influenza nucleoprotein in an RMA.S induction assay (data not shown). The new H2-Kb ligands were identified as described previously 31 by immunoprecipitation of peptide–MHC complexes, treatment with 0.1% trifluoric acid, ultrafiltration, fractionation by HPLC, and subsequent analysis by automated Edman degradation (sequencer model Procise 494A; Applied Bio-systems).
Table 1.
H2-Db Binding Peptide Library DbL (n = 2,000)
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|
| F | Q | N | D | N | G | Y | T | M |
| I | A | V | A | V | ||||
| N | M | Q | L | T | ||||
| S | S | K | M | Q | ||||
| A | E | E |
Generation of Effector Cells and CTL Assays.
Splenocytes were plated in limiting dilution in round-bottomed 96-well plates (Greiner), starting with 1–2 × 105 cells/well and diluting 1:2. Each well received 5 × 105 irradiated (33 Gy) stimulators in 200 μl of α-MEM (GIBCO BRL) with the above-listed supplements, including 5% Con A–induced rat splenocyte supernatant, 50 μM α-methylmannoside (CTL medium; Roth), and, where applicable, a peptide library (500 ng/ml) or a peptide mixture. After 7 d, the cultures were stimulated as before with peptides at 50 ng/ml. The cultures were tested 4–6 d later in a split-well 51Cr-release assay, as described previously 27. For expansion, CTLs were restimulated in 48- and 24-well plates with 250 or 5 × 105 irradiated TAP-1−/− splenocytes, respectively.
Results
Differential Requirement for Synthetic Peptides in Alloreactivity.
For the generation of tumor-specific CTLs, the holes in the self-restricted T cell repertoire led us to raise CTLs against peptide libraries presented on MHC molecules foreign to the T cells 27 28. In the course of such experiments, we noticed difficulties in raising H2-Kb– or H2-Db–restricted CTLs from H2d and H2k animals against several individual peptides (data not shown) and investigated the influence of the responder's haplotype.
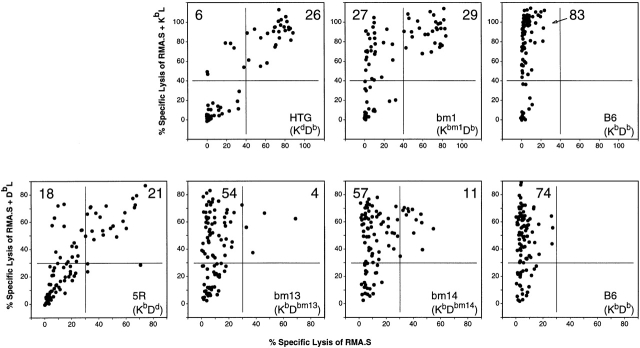
Splenocytes from HTG, bm1, and B6 mice were stimulated with TAP-1−/− (H2b) splenocytes in the presence of an H2 KbL under limiting dilution conditions. HTG animals express the Kd allele with 35 polymorphic amino acids within the α1 and α2 domains, whereas the Kbm1 allele carries merely 3 amino acid exchanges, both compared with Kb (Table ). After one restimulation, the cultures were tested against TAP− RMA.S cells (H2b) in the presence or absence of KbL. H2 expression was not increased under our experimental conditions (data not shown). The library is a strong antigen for B6 responders, as it contains numerous nonself-peptides (Fig. 1, top right). Previously, we have shown that such CTLs can be of high avidity towards the particular peptide or peptides recognized 27. For the allogeneic responders, it turned out that the HTG splenocytes generated less allorestricted, i.e., Kb-restricted, and peptide-specific/dependent cultures (ratio of peptide-specific [P] to structure-specific [S] cultures 1:4) than bm1 splenocytes (P/S = 1:1; Fig. 1, top). These results suggest that the bm1 repertoire is more prone to react in a peptide-dependent way towards Kb than the HTG repertoire. Out of the 35 amino acids polymorphic between Kd and Kb, 13 make contact with the bound peptide 32 33, whereas 2 (residues 79 and 155) are bound by the 2C TCR directly 34. However, one of the three amino acids that are polymorphic between Kb and Kbm1 contacts the 2C TCR directly (residue 155; Table ). It is tempting to speculate that this residue is responsible for the finding that about half of the cultures recognize RMA.S cells in the absence of KbL. These data show that allogeneic MHC molecules, which carry amino acid exchanges at positions that are responsible for peptide binding, preferentially elicit an alloresponse against the bound peptides, as one would expect from considering the structure of the MHC–peptide complex.
Table 2.
| Mutation | Position | Exchanges | Pocket (peptide res.) | TCR interaction |
|---|---|---|---|---|
| Kbm1 | 152 | Glu→Ala | D (3), C (6), E (7) | – |
| 155 | Arg→Tyr | D (3), C (6), E (7) | CDR1α, 2α, 3β | |
| 156 | Leu→Tyr | D (3), C (6), E (7) | – | |
| Dbm13 | 114 | Leu→Gln | D (3), C (6), E (7), F (9/10) | – |
| 116 | Phe→Tyr | C (6), E (7), F (9/10) | – | |
| 119 | Glu→Asp | |||
| Dbm14 | 70 | Gln→His | B (2), D (3), C (6), F (9/10) | – |
Figure 1.
The responder H2 haplotype influences the role of peptide libraries in CTL alloreactivity. Splenocytes of the given strains were plated in limiting dilution and stimulated with irradiated TAP-1−/− (H2b) splenocytes in the presence of KbL (top) or DbL (bottom). The 96 cultures were assayed for CTL activity against RMA.S cells 51Cr labeled in the absence or presence of the respective library. Numbers of cultures within the arbitrarily chosen regions are indicated to allow for better comparison. Spontaneous release was <10%. The experiments were repeated with similar results.
None of the polymorphic amino acids of H2-Dbm13 and H2-Dbm14 (compared with Db) are directly involved in TCR contacts (Table ). Consequently, after stimulation of splenocytes with TAP-1−/− cells and a DbL, the majority of cytotoxicity was directed towards DbL-coated RMA.S cells (Fig. 1, bottom). Amino acid 70, which is exchanged in Dbm14, is positioned at the inner rim of the α1 helix and seems to cause some structural change that is detected by several cultures reacting to both DbL-coated and untreated RMA.S cells (P/S = 5:1). The contribution of cultures specific for structural elements to overall alloreactivity is even further reduced in the case of bm13 responders, which carry polymorphic residues primarily determining the chemical environment of the binding groove's F pocket (P/S = 10:1). Our data indicate that increasing similarity between α-helical residues of the responder's and the stimulator's MHC increases the contribution of peptide-specific or -dependent T cells to an alloresponse.
Differential Requirement for Natural Peptides in Alloreactivity.
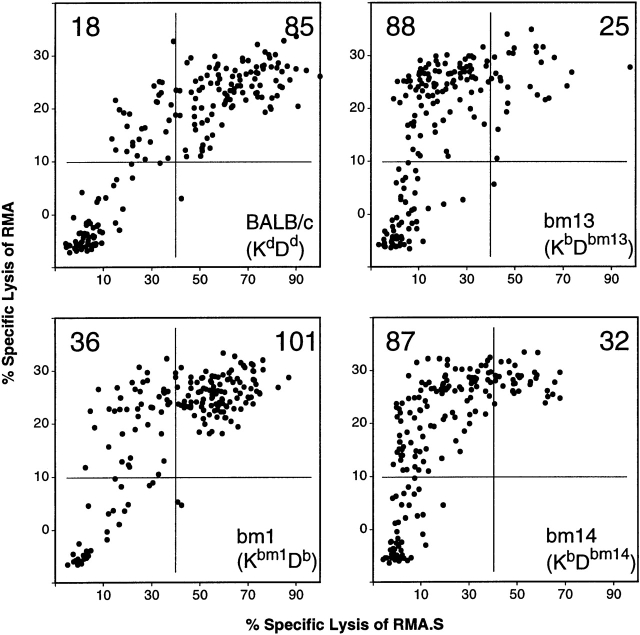
In the experiments described above, we made use of peptide libraries of limited complexity to replace the more diverse set of self-peptides on TAP+ cells. Under our loading conditions (1 h of peptide incubation at 37°C), no increase in the expression of H2 molecules on RMA.S cells could be detected. This was true both for incubation with the peptide library and with individual peptides (data not shown). We wondered whether the above observations also apply to alloreactivity against cells expressing TAP, the wild-type complement of peptides and, therefore, the natural density of class I molecules on the cell surface. BALB/c, bm1, bm13, and bm14 splenocytes were stimulated with B6 cells under limiting dilution conditions and tested against RMA versus RMA.S target cells. In general, a higher proportion of microcultures reacting with the foreign MHC was detected irrespective of TAP expression (Fig. 2). This result suggests that TAP+ cells could stimulate more T cells specific for structural elements of Kb present on both TAP+ and TAP− cells, because they express a higher density of H2 molecules on the cell surface. But again, bm1 responds with more peptide-specific cultures (P/S = 1:3) than the completely allogeneic responder BALB/c (P/S = 1:5), and bm13 and bm14 respond with even more (P/S = 3:1) than bm1 (Fig. 2). These results extend our conclusion that the particular responder–stimulator combination determines the role of peptides to alloresponses among wild-type stimulators and targets.
Figure 2.
The responder H2 haplotype influences the role of endogenous peptides in CTL alloreactivity. Splenocytes of the given strains were stimulated under limiting dilution conditions with irradiated B6 splenocytes. 192 cultures were assayed against RMA and RMA.S. Numbers of cultures within the arbitrarily chosen areas are indicated to allow for better comparison. Spontaneous release was <21%.
Allorestricted CTLs against Defined Self-Peptides.
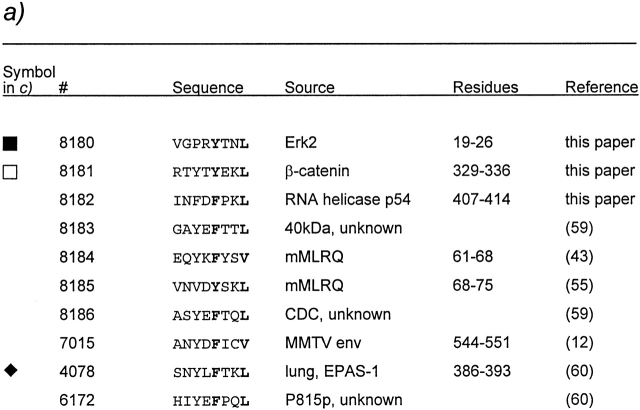
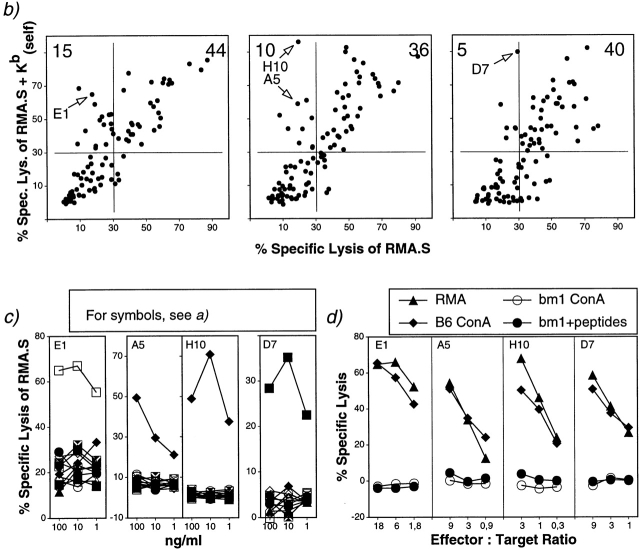
To further characterize alloreactive CTLs raised from a responder expressing an H2 molecule closely related to the stimulating one, we cultured bm1 splenocytes with irradiated TAP-1−/− cells in the presence of 10 self-peptides eluted from Kb (Kb [self]) by us and other laboratories (Fig. 3, a and b). Peptide-specific microcultures were expanded, and the specific peptide recognized was determined. We detected reactivities against determinants from the transcription factor Erk2, the cytosolic protein β-catenin, and the endothelial pas domain protein (EPAS-1; Fig. 3 c) as well as from RNA helicase p54 and the mouse mammary tumor virus envelope protein (data not shown). The specificity of the “aberrant” CTL lines recognizing RMA-S in the absence but not presence of added peptides (see Fig. 3 b, right) could not be elucidated. Importantly, all peptide-specific CTL lines recognized RMA tumor cells as well as B6 Con A blasts, but not bm1 Con A blasts, not even in the presence of peptides (Fig. 3 d). These results demonstrate that allorestricted and peptide-specific CTLs can be generated easily without depletion or extensive cloning from a related mouse strain. The lines that were raised against synthetic self-peptides can efficiently lyse target cells expressing physiological levels of the self-antigen and thereby circumvent peptide-specific self-tolerance.
Figure 3.
Allorestricted CTL lines from bm1 mice against Kb binding self-peptides. (a) Self-peptides eluted from Kb and used in this study as mixture Kb (self) (references 12, 43, 55, 59, 60; and this study). (b) Splenocytes from bm1 animals were plated in limiting dilution and stimulated with TAP-1−/− cells in the presence of a mixture of self-peptides at 500 (left), 100 (middle), and 50 ng/ml (right). After 10 d, the cultures were assayed against RMA.S in the absence or presence of the mixture. Spontaneous release was <10%. (c) Peptide specificity of expanded CTL lines. All peptides listed in a were titrated on 51Cr-labeled RMA.S cells. The indicated effector cells were added at effector to target ratios between 2 and 5. (d) The expanded CTL lines recognize tumor cells and Con A–induced blasts from B6, but not from bm1 mice. Spontaneous release was <10% for tumor cells and <18% for blasts.
Allorestricted CTLs against Defined Viral Peptides.
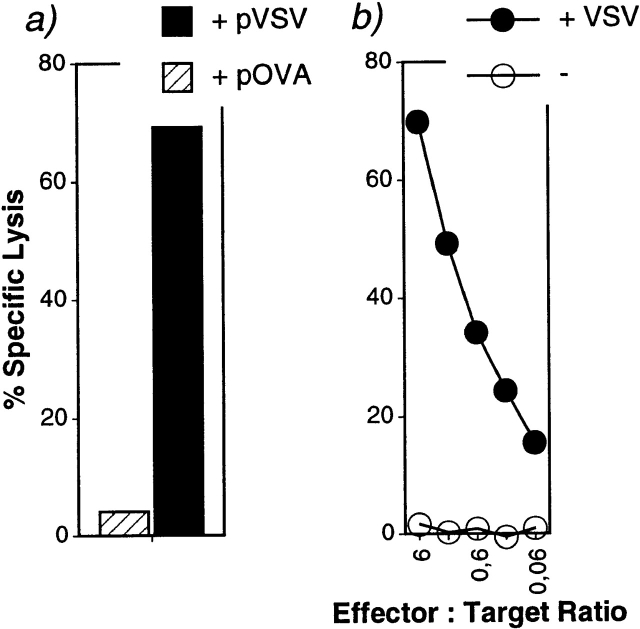
To demonstrate more clearly the exclusive specificity of allorestricted CTLs from a related strain, a line was generated from bm1 against Kb and the immunodominant peptide from VSV (pVSV). The CTL line B6E10 was specific for pVSV, since it did not lyse RMA.S cells coated with the OVA257–264 peptide (Fig. 4 a) or any other of 38 Kb binding peptides tested (data not shown). The line also does not cross-react with self-peptides presented by Kb, since the TAP+ cell line EL4 was not recognized. In contrast, however, EL4 cells infected with VSV were lysed efficiently (Fig. 4 b). Thus we show, to our knowledge for the first time, that allorestricted and highly specific CTLs can be generated in vitro with synthetic peptides to lyse virus-infected targets.
Figure 4.
Allorestricted CTLs can recognize VSV-infected cells. (a) The bm1-derived CTL line B6E10 raised against TAP-1−/− and the VSV-N52-59 peptide (pVSV) lyses RMA.S cells after incubation with the stimulating peptide, but not with the OVA257–264 peptide (pOVA). Effector to target ratio was 2. (b) The same line recognizes EL4 cells after infection with 2 × 107 PFU VSV only. Spontaneous release was <20%.
Discussion
We have previously shown that Kb-restricted and peptide-specific CTLs can be detected among BALB/c splenocytes. Such cells were very rare and were obscured by many alloreactive cells recognizing the target cells in a peptide-independent manner. We had to deplete such cells in order to generate several peptide-specific CTL lines 27. Similar protocols have been successfully used to deplete graft-versus-host activity without compromising the graft-versus-leukemia effect of allogeneic PBLs in bone marrow transplantation 35 36 37. However, since we (data not shown) and others 38 had difficulty generating allorestricted CTLs against individual peptides when CTLs and target cells were completely allogeneic to each other, we wished to analyze the spectrum of CTL precursors specific for allogeneic MHC–peptide complexes in closely related versus unrelated MHC combinations. Our results show that an alloresponse against a related MHC molecule contains more peptide-specific T cells than a response against an unrelated one: alloreactions against an MHC molecule carrying groove mutations only (bm13 anti-B6, bm14 anti-B6) were clearly dominated by peptide-specific cells compared with a response against a molecule with both groove and α-helical replacements (bm1 anti-B6). Among responses against molecules with several groove and α-helical exchanges, peptide-specific cultures were further reduced (HTG anti-B6, 5R anti-B6, BALB/c anti-B6; Fig. 1 and Fig. 2). In addition, Kb-restricted CTLs from bm1 against self- as well as viral peptides showed high specificity and efficiently recognized naturally presented peptides (Fig. 3 and Fig. 4). We take these findings as evidence for an increased contribution of molecular mimicry due to partial identity to the alloresponse against similar MHC molecules, as first suggested by Lechler et al. 39: the self haplotype determines whether an alloresponse is dominated by cells recognizing nonself via multiple binary interactions or rather by cells seeing the allelic MHC molecules regardless of bound peptides. The mode of allorecognition, however, seems unrelated to the “antigenic strength,” as skin grafts done in the mutant strain combinations used in our study are rejected in a similar time frame 40 41.
The importance of the peptide-specific mode of alloreaction was not obvious from structural information published to date. Studies on molecules recognized by the 2C allo-TCR 4 34 42 43 and the xenoreactive clone AHIII12.2 5 44 do not explain the clones' cross-reactivities by structural molecular mimicry. For 2C, only a critical negative charge on the self-MHC–peptide complex has been mimicked by the allogeneic ligand, as suggested in reference 4. We speculate that T cells responding to foreign MHC molecules carrying groove mutations interact with them in the same way as T cells interacting with their respective self-molecules. Such alloreactive T cells respond to the bound peptides, as these peptides had not been present during negative selection.
The T cell repertoire is shaped by selection events in the thymus and the periphery 45 46 47 48. It is currently assumed that a certain MHC haplotype selects 15–20% of CD4+ CD8+ thymocytes 49, of which 50% or more undergo clonal deletion by bone marrow–derived APCs 50 51. Despite this profound imprint of the self-MHC on the T cell repertoire, an influence of selection events on peptide requirements of alloreactivity has not been shown. Our detailed analysis of alloreactive cells on a population level now shows that allorecognition is readily influenced by the selecting MHC molecule—rather than “hard-wired into the TCR genes” 45 52—and mirrors the resemblance between self and foreign.
It is not clear whether negative or positive selection, peripheral survival, or all three are skewing the repertoire towards peptide-specific/dependent recognition of related MHC molecules. Favoring negative selection, one could argue that, for example, the bm13 repertoire, due to its similarity to B6, is simply purged of T cells reactive against structural determinants of Db and contains peptide-specific cells only. However, negative selection may not completely explain the skewing, since the antigenic strength of the pocket mutants bm13 and bm14 is almost as high as that of bm1, which carries a polymorphic amino acid at a position in direct contact with the TCR 40 41. It is tempting to speculate that positive selection and peripheral survival enrich for cells able to recognize similar MHC molecules in a peptide-specific way and thereby modify the alloresponse as well.
The fact that tolerance is MHC restricted 20 23 24 25 permits the generation of high avidity CTLs against MHC–self-peptide complexes for adoptive tumor therapy in three ways. First, a responder's T cell can react towards a peptide from its own MHC groove if it is presented in a new context (same groove, new α helices; references 3, 53, 54). Second, CTLs have been raised against peptides presented by allogeneic MHC molecules carrying several α-helical and groove mutations (new groove, new helices; references 21, 27). Third, alloreactive CTLs can recognize an MHC molecule with groove mutations only because of the many new peptides it carries, as in a self-restricted response (new groove, same helices [reference 55, and this study]). We show here that the latter possibility allows for the easy generation of allorestricted CTLs against defined self- and viral peptides. We therefore suggest that this approach is more likely to yield high avidity human CTLs against self- and other peptides, including tumor-associated antigens. For example, to raise HLA-A*0201–restricted CTLs, it may be useful to start with T cells from a donor expressing A*0206 (1 mutation in pocket B) rather than from a donor expressing, e.g., A*0301 (3 pocket mutations and 11 exchanges outside the groove). In fact, 9 out of 22 HLA-A2 subtypes carry exclusively pocket mutations relative to A*0201 56. Which of these subtypes is best suited to select T cells that respond to A*0201-bound self-peptides remains to be determined.
Taken together, we have shown that self-MHC shapes the repertoire not only of self-restricted, but also of alloreactive T cells. A practical consequence of our data bears on the generation of allorestricted, peptide-specific T cells, especially for those directed against tumor-associated peptides: the success rate for getting such T cells should be higher if T cell donor and stimulating APCs express MHC molecules with similar α helices but different peptide binding grooves.
Acknowledgments
We thank Drs. M. van Roon, H. ter Rile, A. Berns, R. Brandt, and C.J.M. Melief for mice, Dr. R. Zawatzky for virus aliquots, Patricia Hrstic for expert technical assistance, and Drs. L. Antón, P. Blader, M. Correia-Neves, N. Martin-Orozco, D. Mathis, C. Münz, S. Rojo, and H.M. van Santen for discussion and comments on the manuscript.
This work was supported by grants from the Deutsche For-schungsgemeinschaft (Leibnizprogramm, Ra 369/4-1), the European Union (Biomed CT 95-1627), the Deutsche Krebshilfe (10-1258-St1), and Merck KGaA.
Footnotes
R. Obst's present address is Joslin Diabetes Center, One Joslin Place, Boston, MA 02215. E-mail: reinhard.obst@joslin.harvard.edu
Abbreviations used in this paper: B6, C57BL/6; KbL or DbL, Kb or Db binding peptide library, respectively; Kb (self), mixture of 10 Kb binding self-peptides; TAP, transporter associated with antigen processing; VSV, vesicular stomatitis virus.
References
- Klein J. Natural History of the Major Histocompatibility Complex 1986. John Wiley & Sons, Inc; New York: pp. 291–422 [Google Scholar]
- Sherman L.A., Chattopadhyay S. The molecular basis of allorecognition. Annu. Rev. Immunol. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- Rötzschke O., Falk K., Faath S., Rammensee H.-G. On the nature of peptides involved in T cell alloreactivity. J. Exp. Med. 1991;174:1059–1071. doi: 10.1084/jem.174.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir J.A., Garcia K.C., Brunmark A., Degano M., Peterson P.A., Teyton L., Wilson I.A. Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes. Immunity. 1998;8:553–562. doi: 10.1016/s1074-7613(00)80560-9. [DOI] [PubMed] [Google Scholar]
- Zhao R., Loftus D.J., Appella E., Collins E.J. Structural evidence of T cell xeno-reactivity in the absence of molecular mimicry. J. Exp. Med. 1999;189:359–370. doi: 10.1084/jem.189.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P., Bevan M.J. Hypothesiswhy do so many lymphocytes respond to major histocompatibility antigens? Cell. Immunol. 1977;29:1–5. doi: 10.1016/0008-8749(77)90269-6. [DOI] [PubMed] [Google Scholar]
- Kaye J., Janeway C.A., Jr. The Fab fragment of a directly activating monoclonal antibody that precipitates a disulfide-linked heterodimer from a helper T cell clone blocks activation by either allogeneic Ia or antigen and self-Ia. J. Exp. Med. 1984;159:1397–1412. doi: 10.1084/jem.159.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M.J. High determinant density may explain the phenomenon of alloreactivity. Immunol. Today. 1984;5:128–129. doi: 10.1016/0167-5699(84)90233-0. [DOI] [PubMed] [Google Scholar]
- Panina-Bordignon P., Corradin G., Roosnek E., Sette A., Lanzavecchia A. Recognition by class II alloreactive T cells of processed determinants from human serum proteins. Science. 1991;252:1548–1550. doi: 10.1126/science.1710827. [DOI] [PubMed] [Google Scholar]
- Eisen H.N., Sykulev Y., Tsomides T.J. Antigen-specific T-cell receptors and their reactions with complexes formed by peptides with major histocompatibility complex proteins. Adv. Protein Chem. 1996;49:1–56. doi: 10.1016/s0065-3233(08)60487-8. [DOI] [PubMed] [Google Scholar]
- Malarkannan S., Gonzalez F., Nguyen V., Adair G., Shastri N. Alloreactive CD8+ T cells can recognize unusual, rare, and unique processed peptide/MHC complexes. J. Immunol. 1996;157:4464–4473. [PubMed] [Google Scholar]
- Malarkannan S., Serwold T., Nguyen V., Sherman L.A., Shastri N. The mouse mammary tumor virus env gene is the source of a CD8+ T-cell-stimulating peptide presented by a major histocompatibility complex class I molecule in a murine thymoma. Proc. Natl. Acad. Sci. USA. 1996;93:13991–13996. doi: 10.1073/pnas.93.24.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Gulden P.H., Pierce R.A., Shabanowitz J., Man S.T., Hunt D.F., Engelhard V.H., Shabanowitz J.A. A naturally processed peptide presented by HLA-A*0201 is expressed at low abundance and recognized by an alloreactive CD8+ cytotoxic T cell with apparent high affinity. J. Immunol. 1997;158:5797–5804. [PubMed] [Google Scholar]
- Elliott T.J., Eisen H.N. Cytotoxic T lymphocytes recognize a reconstituted class I histocompatibility antigen (HLA-A2) as an allogeneic target molecule. Proc. Natl. Acad. Sci. USA. 1990;87:5213–5217. doi: 10.1073/pnas.87.13.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.A., Brunmark A., Jackson M., Potter T.A. Peptide-independent recognition by alloreactive T lymphocytes. J. Exp. Med. 1997;185:1023–1034. doi: 10.1084/jem.185.6.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L.A. Recognition of conformational determinants on H-2 by cytolytic T lymphocytes. Nature. 1982;297:511–513. doi: 10.1038/297511a0. [DOI] [PubMed] [Google Scholar]
- Bluestone J.A., Kaliyaperumal A., Jameson S., Miller S., Dick R., II. Peptide-induced changes in class I heavy chains alter allorecognition. J. Immunol. 1993;151:3943–3953. [PubMed] [Google Scholar]
- Chattopadhyay S., Theobald M., Biggs J., Sherman L.A. Conformational differences in major histocompatibility complex–peptide complexes can result in alloreactivity. J. Exp. Med. 1994;179:213–219. doi: 10.1084/jem.179.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C., Horvath S., Allen P.M. A basis for alloreactivityMHC helical residues broaden peptide recognition by the TCR. Immunity. 1998;8:543–552. doi: 10.1016/s1074-7613(00)80559-2. [DOI] [PubMed] [Google Scholar]
- Rammensee H.-G., Bevan M.J. Evidence from in vitro studies that tolerance to self antigens is MHC-restricted. Nature. 1984;308:741–744. doi: 10.1038/308741a0. [DOI] [PubMed] [Google Scholar]
- Sadovnikova E., Stauss H.J. Peptide-specific cytotoxic T lymphocytes restricted by nonself major histocompatibility complex class I moleculesreagents for tumor immunotherapy. Proc. Natl. Acad. Sci. USA. 1996;93:13114–13118. doi: 10.1073/pnas.93.23.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauss H.J. Immunotherapy with CTLs restricted by nonself MHC. Immunol. Today. 1999;20:180–183. doi: 10.1016/s0167-5699(99)01443-7. [DOI] [PubMed] [Google Scholar]
- Dos Reis G.A., Shevach E.M. Antigen-presenting cells from nonresponder strain 2 guinea pigs are fully competent to present bovine insulin B chain to responder strain 13 T cells. Evidence against a determinant selection model and in favor of a clonal deletion model of immune response gene function. J. Exp. Med. 1983;157:1287–1299. doi: 10.1084/jem.157.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves E.S., Singer A. Role of the H-2 complex in the induction of T cell tolerance to self minor histocompatibility antigens. J. Exp. Med. 1983;158:1483–1497. doi: 10.1084/jem.158.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P., Zamoyska R., Waldmann H. Self tolerance is H-2-restricted. Nature. 1984;308:738–741. doi: 10.1038/308738a0. [DOI] [PubMed] [Google Scholar]
- Sadovnikova E., Jopling L.A., Soo K.S., Stauss H.J. Generation of human tumor-reactive cytotoxic T cells against peptides presented by non-self HLA class I molecules. Eur. J. Immunol. 1998;28:193–200. doi: 10.1002/(SICI)1521-4141(199801)28:01<193::AID-IMMU193>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Obst R., Münz C., Stevanovic S., Rammensee H.-G. Allo- and self-restricted cytotoxic T lymphocytes against a peptide libraryevidence for a functionally diverse allorestricted T cell repertoire. Eur. J. Immunol. 1998;28:2432–2443. doi: 10.1002/(SICI)1521-4141(199808)28:08<2432::AID-IMMU2432>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Münz C., Obst R., Osen W., Stevanovic S., Rammensee H.-G. Alloreactivity as a source of high avidity peptide-specific human CTL. J. Immunol. 1999;162:25–34. [PubMed] [Google Scholar]
- Matzinger P. Why positive selection? Immunol. Rev. 1993;135:81–117. doi: 10.1111/j.1600-065x.1993.tb00645.x. [DOI] [PubMed] [Google Scholar]
- Tourne S., van Santen H.M., van Roon M., Berns A., Benoist C., Mathis D., Ploegh H. Biosynthesis of major histocompatibility complex molecules and generation of T cells in Ii TAP1 double-mutant mice. Proc. Natl. Acad. Sci. USA. 1996;93:1464–1469. doi: 10.1073/pnas.93.4.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanovic S., Jung G., Rammensee H.-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Fremont D.H., Peterson P.A., Wilson I.A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- Chelvanayagam G. A roadmap for HLA-A, HLA-B, and HLA-C peptide binding specificities. Immunogenetics. 1996;45:15–26. doi: 10.1007/s002510050162. [DOI] [PubMed] [Google Scholar]
- Garcia K.C., Degano M., Pease L.R., Huang M., Peterson P.A., Teyton L., Wilson I.A. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- Garderet L., Snell V., Przepiorka D., Schenk T., Lu J.G., Marini F., Gluckman E., Andreeff M., Champlin R.E. Effective depletion of alloreactive lymphocytes from peripheral blood mononuclear cell preparations. Transplantation. 1999;67:124–130. doi: 10.1097/00007890-199901150-00021. [DOI] [PubMed] [Google Scholar]
- Harris D.T., Sakiestewa D., Lyons C., Kreitman R.J., Pastan I. Prevention of graft-versus-host disease (GVHD) by elimination of recipient-reactive donor T cells with recombinant toxins that target the interleukin 2 (IL-2) receptor. Bone Marrow Transplant. 1999;23:137–144. doi: 10.1038/sj.bmt.1701535. [DOI] [PubMed] [Google Scholar]
- Montagna D., Yvon E., Calcaterra V., Comoli P., Locatelli F., Maccario R., Fisher A., Cavazzana-Calvo M. Depletion of alloreactive T cells by a specific anti-interleukin-2 receptor p55 chain immunotoxin does not impair in vitro antileukemia and antiviral activity. Blood. 1999;93:3550–3557. [PubMed] [Google Scholar]
- Frelinger J.A., McMillan M. The role of peptide specificity in MHC class I-restricted allogeneic responses. Immunol. Rev. 1996;154:45–58. doi: 10.1111/j.1600-065x.1996.tb00929.x. [DOI] [PubMed] [Google Scholar]
- Lechler R.I., Lombardi G., Batchelor J.R., Reinsmoen N., Bach F.H. The molecular basis of alloreactivity. Immunol. Today. 1990;11:83–88. doi: 10.1016/0167-5699(90)90033-6. [DOI] [PubMed] [Google Scholar]
- Morgan G.M., Dellos H., McKenzie I.F., Melvold R.W., Bailey D.W. Studies of two H-2Db mutantsB6.C-H-2bm13 and B6.C-H-2bm14 . Immunogenetics. 1980;11:341–349. doi: 10.1007/BF01567801. [DOI] [PubMed] [Google Scholar]
- Rosenberg A.S., Mizuochi T., Singer A. Analysis of T-cell subsets in rejection of Kb mutant skin allografts differing at class I MHC. Nature. 1986;322:829–831. doi: 10.1038/322829a0. [DOI] [PubMed] [Google Scholar]
- Brock R., Wiesmüller K.-H., Jung G., Walden P. Molecular basis for the recognition of two structurally different major histocompatibility complex/peptide complexes by a single T-cell receptor. Proc. Natl. Acad. Sci. USA. 1996;93:13108–13113. doi: 10.1073/pnas.93.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist M.D., Weaver A.J., Pease L.R. Degenerate recognition of alloantigenic peptides on a positive-selecting class I molecule. J. Immunol. 1998;160:802–809. [PubMed] [Google Scholar]
- Loftus D.J., Chen Y., Covell D.G., Engelhard V.H., Appella E. Differential contact of disparate class I/peptide complexes as the basis for epitope cross-recognition by a single T cell receptor. J. Immunol. 1997;158:3651–3658. [PubMed] [Google Scholar]
- Jameson S.C., Bevan M.J. T-cell selection. Curr. Opin. Immunol. 1998;10:214–219. doi: 10.1016/s0952-7915(98)80251-3. [DOI] [PubMed] [Google Scholar]
- Ernst B., Lee D.S., Chang J.M., Sprent J., Surh C.D. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 1999;11:173–181. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- Goldrath A.W., Bevan M.J. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity. 1999;11:183–190. doi: 10.1016/s1074-7613(00)80093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel R.M., Althage A. On the role of thymic epithelium vs. bone marrow-derived cells in repertoire selection of T cells. Proc. Natl. Acad. Sci. USA. 1999;96:8092–8097. doi: 10.1073/pnas.96.14.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., Graf D., Lovatt M., Bommhardt U., Zamoyska R., Fisher A.G. How many thymocytes audition for selection? J. Exp. Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meerwijk J.P., Marguerat S., Lees R.K., Germain R.N., Fowlkes B.J., MacDonald H.R. Quantitative impact of thymic clonal deletion on the T cell repertoire. J. Exp. Med. 1997;185:377–383. doi: 10.1084/jem.185.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M.J. In thymic selection, peptide diversity gives and takes away. Immunity. 1997;7:175–178. doi: 10.1016/s1074-7613(00)80520-8. [DOI] [PubMed] [Google Scholar]
- Jerne N.K. The somatic generation of immune recognition. Eur. J. Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- Grandea A.G., III, Bevan M.J. Single-residue changes in class I major histocompatibility complex molecules stimulate responses to self peptides. Proc. Natl. Acad. Sci. USA. 1992;89:2794–2798. doi: 10.1073/pnas.89.7.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzushima K., Sun R., van Bleek G.M., Vegh Z., Nathenson S.G. The role of self peptides in the allogeneic cross-reactivity of CTLs. J. Immunol. 1995;155:594–601. [PubMed] [Google Scholar]
- Hogquist K.A., Tomlinson A.J., Kieper W.C., McGargill M.A., Hart M.C., Naylor S., Jameson S.C. Identification of a naturally occurring ligand for thymic positive selection. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- Rammensee H.-G., Bachmann J., Stevanovic S. MHC Ligands and Peptide Motifs 1997. Landes; Austin, TX: pp. 462 [Google Scholar]
- Nathenson S.G., Geliebter J., Pfaffenbach G.M., Zeff R.A. Murine major histocompatibility complex class I mutantsmolecular analysis and structure-function implications. Annu. Rev. Immunol. 1986;4:471–502. doi: 10.1146/annurev.iy.04.040186.002351. [DOI] [PubMed] [Google Scholar]
- Hemmi S., Geliebter J., Zeff R.A., Melvold R.W., Nathenson S.G. Three spontaneous H-2Db mutants are generated by genetic micro-recombination (gene conversion) events. Impact on the H-2–restricted immune responsiveness. J. Exp. Med. 1988;168:2319–2335. doi: 10.1084/jem.168.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., Carbone F.R. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Rammensee H.-G., Bachmann J., Emmerich N.P.N., Bachor O.A., Stevanovic S. SYFPEITHIdatabase for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]