Abstract
The gonococcal pilus is a major virulence factor that has well-established roles in mediating epithelial cell adherence and DNA transformation. Gonococci expressing four gonococcal pilin variants with distinct piliation properties under control of the lac regulatory system were grown in different levels of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG). These pilin variants expressed various levels of pilin message and pilin protein in response to the level of IPTG in the growth medium. Moreover, posttranslational modifications of the variant pilin proteins were detected, including S-pilin production and glycosylation. The ratio of the modified and unmodified pilin forms did not substantially change with different levels of pilin expression, showing that these modifications are not linked to pilin expression levels. DNA transformation competence was also influenced by IPTG levels in the growth medium. Substantial increases in transformation competence over an isogenic, nonpiliated mutant were observed when limited amounts of three of the pilin variants were expressed. Immunoelectron microscopy showed that when limited amounts of pilin are expressed, pili are rare and do not explain the pilin-dependent transformation competence. This pilin-dependent transformation competence required prepilin processing, the outer membrane secretin PilQ, and the twitching-motility-regulating protein PilT. These requirements show that a fully functional pilus assembly apparatus is required for DNA uptake when limited pilin is produced. We conclude that the pilus assembly apparatus functions to import DNA into the bacterial cell in a pilin-dependent manner but that extended pili are not required for transformation competence.
The gonococcal pilus is a member of the type IV family of pili, a classification based on similarities in the pilin subunit primary amino acid sequence, cleavage of a 5- to 15-amino-acid signal sequence followed by N-methylation of the first amino acid of the mature pilin subunit, and conservation of the genes required for pilus assembly (40). Other bacteria that produce type IV pili include, but are not limited to, Neisseria meningitidis, Pseudomonas aeruginosa, Moraxella lacunata, Dichelobacter nodosus, Vibrio cholerae, enteropathogenic Escherichia coli, Synechocystis, and Myxococcus xanthus (37, 44). Pilin can be modified at a variable residue by glycosylation (29) or at conserved residues by α-glycerophosphate (39), phosphoserine (9), or phosphorylcholine (45). Mature pilin can be further processed by an unknown protease into S-pilin, and S-pilin is found predominately in the growth medium (14). The implications of these posttranslational modifications on pilus function or gonococcal pathogenesis are unknown.
Type IV pili mediate adherence of bacteria to host tissues and are associated with a form of bacterial motility called twitching motility (reviewed in references 40 and 42). Twitching motility may be mediated by assembly and retraction of pili (3), which serves to propel type IV pilus-expressing bacterial cells over a semisolid surface (15). A gene that is dispensable for pilus assembly but is required for DNA transformation and twitching motility is pilT (47). A homologue of pilT is also required for twitching motility in P. aeruginosa (46) and social motility in Myxococcus (51). Due to its consensus nucleotide binding sequences and stretches of hydrophobic residues, PilT is proposed to be a nucleotide binding protein associated with the inner membrane that presumably functions in regulating the disassembly and the retraction of pili (47). PilT has been shown to be required for the movement of gonococci, and the force of PilT-dependent retraction has been measured using optical tweezers (23). By limiting pilin expression using the strain described by Long et al., the force of single pilus retracting was measured at >100 pN (21). These observations suggest that the retraction of pili is not solely due to disassembly but that import machinery must be encoded in the pilus assembly apparatus.
Proteins known to be required for pilus assembly include PilD, PilC, PilQ, PilG, PilF, and PilP. It is assumed that these proteins form a complex that transverses the periplasm to the outer membrane, facilitating pilus assembly. Prepilin is presumed to be targeted to the inner membrane via a Sec-dependent mechanism. The product of the pilD gene processes prepilin into mature pilin by cleaving the seven-amino-acid leader sequence and methylating the N terminus of the pilin protein (6, 27). In the absence of PilD activity, pilin cannot be assembled into pili and is often released as S-pilin (10). The outer membrane pore through which pili are assembled consists of multimers of the PilQ protein (originally called OMPC) (43), which is a member of a family of integral membrane, pore-forming proteins called secretins (reviewed in reference 32). A pilQ loss-of-function mutant is nonpiliated and secretes mostly S-pilin (5). However, in the presence of both pilQ and pilT mutations, assembled pili are detected within the periplasm of gonococci (48). These findings suggest that pilus assembly occurs independent of the PilQ pore and that PilT counteracts pilus assembly by regulating disassembly.
Piliation has been closely associated with natural DNA transformation efficiency of N. gonorrhoeae (38). Gonococci that do not express any pilin protein are greatly reduced in competence (35, 52). Gonococci that produce reduced levels of pilin and a few pili (13, 20, 30) are more competent for transformation than a nonpiliated (ΔpilE) variant. In addition to mutations in pilE, gonococci that are nonpiliated due to a mutation in a pilus assembly gene also have very low transformation efficiencies (5, 10, 31, 41). Mutation of the pilus accessory gene pilT (2, 47) also results in reduced competence. In addition, there are a number of genes that are essential for DNA transformation but are not required for pilus biogenesis or DNA uptake, including comA, comL, tpc, and comP (11, 12, 49). These genes are thought to be involved in transport of DNA across the periplasm and/or inner membrane, prior to recombination into the chromosome. A number of competence proteins that have homology to the type IV pilin proteins but no role in pilus production have been identified in Bacillus subtilis, Haemophilus influenzae, and Acinetobacter sp. (reviewed in reference 4).
The requirement for pilus expression and pilus assembly proteins in gonococcal DNA transformation competence supports the hypothesis that pili and/or the pilus assembly machinery directly mediates DNA transformation. However, this hypothesis has not been proven and, if true, the mechanism used by the pilus to mediate DNA transformation competence is unknown. Our investigators have previously shown that pilus-mediated cell adherence and DNA transformation can be modulated by lac-regulated control of pilin expression (19). Here we examine the effect of regulating pilE expression in four defined pilin variants of strain FA1090. We characterize the effect of different levels of pilin expression on posttranslational modifications of pilin and show that the posttranslational modifications of pilin are independent of the amount of pilin in a cell. We also show that a substantial level of DNA transformation competence is maintained when limited pilin is expressed and few pili are detected. This pilin-dependent transformation competence requires a fully functional pilus assembly apparatus and is therefore genetically indistinguishable from normal DNA transformation in gonococci. These data argue that an extended pilus is not absolutely required for transformation in gonococci.
MATERIALS AND METHODS
Bacterial strains and media.
All N. gonorrhoeae strains were derivatives of FA1090, and E. coli strain DH10B was used for all molecular biology experiments. Gonococci were grown on GC medium base (GCB; Difco) with Kellogg supplements (17) at 37°C in 5% CO2, or in GC liquid (GCBL) medium (1.5% Proteose Peptone no. 3 [Difco], 0.4% K2HPO4, 0.1% KH2PO4, 0.1% NaCl) with Kellogg supplements and 0.042% sodium bicarbonate (25) at 37°C with rotation. For antibiotic selection of gonococci, nalidixic acid (Nal) at 2 μg/ml, erythromycin (Erm) at 1 to 10 μg/ml, and chloramphenicol (Chlor) at 2 μg/ml were used. E. coli cells were grown at 37°C on Luria-Bertani medium (LB agar; Difco) or shaking in LB broth (Difco) at 37°C. Immediately following electroporation, E. coli cells were grown in LB plus 20 mM glucose, 10 mM MgSO4, and 10 mM MgCl2. E. coli cells were selected for antibiotic resistance at 40 μg of kanamycin (Kan)/ml, 20 μg of Chlor/ml, and 200 to 300 μg of Erm (Sigma)/ml.
Construction of regulatable RM11, RM11.2, RM11.6, and RM21.
The pilE coding sequences of FA1090 recA6 variants RM11, RM11.2, RM11.6, and RM21 were PCR amplified from chromosomal DNA using PILRBS (50) and OPAE1REV (16) primers and cloned into pCR-BLUNT (Invitrogen). Kmr transformants were screened for insertions of the appropriate size by PCR with M13FOR-20 (GTAAAACGACGGCCAG) and M13REV (Gibco BRL) primers, and the PCR products were sequenced. A fragment containing lacIOP, ermC, and 4 kb of gonococcal DNA upstream of the pilE2 locus from strain MS11-C9 was released from pCDL1 (19) with EcoRI and inserted into the NotI site 5′ of the pilE coding sequence of variant pilE clones. Erm-Kanr transformants were screened by restriction endonuclease digestion and PCR with LACPFOR (GAGCGGATAACAATTTCAC) and OPAE1REV primers, and the DNA sequence of the pilE gene was sequenced using CONSTF2 (50) to confirm maintenance of the correct pilE sequence. Regulatable pilE clones were then used to transform the wild-type FA1090 variants with the matched pilE sequence, and transformants were selected on GCB-Erm. Transformants of FA1090 RM11, RM11.2, and RM11.6 were checked by Southern blotting and PCR analysis to confirm proper incorporation of the regulatable construct into the chromosome.
Using the protocol described above, Erm-resistant transformants of RM21 were never isolated. A cat gene with an associated gonococcal uptake sequence was excised from pCmUp with EcoRI and inserted into the HindII site of pRM21Reg. Kan-Chlorr transformants were analyzed by PCR and restriction endonuclease digestion and sequenced. pRM21RegCmUp was used to transform FA1090 RM21.20, which has the same HV pilE sequence as FA1090 RM21 but has a more piliated colony morphology. Erm-resistant colonies were screened by Southern and PCR analyses to confirm proper recombination into the chromosome, and the pilE sequence was determined to ensure that the RM21, and not the RM21.20, pilE sequence was present.
Analysis of isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent mRNA levels.
Plasmid-derived standards were linearized by restriction enzyme digestion and were then separated in a 0.8% agarose gel stained with ethidium bromide along with the Low DNA mass ladder (Invitrogen) carrying DNA fragments of known concentrations. Densitometry on the gel was performed using ImageQuant (Molecular Dynamics) to quantify the linear plasmid standards to the Low DNA mass ladder. Copy numbers of pilE transcripts were calculated as previously described (28) using serial dilutions of the standard representing 108 to 101 copies.
Gonococci were swabbed from solid media into ice-cold phosphate-buffered saline (PBS), and either TRIzol (Invitrogen) or RNeasy (Qiagen) extraction of total RNA was performed according to the supplier's instructions. DNase treatment was then performed twice, followed by RNA purification using the RNeasy RNA Cleanup protocol (Qiagen). RNA was run in a 0.8% agarose gel stained with ethidium bromide to assess purity. 28S rRNA bands were compared and normalized by performing densitometry with the ImageQuant software. After RNA levels from all samples were normalized, cDNA synthesis was performed as described by Serkin and Seifert (36) using the primer LC-CONS-rev for the 5′ constant region of pilE or LC-RecA-rev for recA (see below). The reverse transcription reaction was purified using the Qiaquick PCR purification kit (Qiagen). All primers were reverse-phase high-performance liquid chromatography purified (IT Biochem). Primer and probe melting temperatures were calculated using the freeware program TM Utility v0.1.3 (Idaho Technology; http://www.idahotech.com/downloads_up/Tmutility_form.htm).
Hybridization probes were created in accord with previously published guidelines (24). For pilE transcripts we used primers LC-CONS-for (CGAGCTGATGATTGTGATCGC) and LC-CONS-rev (GCTGATTTTTGACCTTCGGCC) for amplification and probes LC-Hyb-CONS-1 (CGCCTACCAAGACTACACCGCCC-F, where F is fluorescein) and LC-Hyb-CONS-2 (LC640-CGCGCAAGTTTCCGAAGCCATCC-P, where LC640 is LightCycler Red 640-N-hydroxysuccinimide ester and P is phosphate) were used for detection. For recA transcripts primers LC-RecA-for (CATGAAAATGGACGGCAGCC) and LC-RecA-rev (GACGGCTTCGAGGCAGAG) were used for amplification, and probes LC-Hyb-recA-1 (CGGGCGCATCGTCGAAATCTTCGG-F) and LC-Hyb-RecA-2 (LC640-CCCGAATCCTCCGGCAAAACCACA-P) were used for detection. For por transcripts primers LC-POR-for (TTTCTTACGCCCACGGCT) and LC-POR-rev (CGCCTTTGCCTTCTTGC) were used for amplification, and probes LC-Hyb-Por-1 (GCGGAATACGACTTCTCCAAACGCA-F) and LC-Hyb-Por-2 (LC640-TTCTGCCTTGGTTTCTGCCGGC-P) were used for detection.
Real-time PCR was performed on a LightCycler instrument (Roche Diagnostics). All assays using hybridization probes contained 2 μl of LightCycler DNA master hybridization probes mix (Roche Diagnostics), 3.2 μl of MgCl2 (for a final concentration of 5 mM), 2 μl of each primer (0.5 mM), 2 μl of donor hybridization probe (0.2 mM), 2 μl of acceptor hybridization probe (0.2 mM), 2 μl of template, and PCR-grade sterile water for a final volume of 20 μl. The annealing temperature was 53°C for pilE and 60°C for both recA and porA. Fluorescence in channels F1 and F2 was acquired at the end of every extension step, and the F2/F1 ratio was analyzed.
Western analysis of pilin and S-pilin production.
Gonococci were grown in GCBL in the presence of 0 to 1 mM IPTG. Cell pellet and culture supernatant protein fractions were isolated as previously described (20). Cell pellet and culture supernatant fractions were separated on sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis gels and either stained with Coomassie brilliant blue R-250 (Sigma) or transferred to nitrocellulose (MSI) using a Trans-Blot cell (Bio-Rad). The membranes were blocked with MegaBlocI (CEL Associates) as recommended by the manufacturer and probed with the pilin monoclonal antibody (MAb) 1E8/G8 (gift from M. Koomey and M. Blake) (7) at a dilution of 1:1,000. Alkaline phosphatase-linked anti-mouse antibody (Promega) was used at a dilution of 1:7,500, and the immunoblots were developed colorimetrically as previously described (20).
Construction of pilT mutants.
FA1090 RM11 recA6, RM11.2 recA6, RM11.6 recA6, RM21 recA6, regulatable RM11, and regulatable RM11.2 were transformed with p2E1 DNA (47), which is a cloned gonococcal pilT gene with an Erm resistance insertion mutation. Erm-resistant transformants were screened by Southern blotting and PCR analysis, and the pilE sequence was determined.
Construction of pilQ mutants.
One-kilobase regions upstream and downstream of the pilQ gene were amplified from FA1090 chromosomal DNA using primers PILQFOR1 (CCCCAACCTCCCGTAGATG), PILQREV1 (GACTTTGACGATTTTCTGTTTG), PILQFOR2 (GTCGGCGGTATTTATGAAG), and PILQREV2 (GAATCCAGTCCGTTCGGTT), and each was cloned into pCR-BLUNT. Kanr transformants were screened by restriction endonuclease digestion. The cat gene from pHSS6cat2-9 was inserted into the KpnI site of p5′pilQ, and the 3′ insert was excised from p3′pilQ with KpnI and MluI and inserted between the KpnI and MluI sites of p5′pilQcat. Kan-Chlorr transformants were screened by restriction endonuclease digestion. pPilQ::cat was used to transform FA1090 RM11 recA6, RM11.2 recA6, regulatable RM11, and regulatable RM11.2, selecting on GCB-Chlor. Transformants were screened to confirm knockout of the pilQ coding sequence and replacement with the cat gene by PCR with primers CATF (ATCGAGATTTTCAGGAGCTAAG), CATMREV (CCATTGGGATATATCAACGGTG), PILQFOR3 (CGCAACCGCCGCCTTTCA), and PILQREV3 (GCATCAATAGCGCAGGCTG), and the 1-kb region 5′ of pilQ was sequenced to confirm that a secondary mutation had not been introduced into pilP. The pilE sequences of Chlor-resistant gonococcal transformants were also determined to confirm maintenance of the proper variant pilE sequence.
DNA sequencing.
Twenty to 50 ng of PCR template or 200 to 500 ng of plasmid DNA was subjected to automated sequencing using either AmpliTaq dye terminator DNA sequencing or Big Dye terminator cycle sequencing (Perkin-Elmer Applied Biosystems) according to the manufacturer's specifications. Reactions were electrophoresed on an Applied Biosystems International ABI 373 or 377 sequencing apparatus. Sequences were analyzed using Gene Runner 3.04 and Chromas 1.43 (Griffith University, Queensland, Australia) DNA analysis software.
Gonococcal chromosomal DNA preparation and Southern analysis.
Gonococcal chromosomal DNA was prepared as described previously (35) and was digested with ClaI (NEB). Southern analysis was performed as described elsewhere (33). pilE gene probe DNA was amplified by PCR from FA1090 chromosomal DNA using primers PILRBS and SP3A (50). ermC gene probe DNA was amplified from FA1090 chromosomal DNA using primers ERMA-IN and ERMB-IN (22). cat gene probe DNA was amplified from pCmUp using primers CMFOR and CMREV. Gene probes were labeled by random-primer labeling (8).
Gonococcal transformation.
For liquid transformations, gonococci were grown on GCB for 16 to 18 h, collected with Dacron swabs, and suspended to a density of 108 CFU/ml in 1 ml of GCBL. Twenty microliters of cells was added to 200 μl of GCBL that contained 5 mM MgSO4 (35), 0 to 1 mM IPTG, and transforming DNA. After 10 to 15 min at 37°C, the transformation mixes were diluted into 2 ml of GCBL plus Kellogg supplements, 1 mM IPTG if necessary, and 5 mM MgSO4 and incubated at 37°C in 5% CO2 for 5 h without agitation. Transformants were selected on GCB containing the appropriate antibiotic(s).
For spot transformations, gonococci were passaged onto GCB medium without antibiotics, and 10 to 15 μl of transforming DNA in a suspension of GCBL plus Kellogg supplements and MgSO4 was applied in a spot on the freshly passaged GCB plate. After overnight incubation, gonococci were collected with a Dacron swab, suspended in GCBL, and plated on the appropriate antibiotic.
Immunoelectron microscopy.
To detect surface-exposed pili, polyclonal antibodies were raised against a synthetic peptide representing a hypervariable loop region (also called mc2) of the pilin variants expressed on strains RM11.2 and RM11 (KRDAGAKTGADDVKADGN). The peptide was synthesized on MAP resin and used to immunize two New Zealand White female rabbits (Research Genetics). The antiserum recognized the pilin protein from RM11 and RM11.2 but did not recognize the pilins from RM11.6 or RM21 in Western blotting (data not shown).
For analysis of pilus expression in liquid medium, RM11.2recA6 was grown for 18 h on GCB plates and swabbed into GCBL containing Kellogg supplements. Formvar-coated grids (Ladd Research Industries, Inc.) were placed into a 12-well microtiter plate into which the resuspended bacteria were added. Grids were incubated with the bacteria for 4 h at 37°C in 5% CO2. For analysis of piliation on bacteria growth on solid medium, Formvar-coated grids were used to lift cells directly from 18-h-old colonies.
Grids from both culture conditions were fixed for 15 min by floating on a drop of 0.2% glutaraldehyde, 4% paraformaldehyde (Fisher Scientific). After fixation, the grids were rinsed three times with Dulbecco's PBS (Gibco) plus 1% bovine serum albumin (BSA; Sigma) and incubated in 0.1% gelatin (Aurion, Inc.) in PBS for 30 min. The grids were rinsed once with PBS plus BSA and incubated with a 1:10 dilution of rabbit α-pilin peptide antisera for 1 h. Grids were rinsed three times with PBS-BSA and incubated with 0.1% gelatin in PBS for 30 min. The grids were rinsed once with PBS plus BSA and incubated with goat anti-rabbit immunoglobulin G antibody conjugated to 12-nm gold particles (1:20 dilution; Jackson Immunolabs) for 1 h. Grids were rinsed five times in water for 3 min each step. Excess water was carefully wicked away, and grids were negatively stained with 1% uranyl acetate for 1 min. All incubations were performed at 25°C. Grids were viewed using a Jeol JEM-1220 transmission electron microscope at 60 kV.
RESULTS
Pilin protein and transcript expression in regulatable pilE variants RM11, RM11.2, RM11.6, and RM21 in response to IPTG.
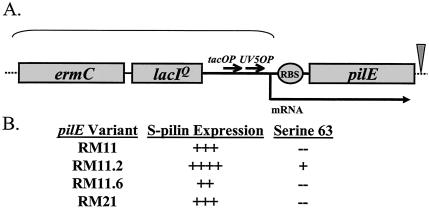
We previously described the construction and characterization of a regulatable pilE strain in the MS11 background that demonstrated IPTG-dependent DNA transformation competence and epithelial cell adherence (19). To extend these analyses, we chose four pilin variants from a panel of FA1090 colony variants previously analyzed (20). These variants were all isolated from a progenitor FA1090 variant but carry variant pilin sequences (variant pilE genes) and express different levels of DNA transformation competence. One variant carries a glycosylation site, one is inefficiently assembled into pili, and each expresses different levels of pilin and S-pilin (Fig. 1). The lac regulatory sequences were introduced into the 5′-untranslated region of the pilE gene of variants RM11.2 and RM11.6 (highly piliated), RM11 (moderately piliated), and RM21 (minimally piliated).
FIG. 1.
Schematic of the regulatable pilE construct and characteristics of regulatable pilE variants. (A) Cartoon of regulatable constructs. Shaded boxes indicate genes and are drawn to scale. Thick arrows indicate operators and promoters. The curved bracket indicates the inserted regulatory sequences. The triangle indicates the site of the cat gene insertion used to create regulatable RM21. (B) Characteristics of the four FA1090 pilin variants. S-pilin expression was determined by densitometric analysis of Coomassie blue-stained gradient gels (20). ++++, high level of S-pilin produced; +, low level of S-pilin produced. Variant pilin residue 63 is glycosylated when a serine is present at this position.
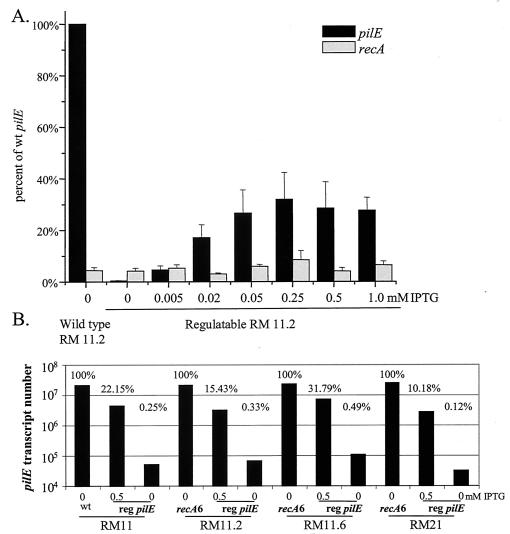
Quantitative reverse transcription-PCR (Q-RT-PCR) analysis of pilE transcript levels was performed on the regulatable RM11.2 strain grown on solid medium with various amounts of IPTG and compared to wild-type recA message levels from the same cultures (Fig. 2A). The pilE message level without IPTG was about 1% of wild-type message levels, and the steady-state levels increased with increasing amounts of IPTG until a plateau was reached at about 0.25 mM IPTG (Fig. 2A). No increase in transcript level was measured when higher levels of IPTG were used (Fig. 2A and data not shown). The recA transcript levels did not respond to IPTG levels but showed the normal variability of bacterial message levels. The pilE transcript levels of the four regulatable pilin variants, grown with or without IPTG in liquid medium, were compared to pilE transcript levels of the four strains with wild-type pilE promoters (Fig. 2B). The four regulatable strains produced from 10 to 32% of wild-type levels of pilE message when induced with 0.5 mM IPTG. Therefore, the dual lac promoters used to construct the regulatable strains were not as strong as the wild-type pilE promoter in gonococci. Since each variant produced similar levels of transcript, we can conclude that the changes in pilE sequence do not substantially alter message stability. Even without IPTG induction, the four regulatable strains produced detectable pilE message, but at levels that were less than 1% of the wild-type pilE message levels and between 1 and 2% of the 0.5 mM IPTG levels. While the uninduced pilE transcripts were reproducibly detected in this assay, they were at levels that were near the minimal detectable level of 2 × 104 transcripts in this Q-RT-PCR assay.
FIG. 2.
Q-RT-PCR analysis of pilE transcript levels. (A) The strains were grown on solid medium with the indicated level of IPTG. Total RNA was isolated with and reverse transcribed with specific primers. Transcript copies were determined by FRET hybridization probe detection in a Roche LightCycler compared to known amounts of cloned DNA standards. Values are the percentage of the wild-type RM11.2 strain value. Shown is an average of three replicates. (B) The strains were grown in liquid medium with the indicated level of IPTG. Q-RT-PCR was performed as described for panel A using por primers and probes, and RNA samples were normalized to contain similar levels of por transcript levels (data not shown). pilE transcript numbers for fully induced and uninduced regulatable (reg) pilE variants were determined and expressed as the percentage of wild-type (recA6) pilE transcript level for each variant.
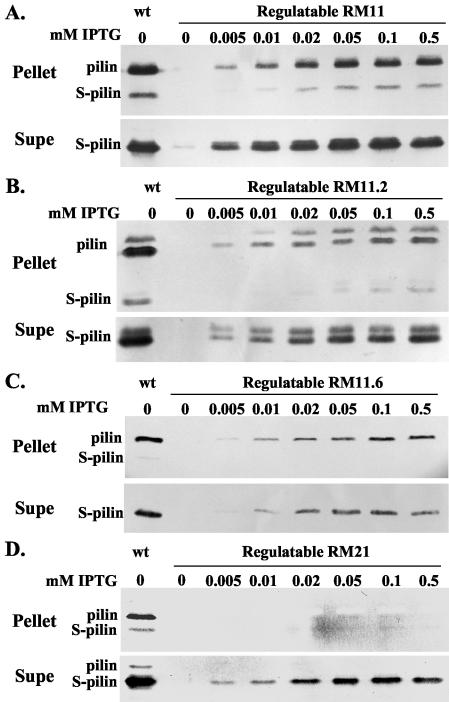
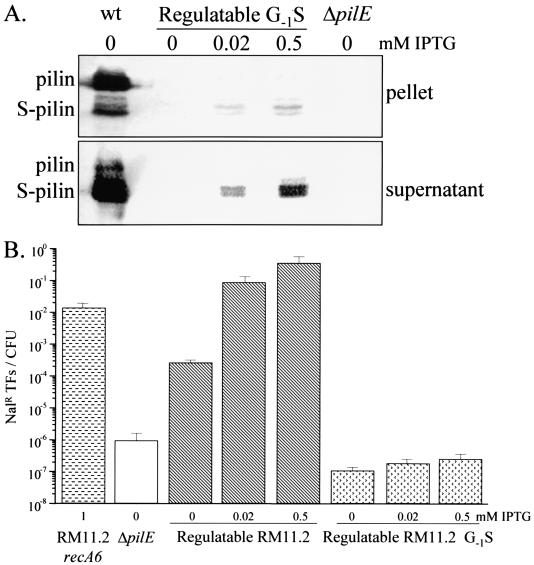
Pilin expression of the pilin variants in response to varying IPTG levels was determined by Western blot analysis. With the exception of the supernatant from regulatable RM11, pilin and S-pilin were never detected by Western blotting when these strains were grown without IPTG. The three piliated colony variants expressed increasing amounts of pilin and S-pilin with increasing concentrations of IPTG (Fig. 3). Consistent with the transcript analysis, the fully induced level of protein never reached wild-type levels in any of the four variants (Fig. 3). At each level of IPTG-induced transcription, the ratio of full-length pilin to S-pilin was diagnostic for each variant and was similar to the ratio observed with the corresponding wild-type variant.
FIG. 3.
Western analysis of regulatable pilE variants. Gonococcal strains were grown in medium containing the indicated amounts of IPTG. Cultures were separated into a cell pellet and culture supernatant, and the supernatant proteins were precipitated with trichloroacetic acid (see Materials and Methods). Pilin and S-pilin were detected with the MAb 1E8/G8. wt, wild-type pilE expressed in a recA6 background. Strains used are as follows: RM11 (A); RM11.2 (B); RM11.6 (C); RM21 (D).
The RM11.2 pilin protein carries a serine residue at position 63 of the mature pilin protein, which is glycosylated (29). Western analysis of cell pellets and culture supernatants of regulatable RM11.2 demonstrated that there was a mixture of glycosylated and nonglycosylated pilin and S-pilin in RM11.2 and that the ratios of glycosylated to nonglycosylated pilin and S-pilin remained the same, independent of the level of pilin production (Fig. 3B).
DNA transformation efficiencies of regulatable pilE variants respond to changes in pilE transcription.
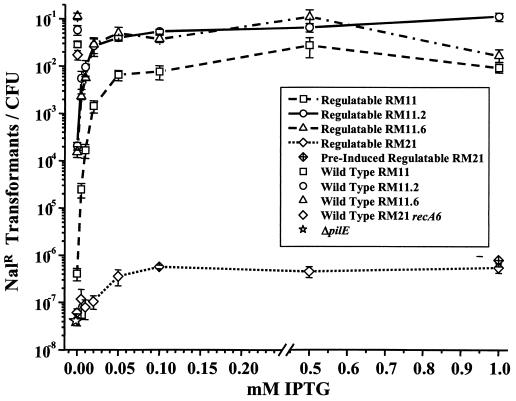
A dose response for IPTG-dependent transformation efficiencies of the four regulatable pilE strains was measured to determine whether all four pilin variants responded similarly to changes in pilE transcription. Transformation efficiencies of the four regulatable pilE variants increased over a range of 0 to 0.05 mM IPTG, at which point transformation competence reached a plateau at a level of transcription that was about 1/6 of maximal expression levels (Fig. 2). These results are similar to those obtained with a single regulatable MS11 pilin variant (19). RM11.2 and RM11.6 expressed similar transformation efficiencies at each IPTG level, RM11 expressed intermediate transformation efficiencies, and RM21 expressed very low transformation efficiencies at each level of IPTG induction (Fig. 4). Therefore, while each pilin variant showed an IPTG-dependent regulation of transformation competence, the differences in primary pilin amino acid sequence still influenced the levels of DNA transformation competence. Even though the transcript and protein levels at 0.05 mM were well below wild-type levels (Fig. 2 and 3), three of the regulatable variants expressed transformation competence close to wild-type levels that did not significantly increase with the further induction of pilE. This finding suggests that wild-type levels of pilin are in excess of that required for full transformation competence. When the regulatable pilin variants were grown without IPTG, three of the regulatable pilE variants expressed transformation competence levels 10- to 1,000-fold higher than that of the nonpiliated, ΔpilE strain (Fig. 4), even though transcript and protein levels were very low in the uninduced cultures (Fig. 2 and 3).
FIG. 4.
IPTG dose response of DNA transformation competence in regulatable and control strains. Strains were induced as indicated during incubation with plasmid DNA that confers resistance to nalidixic acid (Nalr) when recombined into the chromosome. Efficiency was expressed as the number of Nalr transformants per CFU. The mean and standard error of 8 to 10 identical experiments are shown. Each variant with its wild-type promoter is shown without IPTG. The recA6 version of RM21 was used because the pilE sequence in wild-type RM21 was found to be unstable (data not shown).
The regulatable RM21 strain, unlike the three other regulatable strains, exhibited transformation efficiencies well below its matched control strain with a wild-type pilin promoter, even when grown overnight with IPTG prior to transformation. The greater transformation competence of the wild-type RM21 was not due to antigenic variation, since a majority of transformants expressed the same pilE sequence (data not shown). Western analysis of cell pellet fractions of regulatable RM21 induced at 0.5 mM IPTG and RM21 recA6 revealed that even when fully induced, regulatable RM21 makes less than 2% of the full-length pilin produced by RM21 recA6, while the other regulatable variants express between 20 and 50% of the wild-type pilin amounts (data not shown). This reduction in full-length pilin levels accounts for the 10,000-fold difference in transformation competence between these two strains with the same pilE sequence. We cannot explain why this variant expresses such a reduced level of pilin relative to the wild-type promoter but propose that it is likely to be related to the instability of the RM21 pilin protein.
Immunoelectron microscopy revealed that regulatable RM11.2 expresses few pili under conditions of limited pilin expression.
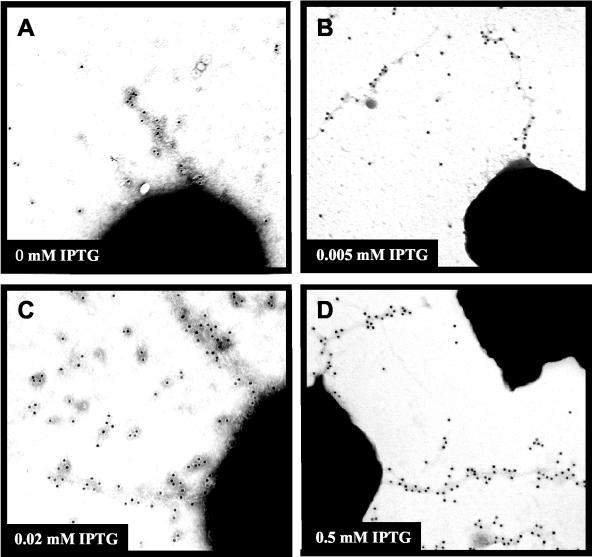
The greater than 1,000-fold increase in DNA transformation competence expressed by RM11.2 and RM11.6 regulatable pilin variants without IPTG over an isogenic nonpiliated control led us to two major hypotheses. An occasional pilus is assembled under limited pilin expression levels, and those rare pili are responsible for the transformation competence observed. Alternatively, it is possible that the small amount of pilin expressed without IPTG is responsible for the increased competence in a mechanism that does not require an extended pilus. To examine pili produced in the absence of IPTG, a polyclonal antiserum was raised against a peptide synthesized from the hypervariable loop sequence of RM11.2 pilin and used in immuno-gold electron microscopy studies of regulatable RM11.2. Cultures of regulatable RM11.2 were grown without IPTG or with 0.005, 0.02, or 0.5 mM IPTG and compared to an FA1090 variant with a deleted pilE (Fig. 5). Gonococcal cells were lifted directly from colonies grown on agar medium or were grown in liquid medium on top of an electron microscopy grid for 4 h to mimic the conditions used in the transformation assays and to ensure that pili were not being broken during disruption of colonies. Figure 5 shows representative electron micrographs, except for the no-IPTG micrograph, which is not representative since it shows the only pilus structure detected on cells grown without IPTG. Quantification of the pili expressed on cells from the different cultures revealed that the liquid-grown cells expressed pili on a lower percentage of cells than those grown on solid medium (Table 1). The regulatable RM11.2 grown with 0.5 mM IPTG expressed the most pili, with over 50% of the cells directly lifted from colonies showing pili, and 5% of liquid-grown bacteria expressed detectable pili. These percentages of cells expressing detectable pili are similar to that of a strain with a wild-type pilin locus (19). Cells grown at 0.005 mM IPTG in liquid medium expressed about 10-fold fewer pili, while cells grown on agar medium expressed about 3-fold fewer pili than those grown in 0.5 mM IPTG. Without IPTG, pili were undetected on 5,185 liquid-grown cells, and a single shortened pilus structure was detected on 1 of 6,522 cells lifted from agar medium (Fig. 5A). Most fields of bacteria grown without IPTG were devoid of specific gold labeling, and the extent of background gold was identical to that of the ΔpilE control strain (data not shown). Therefore, this single pilus was detected on only 0.00015% of the uninduced RM11.2 cells (Table 1). These data strongly suggest that the single pilus detected on a low percentage of cells grown without IPTG cannot account for the large increase in transformation competence expressed by the uninduced cultures relative to that of the ΔpilE strain (see Discussion).
FIG. 5.
Electron micrographs of immuno-gold-labeled pili. Regulatable RM11.2 recA6 was grown on solid medium in the presence of 0, 0.005, or 0.5 mM IPTG and lifted directly onto Formvar-coated grids. Pili were detected with an antipeptide polyclonal antiserum raised against the hypervariable loop sequence of the RM11.2 pilin and detected with gold-labeled secondary antibody. Representative electron micrographs are shown for each growth condition except for 0 mM IPTG, where the single detected pilus is shown. IPTG concentrations were as follows: 0 (A); 0.005 (B); 0.2 (C); 0.5 mM IPTG (D).
TABLE 1.
Summary of immunogold detection of pili expressed by regulatable RM11.2
| Growth condition | Level of induction (mM IPTG) | Total cells examined | % Piliated cells |
|---|---|---|---|
| Liquid medium | 0 | 5,185 | NDa |
| 0.005 | 859 | 0.5 | |
| 0.5 | 247 | 5 | |
| Agar medium | 0 | 6,522 | 0.00015 |
| 0.005 | 704 | 3 | |
| 0.02 | 661 | 8 | |
| 0.5 | 123 | 55 |
ND = not detected.
Prepilin processing is required for pilin-dependent DNA transformation competence.
The substantial transformation competence of three uninduced regulatable pilin variants shows that the small amount of pilin protein expressed in these strains without IPTG is responsible for this transformation competence. The pilin protein is expressed as a prepilin, and the six-amino-acid leader peptide is removed by the PilD prepilin peptidase (10). Therefore, we asked whether DNA transformation competence in regulatable RM11.2 was still observed when an amino acid required for prepilin processing was mutated (18) (RM11.2 G-1S). Western analysis of the regulatable RM11.2 G-1S mutant demonstrated that neither pilin nor S-pilin was detectable when grown in the absence of IPTG (Fig. 6A). When the G-1S RM11.2 strain was grown in the presence of 0.02 or 0.5 mM IPTG, the majority of detectable pilin was in the S-pilin form found in the medium, and there was a drastic reduction in the amount of total pilin detected. This confirms the observations previously made in strain MS11 that the G-1S mutation prevents prepilin processing and results in S-pilin in the medium (18). The RM11.2 G-1S mutant exhibited transformation efficiencies that were not significantly different from that of the ΔpilE nonpiliated control strain, independent of IPTG induction (Fig. 6B). This shows that proper processing of pilin by PilD is required for pilin-dependent transformation competence. However, since the unprocessed prepilin was unstable, we cannot absolutely conclude whether it is the processed form of the protein that is responsible for the transformation competence.
FIG. 6.
Effect of the prepilin processing Gly-1Ser mutation on DNA transformation competence in a regulatable strain. (A) Western blot analysis of pilin. Gonococci were grown overnight in liquid in the presence of 0, 0.02, or 0.5 mM IPTG as indicated. Pilin and S-pilin were detected with MAb 1E8/G8. wt, RM11.2 recA6. (B) Transformation efficiency was determined as described in the legend for Fig. 4. Values shown are the means and standard errors of three to nine experiments.
The pilus assembly apparatus is required for pilin-dependent DNA transformation competence.
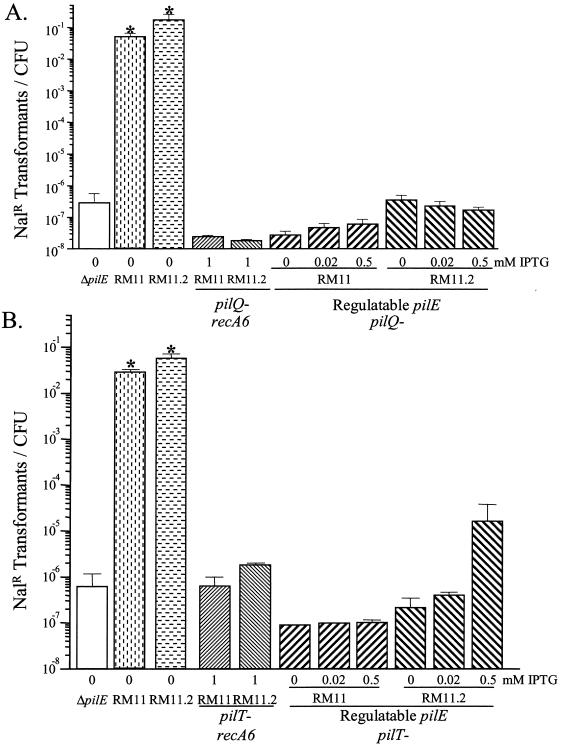
Once processed by PilD, mature pilin is assembled into a fiber in a process that is poorly understood. One hypothesis to explain the relationship between low-level pilin production and transformation competence is that processed pilin interacts with the pilus assembly machinery to allow DNA access to the cytoplasm. An alternative hypothesis is that pilin interacts with unknown components of the cell to allow for transformation competence. To test whether a fully functional pilus assembly apparatus is required for transformation when low levels of pilin are expressed, loss-of-function mutations in genes required for pilus biogenesis (pilQ) and function (pilT) were assayed for their effect on pilin-dependent DNA transformation.
A pilQ loss-of-function mutation introduced into strains RM11 and RM11.2 confirmed that PilQ is required for normal DNA transformation competence (5) (Fig. 7A). The same pilQ mutation in the regulatable RM11 and RM11.2 strains demonstrated that the pilin-dependent increase of transformation competence was also dependent on PilQ (Fig. 7A). This finding shows that the PilQ secretin is required for DNA to efficiently enter into the cell regardless of whether pili are produced.
FIG. 7.
DNA transformation competence of pilQ and pilT loss-of-function mutants in regulatable RM11 and RM11.2. Transformation efficiency was determined as described in the legend for Fig. 4. Values shown are the mean and standard error of three to seven experiments. (A) Competence of pilQ mutants and control strains. (B) Competence of pilT mutants and control strains. The asterisk indicates a statistically significant difference from the ΔpilE strain, as determined by Student's independent t test (P < 0.05).
PilT is not required for pilus assembly, but it is required for twitching motility (23) and DNA transformation competence (47). As a predicted cytoplasmic NTPase, PilT is thought to regulate pilus disassembly to allow twitching motility. A pilT mutation was introduced into variants with wild-type pilE promoters (recA6) (34) as well as the regulatable pilE variants RM11.2 and RM11 to determine its effect upon DNA transformation efficiency. The pilT mutants showed DNA transformation competencies similar to that of a ΔpilE strain (Fig. 7B). Moreover, in the IPTG-regulatable strains, the pilT loss-of-function mutation also prevented pilin-dependent increases in transformation competence (Fig. 7B). While the transformation competence of the regulatable RM11.2 pilT mutant grown with 0.5 mM IPTG was higher than that of the ΔpilE strain (Fig. 7B), it was still 1,000-fold lower than that of its matched wild-type control and not statistically different than that of the ΔpilE control strain. Therefore, the putative NTPase, PilT, is required for transformation competence regardless of the level of pilin expressed.
DISCUSSION
Four previously described FA1090 pilE variants (RM11, RM11.2, RM11.6, and RM21) were chosen for this study based on their differences in pilin amino acid sequence, S-pilin expression, glycosylation, and relative level of pilus-mediated function (Fig. 1; see also reference 20). By growing the four regulatable strains in select amounts of IPTG, a series of pilE steady-state expression levels were analyzed for each variant. In both the wild-type and regulatable pilE strains, only a subset of pilin molecules was modified by proteolysis to produce S-pilin. As previously observed, the amount of S-pilin relative to mature pilin was different for each pilin variant. Unexpectedly, the ratio of S-pilin to pilin was similar for each variant regardless of how much pilin was expressed. This finding rules out the possibility that the level of S-pilin produced in each variant is the result of a limited activity of the undefined protease. Since the S-pilin cleavage site is within an absolutely conserved sequence, this suggests that secondary or tertiary structural determinants of the pilin protein, dictated by the primary sequence of each pilin variant, modulate the accessibility of the cleavage site to the protease. This differential processing could occur by changes in accessibility of the cleavage site to the protease in the folded polypeptide, by different rates of pilus assembly or disassembly modulating protease efficiency, or by pilin and S-pilin forms resulting from divergent transport pathways. We cannot distinguish between these possibilities at this time. Two different molecular weight forms of variant RM11.2 pilin and S-pilin proteins were also detected, and the ratio of the forms was similar regardless of the level of pilE transcription. These different-mobility pilin proteins are most likely the result of differential glycosylation of serine 63 (29). The constant ratio of the forms shows that incomplete pilin glycosylation is not due to limiting enzyme. Whether or not the mechanisms behind partial glycosylation are similar to those for partial proteolysis is unknown at this time. The observation that a similar ratio of the two modified forms occurs for both mature and S-pilin forms suggests that glycosylation precedes proteolysis. Although the functional outcomes of the extensive posttranslational modifications of pilin are still undefined, our results show that modification is rarely, if ever, enacted on all pilin molecules and confirm that pilin antigenic variation can alter posttranslational processing of pilin.
The IPTG dose response of transformation efficiencies of the three fully piliated, regulatable pilin variants followed the pattern observed previously for a similar construct in strain MS11. The regulatable RM21 variant, which produces mostly S-pilin and very little pilin in the wild-type strain, responded differently. This difference is presumably due to the very limited amount of mature pilin expression in the regulatable RM21. In the other three regulatable variants, transformation efficiencies were 10- to 1,000-fold higher than that of the ΔpilE strain when grown without IPTG, the transformation efficiencies increased with the addition of IPTG to the growth medium, and a plateau was reached at a level of IPTG that is approximately 1/6 of the maximum level of pilin expression. Regulatable RM11 transformed consistently lower than regulatable RM11.2 and RM11.6 at each level of IPTG, and the plateau level of transformation competence was also lower. RM11 with a wild-type pilE promoter produces less pilin and fewer pili than the other two variants (20) and, when fully induced, the regulatable RM11 also produced fewer pili than the other two regulatable strains (data not shown). The transcript level analysis shows that this was not due to differences at the message level. Therefore, the different levels of piliation and transformation competence of these variants are the result of conditions set by the pilin protein amino acid sequence that appears to alter pilin stability. It is possible that some pilin variants are turned over at different rates and that similar signals, defined by the variant amino acid sequences, allow for differential processing to S-pilin, glycosylation, and degradation.
The dose response of transformation competence versus IPTG revealed that even without IPTG induction, high levels of transformation competence could be observed compared to that in the pilin null mutant. There are several reasons to propose that extended pili are not responsible for the transformation competence observed when the regulatable strains were grown without IPTG. First, we observed one short pilus on 1 cell out of 6,522 examined from agar-grown cultures, and 0 pili on 5,182 examined from liquid medium cultures (Table 1). This paucity of pili is consistent with the barely detectable levels of pilin observed by Western blot analysis (Fig. 3). Secondly, regulatable RM11.2 grown without IPTG transforms with about 5,000-fold greater efficiency than the isogenic strain with ΔpilE (Fig. 4). If the single pilus expressed on 6,522 cells were to be responsible for this transformation competence, it would have to increase the frequency by 5,000-fold. Moreover, when regulatable RM11.2 was grown with 0.005 mM IPTG, 3% of the cells expressed detectable pili, which is a 20,000-fold increase from the identical strain grown without IPTG. However, the transformation competence only increased by 40-fold between 0 and 0.005 mM IPTG. These comparisons clearly show that there is no direct correlation between the number of cells expressing pili and the transformation competence of the culture and strongly support the hypothesis that the pilin dependence of transformation competence does not require extended, assembled pili.
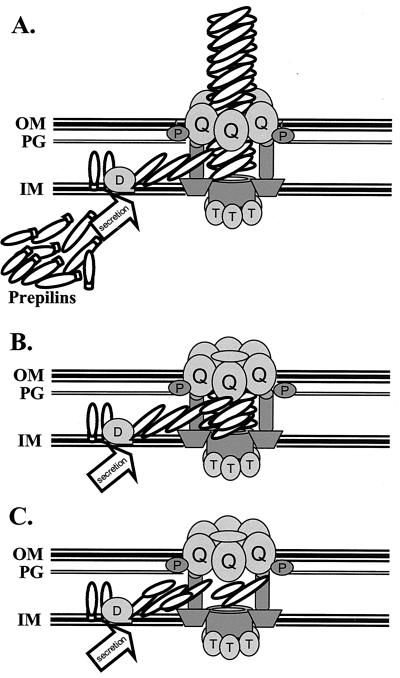
From the data presented here, we propose models for how pilin and the pilus assembly machinery promote DNA transformation competence (Fig. 8). When there is no mature pilin present in the bacterial cell, as in a ΔpilE or G-1S mutant, the pilus assembly machinery is in an inactive state. However, when a small amount of pilin is secreted into the periplasm and processed by PilD, the processed pilin provides a signal that allows DNA transport into the cell for transformation. There are two different ways this pilin dependence could be expressed. The most likely model is that the mature pilin is shuttled into its normal assembly pathway and associates with pilin-like proteins and possibly other proteins of the assembly apparatus to begin the process of pilus assembly (Fig. 8B). However, since a limited amount of pilin is expressed in the regulatable strains without IPTG, the pilus is not assembled to a point where it can be detected by immunoelectron microscopy. It is unknown if pilin associates with the minor pilin-like proteins to form a structure that is functionally distinct from the pilus. The major alternative model is that transport of pilin into the periplasm can activate some pilus assembly apparatus to become transformation competent without ever forming a pilus structure (Fig. 8C). This model suggests that either the pilin protein can interact with the assembly apparatus without forming a pilus-like structure or that an assembly apparatus with residual pili can communicate with other assembly apparatus without pili to become competent for DNA uptake. In support of this view is the fact that the PilQ secretin makes up 10% of the outer membrane proteins in gonococci (26), which would be in excess of the amount required for pilus expression. We have also observed that the level of PilQ does not change with varying levels of pilin expression (data not shown). These observations present the possibility that in wild-type cells, the pilus assembly apparatus that transports DNA could be distinct from the ones that express pili, a hypothesis that awaits experimental testing.
FIG. 8.
Models for pilin-dependent DNA transformation competence. (A) Hypothetical model of the pilus assembly apparatus. This cartoon shows production of the pilin and pilin-like subunits in the cytoplasm and processing by the PilD peptidase (D), followed by polymerization at an unknown location and extrusion through the PilQ secretin (Q), which is associated with PilP (P). PilT is shown interacting with unknown proteins in the cytoplasmic membrane, but this has not been demonstrated. The presence of pilin-like proteins in the exposed fiber is speculative. Not all proteins known to be required for pilus assembly or DNA transformation competence are shown. (B) The residual pilus model. When limited pilin is expressed in the regulatable strain grown without IPTG, a small amount of pilin is produced. These pilin subunits are transported into the periplasm, where the PilD peptidase cleaves the signal sequence and the mature pilin interacts with pilin-like proteins and other members of the assembly apparatus to produce a residual pilus. While the residual pilus is drawn as not being exposed on the cell surface, the residual pili would be predicted to be of different lengths with some protruding from the cell surface but too short to be detected frequently by negatively stained electron microscopy. (C) The pilus-independent transport model. In this model the pilus assembly apparatus assumes a transport-competent form which is dependent on pilin but does not require a polymerized pilus to be within the assembly apparatus that transports DNA. In this model, pilus assembly and DNA transport machinery are separate, but each requires the presence of pilin.
The PilT requirement for pilin-dependent transformation competence could be used to argue that this cytoplasmically localized NTPase functions to allow pilus retraction and that this results in DNA transformation. However, we propose that PilT may act to regulate an import function of the pilus assembly apparatus and that both the pilus and DNA are potential substrates of this apparatus, but only when pilin is expressed. In support of this view is the observation that overexpression of the pilin-like protein ComP produces increased transformation competence when minimal pilin is expressed (1, 49). Moreover, the well-established relationship between transformation competence and type IV pilus systems in other species and the essential role of pilus-like proteins in B. subtilis transformation, where no pilus exists, favor a role for the transport apparatus independent of the pilus but absolutely dependent on the pilin protein (4). Further elucidation of the precise roles of each pilus assembly apparatus component is necessary to test these models, and it will be interesting to determine whether this system can export or import other molecules important for pathogenesis.
Acknowledgments
We thank M. Wolfgang and M. Koomey for the pilT mutant, M. Blake for antibody reagents, and E. Stohl and E. Skaar for critical reviews of the manuscript.
These studies were supported by Public Health Service grants U01 AI31494, R01 AI44239, and R01 AI33493. C.D.L. and K.A.K. were partially supported by Public Health Service Training Grant T32GM08061, and D.M.T was supported by T32 AI07476 and NSRA F32 AI10568.
Editor: D. L. Burns
REFERENCES
- 1.Aas, F. E., C. Lovold, and M. Koomey. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46:1441-1450. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, G. D., S. A. Lacks, and P. F. Sparling. 1989. Transformation-deficient mutants of piliated Neisseria gonorrhoeae. J. Bacteriol. 171:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 4.Chen, I., and D. Dubnau. 2003. DNA transport during transformation. Front Biosci. 8:S544-S556. [DOI] [PubMed] [Google Scholar]
- 5.Drake, S. L., and M. Koomey. 1995. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol. 18:975-986. [DOI] [PubMed] [Google Scholar]
- 6.Dupuy, B., and A. P. Pugsley. 1994. Type IV prepilin peptidase gene of Neisseria gonorrhoeae MS11: presence of a related gene in other piliated and nonpiliated Neisseria strains. J. Bacteriol. 176:1323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards, M., R. L. McDade, G. Schoolnik, J. B. Rothbard, and E. C. Gotschlich. 1984. Antigenic analysis of gonococcal pili using monoclonal antibodies. J. Exp. Med. 160:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 9.Forest, K. T., S. A. Dunham, M. Koomey, and J. A. Tainer. 1999. Crystallographic structure reveals phosphorylated pilin from Neisseria: phosphoserine sites modify type IV pilus surface chemistry and fibre morphology. Mol. Microbiol. 31:743-752. [DOI] [PubMed] [Google Scholar]
- 10.Freitag, N. E., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 95:575-586. [DOI] [PubMed] [Google Scholar]
- 11.Fussenegger, M., D. Facius, J. Meier, and T. F. Meyer. 1996. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol. Microbiol. 19:1095-1105. [DOI] [PubMed] [Google Scholar]
- 12.Fussenegger, M., A. F. Kahrs, D. Facius, and T. F. Meyer. 1996. Tetrapac (tpc), a novel genotype of Neisseria gonorrhoeae affecting epithelial cell invasion, natural transformation competence and cell separation. Mol. Microbiol. 19:1357-1372. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs, C. P., B. Y. Reimann, E. Schultz, A. Kaufmann, R. Haas, and T. F. Meyer. 1989. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature 338:651-652. [DOI] [PubMed] [Google Scholar]
- 14.Haas, R., H. Schwarz, and T. F. Meyer. 1987. Release of soluble pilin antigen coupled with gene conversion in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 84:9079-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrichsen, J. 1975. The occurrence of twitching motility among gram-negative bacteria. Acta Pathol. Microbiol. Scand. B 83:171-178. [DOI] [PubMed] [Google Scholar]
- 16.Howell-Adams, B., and H. S. Seifert. 1999. Insertion mutations in pilE differentially alter gonococcal pilin antigenic variation. J. Bacteriol. 181:6133-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellogg, D. S., Jr., W. L. Peacock, W. E. Deacon, L. Brown, and C. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koomey, M., S. Bergstrom, M. Blake, and J. Swanson. 1991. Pilin expression and processing in pilus mutants of Neisseria gonorrhoeae: critical role of Gly-1 in assembly. Mol. Microbiol. 5:279-287. [DOI] [PubMed] [Google Scholar]
- 19.Long, C. D., S. F. Hayes, J. P. van Putten, H. A. Harvey, M. A. Apicella, and H. S. Seifert. 2001. Modulation of gonococcal piliation by regulatable transcription of pilE. J. Bacteriol. 183:1600-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long, C. D., R. N. Madraswala, and H. S. Seifert. 1998. Comparisons between colony phase variation of Neisseria gonorrhoeae FA1090 and pilus, pilin, and S-pilin expression. Infect. Immun. 66:1918-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier, B., L. Potter, M. So, C. D. Long, H. S. Seifert, and M. P. Sheetz. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA 99:16012-16017. (Erratum, 100:6287, 2003.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation, and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 23.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 24.Meuer, S. C., C. Wittwer, and K. Nakagawara. 2001. Rapid cycle real-time PCR: methods and applications. Springer, Berlin, Germany.
- 25.Morse, S. A., and L. Bartenstein. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc. Soc. Exp. Biol. Med. 145:1418-1421. [DOI] [PubMed] [Google Scholar]
- 26.Newhall, W. J., C. E. Wilde, W. D. Sawyer, and R. A. Haak. 1980. High-molecular-weight antigenic protein complex in the outer membrane of Neisseria gonorrhoeae. Infect. Immun. 27:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okeke, C. N., R. Tsuboi, and H. Ogawa. 2001. Quantification of Candida albicans actin mRNA by the LightCycler system as a means of assessing viability in a model of cutaneous candidiasis. J. Clin. Microbiol. 39:3491-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 30.Rudel, T., D. Facius, R. Barten, I. Scheuerpflug, E. Nonnenmacher, and T. F. Meyer. 1995. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 92:7986-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudel, T., I. Scheurerpflug, and T. F. Meyer. 1995. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373:357-359. [DOI] [PubMed] [Google Scholar]
- 32.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Seifert, H. S. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215-220. [DOI] [PubMed] [Google Scholar]
- 35.Seifert, H. S., R. S. Ajioka, D. Paruchuri, F. Heffron, and M. So. 1990. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J. Bacteriol. 172:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serkin, C. D., and H. S. Seifert. 1998. Frequency of pilin antigenic variation in Neisseria gonorrhoeae. J. Bacteriol. 180:1955-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, W., and H. Sun. 2002. Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect. Immun. 70:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparling, P. F. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stimson, E., M. Virji, S. Barker, M. Panico, I. Blench, J. Saunders, G. Payne, E. R. Moxon, A. Dell, and H. R. Morris. 1996. Discovery of a novel protein modification: alpha-glycerophosphate is a substituent of meningococcal pilin. Biochem. J. 316:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 41.Tonjum, T., N. E. Freitag, E. Namork, and M. Koomey. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 95:451-464. [DOI] [PubMed] [Google Scholar]
- 42.Tonjum, T., and M. Koomey. 1997. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships—a review. Gene 192:155-163. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, W. M., S. H. Larsen, and C. E. Wilde. 1989. Cloning and DNA sequence of the omc gene encoding the outer membrane protein-macromolecular complex from Neisseria gonorrhoeae. Infect. Immun. 57:2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wall, D., P. E. Kolenbrander, and D. Kaiser. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiser, J. N., J. B. Goldberg, N. Pan, L. Wilson, and M. Virji. 1998. The phosphorylcholine epitope undergoes phase variation on a 43-kilodalton protein in Pseudomonas aeruginosa and on pili of Neisseria meningitidis and Neisseria gonorrhoeae. Infect. Immun. 66:4263-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 47.Wolfgang, M., P. Lauer, H.-S. Park, L. Brossay, J. Hébert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:312-330. [DOI] [PubMed] [Google Scholar]
- 48.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfgang, M., J. P. van Putten, S. F. Hayes, and M. Koomey. 1999. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31:1345-1357. [DOI] [PubMed] [Google Scholar]
- 50.Wright, C. J., A. E. Jerse, M. S. Cohen, J. G. Cannon, and H. S. Seifert. 1994. Nonrepresentative PCR amplification of variable gene sequences in clinical specimens containing dilute, complex mixtures of microorganisms. J. Clin. Microbiol. 32:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Q. Y., D. DeRyckere, P. Lauer, and M. Koomey. 1992. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc. Natl. Acad. Sci. USA 89:5366-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]