Dynamic covalent chemistry (DCC) is a reversible exchange process which allows noncovalent interactions to template covalent-bond formation.[1] A dynamic combinatorial library (DCL) is an equilibrating mixture, under thermodynamic control, in which each of the species is represented in proportion to its free energy.[2] The introduction of a molecular template to a DCL will shift the equilibrium to favor individual library members that bind the template and are thus stabilized. Since its conception, DCC has been applied to both small-molecule[2] and macromolecular targets.[3] Examples of DCC that involve biomolecules include the work of Ramström and Lehn[3b] who generated a small disulphide-based carbohydrate library and screened in situ against the common jack bean-lectin Concanavalin A, and of Miller and co-workers[4] who targeted oligonucleotides with non-nucleotide building blocks and selected metal–ligand complexes that bind nucleic acid hairpins with high affinity and selectivity. An experimentally challenging aspect of employing biomolecules as templates in DCLs is the analysis of the reversibility of the dynamic chemistry and quantitation of ligand amplification. This analysis requires detailed investigations on a relatively simple DCL, and has been a key objective of the work we report in this paper.
The molecular target for our study is a four-stranded G-quadruplex formed by DNA sequences with stretches of G-nucleotides under near physiological salt conditions in vitro.[5] Quadruplex structures may be involved in cellular functions that include chromosomal alignment[6] and telomere length regulation.[7] In particular, the latter has been implicated in the control of cellular aging and mechanisms of cancer proliferation.[7] Furthermore, recent studies reported for the oncogene c-myc[8] suggest that quadruplex stabilization might have the potential to control gene expression. Therefore, there is considerable interest in developing quadruplex-stabilizing ligands for novel therapeutic approaches and a desire to understand the molecular basis of quadruplex recognition.[9] The G-quadruplex structural motif has features that distinguish it from double-stranded B-DNA and that can be exploited in the design of quadruplex specific ligands. A number of promising small-molecule ligands have been reported based on recognition of the terminal G-tetrad through hydrophobic and π–π interactions.[9] There is potential for specific ligands that make contacts with loop and groove regions of quadruplex DNA.[10] The philosophy of combining distinct binding elements to generate improved ligands has been effective for nucleic acids.[11] Herein, we investigate the potential of DCC to generate quadruplex ligands through a reversible, covalent combination of two distinct recognition elements: a hydrophobic acridone unit, designed to interact with the terminal G tetrad of a parallel quadruplex,[12] and a tetrapeptide sequence (FRHR), which was recently shown by us to have quadruplex-recognition properties.[13]
Disulphide chemistry was chosen because it is water compatible, relatively fast, and thiol exchange operates at mild pH. It allows the mixture of DCL components to interconvert and reach equilibrium, and can be switched off by lowering the pH value to ≤5.[14] The peptide fragment, FRHR, was derivatized with sulfanylacetic acid at the N terminus to give monomer P, which has an N-terminal thiol group. P was synthesized by using standard solid-phase Fmoc-peptide chemistry (Fmoc = 9-fluorenylmethoxycarbonyl) on Rink amide resin. Acridone A was prepared by coupling of 3,6-bis[(2-carbamoylethyl)methylamino]acetic acid-substituted acridone[15] onto cysteine-loaded Wang resin. Based on preliminary modeling studies, the sarcosine linker was shown to position the cysteine residue at the top of a quadruplex groove (data not shown). In our DCC experiments we employed a human telomeric quadruplex, formed from the deoxyoligonucleotide 5′-biotin(GTTAGG)5.[16]
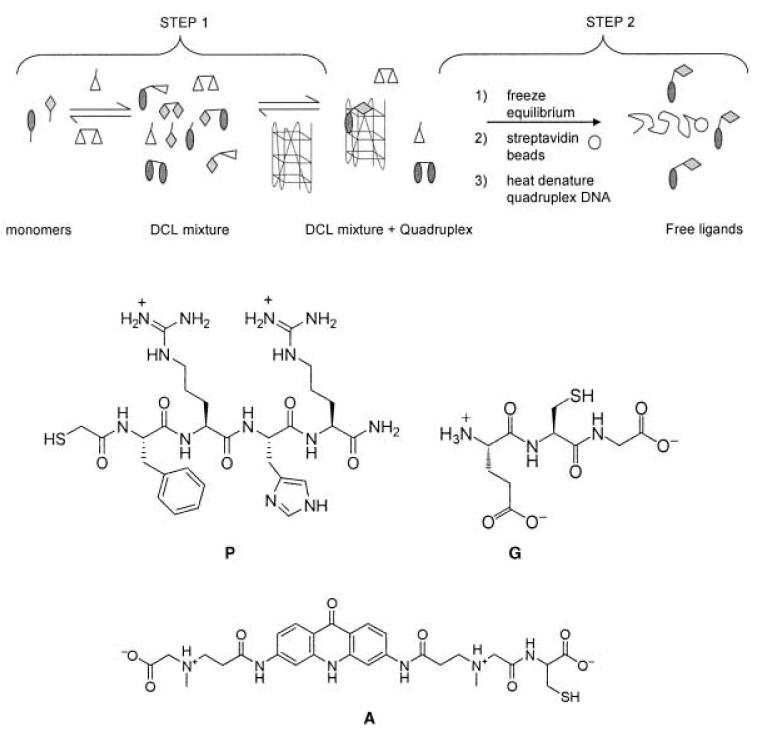
The experimental design principles are detailed in Scheme 1. An exchange buffer containing an excess of both oxidized (G-G) and reduced glutathione (G) mediates exchange between the components of the DCL, which enables the use of relatively low concentrations of A and P. Furthermore, both, G and G-G, have the potential to interact with the quadruplex and thus act as competitive library members. When equilibrium is reached, the exchange reaction is stopped by lowering the pH from 7.4 to 2 and the biotinylated quadruplex target, with bound ligands, can be isolated from the solution by immobilization onto streptavidin beads. The quadruplex is heat-denatured at 85°C to release any bound ligand and HPLC is used to identify and quantify all members of the DCL. A parallel control experiment, without quadruplex target, enabled us to make a comparison that identified species amplified by the quadruplex-DNA template.
Scheme 1.
Schematic experimental method. Step 1: Monomers P and A are used in the presence of glutathione (oxidized: G-G (375 μm), reduced: G (1.5 mm)) in redox exchange buffer to equilibrate in either the presence or absence of the target quadruplex DNA. Step 2: The exchange is frozen by reducing the pH to 2 with 0.1% TFA in water, the quadruplex (+ ligand) is removed from solution by streptavidin immobilization and heat denatured to dissociate bound ligands. All ligands are analyzed by HPLC. TFA = trifluoroacetic acid.
Critical features of any DCC experiment are the need to prove thermodynamic control and demonstrate an equilibrium shift induced by a template. To investigate disulfide-exchange reactions of A and P under thermodynamic control, a glutathione-containing buffer was used. The glutathione was utilized in a large excess to mediate fast exchange at relatively low levels of A and P.[14, 17]
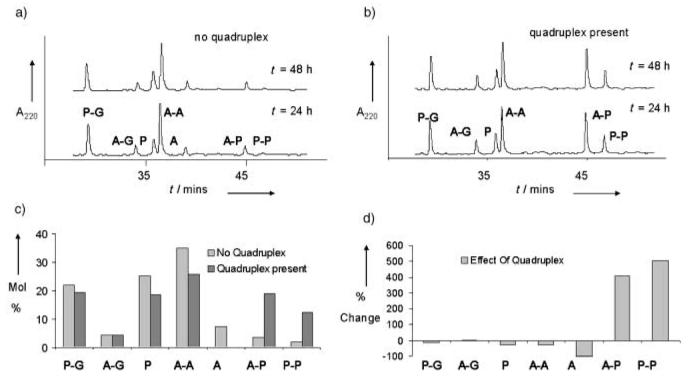
In the absence of target quadruplex DNA, the system equilibrated within 22hours and remained unchanged over the next 26 hours (Figure 1 a). The same distribution of species was reached in 48 hours but starting from homodisulfides A-A and P-P (data not shown), which showed that the system is truly reversible and under thermodynamic control. At equilibrium, building block P was predominantly found in the form of a heterodisulfide with glutathione, P-G. In contrast, building block A was predominantly found as the homodisulfide A-A, despite the excess of competing G present. This observation is in accordance with air oxidation experiments (see Supporting Information) and suggests self-recognition of the acridone.[14]
Figure 1.
a) HPLC traces showing the component composition for exchange experiments in glutathione-containing buffer with use of A and P (200 μm each) in the absence of quadruplex DNA, at times denoted. b) HPLC traces showing the component composition for exchange experiments in glutathione-containing buffer with use of A and P (200 μm each) in the presence of quadruplex DNA, at times denoted. c) Histogram showing the change in equilibrium mixture composition on introduction of quadruplex DNA. Values were measured by HPLC-peak area, taking into account differences in extinction coefficients. d) Histogram showing the percentage changes in each species of the equilibrium mixture upon introduction of quadruplex DNA.
Parallel experiments were carried out in the presence of 200 μm 5′-biotinylated, folded[16] human telomeric quadruplex, under otherwise identical conditions. Equilibrium was achieved within 48 hours (Figure 1 b) and the same distribution of species was reached starting with A and P as either free thiols or as homodisulfides A-A and P-P, which confirmed a true equilibration of the system in the presence of quadruplex DNA. Analysis of the reaction stopped at 12, 24, 48, and 100 hours revealed an increase in the proportion of both, acridone–peptide heterodisulfide (A-P) and peptide homodisulfide (P-P), until equilibrium was reached within 48 hours with a constant A-P/P-P ratio of 1.4:1. The formation of A-P and P-P was at the expense of disulfides P-G and A-A and thiols A and P. At equilibrium the acridone–peptide heterodisulfide A-P and peptide homodisulfide P-P are 19 and 12%, respectively, of the library's mole composition (Figure 1 c). This is a four and fivefold amplification of A-P and P-P, respectively, induced by the presence of the quadruplex DNA (Figure 1 d).
The amplification of P-P was particularly surprising since there are no reports of short peptide ligands that bind quadruplex DNA with high affinity. To confirm this result, exchange experiments were carried out by using P alone in the G/G-G-containing buffer. In the absence of quadruplex DNA the major species observed was P-G, as expected. Upon introduction of the quadruplex DNA, the proportion of P-P in the exchange mixture rose from 2 to 12%, a sixfold increase at the expense of P-G (see Supporting Information).
Quantitative-binding studies of species A-P, P-P, and A-A were carried out by using surface plasmon resonance (SPR) with the same quadruplex-DNA target. SPR is a valuable and extensively used method for the study of DNA small molecule interactions.[13, 18] Figure 2 shows the binding curves derived from the SPR data. A-P and P-P bind the human telomeric quadruplex DNA with dissociation constants of 30 ± 1.5 and 22.5 ± 1.1 μm, respectively, consistent with their selection in the DCC experiments. No binding of A-A was detected at 88 μm indicative of a lower limit for the KD of 2.5 mm (see Supporting Information).
Figure 2.
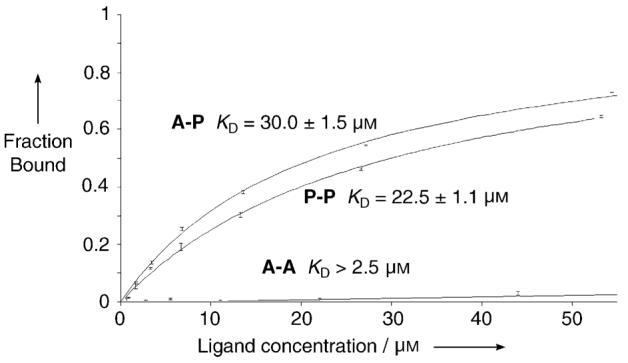
Binding curves obtained from SPR measurements for the determination of KD values for A-P, P-P, and A-A with quadruplex DNA.
This study shows that dynamic covalent chemistry can be used to evolve molecules that bind to quadruplex DNA from a DCL. The critical requirements of thermodynamic control and target-mediated shifting of the position of equilibrium, have been satisfied. The heterodisulfide A-P and, more surprisingly, homodisulfide P-P have been selected from a library of nine species and shown to have good quadruplex affinity. Based on the design principles, we propose that binding of A-P is mediated by acridone–π–π interactions with the top tetrad of the quadruplex and quadruplex-loop/groove interactions with the appended peptide. This result is also suggested by modeling studies of amide-linked acridone–peptide conjugates with human telomeric quadruplexes (data not shown). The mode by which P-P binds quadruplex DNA is presently unknown and will be the subject of future high-resolution NMR spectroscopic studies. Thus, the DCC approach has generated two novel quadruplex-binding ligands and will be exploited to select aromatic cores and peptide/nonpeptide side chains against G-quadruplex targets in future studies.
Footnotes
This work was funded by the BBSRC. We thank Drs. Y. Krishnan-Ghosh and S. Otto for critically reading the manuscript and helpful comments.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.For selected reviews see Ganesan A. Angew. Chem. 1998;110:2989. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2828::AID-ANIE2828>3.0.CO;2-G. Angew. Chem. Int. Ed. 1998;37:2828. Lehn J-M. Chem. Eur. J. 1999;5:2455. Ramstrom O, Bunyapaiboonsri T, Lohmann S, Lehn J-M. Biochim. Biophys. Acta. 2002;1572:178. Rowan SJ, Cantrill SJ, Cousins GRL, Sanders JKM, Stoddart JF. Angew. Chem. 2002;114:1528. doi: 10.1002/1521-3773(20020315)41:6<898::aid-anie898>3.0.co;2-e. Angew. Chem. Int. Ed. 2002;41:898.
- 2.a) Hioki H, Still WC. J. Org. Chem. 1998;63:904. [Google Scholar]; b) Berl V, Huc I, Lehn J-M, DeCian A, Fischer J. Eur. J. Org. Chem. 1999:3089. [Google Scholar]; c) Otto S, Furlan RLE, Sanders JKM. J. Am. Chem. Soc. 2000;122:12063. [Google Scholar]; d) Furlan RLE, Ng Y-F, Otto S, Sanders JKM. J. Am. Chem. Soc. 2001;123:8876. doi: 10.1021/ja0160703. [DOI] [PubMed] [Google Scholar]; e) Furlan RLE, Ng Y-F, Cousins GRL, Redman JE, Sanders JKM. Tetrahedron. 2002;58:771. [Google Scholar]; f) Roberts SL, Furlan RLE, Cousins GRL, Sanders JKM. Chem. Commun. 2002:938. doi: 10.1039/b201465c. [DOI] [PubMed] [Google Scholar]; g) Stulz E, Ng Y-F, Scott SM, Sanders JKM. Chem. Commun. 2002:524. doi: 10.1039/b111019p. [DOI] [PubMed] [Google Scholar]; h) Otto S, Furlan RLE, Sanders JKM. Science. 2002;297:590. doi: 10.1126/science.1072361. [DOI] [PubMed] [Google Scholar]; i) Brisig B, Sanders JKM, Otto S. Angew. Chem. 2003;115:1308. doi: 10.1002/anie.200390326. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2003;42:1270. [Google Scholar]; j) Otto S, Kubik S. J. Am. Chem. Soc. 2003;125:7804. doi: 10.1021/ja0351589. [DOI] [PubMed] [Google Scholar]
- 3.a) Huc I, Lehn J-M. Proc. Natl. Acad. Sci. USA. 1997;94:2106. doi: 10.1073/pnas.94.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ramström O, Lehn J-M. ChemBioChem. 2000;1:41. doi: 10.1002/1439-7633(20000703)1:1<41::AID-CBIC41>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]; c) Hochgürtel M, Kroth H, Piecha D, Hofmann MW, Nicolau C, Krause S, Schaaf O, Sonnenmoser G, Eliseev AV. Proc. Natl. Acad. Sci. USA. 2002;99:3382. doi: 10.1073/pnas.052703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Klekota B, Miller BL. Tetrahedron. 1999;55:11 687. [Google Scholar]; b) Karan C, Miller BL. J. Am. Chem. Soc. 2001;123:7455. doi: 10.1021/ja010325v. [DOI] [PubMed] [Google Scholar]
- 5.For selected reviews see Neidle S, Parkinson GN. Curr. Opin. Struct. Biol. 2003;13:275. doi: 10.1016/s0959-440x(03)00072-1. Simonsson T. Biol. Chem. 2001;382:621. doi: 10.1515/BC.2001.073.
- 6.Sen D, Gilbert W. Nature. 1988;334:364. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 7.a) Blackburn E. Nature. 2000;408:53. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]; b) Cech TR. Angew. Chem. 2000;112:34. doi: 10.1002/(sici)1521-3773(20000103)39:1<34::aid-anie34>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2000;39:34. [Google Scholar]
- 8.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. USA. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin S, Trent JO, Jenkins TC, Neidle S, Hurley LH. J. Med. Chem. 1997;40:2113. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson GN, Lee MP, Neidle S. Nature. 2002;417:876. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 11.For selected examples, see: Bentin T, Nielsen PE. J. Am. Chem. Soc. 2003;125:6378. doi: 10.1021/ja029936t. Carrasaco C, Helissey P, Haroun M, Baldeyrou B, Lansiaux A, Colson P, Houssier C, Giorgi-Renault S, Bailly C. ChemBioChem. 2003;4:50. doi: 10.1002/cbic.200390014. Carlson CB, Stephens OM, Beal PA. Biopolymers. 2003;70:86. doi: 10.1002/bip.10413.
- 12.a) Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR. Mol. Pharmacol. 2002;61:1154. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]; b) Clark GR, Pytel PD, Squire CJ, Neidle S. J. Am. Chem. Soc. 2003;125:4066. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]; c) Haider SM, Parkinson GN, Neidle SJ. J. Mol. Biol. 2003;326:117. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 13.Schouten JA, Ladame S, Mason SJ, Cooper MA, Balasubramanian S. J. Am. Chem. Soc. 2003;125:5594. doi: 10.1021/ja029356w. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan-Ghosh Y, Balasubramanian S. Angew. Chem. 2003;115:2221. doi: 10.1002/anie.200250551. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2003;42:2171. [Google Scholar]
- 15.Ladame S, Harrison RJ, Neidle S, Balasubramanian S. Org. Lett. 2002;4:2509. doi: 10.1021/ol026130p. [DOI] [PubMed] [Google Scholar]
- 16.The G-quadruplex was folded under standard conditions. The single-stranded oligonucleotide, 5′-biotinylated (GTTAGG)5, was heated in buffer to 90°C and slowly cooled to room temperature overnight. Its fully folded structure was confirmed by circular dichroism spectroscopy and UV melting experiments.
- 17.Saghatelian A, Yokobayashi Y, Soltani K, Ghadiri MR. Nature. 2001;409:797. doi: 10.1038/35057238. [DOI] [PubMed] [Google Scholar]
- 18.a) Lacy ER, Le NM, Price CA, Lee M, Wilson WD. J. Am. Chem. Soc. 2002;124:2153. doi: 10.1021/ja016154b. [DOI] [PubMed] [Google Scholar]; b) Carrasco C, Facompre M, Chisholm JD, Van Vranken DL, Wilson WD, Bailly C. Nucleic Acids Res. 2002;30:1774. doi: 10.1093/nar/30.8.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Carrasco C, Rosu F, Gabelica V, Houssier C, De Pauw E, Garbay-Jaureguiberry C, Roques B, Wilson WD, Chaires JB, Waring MJ, Bailly C. ChemBioChem. 2002;3:1235. doi: 10.1002/1439-7633(20021202)3:12<1235::AID-CBIC1235>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]