Abstract
We report the efficient identification of four human histocompatibility leukocyte antigen (HLA)-A*0201–presented cytotoxic T lymphocyte (CTL) epitopes in the tumor-associated antigen PRAME using an improved “reverse immunology” strategy. Next to motif-based HLA-A*0201 binding prediction and actual binding and stability assays, analysis of in vitro proteasome-mediated digestions of polypeptides encompassing candidate epitopes was incorporated in the epitope prediction procedure. Proteasome cleavage pattern analysis, in particular determination of correct COOH-terminal cleavage of the putative epitope, allows a far more accurate and selective prediction of CTL epitopes. Only 4 of 19 high affinity HLA-A*0201 binding peptides (21%) were found to be efficiently generated by the proteasome in vitro. This approach avoids laborious CTL response inductions against high affinity binding peptides that are not processed and limits the number of peptides to be assayed for binding. CTL clones induced against the four identified epitopes (VLDGLDVLL, PRA100–108; SLYSFPEPEA, PRA142–151; ALYVDSLFFL, PRA300–309; and SLLQHLIGL, PRA425–433) lysed melanoma, renal cell carcinoma, lung carcinoma, and mammary carcinoma cell lines expressing PRAME and HLA-A*0201. This indicates that these epitopes are expressed on cancer cells of diverse histologic origin, making them attractive targets for immunotherapy of cancer.
Keywords: antigen presentation, antigen processing, cytotoxic T lymphocyte induction, human histocompatibility leukocyte antigen class I binding, tumor immunotherapy
Introduction
T cell–based immunotherapy of cancer has been successful in numerous mouse tumor model systems 1 and so far efficacious in a limited number of clinical conditions 2 3 4 5 6. Specific T cell–mediated immunotherapy requires the identification of tumor-specific antigens carrying T cell epitopes presented in the context of HLA class I and/or HLA class II molecules (for reviews, see references 1 and 7 8 9). The strategy, pioneered by Boon and coworkers, of screening melanoma cDNA libraries with CTLs derived from tumor infiltrating lymphocytes, has been successful in the identification of CTL epitopes in unknown tumor-associated proteins. The MAGE, BAGE, and GAGE families of tumor-associated testis-specific antigens 10 11 12 13, as well as the melanocyte differentiation antigens overexpressed in tumors like tyrosinase, Melan-A/MART-1, and gp100 14 15 16 17 have been identified in this manner. This strategy requires the availability of CTL clones from mixed leukocyte tumor cultures of cancer patients. Consequently, any CTL epitope that is not successful in activating CTLs that can be expanded in vitro (e.g., subdominant epitopes) will be missed. For the systematic detection of CTL epitopes presented in a broad range of HLA class I molecules, there is a great need for an efficient strategy. The strategy of predicting potential CTL epitopes in already identified tumor-associated proteins followed by in vitro sensitization of CTLs against these putative epitopes (also designated “reverse immunology”) has the advantage that it does not utilize patient-derived T cells as a primary screen and therefore allows a more systematic search for new CTL epitopes. Reverse immunology has been used to identify new CTL epitopes in MAGE-1 18, MAGE-2 19, MAGE-3 20 21 22 23, TRP2 24, gp100 25, and HER-2/neu 19. However, so far this strategy has been rather inefficient, mainly because CTLs raised against putative epitopes were often unable to recognize tumor cells expressing the source protein 26 27 28 29 30. Explanations for these failures are both an insufficient affinity of the induced CTLs for their MHC–peptide complex but more often the lack of processing of the presumed epitope 9. Recently, Chaux et al. successfully applied an alternative strategy of in vitro CTL inductions against dendritic cells (DCs) transduced with MAGE-1, abandoning the strategy of epitope prediction and allowing processing of the relevant epitopes to take place by the APC 31.
In this study, we chose to improve the epitope prediction strategy by verifying the proteasome-mediated generation of peptides in order to identify HLA-A*0201–presented CTL epitopes in the so-called PRAME protein. The main intracellular mechanisms that define the exact amino acid (aa) sequence of a CTL epitope include enzymatic breakdown of the protein by the proteasome, transporter-associated with antigen processing (TAP)-mediated translocation into the endoplasmic reticulum (ER) and binding of the peptide with sufficient affinity in the groove of an MHC class I molecule (for reviews, see references 32 and 33). The COOH terminus of CTL epitopes requires exact cleavage by the proteasome 34 35 36 37, whereas NH2-terminal extensions of the epitope can apparently be trimmed by putative aminopeptidase activity mainly in the ER 36 38 39 40 41 or in the cytosol 34 42. In vitro proteasome-mediated digestions are known to reliably yield MHC class I ligands from viral and model protein-derived polypeptides 43 44 45 46 47. Therefore, after identification of HLA-A*0201 binding peptides, we now incorporated in vitro proteasome-mediated digestions of 27-mer polypeptides encompassing high affinity binding peptides in the epitope prediction procedure. Digestion pattern analysis permitted assessment of efficient COOH-terminal generation of putative epitopes and, in addition, enabled evaluation of possible premature destruction by major cleavage sites within the epitope, as observed by us in a variant viral sequence 47.
The tumor-associated PRAME protein 48 is a particularly attractive target of T cell–based immunotherapy of cancer because of its expression in a wide variety of tumors, including melanoma (95% of patients), renal cell cancer (41%), lung cancer (50%), mammary cancer (27%), acute leukemias (30%), and multiple myeloma (52%; references 49 50 51), and because of its absence from normal tissues, except testis, and its very low levels in endometrium, ovaries and adrenals. We focussed on HLA-A*0201 as a restriction element because of its high prevalence (45.8%) among the Caucasian population 52. Via the improved multistep epitope prediction procedure, we report here the identification of four naturally processed HLA-A*0201–presented CTL epitopes in PRAME that are recognized by CTLs on cell lines derived from tumors of various histologic origins. This study underscores the importance of incorporating processing criteria for accurate identification of CTL epitopes.
Materials and Methods
Cell Lines and Culture Conditions.
The EBV-transformed B lymphoblastoid cell line (B-LCL) JY was cultured in complete culture medium consisting of IMDM (BioWhittaker) supplemented with 8% FCS (Greiner), 100 IU/ml penicillin, and 2 mM l-glutamine. The processing-defective T2 cell line was a gift from Dr. P. Cresswell (Yale University, New Haven, CT). Melanoma cell lines (Mel603, M453, and FM3), renal cell carcinoma cell lines (MZ1851, MZ1774, and MZ1257), and mammary carcinoma cell line MCF7 were provided by Dr. P. Schrier (Leiden University Medical Center). Mammary carcinoma cell line ZR-75-1 was obtained from the American Type Culture Collection. Lung carcinoma cell lines GLC02 and GLC36 were provided by Dr. L. de Leij (University of Groningen, Groningen, The Netherlands). The CD40 ligand–transfected mouse L cell line 53 that was used for generation of activated B cells was donated by Dr. C. van Kooten (Leiden University Medical Center). PRAME cDNA was provided by Dr. P. Coulie (Ludwig Institute for Cancer Research, Brussels, Belgium). The PRAME-encoding insert was cloned into vector pDR2 (Invitrogen), conferring hygromycine resistance. The renal cell carcinoma cell line MZ1851 was transfected with pDR2-PRAME using Fugene (Boehringer) as transfection reagent. After 48 h, hygromycine (100 μg/ml) was added to select transfected cells. Hygromycine-resistant cells were tested by reverse transcription (RT)-PCR for PRAME expression.
Peptides.
Peptides were synthesized by solid phase strategies on an automated multiple peptide synthesizer (Abimed AMS 422) using 9-fluorenylmethyloxycarbonyl (Fmoc) chemistry. Short peptides for CTL inductions were dissolved in 20 μl DMSO, diluted in 0.9% NaCl to a peptide concentration of 1 mg/ml, and stored at −20°C before usage. The fluorescein (FL)-labeled reference peptide as used in the competition-based HLA-A*0201 binding assay was synthesized, labeled, and characterized as described earlier 54. The sequence of the FL-labeled peptide was FLPSDYFPSV (hepatitis B virus [HBV] nucleocapsid 18–27; reference 55) wherein we substituted the tyrosine with a cysteine to tag an FL group to the peptide: FLPSDC(FL)FPSV 54. The 27-mer polypeptides used for in vitro proteasome digestion were synthesized as described above, purified by reversed phase-HPLC in an acetonitrile–water gradient and lyophilized from acetonitrile–water overnight. Purity was confirmed by mass spectrometry.
Cellular Competition–based HLA-A*0201 Peptide Binding Assay.
The affinity of peptides for HLA-A*0201 was analyzed using the homozygous HLA-A*0201+ B-LCL JY as described previously 54, with minor adaptations. In brief, naturally bound peptides were stripped from the HLA-A*0201 molecules by exposing the JY cells for 90 s to ice-cold citric acid buffer with pH 3.1 (1:1 mixture of 0.263 M citric acid and 0.123 M Na2HPO4). Cells were immediately buffered with ice-cold IMDM containing 2% FCS, washed twice in the same medium, and resuspended in 2% FCS/IMDM containing 2 μg/ml human β2-microglublin (Sigma-Aldrich). Subsequently, the stripped JY cells were plated at 4 × 104/well in a 96-well V-bottomed plate together with 150 nM of a known high affinity HLA-A2*0201 binding FL-labeled reference peptide 55 and titrated concentrations of competitor test peptide. After incubation for 24 h at 4°C, cells were washed three times in PBS containing 1% BSA, fixed with 0.5% paraformaldehyde, and analyzed on a FACScan™ flow cytometer (Becton Dickinson). The percentage inhibition of FL-labeled reference peptide binding was calculated using the following formula: [1 − (MFreference and competitor peptide − MFno reference peptide)/(MFreference peptide − MFno reference peptide)] × 100%. The binding affinity of competitor peptide is expressed as the concentration needed to inhibit 50% binding of the FL-labeled reference peptide (IC50). An IC50 ≤ 5 μM was considered high affinity binding, 5 μM < IC50 ≤ 15 μM was considered intermediate affinity binding, 15 μM < IC50 ≤ 100 μM was judged low affinity binding, and IC50 > 100 μM was considered not binding.
Peptide–MHC Complex Dissociation Assay.
Binding stability at 37°C of peptides complexed with HLA-A*0201 was measured as described previously 56. In short, JY cells were treated with 10−4 M emetine (Sigma-Aldrich) for 1 h at 37°C to stop de novo synthesis of MHC class I molecules. Subsequently, endogenous bound peptides in HLA-A*0201 were removed by mild acid treatment (see above) and reconstituted with the peptide of interest at 200 μM in 2% FCS/IMDM containing 2 μg/ml human β2-microglublin (Sigma-Aldrich) for 1.5 h at room temperature. Hereafter, cells were washed twice to remove free peptide and incubated at 37°C for 0, 2, 4, and 6 h. Subsequently, expressed HLA-A*0201–peptide complexes on JY cells were stained using the conformation-specific Moab BB7.2 and goat anti–mouse IgG–FITC and analyzed on a FACScan™ flow cytometer. The fluorescence index (FI) was calculated for each sample as: (MFsample − MFbackground)/MFbackground, where MFbackground is the value without peptide. The percentage of residual HLA-A*0201 molecules was calculated by equating for each peptide the FI of t = 2 h to 100% and then using the formula: % remaining = (FIt=n/FIt=2) × 100%. Because the dissociation of peptides from MHC is a linear process, the stability of the peptide–MHC complexes was measured as the time required for 50% of the molecules to decay (DT50), starting from t = 2. By linear regression analysis of the sequential measurements plotted against the percentage of remaining HLA-A*0201 molecules, the DT50 was calculated. As a positive control, the known highly stable HBV nucleocapsid 18–27 epitope was used.
In Vitro Proteasome-mediated Digestions.
20S proteasomes were purified from a B-LCL cell line as described 57. This cell type is known to contain immunoproteasomes 58. High LMP2 and LMP7 content was confirmed by two-dimensional immunoblotting (data not shown). To assess kinetics, digestions were performed with different incubation periods. Peptides (27 mer, 20 μg) were incubated with 1 μg of purified proteasome at 37°C for 1, 4, and 24 h in 300 μl proteasome digestion buffer as described 44. TFA (30 μl) was added to stop the digestion and samples were stored at −20°C before mass spectrometric analysis.
Mass Spectrometry.
Electrospray ionization mass spectrometry was performed on a hybrid quadrupole time of flight mass spectrometer, a Q-TOF (Micromass), equipped with an on-line nanoelectrospray interface (capillary tip, 20-μm internal diameter × 90-μm outer diameter) with an approximate flow rate of 250 nl/min. This flow was obtained by splitting of the 0.4 ml/min flow of a conventional high pressure gradient system, using an Acurate flow splitter (LC Packings). Injections were done with a dedicated micro/nano HPLC autosampler, the FAMOS (LC Packings), equipped with two additional valves for phase system switching experiments. Digestion solutions were diluted five times in water/methanol/acetic acid (95:5:1, vol/vol/vol), and 1 μl was trapped on the precolumn (MCA-300-05-C8; LC Packings) in water/methanol/acetic acid (95:5:1, vol/vol/vol). Washing of the precolumn was done for 3 min to remove the buffers present in the digests. Subsequently, the trapped analytes were eluted with a steep gradient going from 70% B to 90% B in 10 min, with a flow of 250 nl/min (A, water/methanol/acetic acid [95:5:1, vol/vol/vol]; B, water/methanol/acetic acid [10:90:1, vol/vol/vol]). This low elution rate allows for a few additional mass spectrometry (MS)/MS experiments if necessary during the same elution. Mass spectra were recorded from mass 50–2,000 daltons every second with a resolution of 5,000 full width/half maximum (FWHM). The resolution allows direct determination of the monoisotopic mass, also from multiple charged ions. In MS/MS mode, ions were selected with a window of 2 daltons with the first quadrupole and fragments were collected with high efficiency with the orthogonal time of flight mass spectrometer. The collision gas applied was argon (4 × 10−5 mbar), and the collision voltage ∼30 V. The peaks in the mass spectrum were searched in the digested precursor peptide using the Biolynx/proteins software (Micromass) supplied with the mass spectrometer. The intensity of the peaks in the mass spectra was used to establish the relative amounts of peptides generated after proteasome digestion. The relative amounts of the peptides are given as a percentage of the total amount of peptide digested by the proteasome at the indicated incubation time.
RT-PCR Assay for PRAME Expression.
Analysis of PRAME mRNA expression was determined by RT-PCR. Total cellular RNA was isolated with Trizol (GIBCO BRL) according to the manufacturer's procedure. RT reaction was performed on 5 μg of total RNA in a reaction volume of 25 μl with 5 μl of 5× reverse transcriptase buffer (Promega), 2.5 μl each of 10 mM deoxynucleotides (Amersham Pharmacia Biotech), 0.5 μg oligo dT15 primer, 25 U of RNAsin (Promega), and 15 U avian myeloblastosis virus (AMV) reverse transcriptase (Promega). The RT reaction was incubated at 42°C for 60 min, heat inactivated for 10 min at 70°C, and diluted two times with water. For PCR amplification, 1 μl of reverse transcribed cDNA reaction mixture was used as a template. PCR primers used for the analysis of PRAME expression were OPC 189 (sense primer, 5′-CTGTACTCATTTCCAGAGCCAGA-3′) and OPC 190 (antisense primer, 5′-TATTGAGAGGGTTTCCAAGGGGTT-3′; reference 48). PCR conditions were 5 min at 94°C followed by 34 cycles consisting of 30 s at 94°C, 2 min at 64°C, and 3 min at 72°C.
In Vitro CTL Response Induction and Generation of CTL Clones.
PBMCs of two HLA-A*0201+ healthy donors (one donor for induction against PRA300–309 and the other donor for inductions against PRA100–108, PRA142–151, PRA425–433, and PRA47–56) were obtained by the Ficoll-Paque method and used for CTL inductions. To optimally use all APCs present in PBMCs, we developed a culture system that yields a mix of activated B cells and mature DCs to be used as APCs during the primary induction step. PBMCs were separated in a T cell fraction and a fraction containing B cells and monocytes by SRBC rosetting. The T cell fraction was cryopreserved. The mixture of monocytes and B cells was cultured in 24-well plates at a concentration of 106 cells/well in complete culture medium containing 800 U/ml GM-CSF (provided by S. Osanto, Leiden University Medical Center), 500 U/ml IL-4 (PeproTech), and 500 ng/ml CD40 mAb (clone B-B20; Serotec) for 6 d. This culture system achieved a threefold effect: (a) GM-CSF and IL-4 induced differentiation of monocytes into immature DCs 59, (b) IL-4 and CD40 mAbs caused activation and proliferation of B cells 60, and (c) CD40 mAb mediated maturation of immature DCs 61. At day 3, cytokines and CD40 mAb were replenished. To further promote CTL-inducing capacity, the APC mix was cultured for an additional 2 d with 0.4 ng/ml LPS (Difco Labs), 500 U/ml IFN-γ (Boehringer Mannheim), and 500 ng/ml CD40 mAb. At day 8 the APC mix was pulsed with 50 μg/ml peptide (each peptide separately) for 4 h at room temperature, irradiated (30 Gy), and washed to remove free peptide. The cryopreserved autologous T cell fraction was thawed and depleted from CD4+ T cells using magnetic beads (Dynal). The primary induction was performed in 96-well U-bottomed plates. APCs at a concentration of 10,000/well were cocultured with 50,000 CD8+ T cells/well in culture medium containing 10% human pooled serum (HPS), 5 ng/ml IL-7 (PeproTech), and 0.1 ng/ml IL-12 (Sigma-Aldrich). At day 7 after initiation of induction, the CTL microcultures were harvested (pooled), washed, and restimulated at a concentration of 40,000 responder cells/well of 96-well U-bottomed plates in culture medium containing 10% HPS, 5 ng/ml IL-7, and 0.1 ng/ml IL-12. Autologous-activated B cells, generated via the protocol described by Schultze et al. 60, irradiated (75 Gy), and peptide pulsed (50 μg/ml) for 4 h at room temperature in culture medium containing 2% FCS and 3 μg/ml β2-microglublin (Sigma-Aldrich) after mild acid elution to remove naturally presented peptides from the MHC class I molecules (see Materials and Methods, MHC binding assay), were used at a concentration of 10,000 cells/well as restimulator APCs. Restimulations were repeated at day 14 and 21 in a similar way, with the exception of IL-7 being replaced by 20 IU/ml IL-2 (Chiron Corp.). At day 29, the CTL bulk culture was cloned by standard limiting dilution procedures. CTL clones were maintained by aspecific stimulation every 7 to 12 d using a feeder mixture consisting of allogeneic PBMCs and B-LCL in culture medium containing 10% FCS, 1.5% leucoagglutinin (Sigma-Aldrich), and 240 IU/ml IL-2.
51Cr Cytotoxicity Assay, HLA Class I Blocking, and Proteasome Inhibition.
CTL activity was measured in standard chromium release assays. In brief, after 51Cr labeling (1 h), target cells (2,000/well) were added to various numbers of effector cells in a final volume of 100 μl complete culture medium in 96-well U-bottomed plates. After 4 h incubation at 37°C, supernatants were harvested. The mean percentage of specific lysis of triplicate wells was calculated according to: (experimental release − spontaneous release)/(maximal release − spontaneous release) × 100%. For peptide titration experiments, 51Cr-labeled target cells (2,000/well) were pulsed with titrated amounts of peptide for 1 h at 37°C in 96-well plates. Subsequently, CTLs were added at an E/T ratio of 10. HLA class I blocking was accomplished by treatment of 2 × 105 51Cr-labeled M453 melanoma cells for 1 h with mAb W6.32 or control IgG2a at room temperature before addition to effector cells. Inhibition of proteasome function in melanoma FM3 was performed by treatment with 10 μM of lactacystin (Calbiochem) for 17 h during culture at 37°C. Thereafter, FM3 cells were harvested and 51Cr labeled for use in the cytotoxicity assay. Reconstitution of lysis by peptide was performed by pulsing lactacystin-treated and 51Cr-labeled FM3 cells for 30 min with 5 μM peptide.
Results
Identification of HLA-A*0201 Binding Peptides from PRAME.
To select candidate HLA-A*0201 binding peptides from PRAME, its aa sequence was screened for HLA-A*0201 binding motif containing peptides with a combination of two known binding prediction algorithms 62 63. Only peptides of 9 or 10 aa length were included, taking into account the low prevalence of HLA-A*0201–restricted CTL epitopes of 8 or 11 aa length 64. In total, 128 peptides (65 nonamers and 63 decamers) were synthesized in order to determine their actual binding affinity for HLA-A*0201 using a competition-based cellular binding assay 54. 19 high affinity binding peptides were identified (IC50 ≤ 6 μM), and 27 peptides bound with intermediate affinity (6 μM < IC50 ≤ 15 μM), whereas the other peptides displayed a low (15 μM < IC50 ≤ 100 μM) or undetectable binding capacity (IC50 > 100 μM; Table ). To more precisely define binding characteristics, peptide–MHC stability was assessed by measuring the dissociation rate of high affinity binding peptides complexed with HLA-A*0201 at 37°C 56. Two of the tested high affinity binding peptides (PRA292–301 and PRA190–199) showed a high off rate from HLA-A*0201, because <10% of HLA-A*0201–peptide complexes were detectable after 2 h incubation at 37°C. In previous work we have detected a strong correlation between MHC–peptide binding stability and immunogenicity in vivo 56. Therefore, PRA292–301 and PRA190–199 are, with respect to their binding characteristics, not likely to be efficiently presented in HLA-A*0201. For all other peptides, the 50% decay time (DT50) was 2.5 h or longer (Table ), indicating a stable association with HLA-A*0201.
Table 1.
Binding Affinity for HLA-A0201 of 128 Nonamers and Decamers Derived from PRAME
| Start | Sequence | Length | IC50 | Start | Sequence | Length | IC50 | Start | Sequence | Length | IC50 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 300 | ALYVDSLFFL | 10 | 1.7 | 466 | RLRELLCEL | 9 | 14.0 | 86 | LMKGQHLHL | 9 | 62.3 |
| 142 | SLYSFPEPEA | 10 | 1.9 | 33 | SLLKDEALAI | 10 | 14.0 | 240 | VTCTWKLPTL | 10 | 67.2 |
| 47 | LLPRELFPPL | 10 | 2.1 | 422 | ALQSLLQHL | 9 | 14.2 | 44 | ALELLPREL | 9 | 71.3 |
| 435 | NLTHVLYPV | 9 | 2.5 | 103 | GLDVLLAQEV | 10 | 15.2 | 379 | TLQDLVFDEC | 10 | 71.9 |
| 292 | FLSLQCLQAL | 10 | 2.5 | 231 | QLDSIEDLEV | 10 | 15.5 | 371 | ALLERASAT | 9 | 72.0 |
| 394 | QLLALLPSL | 9 | 2.9 | 312 | RLDQLLRHV | 9 | 15.7 | 353 | VLSLSGVMLT | 10 | 74.6 |
| 182 | FLKEGACDEL | 10 | 3.0 | 493 | RTFYDPEPI | 9 | 15.8 | 305 | SLFFLRGRL | 9 | 79.4 |
| 294 | SLQCLQALYV | 10 | 3.2 | 308 | FLRGRLDQL | 9 | 16.1 | 409 | TTLSFYGNSI | 10 | 80.8 |
| 422 | ALQSLLQHLI | 10 | 3.2 | 429 | HLIGLSNLT | 9 | 16.3 | 93 | HLETFKAVL | 9 | 89.0 |
| 425 | SLLQHLIGL | 9 | 3.7 | 85 | VLMKGQHLHL | 10 | 17.0 | 319 | HVMNPLETL | 9 | 90.3 |
| 258 | QMINLRRLLL | 10 | 4.0 | 316 | LLRHVMNPL | 9 | 17.4 | 18 | SVWTSPRRLV | 10 | >100 |
| 190 | ELFSYLIEKV | 10 | 4.5 | 353 | VLSLSGVML | 9 | 17.4 | 20 | WTSPRRLVEL | 10 | >100 |
| 248 | TLAKFSPYL | 9 | 4.6 | 172 | FIPVEVLVDL | 10 | 17.5 | 26 | LVELAGQSL | 9 | >100 |
| 39 | ALAIAALEL | 9 | 5.1 | 134 | TVWSGNRASL | 10 | 18.4 | 51 | ELFPPLFMA | 9 | >100 |
| 100 | VLDGLDVLL | 9 | 5.2 | 339 | VMHLSQSPSV | 10 | 18.5 | 67 | QTLKAMVQA | 9 | >100 |
| 333 | RLSEGDVMHL | 10 | 5.4 | 72 | MVQAWPFTC | 9 | 18.5 | 70 | KAMVQAWPFT | 10 | >100 |
| 462 | YLHARLREL | 9 | 5.4 | 390 | ITDDQLLALL | 10 | 18.9 | 78 | FTCLPLGVL | 9 | >100 |
| 360 | MLTDVSPEPL | 10 | 5.6 | 18 | SVWTSPRRL | 9 | 19.1 | 84 | GVLMKGQHL | 9 | >100 |
| 419 | SISALQSLL | 9 | 5.7 | 315 | QLLRHVMNPL | 10 | 19.7 | 95 | ETFKAVLDGL | 10 | >100 |
| 432 | GLSNLTHVL | 9 | 6.8 | 71 | AMVQAWPFT | 9 | 20.0 | 133 | WTVWSGNRA | 9 | >100 |
| 214 | KIFAMPMQDI | 10 | 7.2 | 207 | RLCCKKLKI | 9 | 20.8 | 155 | MTKKRKVDGL | 10 | >100 |
| 320 | VMNPLETLSI | 10 | 8.6 | 247 | PTLAKFSPYL | 10 | 21.1 | 165 | STEAEQPFI | 9 | >100 |
| 39 | ALAIAALELL | 10 | 9.0 | 219 | PMQDIKMIL | 9 | 23.9 | 180 | DLFLKEGAC | 9 | >100 |
| 390 | ITDDQLLAL | 9 | 9.2 | 459 | RLAYLHARL | 9 | 24.3 | 198 | KVKRKKNVL | 9 | >100 |
| 242 | CTWKLPTLA | 9 | 9.3 | 264 | RLLLSHIHA | 9 | 24.6 | 205 | VLRLCCKKL | 9 | >100 |
| 99 | AVLDGLDVLL | 10 | 9.4 | 217 | AMPMQDIKMI | 10 | 24.6 | 222 | DIKMILKMV | 9 | >100 |
| 308 | FLRGRLDQLL | 10 | 9.6 | 361 | LTDVSPEPL | 9 | 26.8 | 224 | KMILKMVQL | 9 | >100 |
| 355 | SLSGVMLTDV | 10 | 9.9 | 430 | LIGLSNLTHV | 10 | 27.2 | 234 | SIEDLEVTC | 9 | >100 |
| 34 | LLKDEALAI | 9 | 10.2 | 33 | SLLKDEALA | 9 | 29.2 | 234 | SIEDLEVTCT | 10 | >100 |
| 284 | YIAQFTSQFL | 10 | 10.4 | 258 | QMINLRRLL | 9 | 31.2 | 237 | DLEVTCTWKL | 10 | >100 |
| 71 | AMVQAWPFTC | 10 | 10.4 | 91 | HLHLETFKAV | 10 | 31.8 | 240 | VTCTWKLPT | 9 | >100 |
| 470 | LLCELGRPSM | 10 | 10.5 | 297 | CLQALYVDSL | 10 | 33.8 | 261 | NLRRLLLSHI | 10 | >100 |
| 186 | GACDELFSYL | 10 | 10.6 | 372 | LLERASATL | 9 | 35.0 | 325 | ETLSITNCRL | 10 | >100 |
| 410 | TLSFYGNSI | 9 | 11.0 | 401 | SLSHCSQLT | 9 | 36.9 | 368 | PLQALLERA | 9 | >100 |
| 25 | RLVELAGQSL | 10 | 11.1 | 397 | ALLPSLSHC | 9 | 42.6 | 382 | DLVFDECGI | 9 | >100 |
| 91 | HLHLETFKA | 9 | 11.1 | 389 | GITDDQLLAL | 10 | 47.3 | 382 | DLVFDECGIT | 10 | >100 |
| 100 | VLDGLDVLLA | 10 | 11.9 | 417 | SISISALQSL | 10 | 48.2 | 383 | LVFDECGIT | 9 | >100 |
| 454 | TLHLERLAYL | 10 | 12.2 | 259 | MINLRRLLL | 9 | 48.2 | 389 | GITDDQLLA | 9 | >100 |
| 371 | ALLERASATL | 10 | 12.9 | 479 | MVWLSANPC | 9 | 49.0 | 401 | SLSHCSQLTT | 10 | >100 |
| 326 | TLSITNCRL | 9 | 13.2 | 160 | KVDGLSTEA | 9 | 51.2 | 473 | ELGRPSMVWL | 10 | >100 |
| 462 | YLHARLRELL | 10 | 13.3 | 436 | LTHVLYPVPL | 10 | 53.0 | 481 | WLSANPCPHC | 10 | >100 |
| 350 | QLSVLSLSGV | 10 | 13.3 | 226 | ILKMVQLDSI | 10 | 56.1 | 493 | RTFYDPEPIL | 10 | >100 |
| 99 | AVLDGLDVL | 9 | 13.4 | 292 | FLSLQCLQA | 9 | 58.7 |
Table 2.
Stability of High Affinity Binding Peptides in HLA-A0201
| Start | Sequence | Affinity IC50 | Stability DT50 |
|---|---|---|---|
| μM | h | ||
| 300 | ALYVDSLFFL | 1.7 | >4 |
| 142 | SLYSFPEPEA | 1.9 | 3 |
| 47 | LLPRELFPPL | 2.1 | 2.5 |
| 435 | NLTHVLYPV | 2.5 | 3 |
| 292 | FLSLQCLQAL | 2.5 | N.S. |
| 394 | QLLALLPSL | 2.9 | >4 |
| 182 | FLKEGACDEL | 3.0 | 3 |
| 294 | SLQCLQALYV | 3.2 | >4 |
| 422 | ALQSLLQHLI | 3.2 | 2.5 |
| 425 | SLLQHLIGL | 3.7 | >4 |
| 258 | QMINLRRLLL | 4.0 | >4 |
| 190 | ELFSYLIEKV | 4.5 | N.S. |
| 248 | TLAKFSPYL | 4.6 | >4 |
| 100 | VLDGLDVLL | 5.2 | 2.5 |
| HBV control | >4 |
In Vitro Proteasome-mediated Digestions of 27-mer Polypeptides Encompassing HLA-A*0201 Binding Peptides.
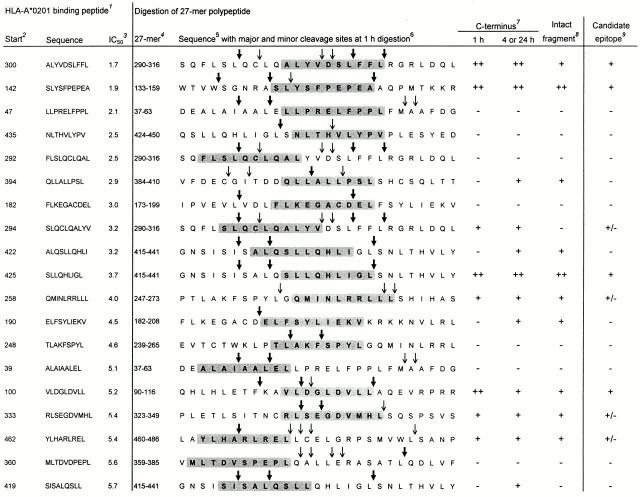
The two most important requirements for a peptide to be naturally presented as CTL epitope are: (a) proper excision from the protein by the proteolytic machinery and (b) sufficient binding affinity for HLA class I molecules. Therefore, we analyzed in vitro proteasome-mediated digestions of 27-mer polypeptides encompassing the 19 identified high affinity HLA-A*0201 binding peptides. Potential epitopes were primarily assessed for efficient liberation (i.e., by a major cleavage site at 1 h incubation) of their precise COOH terminus, which is a first requirement for the generation of most CTL epitopes 34 35 36 37. Intactness of the candidate epitope was evaluated as a secondary factor favoring efficient processing and presentation. 20S proteasomes isolated from a human EBV–transformed B cell line were used for digestions with 1-, 4-, and 24-h incubation periods, and mass spectrometry profiles of the digestion products were analyzed. Digestion patterns of four 27-mer polypeptides, all containing potential high affinity HLA-A*0201 binding epitopes, are shown in Fig. 1.
Figure 1.
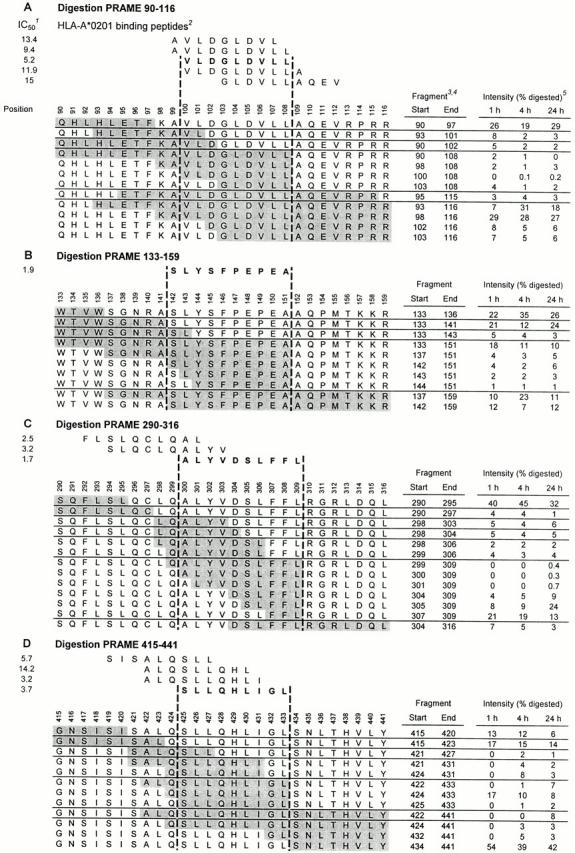
In vitro proteasome-mediated digestions of four 27-mer PRAME polypeptides containing potential HLA-A*0201–restricted epitopes. 20S proteasomes isolated from an EBV-transformed B cell line were coincubated with 27-mer PRAME peptides at 37°C for the indicated time points. Digestion mixtures were analyzed by mass spectrometry as described in Materials and Methods. Generated digestions fragments are depicted as shaded sequences. The digestion of 27-mer PRAME 90–116 is represented in A, digestion of PRAME 133–159 is depicted in B, in C the digestion of PRAME 290–316 is showed, and D represents the digestion of PRAME 415–441. Notes: 1 IC50 as determined in competition binding assay (see Table ); 2 peptides binding with high or intermediate affinity to HLA-A*0201 are shown. Predicted epitopes used for CTL induction are printed in bold; 3 digestion fragments are shaded and ordered according to their COOH terminus; 4 digestion fragments generated for <1% at 1 h digestion or <3% at 4 and 24 h incubation and not relevant for epitope prediction are not shown; 5 intensity is expressed as percentage of total summed mass-peak intensities of digested 27-mer at the indicated incubation time.
Fig. 1 A shows the digestions of PRAME 90–116, which harbors five HLA-A*0201 binding peptides (Table ) with their natural flanking residues. Of the COOH-terminal residues of the five HLA-A*0201 binding peptides, only Leu-108 was generated (fragments containing this COOH terminus added up to 8% at 1 h digestion). Therefore, both the 9-mer VLDGLDVLL (PRA100–108) and the 10-mer AVLDGLDVLL (PRA99–108) represent potential CTL epitopes. The NH2 terminus of the epitope precursor is likely to be Lys-98, because the fragments most frequently generated were aa 90–97 and its complement aa 98–116, indicating an abundantly cleaved site after Phe-97.
Fig. 1 B shows the digestions of 27-mer PRAME 133–159, which contains 10-mer SLYSFPEPEA (PRA142–151), the second best HLA-A*0201 binding peptide (Table ). Fragments sharing Ala-151 as COOH terminus added up to 29% at 1 h digestion, indicating an abundantly cleaved site after this residue. Furthermore, fragment aa 142–159 and the complementary fragment aa 133–141 were abundantly present, pointing to a major cleavage site after Ala-141. Thus, major cleavage sites were present just after and before SLYSFPEPEA, rendering this peptide a potential efficiently generated CTL epitope.
Fig. 1 C depicts digestions of PRAME 290–316 encompassing 10-mer ALYVDSLFFL (PRA300–309), which bound best in HLA-A*0201 (Table ) and has its COOH terminus (Leu-309) in common with the already described HLA-A24–presented 9-mer LYVDSLFFL (PRA301–309; reference 48). As might be expected on that basis, a cleavage site after Leu-309 was observed, because digestion fragments sharing this COOH terminus were abundantly generated after 1 h incubation. However, PRA300–309 itself was found intact only after 24 h incubation at low quantities. This is probably due to cleavage sites within this potential epitope (after Val-303, Asp-304, and Leu-306). HLA-A*0201 binding peptides PRA292–301 and PRA294–303 (also in PRAME 290–316) were, respectively, not COOH-terminally generated and not found as intact fragment, indicating that these peptides are not likely to be naturally generated in the processing pathway.
Fig. 1 D shows the digestion pattern of PRAME 415–441, which harbors four peptides binding in HLA-A*0201 (Table ). The NH2-terminally elongated decameric precursor 424QSLLQHLIGL433 of high affinity binding 9-mer SLLQHLIGL (PRA425–433) was efficiently generated. The abundant generation of the COOH-terminal and NH2-terminal counterparts of this 10-mer precursor (fragments aa 434–441 and aa 415–423, respectively) were also pointing to major cleavage sites just after and before 424QSLLQHLIGL433, indicating PRA425–433, a potential CTL epitope. The three other HLA-A*0201 binding peptides were either not COOH-terminally excised (PRA419–427 and PRA422–430) or the correct COOH terminus was found only after 4 h incubation (PRA422–431).
A concise representation of digestion analysis of 27-mers harboring all 19 high affinity binding peptides, including those discussed above, is shown in Fig. 2. Summarizing, 11 HLA-A*0201 binding peptides were either not COOH-terminally excised (PRA47–56, PRA435–443, PRA292–301, PRA182–191, PRA248–256, PRA100–108, and PRA360–369) or the correct COOH terminus was generated only after 4 h incubation by a minor cleavage site (PRA394–402, PRA422–431, PRA190–199, and PRA419–427). The absence or late appearance of fragments containing the correct COOH terminus render these 11 peptides very unlikely to constitute naturally processed epitopes. Furthermore, three peptides were COOH-terminally liberated at 1 h digestion but only in very low quantities (<1%, data not shown; PRA258–267, PRA333–342, and PRA462–470), whereas PRA294–303 was not found as intact fragment. Consequently, it is doubtful that the latter peptides are efficiently generated in vivo. Only the high affinity binding peptides PRA100–108, PRA142–151, PRA300–309, and PRA425–433 were COOH-terminally excised by a major cleavage site at 1 h incubation and found intact in digestion fragments, indicating possible CTL epitopes (Fig. 1 and Fig. 2). Therefore, these four peptides were chosen for CTL inductions.
Figure 2.
Proteasome-mediated cleavage patterns of 27-mer peptides encompassing 19 high affinity HLA-A*0201 binding PRAME peptides. 20S proteasomes isolated from an EBV-transformed B cell line were coincubated with 27-mer PRAME peptides at 37°C for the indicated time points. Digestion mixtures were analyzed by mass spectrometry as described in Materials and Methods. Major and minor cleavages sites at 1 h digestion are depicted. Notes: 1 all 19 high affinity binding peptides (IC50 < 6 μM) are listed and ranked according to their binding affinity for HLA-A*0201; 2 start aa position in PRAME of the HLA-A*0201 binding peptide. 3 IC50 (in μM) as determined in competition binding assay (see Table ); 4 start and end aa position in PRAME of 27-mer polypeptide encompassing the high affinity binding peptide; 5 aa sequence of 27-mer polypeptide (aa sequence of HLA-A*0201 binding peptide is printed in bold and shaded); 6 major (bold arrows) and minor (thin arrows) cleavage sites at 1 h incubation are depicted, classified according to the following definitions. Major site: fragments containing as COOH terminus the residue NH2-terminal from the cleavage are present for ≥5% at 1 h incubation. Minor site: fragments containing as COOH terminus the residue NH2-terminal from the cleavage are present for <5% at 1 h incubation. 7 Generation by digestion of fragments containing the correct COOH terminus of the HLA-A*0201 binding peptide. Generation at 1 h digestion or after a longer incubation period is separately indicated. Classification: (++) present for ≥5%, (+) present for <5%, (−) no fragments containing the correct COOH terminus were found. 8 Generation by digestion of fragments containing the intact HLA-A*0201 binding peptide and/or NH2-terminal elongated precursors of the peptide. Classification: (++) present for ≥5% at 1 h incubation, (+) present for <5% at 1 h or only detectable after 4 or 24 h, (−) no fragments containing the HLA-A*0201 binding peptide were found. 9 Epitope prediction based on digestion results. Classification: (+) most likely an epitope, (+/−) doubtful epitope, (−) not an epitope.
In Vitro Human CTL Inductions against Four Putative HLA-A*0201–restricted Epitopes.
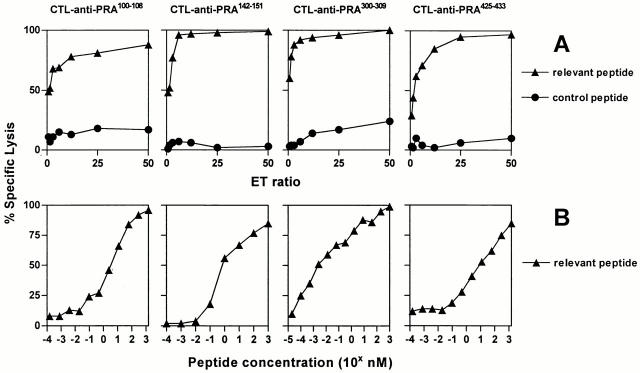
Separate CTL inductions, using PBMCs of healthy donors, were performed against VLDGLDVLL (PRA100–108), SLYSFPEPEA (PRA142–151), ALYVDSLFFL (PRA300–309), and SLLQHLIGL (PRA425–433) according to the protocol described in Materials and Methods. At day 28, the CTL bulk cultures were tested in a 51Cr release assay to asses peptide specificity. The CTL cultures raised against SLYSFPEPEA and SLLQHLIGL showed high specificity for targets loaded with the inducing peptide (at an E/T ratio of 5, both cultures reached 80% lysis compared with 20% lysis of targets loaded with a control peptide), whereas the other two CTL cultures displayed only slightly increased lysis of targets loaded with the relevant peptide (data not shown). The four CTL bulk cultures were cloned by limiting dilution at day 29. The peptide specificity of generated CTL clones was initially assessed in a split-well cytotoxicity assay. Despite low peptide specificity of CTL bulk cultures against VLDGLDVLL and ALYVDSLFFL, CTL clones specific for all four peptides were found. In summary, 51 of 576 (9%) CTL clones induced against VLDGLDVLL (PRA100–108) showed specific lysis of peptide pulsed targets and 19 of 202 (9%) CTL clones raised against ALYVDSLFFL (PRA300–309) displayed peptide specificity. As may be expected, higher percentages, namely 92% (279 of 304), of clones against SLYSFPEPEA (PRA142–151) and 29% (97 of 336) of clones against SLLQHLIGL (PRA425–433) showed peptide specificity. Based on peptide reactivity and growth characteristics, several CTL clones were functionally characterized in detail. For each specificity, one CTL clone is presented in this study. As shown in Fig. 3 A, representative CTL clones raised against the four different peptides all showed highly specific and efficient lysis of T2 cells pulsed with 5 μM of their inducing peptides at low E/T ratios. Peptide sensitivity of the CTL clones was determined in peptide titration experiments (Fig. 3 B). CTL no. 551 (anti-PRA100–108) was able to half-maximally lyse targets loaded with VLDGLDVLL at ∼5 nM peptide. CTL no. 314, raised against SLYSFPEPEA (PRA142–151), lysed T2 cells at half-maximal level when pulsed with <1 nM of the inducing peptide. CTL no. 460 (anti-PRA300–309) was extremely sensitive in lysing T2 cells pulsed with ALYVDSLFFL: half-maximal lysis was reached at ∼3 pM peptide concentration. Finally, CTL no. 1257 (anti-PRA425–433) was able to half-maximally lyse targets loaded with SLLQHLIGL at <12 nM. To analyze clonality of the CTL clones under investigation, we performed RT-PCR analysis with a panel of 24 primers of junctional regions of TCRB transcripts from 22 well-established TCRBV families to determine Vβ usage of the TCR 65. All CTL clones were shown to use a single Vβ, confirming clonality of the clones (data not shown).
Figure 3.
HLA-A*0201–restricted peptide specificity and sensitivity of CTL clones raised against four PRAME peptides. (A) Lysis by CTL clones no. 551 anti-PRA100–108, no. 314 anti-PRA142–151, no. 460 anti-PRA300–309, and no. 1257 anti-PRA425–433 of 51Cr-labeled T2 cells loaded with 5 μM of the relevant peptide (▴) vs. an irrelevant HLA-A*0201 binding peptide (•) at different E/T ratios ranging from 50 to 0.75. (B) Lysis by the same set of CTL clones of 51Cr-labeled T2 cells pulsed for 1 h with titrated concentrations of relevant peptide (▴). The CTL clones were used at an E/T ratio of 10. Results of one representative experiment out of three performed are shown.
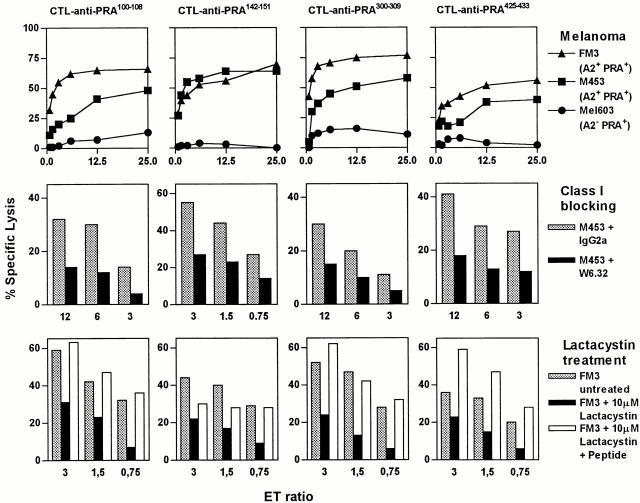
CTLs Raised against Four PRAME Peptides Recognize Melanoma Cell Lines Coexpressing HLA-A*0201 and PRAME in an HLA I–restricted and Proteasome-dependent Fashion.
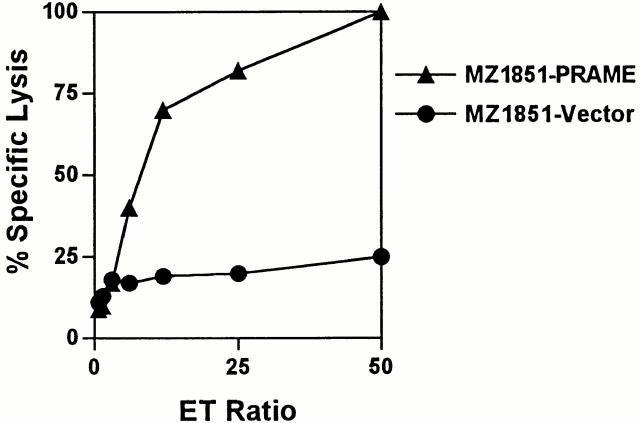
Endogenous presentation of the candidate epitopes PRA100–108, PRA142–151, PRA300–309, and PRA425–433 in HLA-A*0201 was explored by assessing the ability of CTL clones that were raised against these peptides to specifically lyse melanoma cell lines M453 and FM3 expressing HLA-A*0201 (confirmed by flow cytometry, data not shown) and PRAME (determined by Northern blotting, data not shown). Both melanoma cell lines were efficiently lysed by all four CTL clones as measured in a 51Cr release assay, whereas the melanoma Mel603 expressing PRAME (assayed with RT-PCR, data not shown) but lacking HLA-A*0201 was not killed above background level (Fig. 4, top panel). Lysis of M453 was significantly inhibited after treatment of this target with HLA class I blocking mAb W6.32 (Fig. 4, middle panel), indicating that killing of M453 by these CTL clones involved HLA class I–restricted recognition. Furthermore, proteasome inhibition experiments with lactacystin were performed. Lysis of FM3 pretreated for 17 h with lactacystin (10 μM) was significantly diminished (Fig. 4, bottom panel). This indicated, in concordance with our in vitro proteasome digestion data, that generation of the four epitopes is proteasome dependent. To confirm PRAME as source of antigen naturally presented in HLA-A*0201, renal cell carcinoma cell line MZ1851, which is HLA-*0201+ but lacks PRAME expression, was transfected with full-length PRAME cDNA (MZ1851-PRAME). As shown in Fig. 5, PRAME expression (confirmed by RT-PCR) sensitized MZ1851-PRAME for lysis by a CTL clone directed against PRA300–309.
Figure 4.
Recognition by CTL clones of four endogenously processed PRAME epitopes presented on melanoma cell lines in an HLA class I–restricted and proteasome-dependent fashion. (Top) Lysis of 51Cr-labeled melanoma cell line Mel603, expressing PRAME but lacking HLA-A*0201 expression (•), was tested vs. lysis of M453 (▪) and FM3 (▴), both expressing PRAME and HLA-A*0201 together. CTL clones no. 551 anti-PRA100–108, no. 314 anti-PRA142–151, no. 460 anti-PRA300–309, and no. 1257 anti-PRA425–433 were used at E/T ratios ranging from 25 to 0.75. (Middle) Lysis of 51Cr-labeled M453 was tested after 1 h preincubation with HLA class I blocking mAb W6.32 (black bars) or an IgG2a control Ab (gray bars). (Bottom) Lysis of 51Cr-labeled FM3 was tested after 17 h treatment with 10 μM of the proteasome inhibitor lactacystin (black bars) or without treatment (gray bars). As control, the lactacystin-treated cells were loaded with the relevant peptide (white bars). Results of one representative experiment of at least three performed are shown.
Figure 5.
Lysis of PRAME transfected renal cell carcinoma cell line MZ1851 by CTL anti-PRA300–309. CTL no. 460 directed against PRA300–309 was tested on 51Cr-labeled MZ1851 (HLA-A*0201+ but lacking PRAME expression) transfected with PRAME cDNA (▴) vs. MZ1851 transfected with the empty vector (•). CTL no. 460 was used at E/T ratios ranging from 50 to 0.75. Results of one representative experiment of three performed are shown.
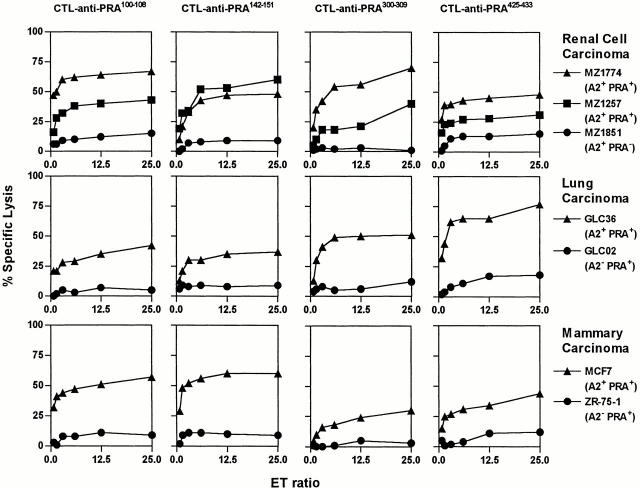
CTLs Reactive with Four PRAME Epitopes Lyse a Broad Array of Tumor Cell Lines Expressing HLA-A*0201 and PRAME.
To investigate HLA-A*0201–restricted presentation of PRA100–108, PRA142–151, PRA300–309, and PRA425–433 on tumor cells from histologic origins other than melanoma, we used panels of cell lines derived from various tumor types which have been reported to express PRAME 48 50.
Lysis by the selected CTL clones of tumor cell lines with or without HLA-A*0201 expression and naturally expressing PRAME or lacking PRAME expression was compared. HLA-A*0201 expression was confirmed by flow cytometry (data not shown) and PRAME expression by RT-PCR or Northern blotting (data not shown). Lysis of the HLA-A*0201+ renal cell carcinoma (RCC) cell line MZ1851, which lacks PRAME expression, was compared with lysis of RCC cell lines MZ1257 and MZ1774, both expressing HLA-A*0201 and PRAME. The CTL clones reactive against the four different PRAME peptides showed significant lysis of the two PRAME+ cell lines but not of MZ1851, again confirming PRAME as the source of target antigens (Fig. 6, top panel). Likewise, lysis of lung carcinoma cell lines was HLA-A*0201 restricted and PRAME specific, because only GLC36 expressing HLA-A*0201+ and PRAME+, and not GLC02, which is PRAME+ but lacks HLA-A*0201 expression, was killed (Fig. 6, middle panel). Mammary carcinoma cell line MCF7 (HLA-A*0201+ and PRAME+) was killed efficiently as well, whereas cell line ZR-75-1, which lacks HLA-A*0201 but expresses PRAME, was not lysed above background level (Fig. 6, bottom panel). Finally, HPV16+ cervix carcinoma cell line C33 and osteosarcoma cell line SAOS, both HLA-A*0201+ and PRAME+, were efficiently killed by the CTL clones (data not shown), underscoring the broad expression pattern of the PRAME epitopes by tumors. Taken together, we observed a consistent lysis of tumor cell lines when both the relevant MHC molecule and the tumor Ag were expressed. These results indicate that PRA100–108, PRA142–151, PRA300–309, and PRA425–433 are presented by HLA-A*0201 on a broad array of tumor cell lines.
Figure 6.
Lysis of tumor cell lines from multiple histologic origins by CTL anti-PRAME in a PRAME-specific and HLA-A*0201–restricted fashion. (Top) Lysis of 51Cr-labeled renal cell carcinoma cell lines MZ1851, expressing HLA-A*0201 but PRAME negative (•), MZ1257 (PRAME+ and HLA-A*0201+) (▪), and MZ1774 (PRAME+ and HLA-A*0201+) (▴) was compared. (Middle) Lysis of 51Cr-labeled lung carcinoma cell lines. GLC02, expressing PRAME but HLA-A*0201 negative (•) and GLC36 (▴) expressing both PRAME and HLA-A*0201 was compared. (Bottom) 51Cr-labeled mammary carcinoma cell lines MCF7 (HLA-*0201+ and PRAME+) (•) and ZR-75-1, expressing PRAME but lacking HLA-A*0201 (▴), were tested. The CTL clones no. 551 anti-PRA100–108, no. 314 anti-PRA142–151, no. 460 anti-PRA300–309, and no. 1257 anti-PRA425–433 were used at E/T ratios ranging from 25 to 0.75. Results of one representative experiment of at least three performed are shown.
CTL Clones Recognizing a Nonprocessed High Affinity Binding Peptide Do Not Lyse PRAME and HLA-A*0201–expressing Tumor Cell Lines.
To further validate our epitope prediction procedure and investigate possible false negative epitope prediction, CTLs were induced against the highest affinity HLA-A*0201 binding peptide that was not generated in vitro by proteasome-mediated breakdown: LLPRELFPPL (PRA47–56; Table and Fig. 2). Digestion of the 27-mer PRAME 37–63 showed that the COOH terminus of LLPRELFPPL was not generated. Instead, cleavage sites were observed after Leu-45, Met-58, and Ala-59 (Fig. 2). Dual cleavage fragments 46ELLPRELFPPLFM58 and 46ELLPRELFPPLFMA59 were abundantly found at 1 h digestion (data not shown). Using the same induction protocol as used for the four identified epitopes, CTLs were generated against LLPRELFPPL. Three CTL clones recognizing this peptide with high affinity were tested for lysis of PRAME and HLA-A*0201–expressing tumor cell lines at different E/T ratios (Table ). For comparison, three CTL clones recognizing the SLYSFPEPEA epitope (PRA142–151) with equal affinity as the affinity of CTL anti-PRA47–56 for LLPRELFPPL were included in the same experiment. The results show that CTLs raised against LLPRELFPPL do not lyse any of the tumor cell lines, whereas the same targets were efficiently killed by the control CTLs directed against SLYSFPEPEA (Table ). These data strongly suggest that LLPRELFPPL is not endogenously generated, supporting the accuracy of our epitope prediction procedure and the relevance of the proteasome digestion analysis. In addition to these data, high affinity CTL clones generated against three different BCR-ABL fusion protein–derived peptides failed to recognize BCR-ABL–expressing target cells (data not shown). Subsequent analysis of in vitro processing of these peptides showed that the proteasome did not generate the COOH terminus of any of these high affinity HLA class I binding peptides. These results confirm the importance of proper proteasomal cleavage for the generation of HLA class I–presented epitopes.
Table 3.
Percent Specific Lysis of PRAME- and HLA-A0201–Expressing Tumor Cell Lines by CTL Clones Specific for LLPRELFPPL (Not Processed) and SLYSFPEPEA (Processed) as Determined in a 51Cr Cytotoxicity Assay
| CTL clonesanti-LLPRELFPPL | CTL clonesanti-SLYSFPEPEA | ||||||
|---|---|---|---|---|---|---|---|
| Tumorcell line | E/Tratio | No. 3 | No. 61 | No. 120 | No. 314 | No. 343 | No. 509 |
| M453 | 6 | 2 | 2 | 10 | 33 | 39 | 26 |
| 3 | 3 | 3 | 11 | 26 | 32 | 16 | |
| 1, 5 | 1 | 2 | 9 | 27 | 17 | 15 | |
| MZ1257 | 6 | 1 | 9 | 0 | 52 | 28 | 27 |
| 3 | 1 | 8 | 4 | 34 | 16 | 26 | |
| 1, 5 | 2 | 8 | 2 | 32 | 12 | 12 | |
| MZ1774 | 6 | 7 | 8 | 10 | 43 | 42 | 31 |
| 3 | 8 | 9 | 10 | 33 | 25 | 16 | |
| 1, 5 | 4 | 4 | 8 | 21 | 13 | 10 | |
| GLC36 | 6 | 2 | 10 | 7 | 26 | 28 | 18 |
| 3 | 7 | 11 | 9 | 30 | 22 | 17 | |
| 1, 5 | 5 | 9 | 7 | 21 | 20 | 16 | |
| MCF7 | 6 | 5 | 8 | 7 | 56 | 26 | 32 |
| 3 | 6 | 9 | 7 | 52 | 24 | 19 | |
| 1, 5 | 4 | 9 | 5 | 48 | 21 | 13 | |
| CTL sensitivity | 0.1–1 | 0.1–1 | 0.1–1 | 0.1–1 | 0.1–1 | 0.1–1 | |
Discussion
In a systematic search for new CTL epitopes in known protein sequences with tumor restricted expression, the strategy of in vitro stimulation of CTLs with predicted epitopes (also coined “reverse immunology”) has successfully led to the identification of several epitopes 18 19 20 21 22 23 24 25, but has met with many failures and is generally inefficient (our unpublished results and references 26 27 28 29 30). The current study reports the identification of four novel HLA-A*0201–restricted CTL epitopes in PRAME (PRA100–108, PRA142–151, PRA300–309, and PRA425–433) by an improved multistep epitope prediction procedure. Using in vitro proteasome-mediated digestion pattern analysis, the four epitopes were chosen for CTL inductions and shown to be naturally presented. In addition, we show that CTL clones with high sensitivity for high affinity binding peptide PRA47–56, which was not produced in vitro by proteasome-mediated digestion (Fig. 2), were unable to lyse PRAME and HLA-A*0201–expressing tumor cell lines (Table ), indicating a lack of endogenous processing of this peptide. Taken together, both findings imply the accuracy and relevance of the proteasome-mediated digestion pattern analysis.
Importantly, only 4 out of the 19 peptides (PRA100–108, PRA142–151, PRA300–309, and PRA425–433) were COOH-terminally excised by a major cleavage site at 1 h incubation and were contained intact in digestion fragments as well, indicating possible abundantly expressed CTL epitopes. This reduction to 21% of high affinity HLA-A*0201 binding peptides being efficiently processed, which is in concordance with an estimation by Yewdell et al. 66, permitted us to avoid laborious and time consuming CTL inductions against unlikely epitopes. Indeed, the four predicted epitopes were all shown to be endogenously processed and presented (Fig. 4 Fig. 5 Fig. 6). For future epitope predictions in other proteins, it will be worthwhile to first systematically characterize proteasome digestion patterns of a complete set of overlapping long (e.g., 30-mer) polypeptides and subsequently determine binding affinities for MHC class I molecules of interest of only those peptides that are shown to be COOH-terminally excised by a major cleavage site. This experimental order reflects the physiological sequence of events, with the primacy of CTL epitope generation at antigen processing and not at MHC binding as indeed has been observed for MHC II epitope presentation 67.
Possibly, in the future, reliable proteasome cleavage prediction algorithms will allow by-passing of experimental digestions. Efforts to develop such algorithms have been reported 68 69. By prediction algorithms of Kuttler et al. 69, three of the four epitopes identified in our study (PRA142–151, PRA300–309, and PRA425–433) were predicted to be correctly COOH-terminally liberated. In contrast, the COOH terminus of PRA100–108, which is generated by a major cleavage site in our study, was not predicted. Of the 25 major cleavage sites observed (Fig. 2), 17 sites (68%) were correctly predicted by the most optimal algorithm variant (type II) as well as 10 of the 23 (43%) minor cleavage sites observed at 1 h digestion (by algorithm variant type III). Furthermore, many cleavage sites were falsely predicted by the algorithms, including erroneous internal epitope destruction sites, thus reducing the value of the current algorithms. Although these differences may be partly attributable to the different types of proteasomes used (immunoproteasomes in this study versus constitutive proteasomes in reference 69), we conclude that this first accessible proteasome cleavage prediction algorithm is not yet accurate enough to be used without experimental verification. As suggested by Kuttler et al. 69, a further enlargement of the training data used to educate the algorithm is likely to improve the accuracy of prediction.
With respect to the utilization and interpretation of proteasome-mediated digestion patterns for epitope prediction as reported here, several points must be raised. For proteasome-mediated digestions, we used 20S proteasomes isolated from an EBV-transformed B cell line known to contain mainly so-called immunoproteasomes with LMP2, LMP7, and MECL1 subunits 58. This implies that the four identified epitopes are likely to be presented on full-length PRAME expressing mature DCs containing immunoproteasomes next to their expression on tumor cells (containing constitutive proteasomes). For whole antigen vaccine development, such epitopes are favorable in contrast to a type of CTL epitope of which the presentation has been reported to be abrogated in mature DCs 70. The reported reverse effect, an inefficient processing in cells containing constitutive proteasomes 71 72 73 74, is excluded for our CTL epitopes by the functional data (Fig. 4 Fig. 5 Fig. 6).
The selection of candidate epitopes was mainly based on generation of the correct COOH terminus by an early major cleavage site, which is considered a sine qua none for efficient epitope generation 34 35 36 37. Late emergence (at 4 or 24 h) of the correct COOH terminus or generation by a minor cleavage site is not expected to yield epitopes or only at very low density, respectively. In contrast, cleavage sites within the epitope were less heavily weighed in the epitope prediction. In particular, ALYVDSLFFL (PRA300–309) was found to be cleaved at several sites within the epitope (Fig. 1 C). Although this phenomenon does not exclude epitope formation, as reported for epitopes of murine leukemia virus and CMV 37 75, partial destruction of an epitope can severely hamper its efficient presentation (for a review by Niedermann et al., see reference 76). Despite that, the PRA300–309 peptide was included in our CTL inductions for several reasons: (a) generation of its COOH terminus by a major cleavage site (Fig. 1 C), (b) reported presentation of the 9-mer length variant PRA301–309 (LYVDSLFFL) in HLA-A24 48, and (c) favorable binding capacity (Table ). Furthermore, (d) it can not be excluded that in vitro digestions are prone to a “recharging-effect”: longer epitope precursor fragments (containing the correct COOH terminus) can be further degraded by reentry in the proteasome, a phenomenon which will presumably not occur in vivo, because translocation of polypeptides by TAP to the ER has been shown to occur as soon as 15 min after protein synthesis (and consequently proteasome digestion 77). However, our control experiments did not reveal a significant effect of peptide substrate concentration on the relative digestion fragment kinetics (data not shown), making pronounced effects of recharging unlikely and indicating that the observed kinetics at least partially reflect the primary cleavage pattern and fragment abundancy. Finally, (e) cleavage within this epitope may be diminished when digested with constitutive proteasomes instead of immunoproteasomes, as enhanced cleavages after leucine and valine (hydrophobic residues) by the latter type of proteasomes have been described 76.
Our data support the current notion that a significant proportion of CTL epitopes is produced by the proteasome as NH2-terminally extended precursor 36 38 39 40 41. Digestion analysis of PRAME 415–441 revealed that 9-mer PRA425–433 is abundantly generated as NH2-terminally elongated 10-mer 424QSLLQHLIGL433 (Fig. 1 D), rendering it likely that this fragment is translocated by TAP to the ER and is trimmed there to its final length. Likewise, 9-mer PRA100–108 is presumably formed as 11-mer 98KAVLDGLDVLL108, which also indicates that the intermediate binding 10-mer PRA99–108 may be presented as well (Fig. 1 A). In contrast, SLYSFPEPEA (PRA142–151) is likely available for TAP translocation in its precise length, because this peptide was found in significant quantities as digestion fragment at 1 h digestion (Fig. 1 B). This is in concordance with a recent report showing that the proteasome can generate both COOH and NH2 termini of some epitopes 78.
PRAME is a particularly attractive tumor-associated antigen because it is widely expressed in many different tumor types 48 49 50 51, but not in normal tissues, except testis, and at very low levels in endometrium, ovaries, and adrenals 48. Indeed, CTL clones recognizing the four novel HLA-A*0201–restricted PRAME epitopes specifically lysed melanoma, renal cell, lung, mammary, and cervical carcinoma cell lines (Fig. 4 and Fig. 6). Therefore, and given the high prevalence of HLA-A*0201 among the Caucasian population, these epitopes are expected to be applicable for immunotherapeutic purposes (adoptive CTL therapy, vaccine design, and/or immunomonitoring) in a high percentage of cancer patients. Undesirable autoimmune CTL reactivity against the few tissues expressing PRAME at low levels is not to be expected, because expression levels are most likely too low to ensure CTL recognition as shown in vitro with human MAGE-specific CTLs by Lethe et al. 79 and in vivo in a murine p53 model by our group 80. Nevertheless, control recognition studies with normal endometrium, ovary, or adrenal tissues should ascertain absence of harmful responses towards healthy tissues expressing PRAME at low levels (expression level <3–5% of that found in melanoma, with the exception of endometrium, which expresses up to 30% of the melanoma level 48). So far we have been unable to establish sufficient primary cell cultures of those sources for functional analysis.
Although not the principle objective of this study, we found a remarkable immunogenicity in healthy donors of the four epitopes, because CTL inductions against the four peptides (performed with blood of two separate donors, see Materials and Methods) were all successful. Particularly, PRA142–151 and PRA425–433 vigorously induced CTL bulk cultures recognizing these peptides as endogenously expressed PRAME epitopes. Apparently, the low level expression of PRAME in some healthy tissues did not induce irreversible tolerance against the four identified epitopes. Future comparison of CTL frequencies in healthy donors versus cancer patients, as determined by, e.g., tetramer studies or enzyme-linked immunospot analysis, will reveal whether cancer patients are naturally primed against the four epitopes. Furthermore, such experiments may allow an immunodominance ranking of the epitopes. Our digestion data suggest a ranking in efficiencies of proteasome-mediated generation of the four epitopes, which is a major factor determining immunodominance 66 76. Because of the higher abundance of epitope precursor fragments and absence of major cleavage sites within the epitopes, PRA142–151 and PRA425–433 are probably more efficiently generated than PRA100–108 and PRA300–309 (Fig. 1 and Fig. 2).
Finally, we expect that our novel epitope prediction methodology will help to rapidly identify PRAME-derived CTL epitopes presented in HLA class I molecules other than HLA-A*0201 and will boost the reverse immunology approach for other tumor specific proteins as well. Such a systematic identification of new CTL epitopes in different tumor antigens will allow the development of multiantigen (epitope-based) tumor vaccines, covering all HLA class I haplotypes, which is probably needed to circumvent tumor escape by antigen loss variants.
Acknowledgments
We thank Dr. P. Coulie for the gift of the PRAME cDNA and Mrs. W. Benckhuijsen for synthesis of peptides.
Footnotes
Abbreviations used in this paper: aa, amino acid; B-LCL, B lymphoblastoid cell line; DC, dendritic cell; ER, endoplasmic reticulum; FI, fluorescence index; FL, fluorescein; HBV, hepatitis B virus; MS, mass spectrometry; RT, reverse transcription; TAP, transporter-associated with antigen processing.
References
- Melief C.J.M., Toes R.E., Medema J.P., van der Burg S.H., Ossendorp F., Offringa R. Strategies for immunotherapy of cancer. Adv. Immunol. 2000;75:235–281. doi: 10.1016/s0065-2776(00)75006-1. [DOI] [PubMed] [Google Scholar]
- Hsu F.J., Benike C., Fagnoni F., Liles T.M., Czerwinski D., Taidi B., Engleman E.G., Levy R. Vaccination of patients with B-cell lymphoma using autologous antigen-pulsed dendritic cells. Nat. Med. 1996;2:52–58. doi: 10.1038/nm0196-52. [DOI] [PubMed] [Google Scholar]
- Nestle F.O., Alijagic S., Gilliet M., Sun Y., Grabbe S., Dummer R., Burg G., Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat. Med. 1998;4:328–332. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- Marchand M., van Baren N., Weynants P., Brichard V., Dreno B., Tessier M.H., Rankin E., Parmiani G., Arienti F., Humblet Y. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int. J. Cancer. 1999;80:219–230. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Thurner B., Haendle I., Roder C., Dieckmann D., Keikavoussi P., Jonuleit H., Bender A., Maczek C., Schreiner D., von den Driesch P. Vaccination with mage-3A1 peptide-pulsed mature, monocyte-derived dendritic cells expands specific cytotoxic T cells and induces regression of some metastases in advanced stage IV melanoma. J. Exp. Med. 1999;190:1669–1678. doi: 10.1084/jem.190.11.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H.J., Holler E. Adoptive immunotherapy with donor lymphocyte transfusions. Curr. Opin. Oncol. 1997;9:139–145. doi: 10.1097/00001622-199703000-00006. [DOI] [PubMed] [Google Scholar]
- Offringa R., van der Burg S.H., Ossendorp F., Toes R.E., Melief C.J. Design and evaluation of antigen-specific vaccination strategies against cancer. Curr. Opin. Immunol. 2000;12:576–582. doi: 10.1016/s0952-7915(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Rosenberg S.A. A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity. 1999;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- Van den Eynde B.J., van der Bruggen P. T cell defined tumor antigens. Curr. Opin. Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- van der Bruggen P., Traversari C., Chomez P., Lurquin C., De Plaen E., Van den Eynde B.J., Knuth A., Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- Traversari C., van der Bruggen P., Luescher I.F., Lurquin C., Chomez P., Van Pel A., De Plaen E., Amar-Costesec A., Boon T. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J. Exp. Med. 1992;176:1453–1457. doi: 10.1084/jem.176.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eynde B.J., Peeters O., De Backer O., Gaugler B., Lucas S., Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J. Exp. Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel P., Wildmann C., Sensi M.L., Brasseur R., Renauld J.C., Coulie P., Boon T., van der Bruggen P. BAGEa new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- Brichard V., Van Pel A., Wolfel T., Wolfel C., De Plaen E., Lethe B., Coulie P., Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J. Exp. Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulie P.G., Brichard V., Van Pel A., Wolfel T., Schneider J., Traversari C., Mattei S., De Plaen E., Lurquin C., Szikora J.P. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J. Exp. Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Eliyahu S., Delgado C.H., Robbins P.F., Rivoltini L., Topalian S.L., Miki T., Rosenberg S.A. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc. Natl. Acad. Sci. USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A.B., Schreurs M.W., de Boer A.J., Kawakami Y., Rosenberg S.A., Adema G.J., Figdor C.G. Melanocyte lineage–specific antigen gp100 is recognized by melanoma-derived tumor-infiltrating lymphocytes. J. Exp. Med. 1994;179:1005–1009. doi: 10.1084/jem.179.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie T., Tahara K., Tanaka F., Mori M., Takesako K., Akiyoshi T. A MAGE-1-encoded HLA-A24-binding synthetic peptide induces specific anti-tumor cytotoxic T lymphocytes. Int. J. Cancer. 1999;80:169–172. doi: 10.1002/(sici)1097-0215(19990118)80:2<169::aid-ijc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Kawashima I., Hudson S.J., Tsai V., Southwood S., Takesako K., Appella E., Sette A., Celis E. The multi-epitope approach for immunotherapy for canceridentification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum. Immunol. 1998;59:1–14. doi: 10.1016/s0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- Celis E., Tsai V., Crimi C., DeMars R., Wentworth P.A., Chesnut R.W., Grey H.M., Sette A., Serra H.M. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc. Natl. Acad. Sci. USA. 1994;91:2105–2109. doi: 10.1073/pnas.91.6.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen P., Bastin J., Gajewski T., Coulie P.G., Boel P., De Smet C., Traversari C., Townsend A., Boon T. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur. J. Immunol. 1994;24:3038–3043. doi: 10.1002/eji.1830241218. [DOI] [PubMed] [Google Scholar]
- Herman J., van der Bruggen P., Luescher I.F., Mandruzzato S., Romero P., Thonnard J., Fleischhauer K., Boon T., Coulie P.G. A peptide encoded by the human MAGE3 gene and presented by HLA-B44 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE3. Immunogenetics. 1996;43:377–383. doi: 10.1007/BF02199806. [DOI] [PubMed] [Google Scholar]
- Tanaka F., Fujie T., Tahara K., Mori M., Takesako K., Sette A., Celis E., Akiyoshi T. Induction of antitumor cytotoxic T lymphocytes with a MAGE-3-encoded synthetic peptide presented by human leukocytes antigen-A24. Cancer Res. 1997;57:4465–4468. [PubMed] [Google Scholar]
- Parkhurst M.R., Fitzgerald E.B., Southwood S., Sette A., Rosenberg S.A., Kawakami Y. Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2) Cancer Res. 1998;58:4895–4901. [PubMed] [Google Scholar]
- Tsai V., Southwood S., Sidney J., Sakaguchi K., Kawakami Y., Appella E., Sette A., Celis E. Identification of subdominant CTL epitopes of the GP100 melanoma-associated tumor antigen by primary in vitro immunization with peptide-pulsed dendritic cells. J. Immunol. 1997;158:1796–1802. [PubMed] [Google Scholar]
- van Elsas A., Nijman H.W., van der Minne C.E., Mourer J.S., Kast W.M., Melief C.J., Schrier P.I. Induction and characterization of cytotoxic T-lymphocytes recognizing a mutated p21ras peptide presented by HLA-A*0201. Int. J. Cancer. 1995;61:389–396. doi: 10.1002/ijc.2910610319. [DOI] [PubMed] [Google Scholar]
- Nijman H.W., van der Burg S.H., Vierboom M.P., Houbiers J.G., Kast W.M., Melief C.J. p53, a potential target for tumor-directed T cells. Immunol. Lett. 1994;40:171–178. doi: 10.1016/0165-2478(94)90189-9. [DOI] [PubMed] [Google Scholar]
- Gedde-Dahl T., III, Spurkland A., Fossum B., Wittinghofer A., Thorsby E., Gaudernack G. T cell epitopes encompassing the mutational hot spot position 61 of p21 ras. Promiscuity in ras peptide binding to HLA. Eur. J. Immunol. 1994;24:410–414. doi: 10.1002/eji.1830240221. [DOI] [PubMed] [Google Scholar]
- Disis M.L., Smith J.W., Murphy A.E., Chen W., Cheever M.A. In vitro generation of human cytolytic T-cells specific for peptides derived from the HER-2/neu protooncogene protein. Cancer Res. 1994;54:1071–1076. [PubMed] [Google Scholar]
- Zaks T.Z., Rosenberg S.A. Immunization with a peptide epitope (p369-377) from HER-2/neu leads to peptide-specific cytotoxic T lymphocytes that fail to recognize HER- 2/neu1 tumors. Cancer Res. 1998;58:4902–4908. [PubMed] [Google Scholar]
- Chaux P., Luiten R., Demotte N., Vantomme V., Stroobant V., Traversari C., Russo V., Schultz E., Cornelis G.R., Boon T., van der Bruggen B.P. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by In vitro stimulation with dendritic cells transduced with MAGE-A1. J. Immunol. 1999;163:2928–2936. [PubMed] [Google Scholar]
- York I.A., Rock K.L. Antigen processing and presentation by the class I major histocompatibility complex. Annu. Rev. Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- Pamer E., Cresswell P. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- Craiu A., Akopian T., Goldberg A., Rock K.L. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl. Acad. Sci. USA. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder H.L., Bacik I., Yewdell J.W., Behrens T.W., Bennink J.R. Promiscuous liberation of MHC-class I-binding peptides from the C termini of membrane and soluble proteins in the secretory pathway. Eur. J. Immunol. 1998;28:1339–1346. doi: 10.1002/(SICI)1521-4141(199804)28:04<1339::AID-IMMU1339>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Mo X.Y., Cascio P., Lemerise K., Goldberg A.L., Rock K. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J. Immunol. 1999;163:5851–5859. [PubMed] [Google Scholar]
- Beekman N.J., van Veelen P.A., van Hall T., Neisig A., Sijts A., Camps M., Kloetzel P.M., Neefjes J.J., Melief C.J., Ossendorp F. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 2000;164:1898–1905. doi: 10.4049/jimmunol.164.4.1898. [DOI] [PubMed] [Google Scholar]
- Snyder H.L., Yewdell J.W., Bennink J.R. Trimming of antigenic peptides in an early secretory compartment. J. Exp. Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelse J., Gromme M., Momburg F., Hammerling G., Neefjes J. Trimming of TAP-translocated peptides in the endoplasmic reticulum and in the cytosol during recycling. J. Exp. Med. 1994;180:1591–1597. doi: 10.1084/jem.180.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T., Willis A., Cerundolo V., Townsend A. Processing of major histocompatibility class I-restricted antigens in the endoplasmic reticulum. J. Exp. Med. 1995;181:1481–1491. doi: 10.1084/jem.181.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E.A., Ortmann B., Surman M., Cresswell P. The protease inhibitor, N-acetyl-L-leucyl-L-leucyl-leucyl-L-norleucinal, decreases the pool of major histocompatibility complex class I–binding peptides and inhibits peptide trimming in the endoplasmic reticulum. J. Exp. Med. 1996;183:1569–1578. doi: 10.1084/jem.183.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beninga J., Rock K.L., Goldberg A.L. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- Dick L.R., Aldrich C., Jameson S.C., Moomaw C.R., Pramanik B.C., Doyle C.K., Demartino G.N., Bevan M.J., Forman J.M., Slaughter C.A. Proteolytic processing of ovalbumin and beta-galactosidase by the proteasome to a yield antigenic peptides. J. Immunol. 1994;152:3884–3894. [PMC free article] [PubMed] [Google Scholar]
- Eggers M., Boes-Fabian B., Ruppert T., Kloetzel P.M., Koszinowski U.H. The cleavage preference of the proteasome governs the yield of antigenic peptides. J. Exp. Med. 1995;182:1865–1870. doi: 10.1084/jem.182.6.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermann G., Butz S., Ihlenfeldt H.G., Grimm R., Lucchiari M., Hoschutzky H., Jung G., Maier B., Eichmann K. Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity. 1995;2:289–299. doi: 10.1016/1074-7613(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Niedermann G., King G., Butz S., Birsner U., Grimm R., Shabanowitz J., Hunt D.F., Eichmann K. The proteolytic fragments generated by vertebrate proteasomesstructural relationships to major histocompatibility complex class I binding peptides. Proc. Natl. Acad. Sci. USA. 1996;93:8572–8577. doi: 10.1073/pnas.93.16.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossendorp F., Eggers M., Neisig A., Ruppert T., Groettrup M., Sijts A., Mengede E., Kloetzel P.M., Neefjes J., Koszinowski U., Melief C. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Lethe B., Lehmann F., van Baren N., Baurain J.F., De Smet C., Chambost H., Vitale M., Moretta A., Boon T., Coulie P.G. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- van Baren N., Chambost H., Ferrant A., Michaux L., Ikeda H., Millard I., Olive D., Boon T., Coulie P.G. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br. J. Haematol. 1998;102:1376–1379. doi: 10.1046/j.1365-2141.1998.00982.x. [DOI] [PubMed] [Google Scholar]
- Neumann E., Engelsberg A., Decker J., Storkel S., Jaeger E., Huber C., Seliger B. Heterogeneous expression of the tumor-associated antigens RAGE-1, PRAME, and glycoprotein 75 in human renal cell carcinomacandidates for T-cell-based immunotherapies? Cancer Res. 1998;58:4090–4095. [PubMed] [Google Scholar]
- Pellat-Deceunynck C., Mellerin M.P., Labarriere N., Jego G., Moreau-Aubry A., Harousseau J.L., Jotereau F., Bataille R. The cancer germ-line genes MAGE-1, MAGE-3 and PRAME are commonly expressed by human myeloma cells. Eur. J. Immunol. 2000;30:803–809. doi: 10.1002/1521-4141(200003)30:3<803::AID-IMMU803>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Imanishi, T., T. Akaza, A. Kimura, K. Tokunaga, and T. Gojobori. 1992. Allele and haplotype frequencies for HLA and complement loci in various ethnic groups. HLA 1991, Proceedings of the Eleventh International Histocompatibility Workshop and Conference. K. Tjuji, M. Aizawa, and T. Sasazuki, editors. Oxford University Press, Oxford/New York/Tokyo. 1065–1220.
- Garrone P., Neidhardt E.M., Garcia E., Galibert L., van Kooten C., Banchereau J. Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J. Exp. Med. 1995;182:1265–1273. doi: 10.1084/jem.182.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg S.H., Ras E., Drijfhout J.W., Benckhuijsen W.E., Bremers A.J., Melief C.J., Kast W.M. An HLA class I peptide-binding assay based on competition for binding to class I molecules on intact human B cells. Identification of conserved HIV-1 polymerase peptides binding to HLA-A*0301. Hum. Immunol. 1995;44:189–198. doi: 10.1016/0198-8859(95)00105-0. [DOI] [PubMed] [Google Scholar]
- Bertoletti A., Chisari F.V., Penna A., Guilhot S., Galati L., Missale G., Fowler P., Schlicht H.J., Vitiello A., Chesnut R.C. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J. Virol. 1993;67:2376–2380. doi: 10.1128/jvi.67.4.2376-2380.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg S.H., Visseren M.J., Brandt R.M., Kast W.M., Melief C.J. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J. Immunol. 1996;156:3308–3314. [PubMed] [Google Scholar]
- Groettrup M., Ruppert T., Kuehn L., Seeger M., Standera S., Koszinowski U., Kloetzel P.M. The interferon-inducible 11S regulator (PA28) and the LMP2/LMP7 subunits govern the peptide production by the 20S proteasome in vitro. J. Biol. Chem. 1995;270:23808–23815. doi: 10.1074/jbc.270.40.23808. [DOI] [PubMed] [Google Scholar]
- Frisan T., Levitsky V., Polack A., Masucci M.G. Phenotype-dependent differences in proteasome subunit composition and cleavage specificity in B cell lines. J. Immunol. 1998;160:3281–3289. [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony–stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze J.L., Michalak S., Seamon M.J., Dranoff G., Jung K., Daley J., Delgado J.C., Gribben J.G., Nadler L.M. CD40-activated human B cellsan alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. J. Clin. Invest. 1997;100:2757–2765. doi: 10.1172/JCI119822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Scheidegger D., Palmer-Lehmann K., Lane P., Lanzavecchia A., Alber G. Ligation of CD40 on dendritic cells triggers production of high levels of interleukin-12 and enhances T cell stimulatory capacityT-T help via APC activation. J. Exp. Med. 1996;184:747–752. doi: 10.1084/jem.184.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amaro J., Houbiers J.G., Drijfhout J.W., Brandt R.M., Schipper R., Bavinck J.N., Melief C.J., Kast W.M. A computer program for predicting possible cytotoxic T lymphocyte epitopes based on HLA class I peptide-binding motifs. Hum. Immunol. 1995;43:13–18. doi: 10.1016/0198-8859(94)00153-h. [DOI] [PubMed] [Google Scholar]
- Parker K.C., Bednarek M.A., Coligan J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- Rammensee H.G., Bachman J., Stevanovic S. MHC Ligands and Peptide Motifs 1997. Springer-Verlag, ; Heidelberg, Germany: pp. 462 pp [Google Scholar]
- van de Corput L., Kluin-Nelemans H.C., Kester M.G., Willemze R., Falkenburg J.H. Hairy cell leukemia-specific recognition by multiple autologous HLA-DQ or DP-restricted T-cell clones. Blood. 1999;93:251–259. [PubMed] [Google Scholar]
- Yewdell J.W., Bennink J.R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- Phelps R.G., Jones V.L., Coughlan M., Turner A.N., Rees A.J. Presentation of the Goodpasture autoantigen to CD4 T cells is influenced more by processing constraints than by HLA class II peptide binding preferences. J. Biol. Chem. 1998;273:11440–11447. doi: 10.1074/jbc.273.19.11440. [DOI] [PubMed] [Google Scholar]
- Holzhutter H.G., Frommel C., Kloetzel P.M. A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20 S proteasome. J. Mol. Biol. 1999;286:1251–1265. doi: 10.1006/jmbi.1998.2530. [DOI] [PubMed] [Google Scholar]
- Kuttler C., Nussbaum A.K., Dick T.P., Rammensee H.G., Schild H., Hadeler K.P. An algorithm for the prediction of proteasomal cleavages J. Mol. Biol. 298 2000. 417 429Algorithm accessible at www.uni-tuebingen.de/uni/kxi/contents.html [DOI] [PubMed] [Google Scholar]
- Morel S., Levy F., Burlet-Schiltz O., Brasseur F., Probst-Kepper M., Peitrequin A.L., Monsarrat B., Van Velthoven R., Cerottini J.C., Boon T. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- Schwarz K., van den Broek M., Kostka S., Kraft R., Soza A., Schmidtke G., Kloetzel P.M., Groettrup M. Overexpression of the proteasome subunits LMP2, LMP7, and MECL-1, but not PA28alpha/beta, enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J. Immunol. 2000;165:768–778. doi: 10.4049/jimmunol.165.2.768. [DOI] [PubMed] [Google Scholar]
- Sijts A.J., Ruppert T., Rehermann B., Schmidt M., Koszinowski U., Kloetzel P.M. Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J. Exp. Med. 2000;191:503–514. doi: 10.1084/jem.191.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijts A.J., Standera S., Toes R.E., Ruppert T., Beekman N.J., van Veelen P.A., Ossendorp F.A., Melief C.J., Kloetzel P.M. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J. Immunol. 2000;164:4500–4506. doi: 10.4049/jimmunol.164.9.4500. [DOI] [PubMed] [Google Scholar]
- van Hall T., Sijts A., Camps M., Offringa R., Melief C., Kloetzel P.M., Ossendorp F. Differential influence on cytotoxic T lymphocyte epitope presentation by controlled expression of either proteasome immunosubunits or PA28. J. Exp. Med. 2000;192:483–494. doi: 10.1084/jem.192.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes B., Hengel H., Ruppert T., Multhaup G., Koszinowski U.H., Kloetzel P.M. Interferon γ stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J. Exp. Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermann G., Geier E., Lucchiari-Hartz M., Hitziger N., Ramsperger A., Eichmann K. The specificity of proteasomesimpact on MHC class I processing and presentation of antigens. Immunol. Rev. 1999;172:29–48. doi: 10.1111/j.1600-065x.1999.tb01354.x. [DOI] [PubMed] [Google Scholar]
- Reits E.A., Vos J.C., Gromme M., Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- Lucchiari-Hartz M., van Endert P.M., Lauvau G., Maier R., Meyerhans A., Mann D., Eichmann K., Niedermann G. Cytotoxic T lymphocyte epitopes of HIV-1 Nefgeneration of multiple definitive major histocompatibility complex class I ligands by proteasomes. J. Exp. Med. 2000;191:239–252. doi: 10.1084/jem.191.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lethe B., van der Bruggen P., Brasseur F., Boon T. MAGE-1 expression threshold for the lysis of melanoma cell lines by a specific cytotoxic T lymphocyte Melanoma Res. 7Suppl. 21997. S83 S88 [PubMed] [Google Scholar]
- Vierboom M.P., Nijman H.W., Offringa R., van der Voort E.I., van Hall T., van den Broek L., Fleuren G.J., Kenemans P., Kast W.M., Melief C.J. Tumor eradication by wild-type p53-specific cytotoxic T lymphocytes. J. Exp. Med. 1997;186:695–704. doi: 10.1084/jem.186.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]