Abstract
The goal of this study was to develop a new surrogate challenge model for use in evaluating protective cell-mediated immune responses against hepatitis C virus (HCV) antigens. The use of recombinant Listeria monocytogenes organisms which express HCV antigens provides novel tools with which to assay such in vivo protection, as expression of immunity against this hepatotropic bacterial pathogen is dependent on antigen-specific CD8+ T lymphocytes. A plasmid DNA vaccine encoding a ubiquitin-NS3 fusion protein was generated, and its efficacy was confirmed by in vivo induction of NS3-specific, gamma interferon-secreting T cells following vaccination of BALB/c mice. These immunized mice also exhibited specific in vivo protection against subsequent challenge with a recombinant L. monocytogenes strain (TC-LNS3) expressing the NS3 protein. Notably, sublethal infection of naive mice with strain TC-LNS3 induced similar NS3-specific T-cell responses. These findings suggest that recombinant strains of L. monocytogenes expressing HCV antigens should prove useful for evaluating, or even inducing, protective immune responses against HCV antigens.
Infection with hepatitis C virus (HCV) represents a major disease problem in the United States and worldwide. As of 1998, approximately 3.9 million people in the United States were infected with HCV, and it has been estimated that over 40% of chronic liver disease cases in the country are HCV related, resulting in 8,000 to 10,000 deaths annually (1). Clearly, there is a need to develop effective new therapies and vaccines for treating and preventing this viral disease, and the development of an effective HCV vaccine will require a better understanding of the protective immune response to this viral infection.
Progress in characterizing the protective immune response to HCV infection has been slow, reflecting in part the limited host range (i.e., humans and chimpanzees) for this virus. Nevertheless, the importance of a protective cell-mediated immune response in preventing viral disease has been suggested by several studies. For example, antigen-specific CD4+ and CD8+ T-cell responses against HCV antigens were observed in the absence of an apparent humoral response or detectable viremia in individuals who did not develop chronic HCV disease following likely occupational exposure (29), suggesting that a vigorous T-cell response can lead to clearance of HCV infection. Memory CD8+ T-cell responses to HCV structural and nonstructural proteins have also been detected from healthy family members of chronically infected HCV patients (40), indicating that the HCV-specific CD8+ T-cell response may protect these exposed family members. In addition, it has been proposed that a strong cytotoxic T lymphocyte (CTL) response and adequate CD4+ T-cell help, especially Th1 T lymphocytes, are important for resolution of HCV infection (13, 18, 34). Furthermore, analyses of the acute phase of viral infection in both chimpanzees (11) and humans (14, 31, 43) suggest that a broad and vigorous early CD8+ CTL response is necessary for successful clearance of the viral infection. Clinical HCV studies have also indicated that the Th1 CD4+ T-cell response is essential for sustaining the effector function of the protective antiviral CD8+ T cells (27). In summary, these findings suggest that any successful vaccine against HCV infection must generate a strong multispecific CD4+ and CD8+ cellular response.
One potential vaccine strategy that satisfies the requirement for CD4+ and CD8+ T-cell stimulation is genetic immunization. Induction of T-cell responses by plasmid DNA vaccines has been demonstrated in a variety of experimental systems, including that of injecting mice with plasmid DNA constructs that encode different HCV structural and nonstructural proteins (9, 16, 19, 30, 39, 44). Although the in vitro and ex vivo analyses of the induced immune responses have been promising, the lack of a suitable small-animal model for HCV prevents the evaluation of the in vivo efficacies of these experimental DNA vaccines.
To partially address the need for additional surrogate small-animal models for this viral disease, we initiated studies which use recombinant Listeria monocytogenes bacteria that express HCV antigens. Several features of the murine model of listeriosis led us to believe that this would be an informative system in which to generate and evaluate immune responses to HCV antigens. First, L. monocytogenes is an intracellular pathogen that has been shown to infect and replicate within hepatocytes both in vitro and in vivo (20, 26, 37). Second, numerous studies have demonstrated that a strong antigen-specific CD8+ T-cell response is required for expression of protective immunity to L. monocytogenes (4, 33). Third, these antigen-specific CD8+ effector cells can specifically recognize L. monocytogenes-infected hepatocytes (7, 22, 26). And finally, recombinant L. monocytogenes strains have been used previously to express foreign antigens in other experimental viral disease models, including those for lymphocytic choriomeningitis virus (41), influenza virus (24), and human immunodeficiency virus (17). Therefore, we reasoned that infection of experimental mice with recombinant L. monocytogenes expressing HCV antigens would provide a useful animal model for in vivo expression of protective immunity to specific HCV proteins. In these studies, we focused on generating responses to the HCV NS3 protein, as previous data have indicated that NS3-specific T-cell responses are correlated with spontaneous resolution of infection and low viral load (38).
To establish this model, we first generated plasmid DNA vaccines that encoded HCV core or NS3 proteins that would induce an antigen-specific cell-mediated immune response in the mouse. To enhance the T-cell responses to the HCV antigens in this model, we used vectors that express HCV antigens as ubiquitin fusion proteins. Previous studies have shown that immunization with DNA vaccine vectors encoding ubiquitin fusion proteins favors enhanced CTL responses and decreased antibody responses (15, 36, 42, 46). We also produced a recombinant strain of L. monocytogenes that expresses the HCV NS3 protein. These recombinant tools were then used to induce and evaluate NS3-specific immune responses in experimental mice, as indicated by in vitro ELISPOT assays and in vivo protection assays. In our initial findings reported here, we demonstrate that immunization of BALB/c mice with plasmid DNA encoding a ubiquitin-NS3 fusion protein induced NS3-specific CD8+ T-cell responses that provided specific protection against recombinant L. monocytogenes expressing the NS3 protein.
MATERIALS AND METHODS
Plasmid DNA constructs.
Based on genomic sequence data from HCV serotype 1a (GenBank accession number M16321), specific oligonucleotide primers were generated, and regions encoding the core and NS3 proteins (amino acids [aa] 1 to 177 and 1027 to 1656, respectively [relative to the full-length HCV 1a polyprotein]) were amplified by PCR from a cDNA template and were then inserted into the pCR2.1-TOPO cloning vector (Invitrogen Life Technologies Inc., Carlsbad, Calif.). The core and NS3 genes were then inserted separately into the pCMVi(-H3)Ubs plasmid (kindly provided by S. Johnston and K. Sykes, University of Texas Southwestern Medical Center, Dallas) by using standard cloning techniques (3). The resulting plasmids, pUb-core and pUb-NS3, were used for DNA immunizations. Plasmid DNA used for injection was prepared from Escherichia coli transformants by using EndoFree plasmid Giga purification kits (Qiagen Inc., Chatsworth, Calif.) according to the manufacturer’s instructions. The NS3 insert of our specific PCR clone was fully sequenced (VA Medical Center Molecular Biology Core Lab, Portland, Oreg.) and differed somewhat from the published sequence, resulting in what we deduced to be several amino acid changes (Fig. 1).
FIG. 1.
Deduced amino acid sequence of the NS3 fragment of pUb-NS3 and the recombinant L. monocytogenes strain TC-LNS3. The NS3 inserts of pUb-NS3 and TC-LNS3 were fully sequenced (VA Medical Center Molecular Biology Core Lab) and differed somewhat from the published sequence (GenBank accession number M62321), resulting in what were deduced to be several amino acid changes. Shaded boxes indicate sequence differences between pUb-NS3, TC-LNS3, and M62321 (which was used as the reference for synthesis of the 15-mer peptides). Underlined text indicates the positions of peptides NS31407-1421 and NS31535-1549. Note that aa 1632 to 1656 are not expressed from pUb-NS3 or by TC-LNS3 due to the inadvertent introduction of a stop codon (*) at position 1632. Also, aa 1616 to 1656 are not represented in the synthetic peptide pools.
Production and characterization of recombinant L. monocytogenes.
The recombinant L. monocytogenes strain TC-LNS3 was created essentially as described previously by Shen et al. (41). First, the NS3 coding sequence was fused in frame to the promoter and signal sequence of the listeriolysin (LLO) gene in plasmid pEJ140 (kindly provided by J. Miller, University of California, Los Angeles), thus creating an antigen expression cassette that will direct expression and secretion of the fusion protein in L. monocytogenes as driven by the LLO promoter. This antigen expression cassette, which also carries the aphA3 gene conferring kanamycin resistance, was then spliced into the NotI site of the homologous recombination vector pLMD3 (J. Miller) to create pLMD3-NS3. In addition to a 5.6-kb fragment of L. monocytogenes genomic DNA, pLMD3 also carries a temperature-sensitive origin of replication which allows for conditional replication in gram-positive bacteria. After selecting for bacteria that had undergone allelic exchange (41), putative recombinants were screened by PCR and Southern blot analysis to confirm the genomic insertion of the antigen expression cassette and the loss of the pLMD3 vector backbone. Reverse transcriptase PCR and Western blot analyses were also performed to confirm NS3 mRNA and protein production (data not shown).
Cell lines, synthetic peptide reagents, and recombinant antigens.
The H2-Kd-transfected RMAS cell line (RMAS-Kd; originally obtained from M. Bevan, University of Washington, Seattle) was maintained in antibiotic-free RPMI 1640 medium (Invitrogen) that was supplemented with 10% fetal bovine serum (FBS) and 200 μg of Geneticin (Invitrogen) per ml. Overlapping 15-mer NS3 peptides spanning the HCV NS3 protein (aa 1007 to 1615) derived from HCV-1 (genotype 1a; accession number M62321) were synthesized by the Natural and Medical Sciences Institute at the University of Tuebingen (Ruetlingen, Germany). These 15-mer peptides (overlapping by 11 aa) were grouped into 15 pools of 10 consecutive peptides each or were used individually. The individual peptide, designated NS31540-1548 (RAYMNTPGL, representing aa 1540 to 1548 of the full-length HCV polyprotein), was synthesized at the Portland Veterans Affairs Medical Center with a Synergy apparatus (Applied Biosystems, Foster City, Calif.) by using standard 9-fluorenylmethoxy carbonyl chemistry. To generate recombinant antigens for immunization, the core and NS3 genes (described above) were inserted into the pProExHT prokaryotic expression system (Invitrogen Life Technologies), and histidine-tagged core or NS3 antigens were produced and purified according to the manufacturer's instructions.
Mice and immunizations.
Four- to five-week-old female BALB/cJ mice purchased from The Jackson Laboratory (Bar Harbor, Maine) were housed with unrestricted access to food and water and were treated in accordance with the animal care policies of the Institutional Animal Care and Use Committee and the Veterinary Medical Unit of the Department of Veterans Affairs Medical Center. For immunization with the plasmid constructs, 6- to 8-week-old mice received the first of a series of three intramuscular (i.m.) immunizations (via the tibialis anterior muscles) at 3- to 4-week intervals with 125 to 150 μg of plasmid DNA in 100 μl of normal saline (50 μl per leg). For immunization with recombinant histidine-tagged core and NS3 antigens, each mouse received, by i.m. injection (50 μl per quadriceps muscle), 25 μg of recombinant antigen that was emulsified 1:1 with Titermax adjuvant (CytRx Corporation, Norcross, Ga.) in a 100-μl total volume. For active immunization with viable L. monocytogenes, 6- to 8-week-old mice received intravenous (i.v.) injections (via the tail vein) with a 0.05 to 0.1 50% lethal dose (LD50) of bacteria (500 CFU for the wild-type strain, Lm10403, and [2 to 4] × 106 CFU for strain TC-LNS3) in 0.2 ml of phosphate-buffered saline (PBS). For all experiments, normal control mice were injected with 0.2 ml of PBS (i.v.) or 100 μl of saline (i.m.) or were not injected. Some groups of mice received a second bacterial immunization 10 to 14 days after the primary injection.
Evaluation of humoral immune responses.
Blood was collected from immunized and control mice with a heparinized capillary tube via retro-orbital puncture. The blood was centrifuged, and the plasma was used in a standard enzyme-linked immunosorbent assay (ELISA) (3). The ELISA plate wells were coated with 0.1 to 0.5 μg of recombinant HCV-NS3 (Mikrogen, Martinsried, Germany) or HCV core proteins (Chiron, Emeryville, Calif.). Horseradish peroxidase-conjugated goat anti-mouse was used as the detection antibody, and o-phenylenediamine was used as the colorimetric detection reagent. Absorbance was measured at 490 nm by using a EL309 ELISA plate reader (Bio-Tek Instruments, Winooski, Vt.).
Depletion of CD4+ or CD8+ T cells.
CD4+ or CD8+ T-cell subsets were depleted from total splenocytes by antibody and complement treatment or by magnetic bead cell sorting (MACS; Miltenyi Biotec, Auburn, Calif.). For antibody and complement lysis, the spleen cells were incubated with anti-CD4 (GK1.5) or anti-CD8 (19/178C1) monoclonal antibody followed by Low-tox M rabbit complement (Cedarlane Laboratories, Ltd., Hornby, Ontario, Canada). For magnetic cell sorting, total splenocytes were labeled with either R phycoerythrin-conjugated anti-CD4 (H129.19) or anti-CD8b.2 (53-5.8) (PharMingen, San Diego, Calif.). After being labeled with primary antibody, the cells were tagged with paramagnetic beads conjugated to anti-R phycoerythrin antibody and sorted on a magnetized column (Miltenyi Biotec). The percentages of CD4+ and CD8+ cells were assessed by flow cytometry (FACScan; Becton Dickinson, San Jose, Calif.) before and after magnetic separation, demonstrating a ≥93% depletion of the specific T-cell subset from the total splenocyte population (data not shown). The CD4+- or CD8+-depleted cell populations were then used for detection of gamma interferon (IFN-γ)-secreting cells by ELISPOT assay. Typically, two spleens from each experimental group were pooled to prepare the cell suspensions.
ELISPOT assays.
ELISPOT assays were performed 21 to 35 days after the final plasmid immunization or 7 to 9 days following infection with L. monocytogenes. The enumeration of IFN-γ-secreting cells was performed essentially as described previously (8), with some minor changes. RMAS-Kd cells were pulsed with a 10−5 molar concentration of the individual peptides or peptide pools. After peptide pulsing and washing, the target cells were placed in the ELISPOT plates (Multiscreen-HA; Millipore Corp., Bedford, Mass.) at a concentration of 105 cells/well in RPMI 1640 medium. Single cell suspensions of immune cells from previously immunized mice were prepared in RPMI 1640 (supplemented with 10% fetal calf serum, 200 U of penicillin per ml, 200 μg of streptomycin per ml, and 60 U of human recombinant interleukin 2 per ml) and added to the ELISPOT plates at concentrations of 100,000 to 250,000 splenocytes per well. After 24 h of incubation, IFN-γ secretion by individual cells was detected by sequential incubation with a biotinylated anti-mouse IFN-γ detection antibody (PharMingen), streptavidin AKP (PharMingen), and BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (KPL, Gaithersburg, Md.), and IFN-γ positive spots were then enumerated with a Zeiss microscopy unit equipped with KS ELISPOT software (Carl Zeiss MicroImaging, Inc., Thornwood, N.Y.).
Culture activation and chromium release cytotoxicity assays.
Spleen cells obtained from normal mice, Listeria-immunized mice (at 2 to 8 weeks following sublethal infection), or plasmid DNA-immunized mice (at 3 to 4 weeks following the final i.m. injection) were cocultured with peptide-pulsed irradiated naive spleen cells (to function as stimulator cells). These stimulator cells were irradiated (3,000 rad from a 137Cs source) and pulsed for 1 to 2 h with peptide NS31407-1421 or NS31535-1549 at a concentration of 10−5 M. After peptide pulse, the stimulator cells were combined, washed once, and then cocultured at 37°C with the donor spleen cell populations (i.e., those from immunized mice) for 6 days at a donor-to-stimulator-cell ratio of 1/1 in RPMI 1640 (supplemented with 10% FBS, 100 U of penicillin per ml, 100 μg of streptomycin sulfate [Sigma, St. Louis, Mo.], and 30 U of recombinant human interleukin 2 [Tecin, Biological Response Modifiers Program; National Cancer Institute, Frederick, Md.] per ml). Typically, two to four spleens from each group were pooled for culture activation. After 6 days, the culture-activated cells were washed twice and then used for chromium release cytotoxicity assays. P815 target cells were labeled with Na51CrO4 (Perkin-Elmer Life Sciences, Inc., Boston, Mass.) for 60 min, washed, and then pulsed with a 5 × 10−8 M concentration of peptide (either LLO91-99, NS31407-1421, or NS31535-1549) in RPMI 1640 medium plus 2% FBS for 60 min. After the peptide pulse, target cells were washed once, resuspended in RPMI 1640 plus 10% FBS, and added in 100-μl volumes to 96-well round-bottom microtiter plates at a concentration of 104 cells/well. Effector cells were added in 100-μl volumes at a concentration of 5 × 105 cells/well. Following a 4-h incubation at 37°C, supernatant from each well was collected, relative radioactivity (counts per minute) was determined (MicroBeta Trilux liquid scintillation counter; Perkin-Elmer), and the percent lysis was calculated with the following equation: 100 × (experimental counts per minute − spontaneous counts per minute)/(total counts per minute − spontaneous counts per minute). The total number of counts per minute was determined following lysis of the target cells with 5% Triton X-100 (Bio-Rad, Redmond, Calif.).
In vivo protection.
Levels of in vivo protection expressed by immunized mice were determined as previously described (4). Briefly, groups of normal or immunized mice received an i.v. injection with 1 to 2 LD50s of L. monocytogenes in 0.2 ml of PBS at 3 to 4 weeks following the final immunization with plasmid DNA. For BALB/c mice, the i.v. LD50s for the Lm10403 wild-type and TC-LNS3 recombinant strains of L. monocytogenes are 104 CFU (5) and 3 × 107 CFU (data not shown), respectively. Control groups consisted of normal (nonimmunized) mice and mice that were previously immunized (4 to 12 weeks earlier) with a sublethal injection of viable L. monocytogenes. Two days after bacterial challenge, spleens and livers were removed from individual mice and separately homogenized in PBS and serially diluted (in PBS); these dilutions were then plated out on brain heart infusion agar. Following overnight culture at 37°C, the log10 CFU per gram of tissue values from individual mice were determined, and the mean and standard error of the mean (SEM) values for each group were calculated. The detection limit of this assay is 1.7 to 1.9 log10 CFU/g.
Statistical analyses.
Standard one-way analysis of variance (ANOVA) with Tukey posttest was performed by using the Prism software package (GraphPad, San Diego, Calif.).
RESULTS
DNA immunization induces little or no humoral response.
BALB/c mice were immunized with the pUb-core or the pUb-NS3 plasmid DNA constructs three times at 4-week intervals, and blood samples were collected at 4 to 6 weeks after the final immunization. We were unable to detect significant levels of antibody to HCV-NS3 following repeated immunization of BALB/c mice with the pUb-NS3 plasmid construct (of nine mice tested, none were positive). In contrast, pUb-NS3 plasmid DNA immunization followed by protein antigen boost did result in the development of low titers of NS3-specific antibody (in two of two mice). Similarly, mice immunized with pUb-core had undetectable or very low antibody titers against HCV core (2 of 14 mice), while analogous plasmid immunization followed by recombinant antigen boost did induce modest anticore antibody responses (2 of 2 mice).
DNA immunization induces a CD8+ T-cell response to HCV NS3 peptides.
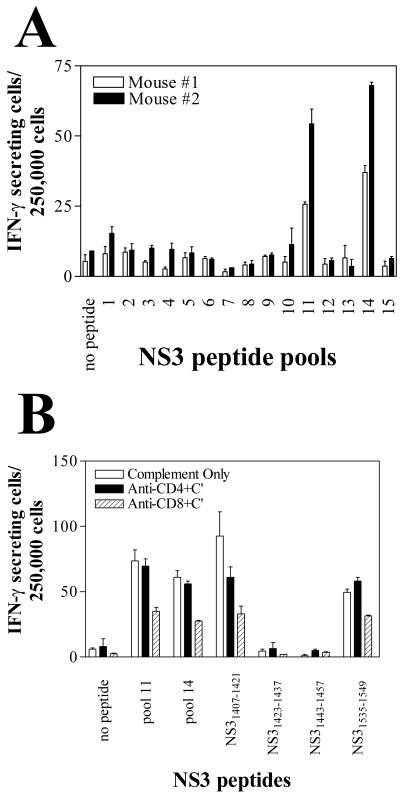
The cellular response observed in two pUb-NS3 plasmid DNA-immunized mice was measured initially by direct ex vivo ELISPOT assays against pools of 10 overlapping 15-aa peptides covering the entire sequence of the NS3 protein (Fig. 1). Using additional pUb-NS3-immunized mice, we detected enhanced numbers of IFN-γ-secreting cells that were reactive to peptide pools 11 and 14, corresponding to aa 1407 to 1457 and 1527 to 1577, respectively, of the full-length HCV polypeptide (Fig. 2A). Further testing of individual peptides from these two pools revealed that the response was restricted to peptides NS31407-1421 and NS31411-1425 from pool 11 and peptides NS31535-1549 and NS31539-1553 from pool 14 (data not shown). The individual peptides NS31407-1421 (LVALGINAVAYYRGL) and NS31535-1549 (TTVRLRAYMNTPGLP) were used in subsequent experiments. Based on HLA binding predictions (35), a putative H2-Kd-binding epitope of NS31535-1549 was identified, and the representative 9-mer peptide NS31540-1548 (RAYMNTPGL) was synthesized.
FIG. 2.
Immunization of mice with the pUb-NS3 DNA vaccine induces IFN-γ-secreting cells against two NS3 peptide epitopes. BALB/c mice (H2d haplotype) were immunized three times with the pUb-NS3 plasmid DNA vaccine. At 4 weeks following the final immunization, NS3-specific IFN-γ-secreting spleen cells were enumerated by using the ELISPOT assay. Target cells for this assay consisted of H2-Kd-transfected RMAS cells that were pulsed with synthetic NS3 peptides. (A) Direct ex vivo evidence of IFN-γ-secreting cells against two distinct pools of NS3 peptides (consisting of overlapping 15-aa peptides, 10 peptides/pool), as demonstrated from two representative mice. Pools 11 and 14 correspond to aa 1407 to 1457 and 1527 to 1577, respectively. Error bars represent the SEM of triplicate wells. (B) Treatment of the donor spleen cells with anti-CD8 antibody and complement (C′) markedly reduced the detectable numbers of IFN-γ-secreting cells. The individual 15-mer peptides from each of the two pools were tested; only peptides NS31407-1421, NS31423-1437, NS31443-1457, and NS31535-1549 are shown here. Error bars represent the SEM of duplicate wells.
To determine whether the HCV-specific response was CD4+- or CD8+-T-cell mediated, we first depleted splenocyte populations of either CD4+ or CD8+ cells by treatment with antibody and complement, and we then compared the depleted populations to the total number of splenocytes in ELISPOT assays. The number of IFN-γ-secreting cells responding to NS31407-1421, NS31535-1549, or peptide pools 11 or 14 was reduced markedly by CD8+ T-cell depletion, whereas depletion of CD4+ T cells had little or no effect (Fig. 2B). In a separate experiment, CD4+ or CD8+ cells from pUb-NS3-immunized mice were depleted by magnetic cell sorting, and ELISPOT assays were performed with target cells that had been pulsed with NS31407-1421. Again, the reactivity (i.e., the number of IFN-γ-secreting cells) to NS31407-1421 was markedly reduced by CD8+ T-cell depletion, but not by CD4+ T-cell depletion, of the total spleen cell population derived from pUb-NS3-immunized mice (data not shown). These data demonstrate that immunization of BALB/c mice with the pUb-NS3 vaccine construct induces activation of CD8+ CTL against two different H2d-restricted NS3 peptide epitopes.
Immunization with recombinant L. monocytogenes induces a specific response to NS3 epitopes.
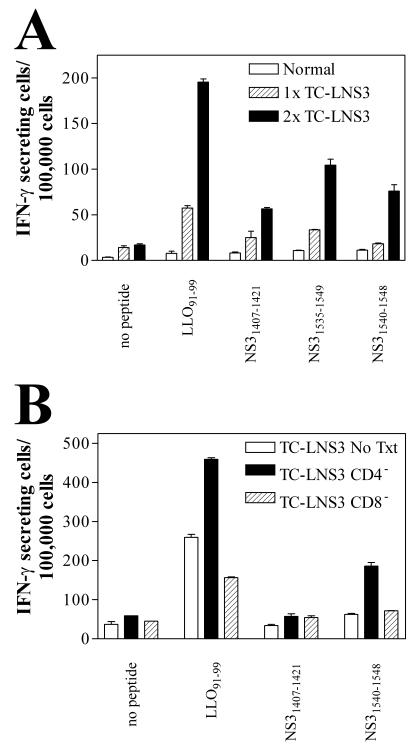
Based on previous observations of other experimental models (17, 24, 41), we suspected that infection of mice with recombinant L. monocytogenes expressing HCV antigens should also induce HCV-specific CTL responses. To examine this possibility, mice were infected with sublethal doses of TC-LNS3 and in vivo induction of NS3 peptide-specific T-cell responses was determined by subsequent ELISPOT assays. After a single immunization with TC-LNS3, an LLO91-99-specific T-cell response was detected but NS3-specific responses were limited or were present only at background levels (Fig. 3A). However, after a second immunization with the recombinant TC-LNS3, donor mice developed a relatively strong response to the 15-mer peptides NS31407-1421 and NS31535-1549 as well as to the shorter NS31540-1548 peptide (Fig. 3A). These results confirm that TC-LNS3 expresses NS3 protein that is available for immune presentation.
FIG. 3.
Immunization with recombinant L. monocytogenes TC-LNS3 induces NS3 peptide-specific T-cell responses in BALB/c mice. Two spleens were pooled for each experimental group, and mean numbers of IFN-γ-secreting cells were determined for duplicate wells by using ELISPOT assays. (A) Mice were immunized once (1×) or twice (2×) with viable TC-LNS3, and NS3 peptide-specific IFN-γ-secreting cells were enumerated. (B) Mice were immunized twice with TC-LNS3, and relative numbers of IFN-γ-secreting cells were measured after in vitro depletion (>90%) of CD4+ and CD8+ T-cell subsets by magnetic cell sorting. Note that the same total number of cells was added to each well regardless of treatment, and therefore the CD4+-T-cell depleted wells were enriched for CD8+ T cells.
In a separate experiment, CD4+ or CD8+ T cells from mice immunized twice with TCL-NS3 were depleted by magnetic cell sorting and ELISPOT assays were performed with target cells that had been pulsed with NS3 peptides. Notably, partial depletion of CD8+ T cells (Fig. 3B) did not totally eliminate the IFN-γ-secreting cells; in contrast, a large proportionate increase in IFN-γ-secreting cells was detected after depletion of CD4+ cells.
Immunization with pUb-NS3 or TC-LNS3 induces NS3-specific cytotoxic T cells.
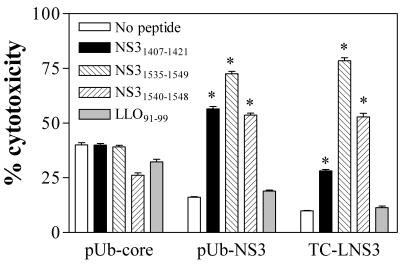
To determine if these NS3-specific IFN-γ-secreting cells detected by ELISPOT assays were in fact cytolytic, we subsequently conducted in vitro chromium release cytotoxicity assays. Spleen cells from naive or immunized mice (i.e., with three immunizations with pUb-NS3 or a single immunization with TC-LNS3) were stimulated in vitro with peptide-pulsed irradiated splenocytes and then used in cytotoxicity assays. Following culture stimulation of these immune donor cells with peptides NS31535-1549 or NS31407-1421, we detected a strong cytotoxic activity against target cells that had been pulsed with peptide NS31407-1421 or the peptides NS31535-1549 and NS31540-1548, respectively (Fig. 4).
FIG. 4.
NS3-specific T cells induced by immunization with pUb-NS3 or TC-LNS3 exhibit cytotoxic activity. Splenocytes from BALB/c mice that were immunized with the pUb-NS3 or pUb core DNA vaccine constructs, or with L. monocytogenes strain TC-LNS3 (expressing the HCV NS3 protein), were culture stimulated for 6 days with peptides NS31407-1421 and NS31535-1549 (as described in Materials and Methods) and were then used as effector cells in chromium release assays. Chromium-labeled P815 cells that had been pulsed with LLO91-99, NS31407-1421, NS31535-1549, or NS31540-1548 peptides served as target cells (effector-to-target ratio, 50/1). Following a 4-h incubation at 37°C, relative radioactivity (counts per minute) in supernatants from each well was determined. Data represent the mean and SEM of triplicate wells. *, significant difference compared to controls (which used no peptide) (P < 0.01) by one-way ANOVA with the Tukey posttest.
DNA immunization protects mice against challenge with recombinant L. monocytogenes.
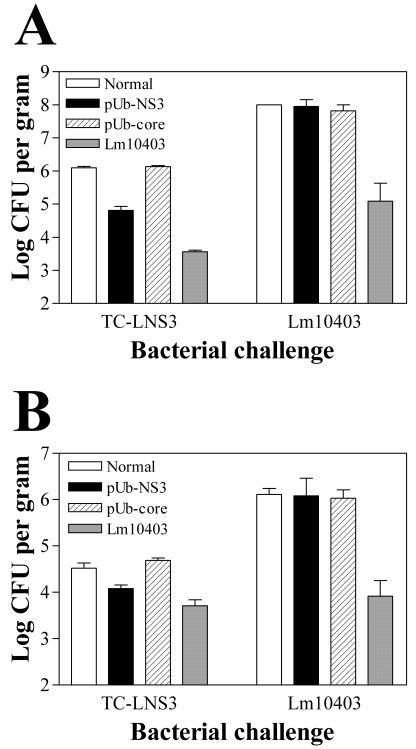
Numerous studies have demonstrated that expression of in vivo protection against challenge with virulent L. monocytogenes is dependent on antigen-specific CTL. To evaluate similar effector function of the NS3-specific T cells induced following DNA vaccination, BALB/c mice were immunized with either the pUb-NS3 or the pUb-core plasmid DNA constructs and then challenged either with the recombinant L. monocytogenes strain TC-LNS3 (producing the HCV NS3 protein) or with wild-type L. monocytogenes (Lm10403). Two days after challenge, the numbers of viable bacteria in the spleens and livers of individual mice were determined by plating organ homogenates on brain heart infusion agar. The results of these assays (Fig. 5) demonstrated that pUb-NS3-immunized mice exhibited significantly (P < 0.001) lower bacterial loads in the spleen (4.81 log CFU/g) following challenge with TC-LNS3 than did naive controls (6.41 log CFU/g) or pUb-core-immunized mice (6.13 log CFU/g). In this experiment, reduced bacterial counts were also seen for the liver homogenates obtained from pUb-NS3-immunized mice compared to similar liver homogenates obtained from naive controls and pUb-core-immunized mice (4.08 log CFU/g versus 4.52 and 4.69 log CFU/g, respectively); however, these differences were not statistically significant (P ≥ 0.05). Notably, none of the pUb-NS3- or pUb-core-immunized mice were protected against challenge with wild-type L. monocytogenes (Fig. 5). In four different experiments, the level of protection (i.e., the difference in the mean log10 CFU values between pUb-NS3-immunized and nonimmune control mice) observed in the spleen ranged from 1.11 to 1.95 log units, whereas the level of protection observed in the liver was always reduced, ranging from 0.44 to 0.84 log units.
FIG. 5.
Mice immunized with the pUbNS3 DNA vaccine exhibit specific protection against the recombinant L. monocytogenes strain TC-LNS3. BALB/c mice that had been immunized with the pUb-NS3 or pUb core DNA vaccine constructs or with wild-type L. monocytogenes were challenged with the recombinant L. monocytogenes strain TC-LNS3 (expressing the HCV NS3 protein) or with the wild-type strain (Lm10403). After 48 h, viable bacterial counts in the spleen (A) and the liver (B) were determined. The pUbNS3-immunized mice exhibited specific protection against challenge with strain TC-LNS3 but not against challenge with wild-type Lm10403. This observed protection was significant (P < 0.001) in the spleen but not the liver (P > 0.05) by one-way ANOVA with the Tukey posttest. In this experiment, four mice were used for the pUb-NS3-immunized group challenged with TC-LNS3, and two mice were used for each of the remaining groups. The data are representative of four separate experiments.
DISCUSSION
In the absence of a suitable small-animal model for HCV infection, experimental studies evaluating protective immunity against HCV have been limited to those conducted with the chimpanzee model (10). Nevertheless, novel strategies have been proposed in attempts to develop small-animal models of in vivo HCV replication and pathogenesis (25, 32), and it has been reported that HCV can infect and replicate within tree shrews (47). Although such models may prove suitable for testing antiviral compounds, they will probably be of little use for immunological and vaccination studies. The protective efficacy of any experimental vaccine formulation can best be assessed in an in vivo bioassay model, and to date both stably transfected cell lines and recombinant vaccinia virus expressing HCV antigens have been used to simulate a challenge with HCV (16, 23, 44, 45). Since L. monocytogenes exhibits a hepatotropic intracellular phenotype, we propose that recombinant L. monocytogenes expressing HCV antigens provides another useful challenge model for the evaluation of HCV-specific CTL responses. While early innate protection against L. monocytogenes infection is largely dependent on natural killer cells and macrophages, long-lived protective immunity to L. monocytogenes requires the expression of effector function by CD8+ T cells (4, 33). Interestingly, IFN-γ secretion by CD8+ T cells may only represent an in vitro correlate of the protective immune response to L. monocytogenes, as IFN-γ-deficient mice exhibit CD8+ T-cell-dependent protective immunity against challenge with L. monocytogenes after immunization with plasmid DNA vaccines encoding LLO (6). In the studies reported here, we have used recombinant bacteria expressing the HCV NS3 protein for the in vivo assay of protective immunity following immunization of mice with DNA vaccines encoding HCV antigens. The data provided demonstrate that vaccination of BALB/c mice with the pUb-NS3 plasmid DNA construct (encoding a ubiquitin-NS3 fusion protein) induces an NS3-specific T-cell response in the absence of significant antibody production. Furthermore, this T-cell response confers immune protection against subsequent challenge with recombinant L. monocytogenes (TC-LNS3) which secretes the HCV NS3 protein.
Since CD8+ T-cell responses presumably play a major role in protective immunity to HCV infection (11, 14, 31, 40), we employed plasmid DNA vectors that were designed to enhance CD8+ T-cell stimulation through efficient major histocompatibility complex (MHC) class I presentation via ubiquitination and proteosome processing. This enhancement of MHC class I presentation can result in a concomitant reduction of the humoral response, as reported here, and this observation is consistent with previous findings which indicated that immunization with DNA vaccine vectors encoding ubiquitin fusion proteins favors enhanced CTL responses and decreased antibody responses (15, 36, 42, 46). Indeed, the immune cell populations induced following pUb-NS3 vaccination of BALB/c mice included peptide-specific CD8+ T lymphocytes that secrete IFN-γ following coculture with target cells pulsed with two NS3 peptides, designated NS31407-1421 and NS31535-1549 (amino acid sequences, LVALGINAVAYYRGL and TTVRLRAYMNTPGLP, respectively). We also demonstrated that NS31540-1548, a shorter peptide containing a predicted H2-Kd-binding epitope (RAYMNTPGL), was recognized by these CD8+ T lymphocytes. We have not yet identified a shorter H2-Kd-binding peptide motif within NS31407-1421. Since we screened the NS3-specific response on peptide-pulsed RMAS-Kd cells that do not express H2-Dd or -Ld, we identified only NS3-specific CTL populations that recognized H2-Kd-restricted peptides. Also, the minor differences between the deduced amino acid sequences of pUb-NS3 and the 15-mer peptides (Fig. 1) might have precluded us from identifying other potential epitopes encoded by these divergent sequences. A search of National Center for Biotechnology Information's GenBank reveals that most of the amino acid changes observed in our sequence have been reported in genotype 1a or 1b clinical HCV isolates. However, the serine-to-glycine change at position 1465 (S1465G) is likely a PCR or cloning error, as this position is invariant among the described isolates. Likewise, the K1094T and Q1106K substitutions, as well as the stop codon at position 1632, are not seen in genotype 1a or 1b isolates and probably also represent PCR or cloning errors. Thus, our findings indicate that at least two H2d MHC class I-restricted peptide epitopes for the HCV NS3 protein exist. In a recent report, Arribillaga et al. (2) showed that NS3-immunized BALB/c mice generated T-cell responses to several NS3 peptides (identified from H-2d-binding predictions). One peptide identified by Arribillaga et al. overlaps with one epitope (peptides NS31535-1549 and NS31540-1548) that we have identified in this paper. Interestingly, the overlapping genotype 1b-derived peptide AYLNTPGLP (corresponding to amino acids 1541 to 1549 of our genotype 1a isolate) identified by Arribillaga et al. stimulated only IFN-γ secretion and not cytotoxic activity in vitro (2), whereas our genotype 1a-derived peptides induced both IFN-γ secretion and target-cell lysis by NS3-specific T cells.
The induction of these immune NS3-specific CD8+ T cells correlated with the expression of in vivo protection against the NS3 recombinant (TC-LNS3), but not the wild-type (Lm10403), L. monocytogenes strains. As indicated by the reduced numbers of bacteria recovered from spleen homogenates, this level of protection was highly significant (P < 0.001) in pUb-NS3-immunized mice relative to that observed in naive or pUb-core-immunized mice. In contrast, although TC-LNS3 bacterial counts in the livers of immunized mice were reduced nearly three- to fourfold (a log unit difference of 0.44 to 0.84) compared to those in the livers of naive mice, this difference was not significant (P ≥ 0.05). At this point, it is unclear whether the differential response is due to variable levels of infection or bacterial clearance of L. monocytogenes relative to the spleen and liver or to actual differences in the cellular response to or expression of HCV antigens in the spleen versus the liver. Clearly, HCV-specific effector cells are being induced by plasmid immunization; however, expression of their in vivo function may be blunted by the reported tolerogenic environment of the liver tissue (28). In contrast, evidence suggests that for some pathogens, including L. monocytogenes, effective immune responses to infected hepatocytes can be generated and maintained (21, 22, 26). We have also extensively evaluated the induction of protective immunity to challenge with L. monocytogenes following immunization of mice with DNA vaccines which encode the bacterial hemolysin LLO (12), and recent observations suggest that the level of protection (as indicated by reduced numbers of bacteria that are present in tissue homogenates 48 h postinfection) is not as pronounced in the liver as in the spleen (our unpublished observations). Additional studies evaluating the kinetics of the relative bacterial loads in these tissues following the challenge of naive versus immunized mice should help clarify this issue.
In the data reported here, mice immunized with viable wild-type L. monocytogenes (Lm10403) were more protected from challenge with TC-LNS3 than were the pUb-NS3-immunized mice. Presumably, this finding reflects the fact that the L. monocytogenes strains Lm10403 and TC-LNS3 share numerous antigens, including the immunodominant LLO antigen, that are recognized by protective CD8+ T cells following sublethal bacterial infection. In contrast, the pUb-NS3-immunized mice have been exposed to only one antigen (i.e., NS3) expressed by the NS3 recombinant strain L. monocytogenes TC-LNS3. Thus, competition with other L. monocytogenes-derived peptides for MHC class I molecules may limit, to some degree, the level of in vivo NS3 peptide presentation by the TC-LNS3-infected host cells to the NS3-specific T cells.
Since other investigators have previously demonstrated induction of CTL responses to heterologous antigens expressed by recombinant L. monocytogenes, we were interested in comparing the CD8+ T-cell responses induced in mice that had been immunized with either the pUb-NS3 plasmid or the NS3 recombinant L. monocytogenes strain. As indicated by ELISPOT assays, naive mice infected with recombinant L. monocytogenes TC-LNS3 appeared to respond more strongly to NS31535-1549 than to NS31407-1421, whereas the strength of the response to these two peptides was more similar in pUb-NS3-immunized mice. Further characterization of the immune responses generated by the plasmid DNA and recombinant bacterial constructs will be necessary to verify any qualitative differences that may develop in these experimental mice.
In conclusion, this experimental model of challenge with recombinant strains of L. monocytogenes offers a novel in vivo method for the evaluation of HCV vaccine efficacy. Notably, this model will allow us to further evaluate cellular immune responses to HCV antigen expression in murine liver. We hope that extending these studies to other new, unique small-animal models of clinical significance will facilitate the evaluation of effective vaccines that will eventually provide protection against this chronic infectious disease.
Acknowledgments
This work was supported by the Department of Veterans Affairs Merit Review and Research Enhancement Award Program (grant numbers VAMRG004 and VAREAP006) and by the Collins Medical Trust.
Editor: B. B. Finlay
REFERENCES
- 1.Alter, M. J., and H. S. Margolis (ed.). 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Morb. Mortal. Wkly. Rep. 47(RR-19):1-14. [PubMed]
- 2.Arribillaga, L., A. L. D. de Cerio, P. Sarobe, N. Casares, M. Gorraiz, A. Vales, O. Bruna-Romero, F. Borras-Cuesta, G. Paranhos-Baccala, and J. Prieto. 2002. Vaccination with an adenoviral vector encoding hepatitis C virus (HCV) NS3 protein protects against infection with HCV-recombinant vaccinia virus. Vaccine 21:202-210. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.) 2002. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Baldridge, J. R., R. A. Barry, and D. J. Hinrichs. 1990. Expression of systemic protection and delayed-type hypersensitivity to Listeria monocytogenes is mediated by different T-cell subsets. Infect. Immun. 58:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barry, R. A., H. G. Bouwer, D. A. Portnoy, and D. J. Hinrichs. 1992. Pathogenicity and immunogenicity of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 60:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry, R. A., H. G. A. Bouwer, T. R. Clark, K. A. Cornell, and D. J. Hinrichs. 2003. Protection of interferon-γ knockout mice against Listeria monocytogenes challenge following intramuscular immunization with DNA vaccines encoding listeriolysin O. Vaccine 21:2122-2132. [DOI] [PubMed] [Google Scholar]
- 7.Bouwer, H. G., A. Bai, J. Forman, S. H. Gregory, E. J. Wing, R. A. Barry, and D. J. Hinrichs. 1998. Listeria monocytogenes-infected hepatocytes are targets of major histocompatibility complex class Ib-restricted antilisterial cytotoxic T lymphocytes. Infect. Immun. 66:2814-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouwer, H. G., R. A. Barry, and D. J. Hinrichs. 2001. Lack of expansion of major histocompatibility complex class Ib-restricted effector cells following recovery from secondary infection with the intracellular pathogen Listeria monocytogenes. Infect. Immun. 69:2286-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinster, C., S. Muguet, Y. C. Lone, D. Boucreux, N. Renard, A. Fournillier, F. Lemonnier, and G. Inchauspe. 2001. Different hepatitis C virus nonstructural protein 3 (Ns3)-DNA-expressing vaccines induce in HLA-A2.1 transgenic mice stable cytotoxic T lymphocytes that target one major epitope. Hepatology 34:1206-1217. [DOI] [PubMed] [Google Scholar]
- 10.Bukh, J., X. Forns, S. U. Emerson, and R. H. Purcell. 2001. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Intervirology 44:132-142. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 12.Cornell, K. A., H. G. Bouwer, D. J. Hinrichs, and R. A. Barry. 1999. Genetic immunization of mice against Listeria monocytogenes using plasmid DNA encoding listeriolysin O. J. Immunol. 163:322-329. [PubMed] [Google Scholar]
- 13.Cramp, M. E., P. Carucci, S. Rossol, S. Chokshi, G. Maertens, R. Williams, and N. V. Naoumov. 1999. Hepatitis C virus (HCV) specific immune responses in anti-HCV positive patients without hepatitis C viraemia. Gut 44:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cucchiarini, M., A. R. Kammer, B. Grabscheid, H. M. Diepolder, T. J. Gerlach, N. Gruner, T. Santantonio, J. Reichen, G. R. Pape, and A. Cerny. 2000. Vigorous peripheral blood cytotoxic T cell response during the acute phase of hepatitis C virus infection. Cell. Immunol. 203:111-123. [DOI] [PubMed] [Google Scholar]
- 15.Delogu, G., A. Howard, F. M. Collins, and S. L. Morris. 2000. DNA vaccination against tuberculosis: expression of a ubiquitin-conjugated tuberculosis protein enhances antimycobacterial immunity. Infect. Immun. 68:3097-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Encke, J., J. zu Putlitz, M. Geissler, and J. R. Wands. 1998. Genetic immunization generates cellular and humoral immune responses against the nonstructural proteins of the hepatitis C virus in a murine model. J. Immunol. 161:4917-4923. [PubMed] [Google Scholar]
- 17.Friedman, R. S., F. R. Frankel, Z. Xu, and J. Lieberman. 2000. Induction of human immunodeficiency virus (HIV)-specific CD8 T-cell responses by Listeria monocytogenes and a hyperattenuated Listeria strain engineered to express HIV antigens. J. Virol. 74:9987-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlach, J. T., H. M. Diepolder, M. C. Jung, N. H. Gruener, W. W. Schraut, R. Zachoval, R. Hoffmann, C. A. Schirren, T. Santantonio, and G. R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology 117:933-941. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, E. J., R. Bhat, Q. Liu, Y. F. Wang, C. Tackney, and A. M. Prince. 2000. Immune responses to hepatitis C virus structural and nonstructural proteins induced by plasmid DNA immunizations. J. Infect. Dis. 181:42-50. [DOI] [PubMed] [Google Scholar]
- 20.Gregory, S. H., L. K. Barczynski, and E. J. Wing. 1992. Effector function of hepatocytes and Kupffer cells in the resolution of systemic bacterial infections. J. Leukoc. Biol. 51:421-424. [DOI] [PubMed] [Google Scholar]
- 21.Gregory, S. H., and C. C. Liu. 2000. CD8+ T-cell-mediated response to Listeria monocytogenes taken up in the liver and replicating within hepatocytes. Immunol. Rev. 174:112-122. [DOI] [PubMed] [Google Scholar]
- 22.Harty, J. T., and M. J. Bevan. 1996. CD8 T-cell recognition of macrophages and hepatocytes results in immunity to Listeria monocytogenes. Infect. Immun. 64:3632-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiranuma, K., S. Tamaki, Y. Nishimura, S. Kusuki, M. Isogawa, G. Kim, M. Kaito, K. Kuribayashi, Y. Adachi, and Y. Yasutomi. 1999. Helper T cell determinant peptide contributes to induction of cellular immune responses by peptide vaccines against hepatitis C virus. J. Gen. Virol. 80:187-193. [DOI] [PubMed] [Google Scholar]
- 24.Ikonomidis, G., D. A. Portnoy, W. Gerhard, and Y. Paterson. 1997. Influenza-specific immunity induced by recombinant Listeria monocytogenes vaccines. Vaccine 15:433-440. [DOI] [PubMed] [Google Scholar]
- 25.Ilan, E., J. Arazi, O. Nussbaum, A. Zauberman, R. Eren, I. Lubin, L. Neville, O. B. Moshe, A. Kischitzky, A. Litchi, I. Margalit, J. Gopher, S. Mounir, W. Cai, N. Daudi, A. Eid, O. Jurim, A. Czerniak, E. Galun, and S. Dagan. 2002. The hepatitis C virus (HCV)-trimera mouse: a model for evaluation of agents against HCV. J Infect. Dis. 185:153-161. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, X., S. H. Gregory, and E. J. Wing. 1997. Immune CD8+ T lymphocytes lyse Listeria monocytogenes-infected hepatocytes by a classical MHC class I-restricted mechanism. J. Immunol. 158:287-293. [PubMed] [Google Scholar]
- 27.Kalams, S. A., and B. D. Walker. 1998. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J. Exp. Med. 188:2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knolle, P. A., and G. Gerken. 2000. Local control of the immune response in the liver. Immunol. Rev. 174:21-34. [DOI] [PubMed] [Google Scholar]
- 29.Koziel, M. J., D. K. Wong, D. Dudley, M. Houghton, and B. D. Walker. 1997. Hepatitis C virus-specific cytolytic T lymphocyte and T helper cell responses in seronegative persons. J. Infect. Dis. 176:859-866. [DOI] [PubMed] [Google Scholar]
- 30.Lazdina, U., C. Hultgren, L. Frelin, M. Chen, K. Lodin, O. Weiland, G. Leroux-Roels, J. A. Quiroga, D. L. Peterson, D. R. Milich, and M. Sallberg. 2001. Humoral and CD4+ T helper (Th) cell responses to the hepatitis C virus non-structural 3 (NS3) protein: NS3 primes Th1-like responses more effectively as a DNA-based immunogen than as a recombinant protein. J. Gen. Virol. 82:1299-1308. [DOI] [PubMed] [Google Scholar]
- 31.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer, D. F., D. E. Schiller, J. F. Elliott, D. N. Douglas, C. Hao, A. Rinfret, W. R. Addison, K. P. Fischer, T. A. Churchill, J. R. Lakey, D. L. Tyrrell, and N. M. Kneteman. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927-933. [DOI] [PubMed] [Google Scholar]
- 33.Mielke, M. E., S. Ehlers, and H. Hahn. 1988. T-cell subsets in delayed-type hypersensitivity, protection, and granuloma formation in primary and secondary Listeria infection in mice: superior role of Lyt-2+ cells in acquired immunity. Infect. Immun. 56:1920-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Missale, G., R. Bertoni, V. Lamonaca, A. Valli, M. Massari, C. Mori, M. G. Rumi, M. Houghton, F. Fiaccadori, and C. Ferrari. 1996. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J. Clin. Investig. 98:706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker, K. C., M. A. Bednarek, and J. E. Coligan. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152:163-175. [PubMed] [Google Scholar]
- 36.Rodriguez, F., J. Zhang, and J. L. Whitton. 1997. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J. Virol. 71:8497-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosen, H., S. Gordon, and R. J. North. 1989. Exacerbation of murine listeriosis by a monoclonal antibody specific for the type 3 complement receptor of myelomonocytic cells: absence of monocytes at infective foci allows Listeria to multiply in nonphagocytic cells. J. Exp. Med. 170:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen, H., C. Miner, A. Sasaki, D. Lewinsohn, A. Conrad, A. Bakke, H. G. Bouwer, and D. Hinrichs. 2002. Frequencies of HCV-specific effector CD4+ T cells by flow cytometry: correlation with clinical disease stages. Hepatology 35:190-198. [DOI] [PubMed] [Google Scholar]
- 39.Saito, T., G. Sherman, K. Kurokohchi, Z. Guo, M. Donets, M. Yu, J. Berzofsky, T. Akatsuka, and S. Feinstone. 1997. Plasmid DNA-based immunization for hepatitis C virus structural proteins: immune responses in mice. Gastroenterology 112:1321-1330. [DOI] [PubMed] [Google Scholar]
- 40.Scognamiglio, P., D. Accapezzato, M. A. Casciaro, A. Cacciani, M. Artini, G. Bruno, M. L. Chircu, J. Sidney, S. Southwood, S. Abrignani, A. Sette, and V. Barnaba. 1999. Presence of effector CD8+ T cells in hepatitis C virus-exposed healthy seronegative donors. J. Immunol. 162:6681-6689. [PubMed] [Google Scholar]
- 41.Shen, H., M. K. Slifka, M. Matloubian, E. R. Jensen, R. Ahmed, and J. F. Miller. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 92:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sykes, K. F., and S. A. Johnston. 1999. Genetic live vaccines mimic the antigenicity but not pathogenicity of live viruses. DNA Cell Biol. 18:521-531. [DOI] [PubMed] [Google Scholar]
- 43.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokushige, K., T. Wakita, C. Pachuk, D. Moradpour, D. B. Weiner, V. R. Zurawski, Jr., and J. R. Wands. 1996. Expression and immune response to hepatitis C virus core DNA-based vaccine constructs. Hepatology 24:14-20. [DOI] [PubMed] [Google Scholar]
- 45.Wedemeyer, H., S. Gagneten, A. Davis, R. Bartenschlager, S. Feinstone, and B. Rehermann. 2001. Oral immunization with HCV-NS3-transformed Salmonella: induction of HCV-specific CTL in a transgenic mouse model. Gastroenterology 121:1158-1166. [DOI] [PubMed] [Google Scholar]
- 46.Wu, Y., and T. J. Kipps. 1997. Deoxyribonucleic acid vaccines encoding antigens with rapid proteasome-dependent degradation are highly efficient inducers of cytolytic T lymphocytes. J. Immunol. 159:6037-6043. [PubMed] [Google Scholar]
- 47.Xie, Z. C., J. I. Riezu-Boj, J. J. Lasarte, J. Guillen, J. H. Su, M. P. Civeira, and J. Prieto. 1998. Transmission of hepatitis C virus infection to tree shrews. Virology 244:513-520. [DOI] [PubMed] [Google Scholar]