Abstract
A bovine-specific cDNA microarray system was used to compare gene expression profiles of peripheral blood mononuclear cells (PBMCs) from control uninfected (n = 4) and Johne's disease-positive (n = 6) Holstein cows. Microarray experiments were designed so that for each animal, a direct comparison was made between PBMCs stimulated in vitro with Mycobacterium avium subsp. paratuberculosis and PBMCs stimulated with phosphate-buffered saline (nil-stimulated PBMCs). As expected, M. avium subsp. paratuberculosis stimulation of infected cow PBMCs enhanced expression of gamma interferon transcripts. In addition, expression of 15 other genes was significantly affected (>1.25-fold change; P < 0.05) by in vitro stimulation with M. avium subsp. paratuberculosis. Similar treatment of control cow PBMCs with M. avium subsp. paratuberculosis resulted in significant changes in expression of 13 genes, only 2 of which were also affected in PBMCs from the infected cow PBMCs. To compare gene expression patterns in the two cow infection groups (infected cows and uninfected cows), a mixed-model analysis was performed with the microarray data. This analysis indicated that there were major differences in the gene expression patterns between cells isolated from the two groups of cows, regardless of in vitro stimulation. A total of 86 genes were significantly differentially expressed (P < 0.01) in M. avium subsp. paratuberculosis-stimulated PBMCs from infected cows compared to expression in similarly treated PBMCs from control cows. Surprisingly, a larger number of genes (110 genes) were also found to be significantly differentially expressed (P < 0.01) in nil-stimulated cells from the two infection groups. The expression patterns of selected genes were substantiated by quantitative real-time reverse transcriptase PCR. Flow cytometric analysis indicated that there were no gross differences in the relative populations of major immune cell types in PBMCs from infected and control cows. Thus, data presented in this report indicate that the gene expression program of PBMCs from M. avium subsp. paratuberculosis-infected cows is inherently different from that of cells from control uninfected cows.
Johne's disease is an infectious disease of ruminants caused by the facultative intracellular bacterium Mycobacterium avium subsp. paratuberculosis. Infections with M. avium subsp. paratuberculosis can persist in a subclinical state for several years with few outward pathological consequences (20, 22, 23, 34). Thus, M. avium subsp. paratuberculosis infections in ruminants can serve as models for other persistent or chronic infectious diseases caused by intracellular bacteria, such as Brucella abortus, Mycobacterium bovis, and Salmonella. The lesions associated with M. avium subsp. paratuberculosis infection are typically restricted to the illeum and particularly to the illeocecal valve region of the small intestine (10, 42). Like the pathogenesis associated with other mycobacterial infections, the pathogenesis associated with M. avium subsp. paratuberculosis infection is in large part due to a severe immune pathology and chronic inflammation (5, 27, 36, 47).
Infections with M. avium subsp. paratuberculosis can be established in utero, presumably by transfer of bacteria or infected cells from the dam. Alternatively, calves may be infected in the first few months of life via the fecal-oral route or through ingestion of infected colostrum (30, 35, 38, 41). Following the initial exposure to M. avium subsp. paratuberculosis, most animals develop an appropriate T-cell response, which is characterized by release of proinflammatory cytokines, such as gamma interferon (IFN-γ) and tumor necrosis factor alpha, as well as by production of interleukin-2 (IL-2), which presumably activates CD4+ T cells (4, 14). The initial proinflammatory cytokine production leads to recruitment and activation of cytotoxic CD8+ T cells and other immune components, including neutrophils and additional activated macrophages, at sites of M. avium subsp. paratuberculosis infection (1, 2). During the long subclinical stage of infection, the initial cytotoxic or Th1-like response is often replaced by an antibody or Th2-like response characterized by production of immunoglobulin G1 antibodies (for a review see reference 14). The reasons for this shift in the immune response in animals that show clinical signs of Johne's disease are unknown, but they may be related to unknown genetic factors or to the constant exposure of immune cells to antigen released from infected macrophages.
Infections with M. avium subsp. paratuberculosis are typically diagnosed by using an absorbed serum enzyme-linked immunosorbent assay (ELISA) or an in vitro IFN-γ stimulation test or by direct fecal culture (3, 12, 16, 33). Because of the early proinflammatory response to M. avium subsp. paratuberculosis infection, IFN-γ testing may detect infections in animals much earlier in the infection cycle than either a serum ELISA or fecal culturing detects them (33). Widespread use of the IFN-γ test has led to a wealth of information concerning production of this cytokine in response to M. avium subsp. paratuberculosis (3, 17, 25, 31-33). Although several recent reports have also detailed expression patterns of other cytokines in peripheral blood mononuclear cells (PBMCs), lesions, and mesenteric lymph nodes of infected cattle and sheep (4, 31), a paucity of information and specific reagents for detecting expression of bovine cytokines and other immune cell genes and proteins important in disease progression has severely limited studies of immune responses to M. avium subsp. paratuberculosis. Thus, while basic immune cell types that respond to M. avium subsp. paratuberculosis and the presence or absence of several well-characterized proinflammatory cytokines at sites of M. avium subsp. paratuberculosis infection are known (4, 5, 8, 9, 24), the molecular mechanisms that ultimately lead to pathological outcomes, including shifts from a beneficial Th1-like immune response to an unprotective Th2-like immune response in cattle with Johne's disease, are unknown.
In humans, a potential link between M. avium subsp. paratuberculosis exposure and Crohn's disease suggests that in addition to having economic consequences for the dairy industry, this pathogen may also be an important food safety concern. When combined, the high incidence of M. avium subsp. paratuberculosis infection in United States dairy herds (19), the grave economic and animal welfare consequences of Johne's disease, and a potential link to human disease make a powerful case for learning more about how M. avium subsp. paratuberculosis infections progress and about the host genomic response to this fastidious pathogen. This was the main objective of the present study.
Recently, development of cDNA microarrays containing 724 unique bovine genes specifically targeted to enhance studies of bovine immunobiology was described (6, 45). This BOTL-2 (bovine total leukocyte, version 2) cDNA microarray was used to demonstrate differences in the gene expression profiles of PBMCs isolated from cattle in various stages of Johne's disease (13). In this report, we describe experiments in which we used an expanded bovine (BOTL-3) cDNA microarray to test the hypothesis that the gene expression profiles of PBMCs from Johne's disease-positive cows are different from those of PBMCs from healthy uninfected cows. Our results demonstrated that while in vitro stimulation of PBMCs with M. avium subsp. paratuberculosis induced gene expression changes in cells from infected cows distinct from those induced in cells from control cows, there were also a surprising number of gene expression differences between infected and control cow PBMCs not exposed to M. avium subsp. paratuberculosis in vitro. The unique patterns of gene expression did not appear to be due to gross differences in the types or relative numbers of cells present in PBMCs from infected and control cows, suggesting that M. avium subsp. paratuberculosis infection in vivo alters the gene expression program of bovine blood mononuclear cells.
MATERIALS AND METHODS
Experimental animals and preparation of PBMCs.
The infected and control cattle used in this study were Holstein cows ranging in age from 24 to 48 months. All animals were housed on the same commercial dairy operation. The immune status of all study animals with regard to M. avium subsp. paratuberculosis infection had been monitored by serum ELISA on a bimonthly basis for over 24 months prior to the initiation of experiments. Additional screening procedures for M. avium subsp. paratuberculosis infection included quarterly IFN-γ testing with a commercial system (BioCor, Inc., DeMoines, Iowa) and quantitative real-time reverse transcriptase PCR (Q-RT-PCR). Periodic fecal culture testing by a U.S. Department of Agriculture-approved testing laboratory (Michigan State University Animal Health Diagnostic Laboratory, East Lansing) was conducted to confirm infection of ELISA- and IFN-γ-positive cows. The control uninfected animals (n = 4) had shown negative responses for over 2 years of testing by all assays used. The M. avium subsp. paratuberculosis-infected animals (n = 6) were strongly positive as determined by serum ELISA (index value, >75) over the entire testing period, exhibited occasional diarrhea, and were fecal culture positive, with 5 to 100 CFU/g of feces (subclinical shedders). Tests for IFN-γ in infected animals were strongly positive when the results were analyzed as recommended by the manufacturer (BioCor, Inc.).
Blood samples were obtained from all animals via the coccygeal (tail) vein by using 2.5-cm 21-gauge multiple-sample needles and a series of four 8-ml Vacutainer tubes containing acid-citrate dextrose as an anticoagulant (BD Vacutainer, Rutherford, N.J.). PBMCs were prepared as previously described (13, 39, 45). Briefly, blood samples were centrifuged at 4°C for 20 min at 1,000 × g, and the resulting buffy coats (approximately 1 to 2 ml) were transferred to new 50-ml conical tubes containing 34 ml of ice-cold sterile phosphate-buffered saline (PBS) overlaid on a 10-ml cushion of Percoll (1.084 g/ml; Sigma Chemical Co., St. Louis, Mo.). Cells were centrifuged at 1,000 × g for 40 min at room temperature to separate erythrocytes and polymorphonuclear leukocytes from mononuclear cells. Following careful aspiration of the PBS, PBMCs at the PBS-Percoll interface were transferred to new 50-ml conical tubes, rinsed once with 20 ml of sterile PBS, and finally suspended in maintenance medium RPMI 1640 containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (complete RPMI 1640).
The PBMCs from each cow were split into two equal aliquots; one aliquot was used for stimulation with M. avium subsp. paratuberculosis, and the other aliquot was used for PBS (nil) treatment. PBMCs were stimulated with the bacterium by using 106 live M. avium subsp. paratuberculosis cells per ml of medium added in 200 μl of PBS. Nil stimulation was conducted by adding 200 μl of PBS. All PBMCs were then incubated at 37°C for 16 to 18 h in complete RPMI 1640 without antibiotics in a humidified atmosphere consisting of 95% air and 5% CO2. This protocol was selected because overnight stimulation with antigen is standard in IFN-γ tests and results in significant production of this cytokine. In a typical protocol, each treatment (nil stimulation and M. avium subsp. paratuberculosis stimulation) consisted of two or three 75-cm2 flasks, with each flask containing approximately 2 × 107 PBMCs.
RNA extraction, preparation of labeled cDNA, and microarray analysis.
RNA was extracted from nil-stimulated and M. avium subsp. paratuberculosis-stimulated PBMCs by using Trizol reagent (Invitrogen Life Technologies Corp., Carlsbad, Calif.) as previously described (13, 45). The quantity and quality of the extracted total RNA were estimated by UV spectrophotometry and electrophoresis on 1.2% native agarose gels. To evaluate gene expression profiles of PBMCs following nil stimulation or stimulation with M. avium subsp. paratuberculosis, total RNA (5 to 10 μg) from treated PBMCs of each animal were used as templates in reverse transcription reactions (Atlas Powerscript labeling system; BD Biosciences Inc., Alameda, Calif.) in which oligo(dT)15-18 was used as the primer. To provide a control for cDNA synthesis and labeling efficiency, as well as for subsequent cDNA microarray hybridization, 650 pg of synthetic lambda Q gene RNA containing an engineered poly(A) tail was spiked into each cDNA synthesis reaction mixture.
Following first-strand cDNA synthesis, cDNAs from nil-stimulated and M. avium subsp. paratuberculosis-stimulated PBMCs from each animal were differentially labeled by using N-hydroxysuccinimide-derivatized Cy3 and Cy5 dyes (Amersham Pharmacia, Ltd., Piscataway, N.J.). The labeled cDNAs were extensively purified to remove unincorporated dyes, combined, and concentrated to 10 μl by using Microcon 30 spin concentrators (Millipore Corp., Bedford, Mass.). Microarray hybridizations for each animal were performed by addition of concentrated Cy3- and Cy5-labeled probe cDNAs to 100 μl of SlideHyb-3 (Ambion, Inc., Alameda, Calif.). Hybridizations were conducted for 4 h in a commercial microarray hybridization station by using a step-down hybridization protocol (GeneTAC; Genomics Solutions, Inc., Ann Arbor, Mich.). Labeling was conducted so that in the infected group (n = 6), three M. avium subsp. paratuberculosis-stimulated PBMC cDNA preparations were labeled with Cy3 and three preparations were labeled with Cy5 (nil-stimulated samples were labeled with the other dye in each case). In the control group (n = 4), two M. avium subsp. paratuberculosis-stimulated samples were labeled with Cy3 and two samples were labeled with Cy5 (nil-stimulated samples were labeled with the other dye in each case).
Following hybridization, cDNA microarrays were washed in the hybridization station, rinsed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and once in double-distilled H2O, and finally dried by centrifugation in a cushioned 50-ml conical centrifuge tube. This process yielded BOTL-3 cDNA microarrays, which allowed direct comparison of PBMC gene expression profiles following stimulation with M. avium subsp. paratuberculosis and nil stimulation blocked by animal.
Final microarrays were scanned by using a GeneTAC LS IV microarray scanner and GeneTAC LS software (Genomic Solutions, Inc.). GeneTAC analyzer software was then used to process microarray images, to find spots, to integrate robot-spotting files with the microarray image, and finally to create reports of raw spot intensities.
BOTL-3 cDNA microarrays.
The cDNA microarrays used in this study (BOTL-3 cDNA microarrays) were an expanded version of those described previously (6, 13, 45). Each microarray contained 3,888 spots consisting of 709 bovine expressed sequence tag (EST) clone inserts and 345 amplicons representing known immune response genes derived from the bovine sequence (15), all spotted in triplicate (3,162 gene spots). The genes represented on the BOTL-3 cDNA microarrays included the genes encoding most commonly studied cytokines, including IL-1, IL-4, IL-5, IL-6, IL-10, IL-12, tumor necrosis factor alpha, IFN-γ, and transforming growth factor β. The complete list of genes on the BOTL-3 cDNA microarray can be found at http://www.nbfgc.msu.edu (15). The control gene spots included 144 spots representing glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes (three spots in each of 48 patches), 75 spots representing β-actin genes, 75 spots representing ribosomal protein L-19 genes, 96 synthetic lambda Q gene spots (two spots per patch), 48 negative control spots (one spot per patch), and 288 blank spots (six spots per patch). The entire array was organized in a 4 × 12 pattern of patches, with each patch containing 81 spots in a 9 × 9 pattern.
Microarray data analysis.
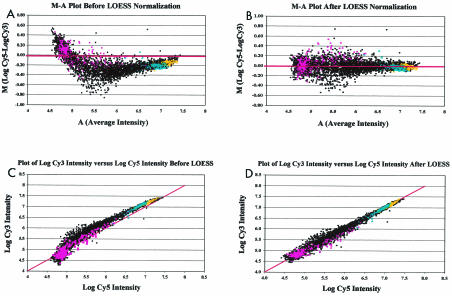
Potential sample and dye intensity biases in microarray data sets were visualized by using log intensity ratio (M)-mean log intensity (A) scatter plots constructed for each array, in which M [M = log (Cy5/Cy3) = (log Cy5 − log Cy3)] was plotted against A [A = (log Cy3 + log Cy5)/2] for each array spot, as described by Yang et al. (44). Array-specific data normalization was then performed by using a robust local regression technique (11) in the LOESS procedure of SAS (28, 29). The efficiency of LOESS normalization was assessed by monitoring M-A plots (Fig. 1A and B) and scatter plots of log Cy3 versus log Cy5 (Fig. 1C and D) for data from each array before and after normalization. Normalized data were then back-transformed prior to further statistical analyses by using the following formulae: log Cy3* = A − M*/2 and log Cy5* = A + M*/2, where log Cy3* and log Cy5* are the normalized log intensities and M* = M − ⁁M represents each of the normalized M values (⁁M is the LOESS-predicted value for each spot).
FIG. 1.
(A) Representative M-A plot of raw cDNA microarray data. In the analysis, data were derived from a cDNA microarray in which RNA from nil (Cy3)-stimulated PBMCs and RNA from M. avium subsp. paratuberculosis (Cy5)-stimulated PBMCs from a Johne's disease-positive cow were compared. Raw cDNA data in the form of relative fluorescence intensity were log transformed and used to calculate M (difference in log intensities) and A (average log intensity) for each spot on the BOTL-3 microarray. Test genes (black circles), blanks and negatives (pink squares), GAPDH (yellow dots), and synthetic lambda Q gene control spots (blue triangles) were plotted as separate series. The red line indicates an M value of zero. (B) M-A plot of LOESS-normalized data. LOESS-corrected values were used to calculate normalized M and A values essentially as described above for panel A. As in panel A, the sample, blank or negative, GAPDH, and lambda Q control genes were plotted as separate series, and the red line indicates an M value of zero. Note the effect of LOESS normalization on the sample M values clustering around the M-0 line (compare this plot with that in panel A). (C) Representative plot of log Cy3 intensity versus log Cy5 intensity before LOESS normalization. Data from panel A were used to calculate log-transformed intensity values for every spot on the BOTL-3 cDNA microarray. Cy3 log intensity values were plotted against Cy5 log intensity values by using a color scheme identical to that described above for panel A. A line of unity was inserted to demonstrate the relationship of sample and control gene log intensity values in an ideal situation, where most points would cluster around the line. (D) Representative plot of log Cy3 intensity versus log Cy5 intensity after LOESS normalization. LOESS-normalized log Cy3 and log Cy5 intensity values were plotted against each other, and a line of unity (log Cy3 intensity = log Cy5 intensity) was inserted to demonstrate the relationship of sample and control gene points in an ideal situation. The colors are as described above for panel A. Note the effect of LOESS normalization both on the clustering of log Cy3-versus-log Cy5 points around the unity line and on the tailing at lower fluorescence intensities observed in the nonnormalized data shown in panel C.
Initial analyses were conducted to explore the effect of treatment (M. avium subsp. paratuberculosis stimulation versus nil stimulation) within each group, and thus we performed a direct comparison of normalized microarray intensities for each BOTL-3 gene in nil-stimulated PBMCs and M. avium subsp. paratuberculosis-stimulated PBMCs blocked by animal. LOESS-normalized values were imported into a Microsoft Excel spreadsheet and combined within infection group. The back-transformed median negative values for each dye within each array were subtracted from the back-transformed LOESS-normalized expression values. The resulting subtracted values were used to calculate mean loge expression differences between treatments for each animal (by using the three gene replicates on each microarray). Finally, the mean loge expression difference values for each animal were combined within infection group and used to calculate an overall mean expression difference (M. avium subsp. paratuberculosis stimulation versus nil stimulation), the standard error, the t statistic, and the t distribution (P value) for each gene.
In our next analysis, we were interested in examining gene expression differences between infected and control cow PBMCs, both with and without in vitro stimulation. This analysis required indirect comparisons across multiple microarrays. To accomplish this, LOESS-adjusted log intensities were analyzed statistically by using a mixed-model approach consisting of two steps (43). The first step involved array-specific spatial variability normalization, and the second step involved gene-specific analyses to test the effects of group (control versus infected cows) and stimulation (nil stimulation versus M. avium subsp. paratuberculosis stimulation) and their interaction on expression profiles for individual genes. The normalization model in the first step included the fixed effects of dye, group, and stimulation and their interaction, as well as the random effects of array (or animal) and patch within array. The second step of the statistical analysis consisted of gene-specific models for estimated residuals obtained from the normalization approach discussed above. Split plot models were considered, having animals as plot (each in one of the groups) and dyes as subplots (with each of the methods of stimulation). These models included gene-specific fixed effects of dye, group, and stimulation and their interaction and random effects of animal within group, stimulation × animal within group, patch within array, and spots within patches. These analyses were performed by using the MIXED procedure of SAS (29).
Q-RT-PCR.
Final validation of selected gene expression changes observed on cDNA microarrays was performed by a Q-RT-PCR procedure by using an Applied Biosystems 7000 DNA sequence detection system (Perkin-Elmer Corp., Foster City, Calif.). Total RNA was extracted from M. avium subsp. paratuberculosis- and nil-stimulated PBMCs, quantified, and quality checked as described above for the microarray analysis. RNA was converted into first-strand cDNA by adding 2 μg of total RNA to a 12-μl reaction mixture containing 10 mM oligo(dT)15-18 primer and each deoxynucleoside triphosphate at a concentration of 1 mM. Following 5 min of incubation at 65°C, the reaction mixture was quick-chilled on ice, and then we added 4 μl of a 5× buffer supplied by the reverse transcriptase manufacturer (the final reagent concentrations were 50 mM Tris-HCl [pH 8.3], 75 mM KCl, and 3 mM MgCl2), 200 U of Superscript II RNase H− reverse transcriptase (Invitrogen Life Technologies), and 10 mM (final concentration) dithiothreitol in a total reaction volume of 20 μl. The reverse transcription reaction was allowed to progress at 42°C for 60 min, and then the reaction mixture was heated to 70°C for 15 min and cooled to 37°C prior to addition of 2 U of DNase-free RNase H (Invitrogen Life Technologies). The preparation was incubated at 37°C for 20 min in the presence of RNase H to remove the original RNA templates. The RNase H was subsequently inactivated by heating the mixture at 70°C for 15 min. First-strand cDNAs were purified by extraction with phenol-chloroform (1:1) and precipitation in ethanol. Final cDNA pellets were suspended in 52 μl of RNase-free double-distilled H2O. The concentration of cDNA in each sample was determined by UV spectrophotometry and was adjusted with RNase-free double-distilled H2O so that the final working concentration was 10 ng per μl. All cDNA dilutions were stored at −80°C until they were used in quantitative real-time PCRs.
Q-RT-PCR was performed by using SYBR Green PCR Master Mix (Perkin-Elmer Corp.), 20 ng of template cDNA, and gene-specific primers. All primers were designed by using Primer Express software (Perkin-Elmer Corp.) and were synthesized at a commercial facility (Operon Technologies, Alameda, Calif.). The primer sequences, the expected melting temperatures of the products, and the appropriate primer concentrations are posted at www.nbfgc.msu.edu. All reactions were performed in duplicate, and Q-RT-PCR data were analyzed by using the 2−ΔΔCt method as described previously (21). To assess the effect of stimulation (M. avium subsp. paratuberculosis stimulation versus nil stimulation) within animal, β-actin served as the control gene and nil-stimulated samples for each animal were used as the calibrators. To assess differential gene expression between infection groups, β-actin was used as the control gene, and the mean control cow value for nil stimulation or M. avium subsp. paratuberculosis stimulation was used as the calibrator.
Flow cytometric analysis of relative immune cell populations in PBMCs from infected and control cows.
Aliquots of PBMCs from cows used in cDNA microarray and Q-RT-PCR analyses were separated and labeled for flow cytometric analysis essentially as described previously (7, 26) by using a Becton Dickinson FACSCalibur flow cytometer. For specific immune cell staining approximately 105 cells from each cow were combined with primary antibodies diluted 1:500 in PBS. The antibodies employed in this study were directed against CD4 (clone CACT138A), CD8 (clone CACT80C), the γδ T-cell receptor (clone GB21A), a B-cell antigen (clone LCT2A), and monocyte antigen (clone BAQ151A). All antibodies were obtained from VMRD, Inc., Pullman, Wash.
Following washing in PBS to remove unbound primary antibodies, a phycoerythrin (PE)-conjugated secondary antibody (goat anti-mouse immunoglobulin G; Caltag Laboratories, Burlingame, Calif.) was diluted 1:500 in PBS and applied to the cells. Cells were incubated for 15 min in secondary antibody and again washed in PBS. The final cell pellets were suspended in 200 μl of sheath fluid (BD Biosciences) for immediate flow cytometric acquisition (FASCalibur flow cytometer and CellQuest software; Becton Dickinson). The percentages of specific cell types in PBMC preparations were determined by using density dot plots, with side scatter plotted on the y axis versus log PE fluorescence intensity (log FL-2) on the x axis. For each primary antibody, an aliquot of cells was stained with an appropriate nonspecific isotype control antibody. Cells stained with isotype controls were also labeled with PE-conjugated secondary antibodies and used to establish boundaries for quadrants on density dot plots based on background fluorescence. The various leukocyte populations present were then recorded as the percentages of immunostained cells falling within the upper or lower right quadrants of the density dot plots.
RESULTS
Microarray data normalization.
As described in Materials and Methods, we adopted a normalization procedure to minimize systematic variations, such as dye biases and variations in sample RNA (cDNA) concentration, in measured gene expression levels before proceeding with significance testing. The systematic biases in data sets were corrected by using the LOESS procedure of SAS. The efficiency of LOESS normalization is illustrated for a typical cDNA microarray experiment in Fig. 1. In the absence of sample and dye intensity bias, observations should have been distributed around the horizontal line M = 0 on M-A plots and around the diagonal line log Cy3 = log Cy5 on scatter plots of log Cy3 versus Log Cy5, respectively (Fig. 1A and C).
In general, most cDNA microarray data display bias at both the high and low ends of the intensity spectrum, even in the absence of cDNA loading differences (Fig. 1A). LOESS normalization effectively removed this bias (Fig. 1B and D). Data in Fig. 1 also show that values for common control genes, such as the GAPDH, β-actin, and synthetic lambda Q genes, clustered at high-intensity mean values, while blank values clustered at low-intensity mean values (as expected). Importantly, LOESS normalization not only improved direct within-microarray comparisons but also allowed data to be compared across multiple microarrays, given appropriate corrections for fixed and random effects. Both of these comparisons were utilized in the analysis of data in this study.
Gene expression changes induced by M. avium subsp. paratuberculosis stimulation of PBMCs from Johne's disease-positive cows.
Mycobacterial proteins and other components are potent immunogens, and several cell types have been proposed to react strongly to these antigens without prior exposure and with or without stimulation by antigen-presenting cells (18, 40, 46, 48). We therefore hypothesized that PBMCs from control and infected cows would both respond to M. avium subsp. paratuberculosis and that the responses of cells from the two groups would be different. To begin testing this hypothesis, we first compared normalized data from BOTL-3 microarrays in which PBMCs were stimulated with PBS (nil stimulation) or M. avium subsp. paratuberculosis, and RNA transcripts from these stimulated cells were directly compared on cDNA microarrays.
Within the infected group of cows (n = 6), the expression levels of 49 PBMC genes were significantly affected by M. avium subsp. paratuberculosis stimulation compared to the effect of nil stimulation (P < 0.05). However, the biological relevance of statistically significant expression differences could be questioned, since the actual changes in expression were quite small (between 0.8- and 1.25-fold decreases or increases). When an additional criterion for selection of interesting genes (>1.25-fold increase or decrease and P < 0.05) was imposed, only 16 genes were highlighted as genes that were differentially expressed in M. avium subsp. paratuberculosis-stimulated PBMCs and nil-stimulated cells (Table 1). Of these 16 genes, 6 were expressed at higher levels when PBMCs from infected cows were stimulated with M. avium subsp. paratuberculosis than when cells from the same cows were subjected to the nil stimulation treatment. Thus, 10 genes were expressed at lower levels following exposure of PBMCs to M. avium subsp. paratuberculosis than following exposure of PBMCs to the nil stimulation treatment.
TABLE 1.
Genes with significantly different expression patterns in M. paratuberculosis- and nil-stimulated PBMCs from Johne's disease-positive cows
| Clone or gene | Identity or description | Mean fold change | P value |
|---|---|---|---|
| EMMPRIN | Amplicon representing bovine EMMPRIN mRNA | 1.74 | 0.00026 |
| BOTL0100003XB05R | Bovine EST clone weakly similar to human interferon-inducible guanylate binding protein 1 mRNA | 1.49 | 0.02938 |
| INF-γ | Amplicon representing bovine IFN-γ mRNA | 1.48 | 0.01185 |
| BOTL0100003XH04R | Bovine EST clone weakly similar to human putative LOC138179 mRNA | 1.38 | 0.00745 |
| GM-CSF-R-β | Amplicon representing bovine granulocyte-macrophage colony-stimulating factor receptor β mRNA | 1.29 | 0.00162 |
| BOTL0100003XF09R | Bovine EST clone highly similar to human leukocyte antigen-related protein mRNA | 1.28 | 0.03403 |
| BOTL0100012_C05 | Bovine EST clone weakly similar to human nuclear autoantigen (SP-100) mRNA | −1.27 | 0.02860 |
| BOTL0100010_A07 | Bovine EST clone highly similar to a gene from COS-1 cells affected by herpes simplex virus type 1 | −1.37 | 0.03951 |
| BOTL0100003XD01R | Bovine EST clone similar to human nucleoside diphosphate-linked moiety X-type 3 (NUDT3) mRNA | −1.37 | 0.02034 |
| Inhibin-A | Amplicon representing bovine inhibin A subunit mRNA | −1.39 | 0.00574 |
| CD4 antigen (p55) | Amplicon representing bovine CD4 antigen p55 subunit mRNA | −1.46 | 0.04854 |
| Flk-1 | Amplicon representing bovine bovine Flk-1 tyrosine kinase receptor mRNA | −1.65 | 0.01075 |
| IL-6 Rα | Amplicon representing bovine IL-6 receptor α-chain mRNA | −1.71 | 0.02903 |
| CYCB2 | Amplicon representing bovine cyclin B2 mRNA | −2.30 | 0.00124 |
| MMP9 | Amplicon representing bovine MMP 9 mRNA | −2.41 | 0.02458 |
| Sentrin | Amplicon representing bovine sentrin mRNA | −3.20 | 0.04328 |
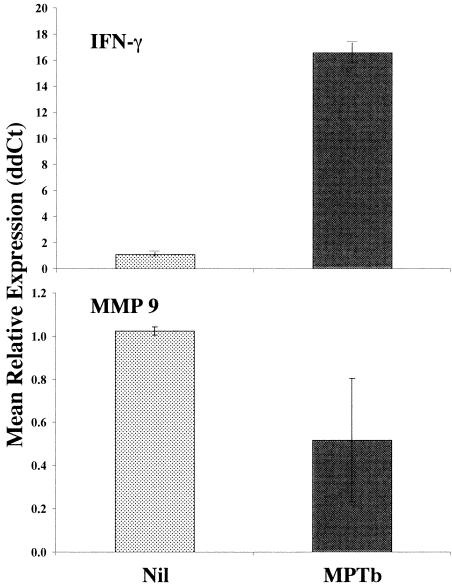
Importantly, the IFN-γ gene was among the genes that were significantly (>1.25-fold change; P < 0.05) activated by stimulation of infected cow PBMCs with M. avium subsp. paratuberculosis compared to the expression in nil-stimulated cells (Table 1). In addition, down-regulation of matrix metalloproteinase 9 (MMP 9) gene expression was consistent with our previous microarray observations for cattle with later-stage Johne's disease (13). Thus, these two genes were selected for Q-RT-PCR validation as representatives of the genes whose expression was affected either positively (IFN-γ) or negatively (MMP 9) following exposure of infected cow PBMCs to M. avium subsp. paratuberculosis compared to the expression in nil-stimulated cells.
Enhanced expression of IFN-γ transcripts in M. avium subsp. paratuberculosis-stimulated PBMCs from infected cows compared to the expression in similar cells treated with PBS was verified by Q-RT-PCR (Fig. 2), although the magnitude of activation (mean, 16.5-fold) was much larger (as measured by Q-RT-PCR) than that observed in cDNA microarray experiments. Down-regulation of MMP 9 gene expression following exposure of infected cow PBMCs to M. avium subsp. paratuberculosis was also verified by Q-RT-PCR (Fig. 2), and the extent of down-regulation observed was similar in Q-RT-PCR analysis (2-fold) and cDNA microarray analysis (2.4-fold).
FIG. 2.
Q-RT-PCR validation of cDNA microarray results for PBMCs from infected cows. Genes encoding IFN-γ and MMP 9 were selected for validation of cDNA microarray results by Q-RT-PCR. The IFN-γ gene was selected because enhanced expression of this cytokine is a well-documented effect following M. avium subsp. paratuberculosis stimulation of immune cells from Johne's disease-positive cows. The MMP 9 gene was selected as a representative of genes that exhibit repressed expression in M. avium subsp. paratuberculosis-stimulated PBMCs compared to the expression in nil-stimulated cells and because repression of MMP 9 gene expression was observed in previous studies. Q-RT-PCR was conducted as described in Materials and Methods by using gene-specific primers. Data were analyzed by using the 2−ΔΔCt method essentially as described previously (21) with β-actin as the control gene and nil stimulation within animal as the calibrator. The data are the means ± standard errors of the means for independent results from four infected cows. MPTb, M. avium subsp. paratuberculosis-stimulated PBMCs.
Gene expression changes induced by M. avium subsp. paratuberculosis stimulation of PBMCs from Johne's disease-negative control cows.
When microarray data for control cow PBMCs were analyzed, 23 genes were expressed significantly different following stimulation with M. avium subsp. paratuberculosis than following nil stimulation (P < 0.05). However, as with the infected group of cows, only 13 of these genes exhibited expression differences greater than 1.25-fold (Table 2). The responses of control cow PBMCs to in vitro M. avium subsp. paratuberculosis stimulation were balanced, with six genes up-regulated and seven genes down-regulated (Table 2).
TABLE 2.
Genes with significantly different expression patterns in M. paratuberculosis- and nil-stimulated PBMCs from control uninfected cows
| Clone or gene | Identity or description | Mean fold change | P value |
|---|---|---|---|
| Sentrin | Amplicon representing bovine sentrin mRNA | 1.901 | 0.03464 |
| PIGF | Amplicon representing bovine placenta growth factor mRNA | 1.671 | 0.00396 |
| IL-11 Rα | Amplicon representing bovine IL-11 receptor mRNA | 1.476 | 0.02818 |
| Endoglin | Amplicon representing bovine endoglin mRNA | 1.296 | 0.02343 |
| BOTL0100005XH03R | Bovine EST clone similar to human CD164 mRNA | 1.292 | 0.03027 |
| BOTL0100012_E10 | Bovine EST clone highly similar to human Wiskott-Aldrich syndrome protein-interacting protein mRNA | 1.261 | 0.02219 |
| uPA | Amplicon representing bovine urokinase plasminogen activator mRNA | −1.274 | 0.02754 |
| BOTL0100013_F11 | Bovine EST clone highly similar to human echinoderm microtubule-associated protein-like EMAP2 mRNA | −1.275 | 0.01496 |
| IL-1 | Amplicon representing bovine IL-1 mRNA | −1.294 | 0.00541 |
| BOTL0100008_G05 | Bovine EST clone not similar to any known gene | −1.358 | 0.04846 |
| MMP1 up | Amplicon representing bovine MMP 1 mRNA | −1.374 | 0.02464 |
| MMP23 | Amplicon representing bovine MMP 23 mRNA | −1.661 | 0.00653 |
| MMP9 | Amplicon representing bovine MMP 9 mRNA | −2.512 | 0.04382 |
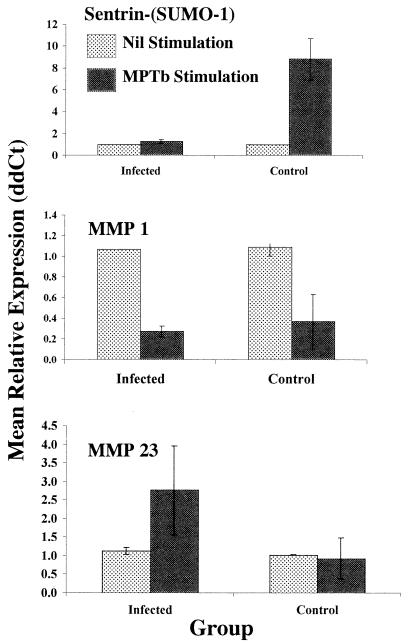
Similar to our study of infected cow PBMC gene expression, we selected genes representing the genes that were apparently up- or down-regulated following exposure of control cow PBMCs to M. avium subsp. paratuberculosis for validation by Q-RT-PCR. The gene encoding Sentrin-(SUMO-1) was selected because it was the gene that was most highly activated by M. avium subsp. paratuberculosis (1.9-fold) and because sentrin has an important role as a ubiquitin-related regulatory protein. As predicted by cDNA microarray analysis, the levels of transcripts encoding Sentrin-(SUMO-1) were significantly elevated (P < 0.05) following exposure of control cow PBMCs to M. avium subsp. paratuberculosis (compared to the levels following nil stimulation), as monitored by Q-RT-PCR (Fig. 3). The genes encoding MMP 1 and MMP 23 were selected to represent the genes that are down-regulated in control cow PBMCs exposed to M. avium subsp. paratuberculosis because previous data suggested that MMPs play an important, albiet unknown, role in PBMC responses to M. avium subsp. paratuberculosis (13). Although Q-RT-PCR analysis indicated that the gene encoding MMP 23 tended to be expressed at higher levels following exposure of PBMCs from infected cows to M. avium subsp. paratuberculosis, the mean down-regulation of MMP 23 gene expression in PBMCs from control cows was less than 1.25-fold and was highly variable when it was measured by Q-RT-PCR (Fig. 3). In contrast, down-regulation of MMP 1 appeared to be a consistent and common response of PBMCs to M. avium subsp. paratuberculosis compared to expression in unstimulated cells (Fig. 3).
FIG. 3.
Q-RT-PCR validation of cDNA microarray results for PBMCs from control cows. Genes encoding Sentrin-(SUMO-1), MMP 1, and MMP 23 were selected for Q-RT-PCR validation from among the genes that exhibit differential expression in nil-stimulated and M. avium subsp. paratuberculosis-stimulated PBMCs from control uninfected cows. The gene encoding Sentrin-(SUMO-1) was selected as a representative of genes that exhibit up-regulation in control cow PBMCs stimulated with M. avium subsp. paratuberculosis compared to expression in nil-stimulated cells. The genes encoding MMP 1 and MMP 23 were selected because each was apparently down-regulated by M. avium subsp. paratuberculosis stimulation of control cow PBMCs on cDNA microarrays and because of previous data suggesting that MMP gene regulation is a major and consistent effect of M. avium subsp. paratuberculosis on PBMCs from infected cows (13). For these reasons, an analysis of nil-stimulated and M. avium subsp. paratuberculosis-stimulated PBMCs from infected cows was also included in this study. Q-RT-PCR was conducted as described in Materials and Methods by using gene-specific primers. Data were analyzed by using the 2−ΔΔCt method essentially as described previously (21) with β-actin as the control gene and nil stimulation within animal as the calibrator. The data are the means ± standard errors of the means for independent results from four infected cows and three control cows. MPTb, M. avium subsp. paratuberculosis.
Many gene expression differences in PBMCs from Johne's disease-positive and control cows do not depend upon antigen stimulation.
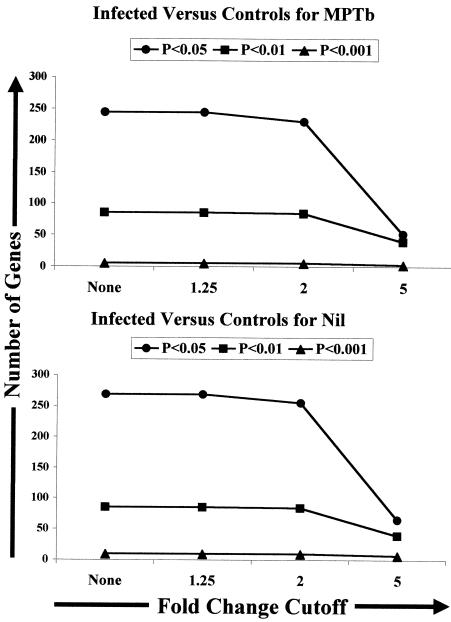
To explore potential gene expression differences in PBMCs from the two groups of cows (infected and control uninfected), a mixed-model analysis was conducted as described in Materials and Methods. The results of this analysis supported our hypothesis and demonstrated that there were major differences in gene expression when the expression patterns of M. avium subsp. paratuberculosis-stimulated PBMCs from infected and control cows were compared. In fact, employing our previous selection criteria, >1.25-fold expression difference and P < 0.05, resulted in identification of 244 differentially expressed genes in M. avium subsp. paratuberculosis-stimulated PBMCs from the infected and control groups. Even with more stringent selection criteria, a twofold expression difference and P < 0.01, 86 genes were identified as genes that were expressed differently by the two groups of M. avium subsp. paratuberculosis-stimulated PBMCs (Fig. 4).
FIG. 4.
Identification of numerous gene expression changes in both nil-stimulated and M. avium subsp. paratuberculosis-stimulated PBMCs when expression levels were compared across infection groups by using a mixed-model analysis. Data from microarray analysis of PBMCs from six infected cows and four control cows were combined and analyzed as described in Materials and Methods by using a two-stage mixed model in SAS. The resulting least square (LS) means were used to construct interaction tables containing relative expression information and confidence intervals for each gene on the BOTL-3 cDNA microarray. Data were imported into Excel, and the Data Filter command was used to select genes with various expression differences (fold changes) and significance values (P < 0.05, P < 0.01, and P < 0.001). The number of genes in each category was tabulated and used to construct plots. MPTb, M. avium subsp. paratuberculosis.
To ascertain the importance of M. avium subsp. paratuberculosis stimulation in identifying gene expression differences in PBMCs from infected and control cows, we next performed an analysis in which we compared gene expression in nil-stimulated cell populations from the two infection groups. Surprisingly, 269 genes that were expressed significantly differently (P < 0.05) were identified in a comparison of nil-stimulated samples from infected and control cows (Fig. 4), suggesting that differential gene expression in PBMCs from infected and control cows was not entirely dependent upon in vitro stimulation with M. avium subsp. paratuberculosis. Use of the more stringent criteria (twofold change and P < 0.01) resulted in identification of 108 genes that were expressed differently in nil-stimulated PBMCs from the infected and control groups (Fig. 4).
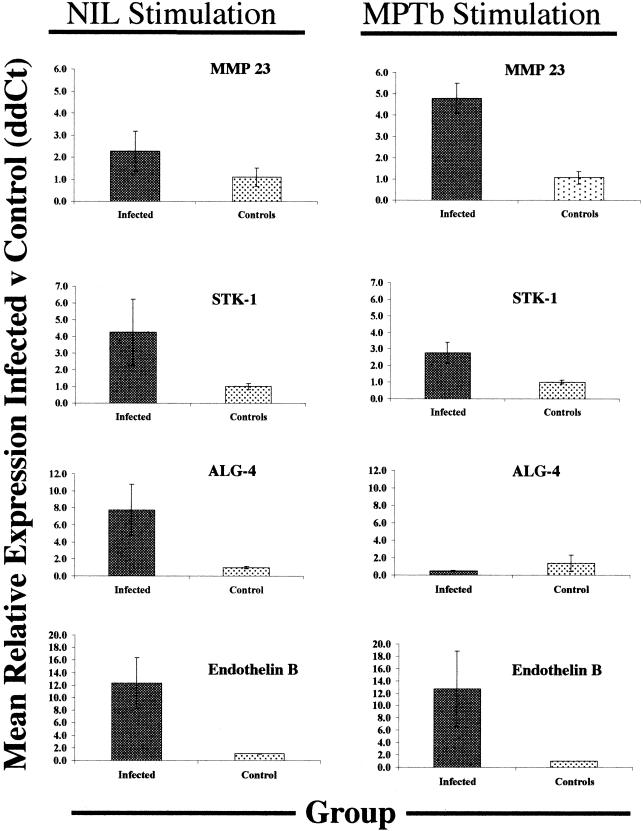
To confirm our observations from the mixed-model analyses, we performed Q-RT-PCR and conducted within- and across-group comparisons of selected genes, as described in Materials and Methods. The genes selected for Q-RT-PCR were a subset of the genes suggested to have the most significant differences between groups and represented a composite of ontological classes. These genes (the MMP 23, STK-1, ALG-4, and endothelin B genes) also represent three common themes observed in expression patterns across groups: (i) reduced expression following exposure to M. avium subsp. paratuberculosis (ALG-4 gene); (ii) enhanced expression following exposure to M. avium subsp. paratuberculosis (MMP 23 gene); and (iii) no change in expression following exposure to M. avium subsp. paratuberculosis (STK-1 and endothelin B genes).
The mean relative level of expression of the MMP 23 gene as measured by Q-RT-PCR tended to be higher in nil-stimulated PBMCs from infected cows than in nil-stimulated PBMCs from control cows (2.4-fold difference) (Fig. 5). Stimulation with M. avium subsp. paratuberculosis enhanced the difference in expression between the infection groups (which is consistent with results shown in Fig. 3), which led to a significantly higher level of expression of the MMP 23 gene in infected cow PBMCs than in similarly treated cells from controls (4.9-fold difference; P < 0.05) (Fig. 5).
FIG. 5.
Q-RT-PCR validation of gene expression changes observed following microarray analysis of PBMCs from control and infected cows. Genes to be validated were selected from a list of the genes whose expression was most significantly different following nil stimulation of PBMCs from infected and control cows. Q-RT-PCR was performed as described in Materials and Methods, and data were analyzed by using the 2−ΔΔCt method with β-actin as the control gene. Mean values for control cow PBMCs with nil stimulation or M. avium subsp. paratuberculosis (MPTb) stimulation were used as calibrators for calculation of all 2−ΔΔCt values, so that the values for control cows always bracketed 1.0. For Q-RT-PCR analysis, samples were arranged in 96-well PCR plates so that comparisons could be made between PBMCs from infected cows and PBMCs from control cows, each stimulated with M. avium subsp. paratuberculosis or PBS (nil stimulation), on the same plate. The data are means ± standard errors of the means for 2−ΔΔCt values for three or four infected cows and three or four control cows for each gene.
In contrast, the ALG-4 gene represents a class of genes whose mean level of expression in nil-stimulated PBMCs from infected cows was significantly higher (eightfold higher; P < 0.05) than the mean level of expression in similarly treated cells from control cows (Fig. 5). However, stimulation of PBMCs with M. avium subsp. paratuberculosis caused a marked reduction in ALG-4 gene expression in infected cow PBMCs, resulting in a mean level of expression that was twofold lower than that in similarly treated control cow PBMCs (Fig. 5). Thus, the overall effect of M. avium subsp. paratuberculosis on ALG-4 gene expression in infected cow PBMCs compared to expression in nil-stimulated cells from the same cows was a 16-fold reduction in expression, as measured by Q-RT-PCR. This rather dramatic difference in expression of the ALG-4 gene was not reflected on cDNA microarrays, on which nil- and M. avium subsp. paratuberculosis-stimulated PBMCs from infected cows were directly compared. On cDNA microarrays, the difference in expression of the ALG-4 gene after M. avium subsp. paratuberculosis stimulation and Nil stimulation was highly significant (P < 0.01), but the mean difference across all infected cows was less than 1.25-fold.
The mean levels of expression of the STK-1 and endothelin B genes measured by Q-RT-PCR were 4.2- and 12-fold higher, respectively, in nil-stimulated PBMCs from infected cows than in similarly treated control cow PBMCs (Fig. 5). The relative levels of expression of the STK-1 and endothelin B genes were not significantly altered by stimulation of PBMCs with M. avium subsp. paratuberculosis (Fig. 5).
Differences in the gene expression programs of PBMCs from infected and control cows are not due to gross changes in the relative populations of immune cells.
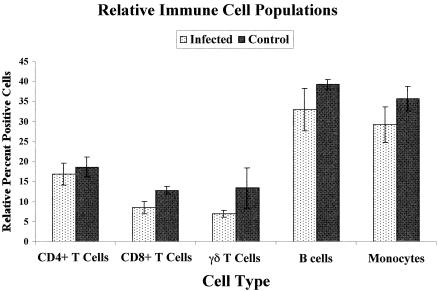
One possible explanation for the dramatic differences in the PBMC gene expression patterns observed between the infected and control cows used in this study could be that the relative populations of specific cell types comprising PBMCs were different in the infection groups. To assess this possibility, a flow cytometric analysis of several major cell types was conducted as described in Materials and Methods. Figure 6 shows that despite a tendency for slightly reduced cell numbers in the infected group, there were no significant differences (P > 0.05 for all cell types) in the relative populations of CD4+ or CD8+ T cells, γδ T cells, B cells, or monocytes in PBMCs from infected and control cows. Although it is possible that another cell type not monitored in our study might account for some of the observed gene expression differences, the total population of cells monitored in this study represented at least 84% of all PBMCs. Together with gene expression data presented in this report, these results suggest that the internal gene expression programs of cells comprising Percoll-isolated PBMC populations are different in cows suffering from Johne's disease and in healthy uninfected control cows.
FIG. 6.
Flow cytometric analysis of major immune cell types in PBMCs from infected and control cows. Aliquots of PBMCs from infected and control cows used in a cDNA microarray analysis were immunostained as described in Materials and Methods to label specific immune cell types, and the relative percentages of each cell type were determined by flow cytometry. The percentage of each immune cell population was determined from density dot plots with side scatter as the y axis and FL-2 (fluorescence of PE) as the x axis. Quadrants were established based on similar plots of cells stained with irrelevant isotype control antibodies to determine the background fluorescence of the cells. The percentage of PE-positive cells was then determined as the percentage of all PBMCs in the upper or lower right quadrants. The data are the means ± standard errors of the means for six infected cow PBMC preparations and four control cow preparations for each cell type.
DISCUSSION
Previously, it was demonstrated that gene expression profiling could detect significant differences in the responses of PBMCs from clinical and subclinical Johne's disease-positive cows to stimulation with M. avium subsp. paratuberculosis (13). Although important in beginning to define critical changes in the immune response during the long course of M. avium subsp. paratuberculosis infection, these studies did not help define critical and consistent differences between the responses of PBMCs from uninfected and infected cows.
In the present study, we focused on comparing the gene expression programs of PBMCs from Johne's disease-positive cows with those of PBMCs from control uninfected cows. Our results are important in that they go well beyond data for the typically monitored cytokines and thus may help to better define how M. avium subsp. paratuberculosis infection influences the host peripheral immune system and affects a variety of cell surface receptors, intracellular signaling mechanisms, apoptosis regulators, and transcription factors. In addition, results presented in this report suggest several excellent potential targets for diagnosis of Johne's disease infection status, particularly when systems such as Q- RT-PCR are used.
Despite the rather profound effect of M. avium subsp. paratuberculosis stimulation on production of IFN-γ by PBMCs from infected cows, there were surprisingly few other genes (a total of 15 genes) in cells from these Johne's disease-positive cows whose mean levels of expression were significantly different when PBMCs were exposed to M. parartuberculosis stimulation than when PBMCs were exposed to nil stimulation. As revealed by IFN-γ transcript production and MMP 9 gene down-regulation, the paucity of significantly affected genes did not appear to be due to a failure of infected cow PBMCs to respond in vitro to M. avium subsp. paratuberculosis. The variability in individual cow responses led to the loss of many profound gene expression differences when our rather stringent selection criteria (>1.25-fold change and P < 0.05 across six infected cows) were used. Thus, the genes identified in Table 1 represent only those genes whose expression in PBMCs from infected cows is most consistently (i.e., statistically significantly) affected by in vitro stimulation with M. avium subsp. paratuberculosis. The potential sources of variability in individual infected cow responses to M. avium subsp. paratuberculosis are likely related to the fact that in naturally infected cows it is difficult to precisely determine the initial exposure dose, the route of entry, and the status of infection. In addition, the genetic factors regulating individual cow responses to M. avium subsp. paratuberculosis are completely unknown.
Although several of the observed gene expression changes (e.g., up-regulation of IFN-γ mRNA) were specific to infected cow PBMCs, other changes (e.g., MMP 9 gene down-regulation) appeared to be more general responses of immune cells to M. avium subsp. paratuberculosis, because they also occurred in PBMCs from control cows. The results of the direct comparison of nil- and M. avium subsp. paratuberculosis-stimulated PBMCs from infected cows reported here support the previous observations that down-regulation of gene expression is a common response to stimulation with M. avium subsp. paratuberculosis and that the MMPs figure prominently in this response (13). Further analysis of the role that MMPs play in PBMC responses to M. avium subsp. paratuberculosis both in vitro and in vivo is clearly warranted.
Mycobacterial cell components are potent immunogens, and thus, it was perhaps not surprising that stimulation of PBMCs from uninfected cattle with M. avium subsp. paratuberculosis significantly altered expression of several immune cell genes. While the consequences of M. avium subsp. paratuberculosis-induced gene expression changes in uninfected cow PBMCs are unclear at present, our results demonstrate that PBMCs from uninfected cows do respond on a genomic level to M. avium subsp. paratuberculosis stimulation in vitro. Our results also indicate that the response of control uninfected cow PBMCs was different from that observed with cells from infected cows, supporting our original hypothesis.
A major goal of the present study was to highlight genes that were differentially expressed in PBMCs from control and Johne's disease-positive cows, in order to both identify potential diagnostic targets and refine our understanding of interactions between M. avium subsp. paratuberculosis and the host immune system. When cDNA microarray data from four control cows and six infected cows were analyzed by using a mixed-model approach, major differences in PBMC gene expression patterns between the two infection groups were apparent. Surprisingly, stimulation of PBMCs with M. avium subsp. paratuberculosis in vitro was not required to detect most of these differences in gene expression (Fig. 4).
Differences in gene expression between nil-stimulated PBMCs from infected cows and nil-stimulated PBMCs from control cows were highly significant, with over 80 genes exhibiting a twofold or greater difference in expression at P < 0.01. Previous studies with similar cDNA microarrays demonstrated that with these selection criteria, the expected number of false positives (genes showing differential expression when none is expected) is almost zero (37). Thus, it is likely that most gene expression differences identified by using these criteria in a cDNA microarray analysis are valid. To demonstrate this, we selected four genes from the list of transcripts that were expressed significantly differently for further evaluation by Q-RT-PCR. These genes were selected without regard to specific ontological class but rather because they represent the three main expression classes observed in cDNA microarray experiments. These classes were (i) genes expressed at higher levels in nil-stimulated cells from infected cows than in controls, where the difference was accentuated by M. avium subsp. paratuberculosis stimulation (MMP 23 gene); (ii) genes for which the expression differences between nil-stimulated cells from infected cows and nil-stimulated cells from control cows were relatively unchanged by M. avium subsp. paratuberculosis stimulation (STK-1 and endothelin B receptor genes); and (iii) genes for which expression in PBMCs from infected cows was reduced by M. avium subsp. paratuberculosis stimulation (ALG-4 gene).
Our results demonstrate that M. avium subsp. paratuberculosis stimulation of PBMCs from infected and control cows does indeed cause changes in the gene expression patterns compared to the patterns observed after nil stimulation, including the IFN-γ response, which can be used to differentiate infected and uninfected animals. However, our results also demonstrate that in vitro stimulation with M. avium subsp. paratuberculosis is not necessary to observe significant differences in the gene expression patterns of PBMCs from infected and control cows. Furthermore, our results suggest that the in vivo infection environment alone may be sufficient to cause a reprogramming of genome usage in PBMCs from infected cows. Differential gene expression observed in the present study was not explained by gross changes in relative immune cell populations comprising PBMCs in infected cows, since most of the major immune cell types present were found to represent similar relative populations in the infected and control groups. However, we cannot exclude the possibility that some observed differences in gene expression were due to altered populations of immune cells not specifically assessed in the present study (i.e., NK T cells). Despite this caveat, our novel results suggest that the global gene expression program in PBMCs from M. avium subsp. paratuberculosis-infected cows is different than that in cells from control uninfected cows.
Based on our results, several significant questions arise. Is overnight culture required to reveal differences in gene expression in PBMCs from infected and control cows? What cell types within PBMC populations are primarily responsible for the observed differences in gene expression programs? What are the pathological and immunological mechanisms involved in developing different gene expression programs in infected and control cows? Can a series of genes be identified that reliably diagnoses infection with M. avium subsp. paratuberculosis (with or without in vitro stimulation), and how early in the infection cycle can these genes provide reliable information? Finally, is the PBMC gene expression reprogramming phenomenon observed in the present study unique to M. avium subsp. paratuberculosis infection, or can our results be extended to different infectious diseases and to other host species? Clearly, further investigation aimed at addressing these important questions is warranted.
Acknowledgments
We acknowledge the outstanding technical support of Sue Sipkovsky and thank Jeanne Burton and George W. Smith for many helpful discussions and critical reviews of the manuscript. We also acknowledge the excellent assistance of Rob Tempelman, Department of Animal Science, Michigan State University, with experimental design issues and analysis of microarray data.
We acknowledge the generous financial support of the College of Agriculture and Natural Science, Michigan State University Agricultural Experiment Station, and the Office of the Vice President for Research and Graduate Studies at Michigan State University. Additional support for this project was provided by the Michigan Animal Industry Coalition and USDA IFAFS grant 2001-52100-11211.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Adams, J. L., and C. J. Czuprynski. 1994. Mycobacterial cell wall components induce the production of TNF-alpha, IL-1, and IL-6 by bovine monocytes and the murine macrophage cell line RAW 264.7. Microb. Pathog. 16:401-411. [DOI] [PubMed] [Google Scholar]
- 2.Alzuherri, H. M., C. J. Woodall, and C. J. Clarke. 1996. Increased intestinal TNF-alpha, IL-1 beta and IL-6 expression in ovine paratuberculosis. Vet. Immunol. Immunopathol. 49:331-345. [DOI] [PubMed] [Google Scholar]
- 3.Billman-Jacobe, H., M. Carrigan, F. Cockram, L. A. Corner, I. J. Gill, J. F. Hill, T. Jessep, A. R. Milner, and P. R. Wood. 1992. A comparison of the interferon gamma assay with the absorbed ELISA for the diagnosis of Johne's disease in cattle. Aust. Vet. J. 69:25-28. [DOI] [PubMed] [Google Scholar]
- 4.Burrells, C., C. J. Clarke, A. Colston, J. M. Kay, J. Porter, D. Little, and J. M. Sharp. 1999. Interferon-gamma and interleukin-2 release by lymphocytes derived from the blood, mesenteric lymph nodes and intestines of normal sheep and those affected with paratuberculosis (Johne's disease). Vet. Immunol. Immunopathol. 68:139-148. [DOI] [PubMed] [Google Scholar]
- 5.Burrells, C., C. J. Clarke, A. Colston, J. M. Kay, J. Porter, D. Little, and J. M. Sharp. 1998. A study of immunological responses of sheep clinically-affected with paratuberculosis (Johne's disease). The relationship of blood, mesenteric lymph node and intestinal lymphocyte responses to gross and microscopic pathology. Vet. Immunol. Immunopathol. 66:343-358. [DOI] [PubMed] [Google Scholar]
- 6.Burton, J. L., S. A. Madsen, J. Yao, S. Sipkovsky, and P. M. Coussens. 2001. An immunogenomics approach to understanding periparturient immunosuppression and mastitis susceptibility in dairy cows. Acta Vet. Scand. 42:407-425. [PubMed] [Google Scholar]
- 7.Burton, J. L., S. A. Madsen, J. Yao, S. S. Sipkovsky, and P. M. Coussens. 2001. An immunogenomics approach to understanding periparturient immunosuppression and mastitis susceptibility in dairy cows. Acta Vet. Scand. 42:407-424. [PubMed] [Google Scholar]
- 8.Chiodini, R. J., and W. C. Davis. 1993. The cellular immunology of bovine paratuberculosis: immunity may be regulated by CD4+ helper and CD8+ immunoregulatory T lymphocytes which down-regulate gamma/delta+ T-cell cytotoxicity. Microb. Pathog. 14:355-367. [DOI] [PubMed] [Google Scholar]
- 9.Chiodini, R. J., and W. C. Davis. 1992. The cellular immunology of bovine paratuberculosis: the predominant response is mediated by cytotoxic gamma/delta T lymphocytes which prevent CD4+ activity. Microb. Pathog. 13:447-463. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, C. J., and D. Little. 1996. The pathology of ovine paratuberculosis: gross and histological changes in the intestine and other tissues. J. Comp. Pathol. 114:419-437. [DOI] [PubMed] [Google Scholar]
- 11.Cleveland, W. S., and E. Grosse. 1991. Computational methods of local regression. Stat. Comput. 1:47-62. [Google Scholar]
- 12.Collins, M. T., D. C. Sockett, S. Ridge, and J. C. Cox. 1991. Evaluation of a commercial enzyme-linked immunosorbent assay for Johne's disease. J. Clin. Microbiol. 29:272-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coussens, P., C. Colvin, A. Abouzied, K. Wiersma, and S. Sipkovsky. 2002. Gene expression profiling of peripheral blood mononuclear cells from cattle infected with Mycobacteriumparatuberculosis. Infect. Immun. 70:5494-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coussens, P. M. 2001. Interactions between Mycobacterium paratuberculosis and the bovine immune system. Anim. Health Res. Rev. 2:141-161. [PubMed] [Google Scholar]
- 15.Coussens, P. M., and W. Nobis. 2002. Bioinformatics and high-throughput approach to create genomic resources for study of bovine immunobiology. Vet. Immunol. Immunopathol. 86:229-244. [DOI] [PubMed] [Google Scholar]
- 16.Dargatz, D. A., B. A. Byrum, L. K. Barber, R. W. Sweeney, R. H. Whitlock, W. P. Shulaw, R. H. Jacobson, and J. R. Stabel. 2001. Evaluation of a commercial ELISA for diagnosis of paratuberculosis in cattle. J. Am. Vet. Med. Assoc. 218:1163-1166. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, H., T. Shirahata, and K. Hasegawa. 1994. Interferon-gamma production of mitogen stimulated peripheral lymphocytes in perinatal cows. J. Vet. Med. Sci. 56:735-738. [DOI] [PubMed] [Google Scholar]
- 18.Jiao, X., R. Lo-Man, P. Guermonprez, L. Fiette, E. Deriaud, S. Burgaud, B. Gicquel, N. Winter, and C. Leclerc. 2002. Dendritic cells are host cells for mycobacteria in vivo that trigger innate and acquired immunity. J. Immunol. 168:1294-1301. [DOI] [PubMed] [Google Scholar]
- 19.Johnson-Ifearulundu, Y., and J. B. Kaneene. 1999. Distribution and environmental risk factors for paratuberculosis in dairy cattle herds in Michigan. Am. J. Vet. Res. 60:589-596. [PubMed] [Google Scholar]
- 20.Kennedy, D. J., and G. Benedictus. 2001. Control of Mycobacterium avium subsp. paratuberculosis infection in agricultural species. Rev. Sci. Tech. 20:151-179. [DOI] [PubMed] [Google Scholar]
- 21.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(t)) method. Methods 4:402-408. [DOI] [PubMed] [Google Scholar]
- 22.Manning, E. J., and M. T. Collins. 2001. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev. Sci Tech. 20:133-150. [DOI] [PubMed] [Google Scholar]
- 23.Merkal, R. S., A. B. Larsen, and G. D. Booth. 1975. Analysis of the effect of inapparent bovine paratuberculosis. Am. J. Vet. Res. 36:837-838. [PubMed] [Google Scholar]
- 24.Navarro, J. A., G. Ramis, J. Seva, F. J. Pallares, and J. Sanchez. 1998. Changes in lymphocyte subsets in the intestine and mesenteric lymph nodes in caprine paratuberculosis. J. Comp. Pathol. 118:109-121. [DOI] [PubMed] [Google Scholar]
- 25.Perez, V., J. Tellechea, J. M. Corpa, M. Gutierrez, and J. F. Garcia Marin. 1999. Relation between pathologic findings and cellular immune responses in sheep with naturally acquired paratuberculosis. Am. J. Vet. Res. 60:123-127. [PubMed] [Google Scholar]
- 26.Preisler, M. T., P. S. Weber, R. J. Tempelman, R. J. Erskine, H. Hunt, and J. L. Burton. 2000. Glucocorticoid receptor down-regulation in neutrophils of periparturient cows. Am. J. Vet. Res. 61:14-19. [DOI] [PubMed] [Google Scholar]
- 27.Rook, G. A., R. A. Attiyah, and N. Foley. 1989. The role of cytokines in the immunopathology of tuberculosis, and the regulation of agalactosyl IgG. Lymphokine Res. 8:323-328. [PubMed] [Google Scholar]
- 28.SAS Institute, Inc. 1990. Procedures guide, version 6, 3rd ed., p. 209-334. SAS Institute, Inc. Cary, N.C.
- 29.SAS Institute, Inc. 2000. SAS/STAT software, 8th ed. SAS Institute, Inc., Cary, N.C.
- 30.Seitz, S. E., L. E. Heider, W. D. Heuston, S. Bech-Nielsen, D. M. Rings, and L. Spangler. 1989. Bovine fetal infection with Mycobacterium paratuberculosis. J. Am. Vet. Med. Assoc. 194:1423-1426. [PubMed] [Google Scholar]
- 31.Stabel, J. R. 2000. Cytokine secretion by peripheral blood mononuclear cells from cows infected with Mycobacterium paratuberculosis. Am. J. Vet. Res. 61:754-760. [DOI] [PubMed] [Google Scholar]
- 32.Stabel, J. R. 2000. Transitions in immune responses to Mycobacterium paratuberculosis. Vet. Microbiol. 77:465-473. [DOI] [PubMed] [Google Scholar]
- 33.Stabel, J. R., and R. H. Whitlock. 2001. An evaluation of a modified interferon-gamma assay for the detection of paratuberculosis in dairy herds. Vet. Immunol. Immunopathol. 79:69-81. [DOI] [PubMed] [Google Scholar]
- 34.Storset, A. K., H. J. Hasvold, M. Valheim, H. Brun-Hansen, G. Berntsen, S. K. Whist, B. Djonne, C. M. Press, G. Holstad, and H. J. Larsen. 2001. Subclinical paratuberculosis in goats following experimental infection. An immunological and microbiological study. Vet. Immunol. Immunopathol. 80:271-287. [DOI] [PubMed] [Google Scholar]
- 35.Streeter, R. N., G. F. Hoffsis, S. Bech-Nielsen, W. P. Shulaw, and D. M. Rings. 1995. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 56:1322-1324. [PubMed] [Google Scholar]
- 36.Strohmeier, G. R., and M. J. Fenton. 1999. Roles of lipoarabinomannan in the pathogenesis of tuberculosis. Microbes Infect. 1:709-717. [DOI] [PubMed] [Google Scholar]
- 37.Suchyta, S., S. Sipkovsky, R. Halgren, R. Kruska, M. Elftman, M. Weber-Nielson, M. Vandeharr, and P. Coussens. Bovine mammary gene expression profiling using a cDNA microarray enhanced for mammary specific transcripts. Physiol. Genomics, in press. [DOI] [PubMed]
- 38.Sweeney, R. W. 1996. Transmission of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 12:305-312. [DOI] [PubMed] [Google Scholar]
- 39.Tooker, B. R., J. L. Burton, and P. M. Coussens. 2002. Survival tactics of M. paratuberculosis in bovine macrophage cells. Vet. Immunol. Immunopathol. 87:443-451. [DOI] [PubMed] [Google Scholar]
- 40.Ulrichs, T., and S. A. Porcelli. 2000. CD1 proteins: targets of T cell recognition in innate and adaptive immunity. Rev. Immunogenet. 2:416-432. [PubMed] [Google Scholar]
- 41.Wells, S. J., and B. A. Wagner. 2000. Herd-level risk factors for infection with Mycobacterium paratuberculosis in US dairies and association between familiarity of the herd manager with the disease or prior diagnosis of the disease in that herd and use of preventive measures. J. Am. Vet. Med. Assoc. 216:1450-1457. [DOI] [PubMed] [Google Scholar]
- 42.Whitlock, R. H., and C. Buergelt. 1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. North Am. Food Anim. Pract. 12:345-356. [DOI] [PubMed] [Google Scholar]
- 43.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 44.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao, J., J. L. Burton, P. Saama, S. Sipkovsky, and P. M. Coussens. 2001. Generation of EST and cDNA microarray resources for the study of bovine immunobiology. Acta Vet. Scand. 42:391-406. [PubMed] [Google Scholar]
- 46.Zhang, Q. H., M. Ye, X. Y. Wu, S. X. Ren, M. Zhao, C. J. Zhao, G. Fu, Y. Shen, H. Y. Fan, G. Lu, M. Zhong, X. R. Xu, Z. G. Han, J. W. Zhang, J. Tao, Q. H. Huang, J. Zhou, G. X. Hu, J. Gu, S. J. Chen, and Z. Chen. 2000. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 10:1546-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., and W. N. Rom. 1993. Regulation of the interleukin-1 beta (IL-1 beta) gene by mycobacterial components and lipopolysaccharide is mediated by two nuclear factor-IL6 motifs. Mol. Cell Biol. 13:3831-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zurbrick, B. G., D. M. Follett, and C. J. Czuprynski. 1988. Cytokine regulation of the intracellular growth of Mycobacterium paratuberculosis in bovine monocytes. Infect. Immun. 56:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]