Particular guanine rich DNA sequences can fold into stable four-stranded G-quadruplex structures, under physiological concentrations of Na+ and K+, in vitro.1 Such sequence motifs are found in the telomeres where they can fold into quadruplexes under the control of specific telomere binding proteins.2 G-quadruplex motifs have been identified throughout the genome3 and concentrate immediately upstream of transcription initiation sites.3a A number of these so-called “promoter quadruplex” sequences have been studied for several proto-oncogenes, including c-MYC,4 BCL2,5 VEGF,6 KRAS7 and two G-quadruplexes in the c-kit promoter8 (c-kit1 and c-kit2). One working hypothesis couples quadruplex formation in promoters to transcription, suggesting an opportunity for chemical intervention of gene expression using small molecule G-quadruplex ligands. Some proof-of-concept has been provided for the case of c-MYC where small molecule ligands, TmPyP4,4 and quindoline9 derivatives have been shown to inhibit gene expression, while KRAS gene expression was inhibited by TmPyP4.7
The c-kit proto-oncogene encodes a tyrosine kinase receptor for the growth-promoting cytokine SCF (stem cell factor) which plays an important biological role in the control of differentiation.10 A small molecule inhibitor of c-kit, Gleevec (imatinib mesylate), is being effectively used in the treatment of gastrointestinal stromal tumors (GIST).11 A small molecule that inhibits c-kit expression at the transcriptional level would provide further evidence to support the promoter-quadruplex hypothesis and might inspire the exploration of quadruplex-based therapeutic approaches to address GIST. Herein, we report the design, synthesis, biophysical evaluation with primary biological data on 3,8,10-trisubstituted isoalloxazines (Figure 1).
Figure 1.
Structures of isoalloxazine ligands 1a–f.
The design of isoalloxazines as potential G-quadruplex ligands was inspired by the observation, arising from SELEX studies, that oxidized riboflavin (7,8-dimethyl-10-ribityl-isoalloxazine) binds to an intramolecular G-quartet with moderate binding affinity (Kd) of 1–5 μM.12 Our design principles maintain the planar isoalloxazine scaffold, to enable interactions with G-quartet.13 Amine side chains were introduced to provide potential for interactions with quadruplex loops and grooves and the negatively charged sugar–phosphate backbone.
We developed and employed a short and robust synthetic route to 3,8,10-trisubstituted isoalloxazines to prepare ligands of general structure 1 (Figure 1). The key isoalloxazine building blocks were synthesized using modified literature procedures.14 An efficient method was developed for the introduction of amino alkyl side chains to afford 1a–f in good overall yields (see Supporting Information).
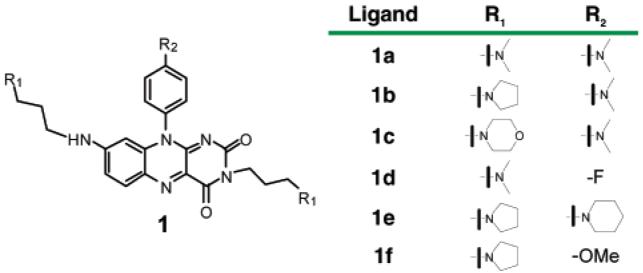
To evaluate the interaction properties of isoalloxazines 1a–f with a number of DNA targets, we employed surface plasmon resonance (SPR) to evaluate equilibrium binding15 and a fluorescence resonance energy transfer (FRET) melting assay16 to evaluate the stabilizing influence of the ligand. We included three distinct G-quadruplex forming sequences in the study, the human telomeric DNA quadruplex sequence htelo [d(AGGG (TTAGGG)3] and the two c-kit promoter G-quadruplex sequences c-kit18b [d(AGGGAGGGCGCTGGGAGGAGGG)] and c-kit28a [d(CGGGCGGGCGCGAGGGAGGGG)]. We have also included a duplex DNA [d(GGCATAGTGCGTGGGCGTTAGC)] hybridized with its complementary strand as a control. The SPR results (Table 1) revealed that ligands 1a–f, bind to the various G-quadruplex DNA targets with measurable dissociation constants ranging from 3 to 69 μM. None of the ligands showed significant binding to the control duplex.17 All ligands tested showed a preference for binding to the c-kit2 quadruplex, with 3-fold selectivity for c-kit2 over c-kit1 for ligand 1d and 14-fold selectivity for c-kit2 over htelo for ligand 1a (Table 1). The discrimination between quadruplex DNA and duplex DNA has now been reported for a number of small molecule ligands, but the discrimination between distinct quadruplexes by a small molecule is perceived to be a greater challenge. Our observations support the potential for small molecules to discriminate between quadruplexes. We do not yet have structural information on the interaction of ligands 1a–f, but we envisage that the observed quadruplex discrimination must, in some part, arise from the distinct loop topology and/or sequences exhibited for the quadruplexes under study.
Table 1.
Dissociation Constants (Kd) Measured by SPR and G-Quadruplex Stabilization (ΔTm) Potential by FRET-Melting
| DNA | 1a | 1b | 1c | 1d | 1e | 1f |
|---|---|---|---|---|---|---|
| Kd [μM] | ||||||
| c-kit2 | 3 ± 0.5 | 5.8 ± 2.3 | 9.2 ± 1.5 | 9.7 ± 0.8 | 2.8 ± 0.5 | 7.7 ± 0.2 |
| c-kit1 | 6.4 ± 0.1 | 9.1 ± 2.3 | 21 ± 2 | 31 ± 1 | 4.9 ± 0.3 | 13.7 ± 1 |
| htelo | 43 ± 3 | 8.6 ± 2 | 69 ± 7 | b | 21 ± 1 | 63 ± 2 |
| ds | nd17 | nd17 | nd17 | nd17 | nd17 | nd17 |
| ΔTm at 1 μM (°C) | ||||||
| c-kit2 | 18 | 17.5 | 5.4 | nda | 17.2 | 14 |
| c-kit1 | 25.3 | 24.6 | 11 | nda | 27.1 | 13.7 |
| htelo | 19.1 | 19.3 | 7 | nda | 15.5 | 15.4 |
| ds | 0 | 0 | 0 | nda | 0 | 0 |
| G-Quadruplex Binding Discrimination (Fold) | ||||||
| c-kit2/ c-kit1/htelo |
1/2/14 | 1/2/2 | 1/2/8 | 1/3 | 1/2/7 | 1/2/9 |
Not determined (nd) due to intrinsic fluorescence of ligand.
No detectable binding was observed; SPR standard deviations are given for two independent experiments; ΔTm ± 1 °C.
The FRET-melting data (Table 1) showed that, with the exception of 1c, all ligands evaluated showed a Tm shift of 14–27 °C, at 1 μM ligand, for the three quadruplexes evaluated and no detectable stabilization of duplex DNA. The FRET melting data is broadly in support of the SPR measurements, although one must exercise caution in the interpretation of FRET-melting since Tm measurements relate to ligand-induced stabilization of folded structure, rather than equilibrium binding (Figure 2).
Figure 2.
(A) SPR binding curve for isoalloxazine 1a to c-kit2 (●), c-kit1 (▲), and htelo (■); running buffer, 50 mM Tris-HCl pH 7.4, 100 mM KCl. (B) FRET-melting assay for c-kit1 (∗), htelo (■), c-kit2 (●), and ds DNA (▲) in the presence of 1a; buffer, 60 mM potassium cacodylate pH 7.4.
On the basis of their selectivity for c-kit2, we carried out a study on the effects of ligands 1a and 1d on c-kit expression. We evaluated the ligands at 5 μM final concentration, near to the measured Kd for both compounds (Table 1). Two human cancer cell lines were used: the epithelial like breast cancer cell line MCF-7 and the gastrointestinal stromal tumor cell line HGC-27. These ligands were not toxic to either cell line at the concentration employed. Experiments were performed in MCF-7 that exhibited relatively low basal levels of c-kit expression. The ligand-induced changes in c-kit expression were measured by quantitation of mRNA using real-time polymerase chain reaction (RT-PCR) (see Supporting Information for details). These changes relative to a control gene β-actin are shown in Figure 3A. The results showed that after 3 h treatment with ligand 1a, expression of c-kit was reduced by 5.4-fold and by 5-fold after 6 h (Figure 3A). For ligand 1d expression of c-kit was reduced by 2.1-fold after 3 h and almost completely suppressed (31-fold reduction) after 6 h (Figure 3A). The strong effect of ligand 1d prompted us to evaluate its influence on HGC-2718 cells, which show much higher basal levels of c-kit oncogene expression (∼16-fold higher expression than MCF-7). Upon treatment with ligand 1d, HGC-27 cells showed that c-kit gene expression was reduced by 1.8-fold, 1.9-fold, and 2.4-fold after 2, 6, and 24 h, respectively (Figure 3B). Overall, ligands show the ability to inhibit c-kit gene expression. The effect of ligand 1d on the expression of c-kit in MCF-7 cells was much more pronounced than in HGC-27 cells, which may be due to the different basal transcriptional levels of c-kit of these two cell lines. It was interesting to note that the inhibitory effects were observed at concentrations near the ligand Kd, which may reflect favorable cellular uptake of such compounds.
Figure 3.
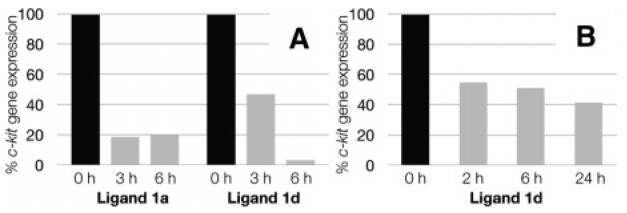
The inhibition of c-kit gene expression by isoalloxazines. Expression of c-kit in control cells treated with 10% DMSO in water is set to 100% (black bars). Expression of c-kit in cells treated with ligands is given as a percentage of untreated controls (gray bars). For all experiments, c-kit expression has been normalized to the expression of the house-keeping gene β-actin: (A) levels of c-kit repression in MCF-7 cells, with indicated ligands and time points at 5 μM final ligand concentration; (B) levels of c-kit repression in HGC-27 cells in response to ligand 1d, after given time points at 5 μM final concentration.
In summary, 3,8,10-trisubstituted isoalloxazines are a promising class of G-quadruplex binding ligands that show selective binding to a c-kit promoter quadruplex and have provided proof of concept for the inhibition of c-kit expression. Detailed investigations on the chemical biology of 3,8,10-trisubstituted isoalloxazines and quadruplexes are now underway.
Acknowledgment
We thank Cancer Research U.K. for programme and project funding, the BBSRC for funding, and COMSATS University faculty development programme funding for a studentship to R.S. S.B. is a BBSRC Career Development Research Fellow. We thank Ashok Venkitaraman for valuable discussion.
Footnotes
Supporting Information Available: Experimental procedures for the synthesis of isoalloxazines 1, SPR binding curves, experimental details, curves of FRET-melting and experimental procedures of cell based experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Phan AT, Modi YS, Patel DJ. J. Am. Chem. Soc. 2004;126:8710. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Parkinson GN, Lee MPH, Neidle S. Nature. 2002;417:876. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 2.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Nat. Struct. Mol. Biol. 2005;12:847. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 3.(a) Huppert JH, Balasubramanian S. Nucleic Acids Res. 2007;35:406. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Todd AK, Johnston M, Neidle S. Nucleic Acids Res. 2005;33:2901. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai J, Chen D, Jones AR, Hurley LH, Yang D. Nucleic Acids Res. 2006;34:5133. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun D, Guo K, Rusche JJ, Hurley HL. Nucleic Acids Res. 2005;33:6070. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogoi S, Xodo LE. Nucleic Acids Res. 2006;34:2536. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman A, Neidle S, Balasubramanian S. Biochemistry. 2006;45:7854. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. J. Am. Chem. Soc. 2005;127:10584. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou TM, Lu YJ, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JC, Chan AS, Gu LQ. J. Med. Chem. 2007;50:1465. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 10.Yarden Y, Kuang WJ, Yang–Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A. EMBO J. 1987;6:3341. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuveson DA, Willis NA, Jacks T, Griffin TD, Singer S, Fletcher CDM, Fletcher JA, Demetri GD. Oncogene. 2001;20:5054. doi: 10.1038/sj.onc.1204704. [DOI] [PubMed] [Google Scholar]
- 12.Lauhon CT, Szostak JW. J. Am. Chem. Soc. 1995;117:1246. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- 13.White EW, Tanious F, Ismail MA, Reszka AP, Neidle S, Boykin DW, Wilson DW. Biophys. Chem. 2007;126:140. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Kipnis F, Weiner N, Spoerri PE. J. Am. Chem. Soc. 1947;69:799. [Google Scholar]
- 15.(a) Teulade–Fichou M-P, Carrasco C, Guittat L, Bailly C, Alberti P, Mergny J-L, David A, Lehn J-M, Wilson WD. J. Am. Chem. Soc. 2003;125:4732. doi: 10.1021/ja021299j. [DOI] [PubMed] [Google Scholar]; (b) Schouten JA, Ladame S, Mason SJ, Cooper MA, Balasubramanian S. J. Am. Chem. Soc. 2003;125:5594. doi: 10.1021/ja029356w. [DOI] [PubMed] [Google Scholar]; (c) Davis TM, Wilson DW. Methods Enzymol. 2001;340:22. doi: 10.1016/s0076-6879(01)40416-2. [DOI] [PubMed] [Google Scholar]
- 16.(a) Harrison RJ, Reszka AP, Haider SM, Ramagnoli B, Morell J, Read MA, Gowan SM, Incles CM, Kelland LR, Neidle S. Bioorg. Med. Chem. Lett. 2004;14:5845. doi: 10.1016/j.bmcl.2004.09.037. [DOI] [PubMed] [Google Scholar]; (b) Darby RAJ, Sollogoub M, Mckeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The isoalloxazines (1a–f) did not show any significant binding with duplex DNA at concentrations up to 200 μM. Ligand aggregation prevented the measurement of binding at higher concentrations.
- 18.Hassan S, Kinoshita Y, Kawanami C, Kishi K, Matsushima Y, Ohashi A, Funasaka Y, Okada A, Maekawa T, He–Yao W, Chiba T. Dig. Dis. Sci. 1998;43:8. doi: 10.1023/a:1018851415704. [DOI] [PubMed] [Google Scholar]


