Abstract
Ly49 receptor genes are expressed by subsets of natural killer (NK) cells in an overlapping fashion, accounting for the capacity of NK subsets to attack host cells that have selectively downregulated self–major histocompatibility complex (MHC) class I molecules. It was shown previously that most NK cells express only one or the other allele of a given Ly49 gene, while a smaller population expresses both alleles. However, the methods used to detect monoallelic and biallelic cells were nonquantitative. Here, new allele-specific antibodies were used to provide the first quantitative examination of biallelic and monoallelic expression of Ly49A and Ly49G2. The results demonstrate conclusively that most Ly49A+ and Ly49G2+ NK cells express the corresponding gene in a monoallelic fashion, with a smaller subset expressing both alleles. Unexpectedly, biallelic Ly49A+ NK cells were more numerous than predicted by completely independent allelic expression, suggesting some heterogeneity among NK progenitors in the potential to express a given Ly49 gene. The data also show that cells expressing one allele of Ly49G2 may express Ly49A from the same or opposite chromosome with equal likelihood, indicating that the expressed allele is chosen independently for different Ly49 genes. Finally, the data demonstrate that biallelic expression of Ly49A or Ly49G2 occurs least frequently in mice that express ligands for these receptors (H-2d mice), and most frequently in class I–deficient mice. Thus, biallelic expression of Ly49 genes is regulated by interactions of NK cell progenitors with MHC class I molecules.
Keywords: major histocompatibility complex, natural killer cells, monoallelic expression, allelic exclusion, Ly49
Introduction
NK cells play an important role in the recognition and lysis of pathogen-infected or transformed target cells (1, 2). NK cells are regulated in part by the various MHC class I–specific receptors expressed on the cell surface 3 4. Most of these receptors transduce an inhibitory signal upon ligand engagement, accounting for the tendency of NK cells to attack class I–deficient target cells while sparing normal cells. Other receptors, including some specific for MHC class I molecules, stimulate NK cells 5 6 7. The balance of inhibitory and stimulatory signaling determines the outcome of NK–target cell interactions.
In mice, two families of MHC class I–specific receptors have been described: the Ly49 family 8 and the NKG2/CD94 family 9 10. The Ly49 family comprises at least nine members, seven of which contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic domains. Functional, genetic, and binding data defined Dd and Dk, among other MHC molecules, as ligands for Ly49A, with no detectable binding to H-2b class I molecules 11 12 13 14 15. Ly49G2 is functionally inhibited by H-2d but not H-2b cells 16, and has been shown to specifically bind to soluble tetrameric Dd molecules 14.
MHC class I–specific inhibitory receptors, including Ly49 receptors, are expressed in a variegated pattern on NK cells, such that each receptor is expressed by 10–50% of NK cells. Receptor expression is stable once initiated, as suggested by experiments in which purified NK subsets are expanded in vitro or transferred in vivo 11 17 18. Coexpression of Ly49 family members occurs frequently 19, with an average of three to four receptors expressed by each NK cell 20. Different receptors are expressed with a considerable degree of independence, as indicated by the finding that the frequency of NK cells coexpressing a given Ly49 receptor pair or trio can be estimated by multiplying the frequencies of cells that express each of the receptors individually (the “product rule” [19]). These findings suggest that the initial choice of which receptors are to be expressed in a developing NK cell is governed by a stochastic mechanism. Additional evidence indicates that different receptors are expressed on developing NK cells in a sequential, cumulative fashion 18 21.
Ly49 genes exhibit limited allelic polymorphism, allowing one to separately monitor the expression of each allele of the same gene using RNA analysis or allele-specific mAbs. Such analyses revealed that Ly49 genes are expressed in a predominantly monoallelic fashion 17 22 23. For example, costaining of NK cells from (B6 × BALB/c)F1 mice with a combination of the A1 mAb, specific for Ly49AB6, and the JR9-318 mAb, reactive with both Ly49AB6 and Ly49ABALB, detected a JR9+A1− population and an equally numerous JR9+A1+ population 17. Analysis of the mRNA in these subsets by reverse transcription (RT)-PCR confirmed that the JR9+A1− population consisted almost exclusively of cells expressing only the Ly49ABALB allele, whereas the JR9+A1+ population consisted predominantly of cells expressing the Ly49AB6 allele. These data suggested that Ly49A+ NK cells in the F1 mice consist primarily of two subsets of approximately equal size, each of which expresses only one of the two Ly49A alleles 17. Monoallelic expression of nonrearranging, non–X–linked genes has been observed in only a few other cases, such as the mouse odorant receptor genes 24, certain cytokine genes 25 26 27, and possibly the Pax-5 transcription factor 27 28. These findings have spurred a great deal of interest in the mechanisms underlying monoallelic gene expression as well as its significance.
The population analysis described above demonstrated that Ly49A is expressed in a predominantly monoallelic fashion, but was insufficiently discriminating to determine whether a minor subset of cells expresses both Ly49A alleles. To address this question, a clonal analysis of Ly49A expression was undertaken. Single Ly49A+ NK cells from F1 mice were expanded in culture into small colonies. RNA samples from the colonies were analyzed by RT-PCR to determine which Ly49A alleles were expressed. The analysis confirmed that most Ly49A+ NK cells expressed only one or the other Ly49A allele. However, a smaller fraction of clones was also detected in which both Ly49A alleles were expressed 22. It appeared that the frequency of biallelic clones could be roughly estimated by multiplying the frequencies of NK cells expressing each allele independently.
Using the clonal system, monoallelic expression was extended to two additional Ly49 genes, Ly49C and Ly49G2. As was observed for Ly49A, biallelic expression in a minority of clones was documented in the case of Ly49G2 23. Consistent with earlier evidence generated with NK cell populations 17, the clonal analysis also suggested that individual NK cells that coexpress two or more Ly49 genes may express alleles from the same or opposite chromosomes 23. Based on these findings, we proposed that the initial choice of NK receptor genes for expression is imposed by mechanisms that act locally to stably regulate cis-acting control sequences associated with each Ly49 allele, independent of neighboring Ly49 genes or of the Ly49 allele on the opposite chromosome 17 19.
Previous data has indicated that the MHC molecules of the host impact the Ly49 receptor repertoire. Specifically, the frequencies of NK cells coexpressing various combinations of Ly49 receptors were lower in class I+ mice than in MHC class I–deficient mice 29. Data suggest that expression by an NK cell of a Ly49 receptor specific for self-MHC reduces the likelihood that the cell will express others. For example, expression of a Ly49A transgene in all NK progenitor cells resulted in a reduced frequency of NK cells expressing endogenous Ly49G2 receptors 30. The effect was observed in mice expressing H-2d, to which the receptors bind, but not in MHC class I–deficient mice. Given that Ly49 receptors are expressed sequentially during development, we proposed that the interaction of an expressed Ly49 receptor with self-MHC ligands could inhibit subsequent expression of additional Ly49 receptors by the same NK cell 19. Why regulate coexpression of multiple Ly49 receptors? Evidence suggests that two Ly49 receptors expressed by the same NK cell independently inhibit the cell when engaged separately 31, and, when both are engaged, function cumulatively to cause greater inhibition than either receptor alone 31a. By regulating the coexpression of multiple self-MHC–specific receptors, the NK cell may optimize its capacity to attack target cells that selectively or only partially downregulate class I molecules. An interesting question is whether this mechanism also limits the coexpression of both alleles of the same Ly49 gene. Consistent with this possibility, clonal analysis suggested that mice lacking MHC class I molecules contained a higher frequency of biallelic Ly49A+ NK cells than normal mice 22.
The clonal analyses suffer from the limited number of clones that could be analyzed, and the fact that the clones were expanded in vitro before analysis, possibly introducing experimental bias. Here we describe new allele-specific mAbs that allow the direct detection of Ly49 monoallelic and biallelic populations in ex vivo NK cells. With the use of these Abs, we provide the first accurate quantitation of monoallelic and biallelic populations, and demonstrate that host MHC molecules determine the extent of biallelic expression of two different Ly49 receptors. Furthermore, the mAbs were used to demonstrate directly that coexpression of two different Ly49 loci can occur from the same or opposite chromosomes with equal likelihood. This finding suggests that each Ly49 locus is independently regulated with respect to which allele is expressed.
Materials and Methods
Mice.
B10.D2nSnJ, C57Bl/6J (B6), BALB.B mice (all from The Jackson Laboratory), and B6-β2m−/− mice backcrossed five times to B6 22 32 were bred in the animal facility at the University of California at Berkeley. BALB/cJ mice were purchased from The Jackson Laboratory. To generate “BALB.B-β2m−/− mice”, B6 β2m−/− mice were crossed to BALB.B mice, and the hybrids were backcrossed twice more to BALB.B. Backcross progeny were selected that harbored one β2m knockout allele (based on Southern blotting with a β2m probe) and that only expressed the NK complex (NKC) of the BALB strain (based on lack of staining of PBLs with PK136 mAb specific for NK1.1 and genomic Southern blotting of EcoRI-digested tail DNA with a NKRP1A partial cDNA probe). Backcrossed animals were intercrossed, selecting β2m−/− progeny. The resulting mice were β2m−/− NKCBALB/BALB and mostly homozygous for BALB alleles at other loci, and were maintained by intercrossing.
The Ly49 heterozygous mice compared in Fig. 5 were generated as follows: β2m−/−-F1 mice by crossing B6-β2m−/− and BALB.B-β2m−/− mice; H-2b-F1 mice by crossing B6 (H-2b) and BALB.B (H-2b) mice; and the H-2d-F1 mice by crossing B10.D2nSnJ (H-2d) and BALB/cJ (H-2d) mice. Comparisons indicated that it made no difference whether one strain or the other was used as the mother.
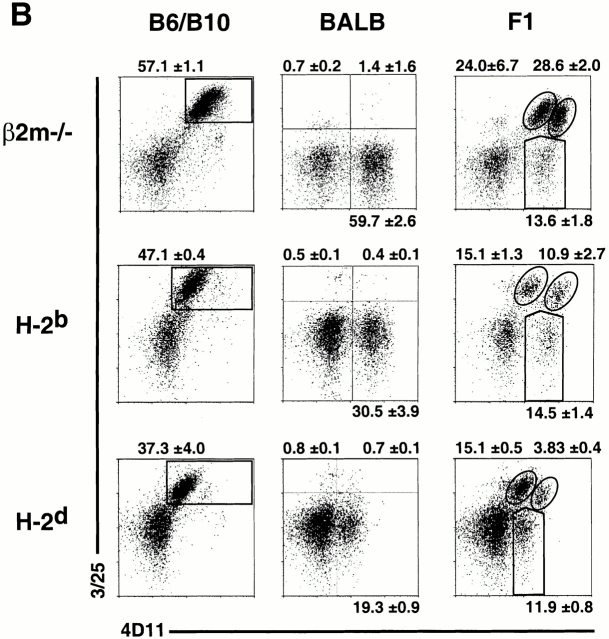
Figure 5.
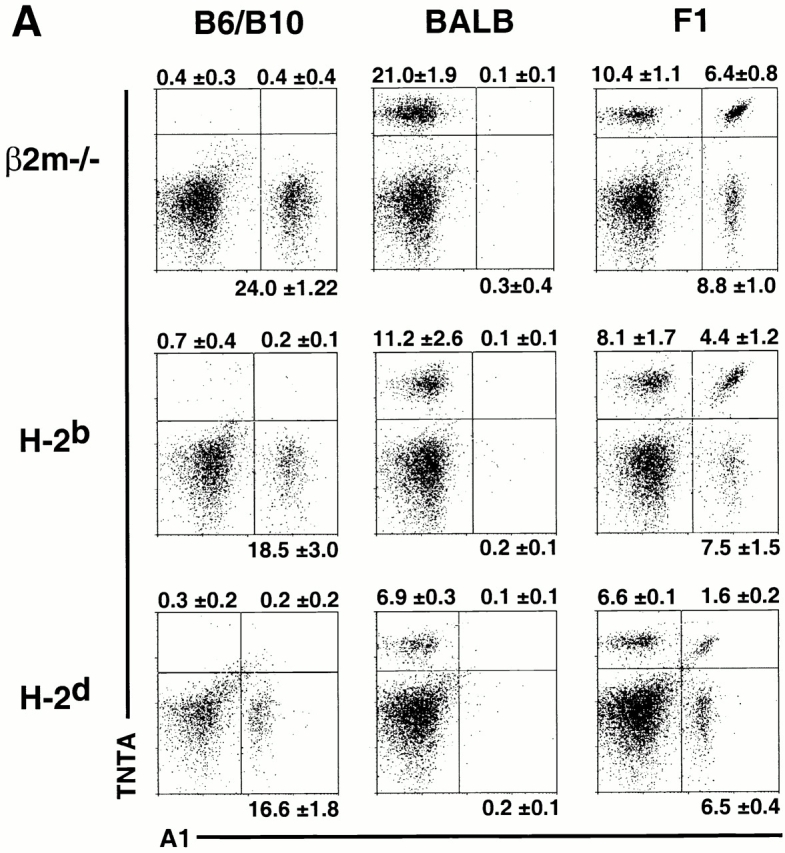
MHC class I molecules determine the extent of biallelic Ly49 expression. Freshly isolated, nylon wool nonadherent, gated DX5+CD3−CD8− NK cells from MHC-different mice were examined for mono- and biallelic expression of (A) Ly49A and (B) Ly49G2. In each case, Ly49 heterozygous (F1) mice were compared with the parental strains. Data represent means ± SD of three or four determinations on cells from individual animals.
Production of Ly49ABALB-specific TNTA mAb.
Ly49ABALB cDNA 17 was cloned into the pIRES vector (CLONTECH Laboratories, Inc.) and stably transfected into Chinese hamster ovary (CHO) cells. B6 mice were immunized intraperitoneally six times with between 1 and 10 × 106 Ly49ABALB-expressing CHO cells in PBS at 2–3-wk intervals. 3 d after the final boost, splenocytes were harvested and fused as described 33, except P3×63-Ag8.653 (American Type Culture Collection) was used as the fusion partner. Hybridoma supernatants were screened by flow cytometry for reactivity with CHO-Ly49ABALB transfectants (but not untransfected CHO cells) and a subset of BALB/c NK cells. Of three Ly49ABALB-specific hybridomas, TNTA (IgG2a, κ) was subcloned and used for this study. TNTA mAb was purified from hybridoma supernatants by affinity chromatography over protein A-agarose columns.
Production of Ly49G2B6-specific 3/25 mAb.
BALB/c mice were immunized repeatedly with CHO cells stably transfected with the Ly49G2B6 cDNA 34 in the pIRES expression vector. Spleen cells were fused to P3×63Ag.8653 cells with polyethylene glycol (PEG), and hybridomas were selected in HAT medium. Supernatants of candidate clones were screened for staining of CHO-Ly49G2B6 (but not CHO-Ly49FB6) transfectants and Ly49G2B6 transgenic thymocytes (Hanke, T., and D.H. Raulet, unpublished observations). Out of two Ly49G2B6-specific mAbs, 3/25 (IgG1, κ) was subcloned and used throughout this study. Depending on the experiment, 3/25 mAb was used directly from hybridoma supernatants, or purified from ascites by ammonium sulfate precipitation followed by size exclusion chromatography.
Other mAbs.
Anti-Ly49AB6 mAb A1 35, anti-Ly49A (nonallele-specific) mAb JR9-318 36, anti-Ly49C mAb 5E6 37, anti-Ly49D mAb SED-85 38, anti-Ly49F mAb HBF-719 38, and the anti-FcR mAb 2.4G2 39 have been described previously. The hybridoma producing 4D11 40 was purchased from American Type Culture Collection. Abs were conjugated to FITC (Boehringer) or biotin (Pierce Chemical Co.). Anti–NK1.1-PE and DX5-PE conjugates were purchased from BD PharMingen. Anti–CD3-Tricolor, anti–CD8-Tricolor, streptavidin-Tricolor, goat anti–mouse IgG, and mouse IgG were purchased from Caltag Labs. Streptavidin-RED613 was purchased from GIBCO BRL. Anti-His tag rabbit polyclonal IgG was purchased from Santa Cruz Biotechnology, Inc. Anti–rabbit–Ig-FITC was purchased from Dako.
Transient Transfection of COS Cells.
Ly49 cDNAs 17 41 cloned into the pME18S expression vector were transiently transfected into COS cells with Lipofectamine Plus (GIBCO BRL). For Ly49D and Ly49H transfections, a DAP12 42 expression vector was cotransfected. 1 d later the cells were trypsinized and replated. On day 2, the COS cells were harvested and stained with TNTA mAb, 3/25 mAb, a positive control Ly49-specific mAb, or an anti-His polyclonal Ab as indicated.
Flow Cytometry and Cell Sorting.
Nylon wool nonadherent splenocytes were preincubated with 2.4G2 mAb to block FcR. To discriminate Ly49A alleles, cells were costained with A1-FITC, TNTA-biotin, DX5-PE, anti–CD3-Tricolor, and anti–CD8-Tricolor followed by straptavidin-613. To discriminate Ly49G2 alleles, cells were stained in four steps: 3/25 mAb; goat anti–mouse IgG-biotin; mouse IgG; and 4D11-FITC, DX5-PE, anti–CD3-Tricolor, anti–CD8-Tricolor, and streptavidin-RED613. In Fig. 3, NK cells were sorted for expression of different Ly49G2 alleles before analysis and subsequent culturing. Nylon wool nonadherent splenocytes from 10–15 B6 mice were stained in four steps: 3/25 mAb; goat anti–mouse-biotin; and mouse IgG; 4D11-FITC, anti–NK1.1-PE, and streptavidin-Tricolor. Stained splenocytes were sorted on an ELITE flow cytometer (Beckman Coulter). Aliquots of the sorted populations were used immediately for RT-PCR analysis and the remaining cells were cultured in RPMI supplemented with 10% FCS, antibiotics, glutamine, βME, and 800 ng/ml IL-2 (Chiron Corp.). After 10 d, an aliquot of cells was used for RT-PCR analysis and the remaining cells were stained for Ly49A (A1-FITC and TNTA-biotin; streptavidin-613) or for Ly49G2 (3/25 mAb; goat anti–mouse IgG-biotin; mouse IgG; 4D11-FITC and streptavidin-613).
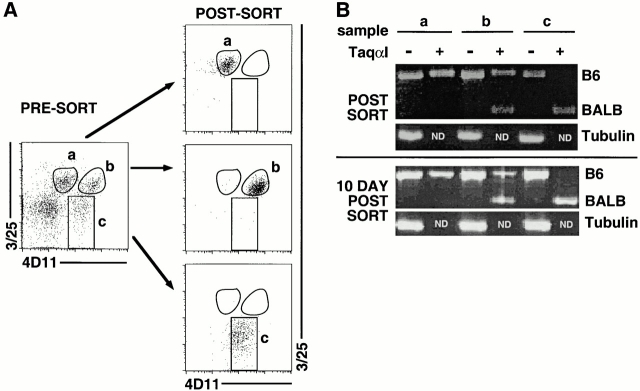
Figure 3.
Cell sorting and identification of Ly49G2 populations. Pooled, freshly isolated nylon wool nonadherent spleen cells from (B6 × BALB.B)F1 mice were stained with NK1.1, 3/25, and 4D11. (A) Three populations of NK1.1+ cells were separated by cell sorting: (a) 3/25+4D11low; (b) 3/25+4D11high; and (c) 3/25−4D11+. As tested by postsort analysis, the populations were each of >90% purity. (B) The Ly49G2 alleles expressed by each population were determined by RT-PCR with Ly49G2 specific primers. Before electrophoresis, PCR products were digested with Taqα1, a diagnostic restriction enzyme that cleaves only the Ly49G2BALB cDNA. RT-PCR analysis was performed on cells immediately postsort (top) and after in vitro expansion for 10 d in the presence of IL-2 (bottom). Because the tubulin PCRs were tested only without digestion, the Taqα1 lanes are empty (ND). The experiment was repeated with NK cells from class I–deficient mice with identical results.
RT-PCR.
RT-PCR was performed on 5–7 × 104 freshly sorted cells or on 10-d IL-2–cultured sorted cells as described previously 23.
Results
Independent Expression of Ly49A Alleles.
The A1 mAb binds Ly49A encoded by the B6 Ly49A allele (Ly49AB6), but not the BALB Ly49A allele (Ly49ABALB; reference 17). A mAb with reciprocal specificity for Ly49ABALB, but not Ly49AB6, was generated by immunizing B6 mice with CHO cell transfectants expressing Ly49ABALB. The resulting allele-specific mAb, TNTA, stained transiently transfected COS cells expressing Ly49ABALB cDNA, but did not stain COS cells expressing any of nine different B6 Ly49 cDNAs, including Ly49AB6 (Fig. 1). Furthermore, TNTA stained 10–15% of freshly isolated NK cells from BALB mice, but did not stain B6 NK cells (Fig. 2 A). It is highly likely that TNTA recognizes only Ly49A in BALB mice, because the staining pattern exactly overlaps with that of the well-characterized nonallele-specific Ly49A mAb, JR9-318. Furthermore, TNTA and JR9-318 reciprocally compete for binding of Ly49ABALB on NK cells from BALB.B mice (data not shown).
Figure 1.
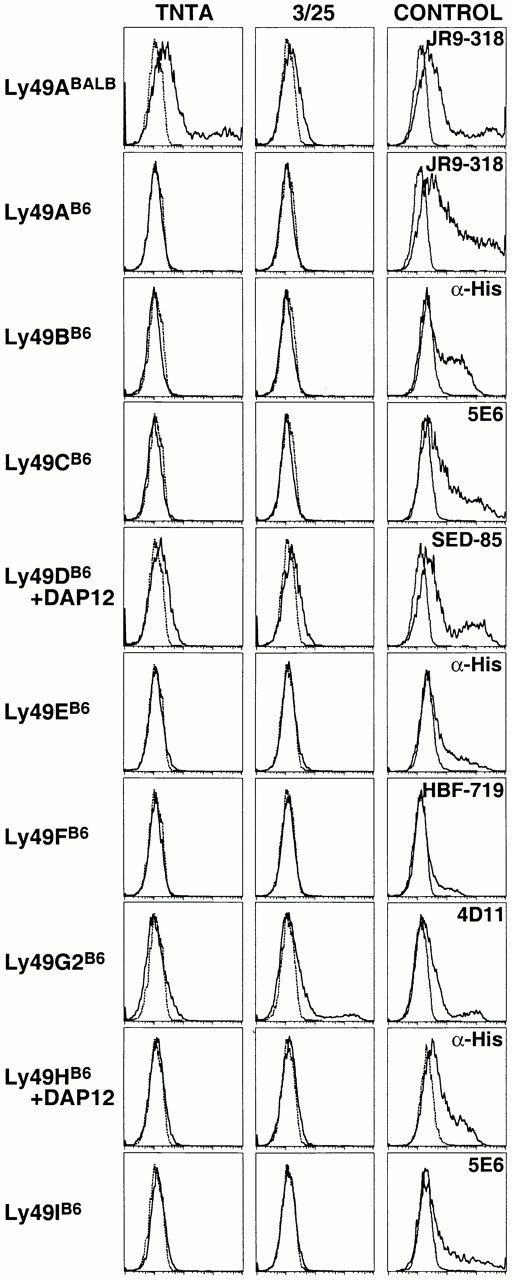
Specificity of TNTA and 3/25 mAbs. TNTA mAb (first column) or 3/25 mAb (second column) was used to stain COS cells transiently transfected with the indicated Ly49 expression constructs (solid lines) or untransfected COS cells (dashed lines). Ly49D and Ly49H constructs were cotransfected with a DAP12 expression construct. Ly49B, Ly49E, and Ly49H cDNAs were epitope (6x-His) tagged. Transiently transfected cells varied in transfection efficiency, as shown by staining with the appropriate Ly49-specific mAb or an anti-his rabbit polyclonal IgG in the case of tagged receptors (third column).
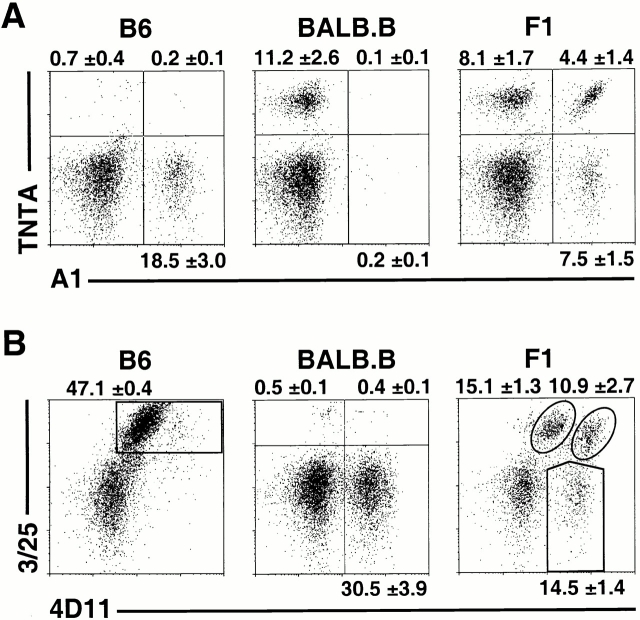
Figure 2.
Predominantly monoallelic expression of Ly49A and Ly49G2. Freshly isolated, nylon wool nonadherent splenic cells from B6, BALB.B, and (B6 × BALB.B)F1 mice (all H-2b) were harvested. Gated DX5+CD3−CD8− NK cells were analyzed with (A) the Ly49AB6 specific mAb, A1, versus the Ly49ABALB specific mAb, TNTA, or (B) with Ly49G2B6-specific mAb 3/25 versus 4D11, which recognizes both Ly49G2B6 and Ly49G2BALB alleles. Data represent means ± SD of three or four determinations on cells from individual animals.
To examine monoallelic versus biallelic expression of Ly49A, (B6 × BALB.B)F1 mice (H-2b/b) were generated. Costaining with A1 and TNTA on freshly isolated, gated NK cells revealed that each mAb stains 10–15% of NK cells (Fig. 2 A). Replicate measurements showed that an average of 8.1% of the (B6 × BALB.B)F1 NK cells expressed Ly49ABALB only, 7.5% expressed Ly49AB6 only, and 4.4% expressed both alleles. Therefore, NK cells express Ly49A predominately from one allele or the other with approximately equal likelihood, but a small subset of cells coexpresses both Ly49A alleles.
Independent Expression of Ly49G2 Alleles.
To distinguish the expression of individual Ly49G2 alleles, a mAb specific for the Ly49G2 receptor expressed on B6 NK cells was generated. BALB/c mice were immunized with CHO cell transfectants expressing the B6 allele of Ly49G2 (Ly49G2B6). A resulting mAb, 3/25, stained Ly49G2B6-transfected COS cells, but not transfectants expressing any of the other eight B6 Ly49 family members (Fig. 1). The 3/25 mAb stained an average of 47.1% of freshly isolated NK cells from B6 mice, and did not stain BALB NK cells (Fig. 2 B). The staining pattern in B6 mice exactly overlapped the staining pattern of the nonallele-specific mAb 4D11, arguing that 3/25 reacts only with Ly49G2B6 in B6 mice (Fig. 2 B).
To examine allelic expression of Ly49G2, NK cells from (B6 × BALB.B)F1 mice were co-stained with 3/25 and the nonallele-specific Ab 4D11 (Fig. 2 B). Because 3/25 recognizes Ly49G2B6, and 4D11 recognizes both the B6 and BALB alleles of Ly49G2, there are no single positive 3/25+ 4D11− NK cells. Unexpectedly, however, two distinct populations were observed in the double-positive 3/25+ 4D11+ quadrant. We hypothesized that the 3/25+4D11high population expressed both Ly49G2B6 and Ly49G2BALB, whereas the 3/25+4D11low population represented NK cells expressing only Ly49G2B6. The dim 4D11 staining of the latter population might occur because 3/25, which was added first in the staining experiments, partially blocks 4D11 binding. Indeed, a control experiment demonstrated that 3/25 blocks binding of Ly49G2B6 by 4D11 (data not shown).
To test allelic expression at the mRNA level, we sorted freshly isolated 3/25+4D11low, 3/25+4D11high, and 3/25− 4D11+ populations to >90% purity (Fig. 3 A). mRNA was prepared directly from these cells, and RT-PCR was performed with primers that amplify the transcripts of both Ly49G2 alleles. The resulting PCR products were then digested with the diagnostic restriction enzyme, TaqαI, which only cleaves the Ly49G2BALB cDNA 23. The analysis confirmed that the 3/25+4D11low population contained only Ly49G2B6 transcripts, the 3/25+4D11high population contained both Ly49G2B6 and Ly49G2BALB transcripts, and the 3/25−4D11+ population contained only Ly49G2BALB transcripts (Fig. 3 B). These expression patterns are stable, as similar results were obtained with sorted cells that had been expanded for 10 d in IL-2 (Fig. 3 B). Based on the staining patterns (Fig. 2 B), we conclude that an average of 15.1% of F1 NK cells express Ly49G2B6 only, 14.5% express Ly49G2BALB only, and 10.9% express both Ly49G2 alleles. Thus, Ly49G2, like Ly49A, is expressed predominantly in a monoallelic fashion but with a distinct population of biallelic cells.
Independent Allelic Expression of Ly49A and Ly49G2.
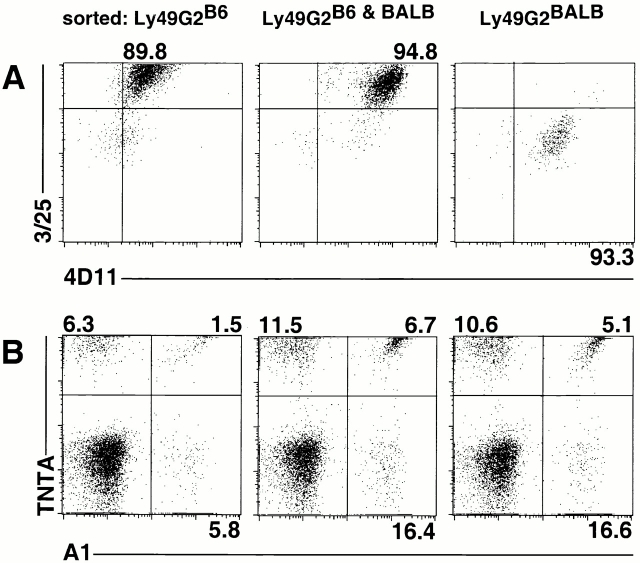
By using the new allele-specific mAbs in conjunction, it was possible to determine directly whether cells that coexpress Ly49A and Ly49G2 do so coordinately from the same chromosome, from opposite chromosomes, or whether there is a bias for one configuration or the other. The three Ly49G2+ populations described above were expanded for 10 d in medium containing IL-2 before reanalysis. Costaining with the 3/25 and 4D11 mAbs confirmed that the populations had largely retained their phenotype with respect to allelic expression of Ly49G2 alleles (Fig. 4 A). Costaining with TNTA and A1 mAbs was employed to determine allelic expression of Ly49A (Fig. 4 B). The analysis revealed that cells expressing Ly49G2B6 were equally likely to express Ly49AB6 or Ly49ABALB, and a small fraction of such cells expressed both Ly49A alleles. Similarly, cells expressing Ly49G2BALB were approximately equally likely to express Ly49AB6 or Ly49ABALB, though there may be a small bias for Ly49AB6 in this instance. Finally, NK cells expressing both Ly49G2 alleles can also express either Ly49A allele, and a small fraction of cells expresses all four alleles. These data suggest that in the case of coexpression of different Ly49 receptor genes, there is little or no coordination in terms of which chromosome is used to direct synthesis of the two receptors.
Figure 4.
Independent allelic expression of Ly49A and Ly49G2. NK cells expressing Ly49G2B6 only, Ly49G2BALB only, or both Ly49G2B6 and Ly49G2BALB were sorted and expanded for 10–15 d in medium containing IL-2. (A) Reanalysis of the sorted populations with Ly49G2 mAbs demonstrated that they remained >89% pure after the culture. (B) Analysis of the sorted populations using allele-specific Ly49A mAbs. The experiment was repeated with NK cells from class I–deficient mice with similar results.
MHC Class I Molecules Control the Frequency of Ly49 Biallelic NK Cells.
The new mAbs were employed to investigate the effect of MHC background on the frequencies of cells expressing Ly49A or Ly49G2 in a monoallelic versus biallelic fashion. For this purpose, we compared three MHC-different Ly49 heterozygous crosses: H-2d-F1, H-2b-F1, or class I–deficient β2m−/−-F1 (see Materials and Methods). All three were hybrids between BALB and B6 or B10 background mice, and therefore were of similar genetic background. In each case we compared F1 mice to the parental Ly49 homozygous strains. Costaining with A1 and TNTA mAbs revealed predominant monoallelic expression of Ly49A in all three F1s, as well as a smaller biallelic population (Fig. 5 A). Similarly, costaining with 3/25 and 4D11 demonstrated predominant monoallelic expression of Ly49G2, with a smaller biallelic population in each case (Fig. 5 B). As reported previously 29, the frequencies of NK cells expressing Ly49A or Ly49G2, regardless of which allele, were marginally lower in H-2b mice than in β2m−/− mice, and lower yet in H-2d mice (which express the strong Dd ligand for both Ly49A and Ly49G2; Fig. 5 and Table ). More significantly, the size of the biallelic population varied substantially depending on MHC expression. Ligand-expressing H-2d mice contained substantially fewer biallelic Ly49A+ cells than either H-2b or class I–deficient mice (Fig. 5 A and Table ). A similar picture emerged when Ly49G2 expression was examined. H-2d F1 mice contained substantially fewer Ly49G2 biallelic cells than H-2b F1 mice, which in turn had significantly fewer biallelic cells than class I–deficient mice (Fig. 5 B, and Table ). These data indicate that expression of a strong Ly49 ligand results in restricted biallelic expression of both Ly49A and Ly49G2.
Table 1.
Monoallelic and Biallelic Ly49 Expression Varies with MHC
| Percentage of NK cells expressing indicated Ly49 alleles | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ly49AB6/BALB | Ly49G2B6/BALB | |||||||
| Heterozygote | Ly49AB6 | Ly49ABALB | Predicted | Observed | Ly49G2B6 | Ly49G2BALB | Predicted | Observed |
| β2m−/−-F1 | 15.2 ± 1.3 | 16.8 ± 1.5 | 2.6 | 6.4 ± 0.8 | 52.6 ± 6.6 | 42.2 ± 3.1 | 22.1 | 28.6 ± 2.0 |
| H-2b-F1 | 11.9 ± 2.6 | 12.4 ± 3.0 | 1.5 | 4.4 ± 1.4 | 26.0 ± 3.9 | 25.4 ± 3.8 | 6.7 | 10.9 ± 2.7 |
| H-2d-F1 | 8.1 ± 0.4 | 8.2 ± 0.3 | 0.7 | 1.6 ± 0.2 | 19.0 ± 0.1 | 15.7 ± 1.2 | 3.0 | 3.8 ± 0.4 |
Discussion
The new mAbs allow the direct detection of NK populations expressing Ly49A and Ly49G2 in a monoallelic or biallelic fashion. The data confirm earlier findings suggesting that Ly49 genes are expressed in a predominantly, but not exclusively monoallelic fashion. Most NK cells that express Ly49A or Ly49G2 express one or the other allele of the respective gene, with approximately equal likelihood. A smaller fraction of cells expresses both alleles. Biallelic Ly49G2+ NK cells are more frequent than biallelic Ly49A+ NK cells, consonant with the higher overall frequency of Ly49G2+ NK cells. Consistent with previous studies of Ly49A, our data suggest that monoallelic expression of Ly49G2 is a stable phenotype that is maintained as NK cells divide in culture.
The mAbs also allow direct cell surface detection of cells that coexpress Ly49A and Ly49G2 alleles in various combinations. The data indicate that every possible assortment of the four Ly49A and Ly49G2 alleles is represented in the NK cell population. Notably, an NK cell that expresses a given Ly49G2 allele can coexpress a Ly49A allele from the same or opposite chromosome with equal likelihood. These data represent strong evidence that the mechanism that chooses a Ly49 allele for expression acts highly locally on the chromosome, without enhancing or diminishing the probability that another gene nearby in the closely linked gene complex will be expressed. It can be concluded that these mechanisms do not “open” or “close” the entire array of Ly49 genes on a given chromosome, but rather act individually on each Ly49 allele. Together with studies suggesting that the average NK cell expresses approximately three Ly49 receptors 20, the data suggest that each cell expresses a patchwork of expressed Ly49 genes selected from one or the other or both chromosomes.
A novel feature of the data is the finding that expression of one Ly49A allele is not completely independent of expression of the other. This conclusion is suggested by deviations between the observed percentages of biallelic NK cells and the percentages calculated under the assumption of allelic independence. The latter value corresponds to the frequency of Ly49AB6+ cells multiplied by the frequency of Ly49ABALB+ cells. Regardless of MHC haplotype, biallelic expression of Ly49A was two- to threefold more common than predicted by the product rule (Table ). Biallelic expression of Ly49G2 also deviated from the predictions of the product rule, but to a much smaller extent (Table ). As noted before 43, the fact that the Ly49 repertoire is controlled to some extent by interactions with host MHC molecules predicts that receptor coexpression is not completely independent, and should lead to deviations from the product rule. However, as deviations from the product rule were observed even in class I–deficient mice (Table ), they are unlikely to result solely from interactions with MHC molecules. The fact that biallelic cells were always more frequent than predicted by the product rule suggests instead that heterogeneity exists in developing NK cells in factors that control expression of Ly49A, with some cells exhibiting a generally higher likelihood of initiating Ly49A expression than others. There are many possible explanations for such variations, but one possibility is that the transcription factor TCF-1, previously reported as necessary for Ly49A expression 44, is not expressed equally in all NK progenitors.
An important finding of the study is that MHC class I genes determine the extent of biallelic Ly49 receptor gene expression. This conclusion was hinted at in a previous clonal analysis, which indicated a greater extent of biallelic expression in class I–deficient mice than in class I+ mice 22. However, the small number of clones analyzed precluded quantitative comparisons, and could not assess whether biallelic expression differs in mice that express different MHC haplotypes. In this study, a strong deficit in biallelic Ly49A and Ly49G2 NK cells was clearly evident in H-2d mice, with H-2b mice intermediate and class I–deficient mice containing the highest percentage of biallelic cells. The control of biallelic expression is thus similar to the control of bigenic Ly49 expression. Previous studies showed that NK cells expressing multiple different Ly49 receptors are less frequent in class I+ mice than in class I–deficient mice 29 45. In the case of Ly49A and Ly49G2, two receptors with strong reactivity with H-2d and not H-2b, the percentage of Ly49A/G2 double-positive cells was lowest in H-2d mice, intermediate in H-2b mice, and highest in class I–deficient mice, similar to the pattern observed in this report with respect to biallelic expression (29; Fig. 5). These data support the conclusion that the MHC-specific education mechanisms that regulate coexpression of different Ly49 genes similarly regulate coexpression of the two alleles of a given Ly49 gene.
The regulation of biallelic expression by MHC molecules is consistent with a previously proposed model of NK cell education 19 21. The regulated sequential expression model proposed that as Ly49 receptor genes are stochastically switched on during NK cell development, engagement of an expressed receptor with host MHC class I molecules initiates feedback inhibition that reduces the likelihood that additional Ly49 receptor genes will be activated. This mechanism should prevent individual NK cells from expressing an excess of self-MHC–specific Ly49 receptors, and thus could optimize the sensitivity of NK cells to changes in host cell class I expression. Our data indicates that this form of regulation controls not only expression of different Ly49 genes, but also different alleles of a given Ly49 gene. A virtue of the model is that it can account for the reduced frequencies of biallelic Ly49A or Ly49G2 populations, as well as of the Ly49A/G2 coexpressing subset, in H-2b mice versus β2m−/− mice, neither of which have strong ligands for these receptors. This is because some NK cells may express NK receptors specific for H-2b class I molecules before they express Ly49A or Ly49G2. The model predicts that in H-2b but not β2m−/− mice, such cells will be inhibited from expressing additional Ly49 receptors, including Ly49A and Ly49G2. The reduced overall frequencies of NK cells expressing Ly49A or Ly49G2 should result in a lower percentage of cells coexpressing these receptor alleles, as was observed (Fig. 5 and Table ).
As the two alleles of a given Ly49 gene do not necessarily exhibit significant differences in specificity 14, it is interesting to ask why these genes are expressed in a predominantly monoallelic fashion. One possibility is based on the findings herein that each Ly49 allele is expressed mostly independently of the opposite allele and of alleles at other Ly49 loci. The mechanism that evolved to switch on a given Ly49 gene in only a fraction of progenitor cells apparently acts on cis-acting elements in each gene, and thus may activate one allele without activating the other. The result is a variegated, overlapping repertoire. A consequence of this is that some NK cells will express receptors specific for one self-MHC molecule and not others. These NK cells can attack self-cells that downregulate that class I molecule selectively. In this view, monoallelic expression is simply a by-product of the mechanism that evolved to ensure variegated expression of Ly49 genes, which happens to act independently on each Ly49 allele. Alternatively, the finding herein that biallelic expression is strongly regulated by MHC class I molecules presents another possibility. By allowing NK cells to express one or two doses of the same gene, the sensitivity of developing NK cells to inhibition by a given class I ligand may be adjusted more precisely. With a weaker ligand, biallelic expression may yield a greater amount of receptor on the cell surface, resulting in a greater inhibitory response in the NK cell, whereas monoallelic expression may be more beneficial in the case of a strong class I ligand. The reagents reported in this paper may be useful in testing the latter model.
Acknowledgments
We thank Hector Nolla for expert flow cytometry and cell sorting assistance, and Scot Liu, Caelin White, and Ann Lazar for excellent technical assistance. We thank Chris McMahon, Deborah Moniot, and Andreas Diefenbach for providing helpful comments on the manuscript.
T. Hanke was a recipient of a Research Fellowship from the Deutsche Forschungsgemeinschaft. This work was supported by National Institutes of Health grant RO1-AI39642 to D.H. Raulet.
Footnotes
Abbreviations used in this paper: CHO, Chinese hamster ovary; RT, reverse transcription.
References
- Trinchieri G. Biology of natural killer cells. Adv. Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron C.A., Nguyen K.B., Pien G.C., Cousens L.P., Salazar-Mather T.P. Natural killer cells in antiviral defensefunction and regulation by innate cytokines. Annu. Rev. Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- Lanier L.L. NK cell receptors. Annu. Rev. Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- Moretta A., Bottino C., Vitale M., Pende D., Biassoni R., Mingari M.C., Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu. Rev. Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- Moretta A., Sivori S., Vitale M., Pende D., Morelli L., Augugliaro R., Bottino C., Moretta L. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J. Exp. Med. 1995;182:875–884. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchins J.P., Lanier L.L., Niemi E.C., Phillips J.H., Ryan J.C. Natural killer cell cytolytic activity is inhibited by NKG2-A and activated by NKG2-C. J. Immunol. 1997;158:3603–3609. [PubMed] [Google Scholar]
- Nakamura M.C., Linnemeyer P.A., Niemi E.C., Mason L.H., Ortaldo J.R., Ryan J.C., Seaman W.E. Mouse Ly-49D recognizes H-2D(d) and activates natural killer cell cytotoxicity. J. Exp. Med. 1999;189:493–500. doi: 10.1084/jem.189.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama W.M., Seaman W.E. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cellsthe NK gene complex. Annu. Rev. Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- Vance R.E., Kraft J.R., Altman J.D., Jensen P.E., Raulet D.H. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical MHC class I molecule Qa-1b . J. Exp. Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R.E., Jamieson A.M., Raulet D.H. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlhofer F.M., Ribaudo R.K., Yokoyama W.M. MHC class I alloantigen specificity of Ly-49+ IL-2 activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- Karlhofer F.M., Hunziker R., Reichlin A., Margulies D.H., Yokoyama W.M. Host MHC class I molecules modulate in vivo expression of a NK cell receptor. J. Immunol. 1994;153:2407–2416. [PubMed] [Google Scholar]
- Brennan J., Mahon G., Mager D.L., Jefferies W.A., Takei F. Recognition of class I major histocompatibility complex molecules by Ly-49specificities and domain interactions. J. Exp. Med. 1996;183:1553–1559. doi: 10.1084/jem.183.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke T., Takizawa H., McMahon C.W., Busch D.H., Pamer E.G., Miller J.D., Altman J.D., Liu Y., Cado D., Lemonnier F.A. Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity. 1999;11:67–77. doi: 10.1016/s1074-7613(00)80082-5. [DOI] [PubMed] [Google Scholar]
- Natarajan K., Boyd L.F., Schuck P., Yokoyama W.M., Eliat D., Margulies D.H. Interaction of the NK cell inhibitory receptor Ly49A with H-2Ddidentification of a site distinct from the TCR site. Immunity. 1999;11:591–601. doi: 10.1016/s1074-7613(00)80134-x. [DOI] [PubMed] [Google Scholar]
- Mason L.H., Ortaldo J.R., Young H.A., Kumar V., Bennett M., Anderson S.K. Cloning and functional characteristics of murine LGL-1a member of the Ly-49 gene family (Ly-49G2) J. Exp. Med. 1995;182:293–303. doi: 10.1084/jem.182.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W., Roland J., Raulet D.H. Allelic exclusion of Ly49 family genes encoding class I-MHC-specific receptors on NK cells. Nature. 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- Dorfman J.R., Raulet D.H. Acquisition of Ly49 receptor expression by developing natural killer cells. J. Exp. Med. 1998;187:609–618. doi: 10.1084/jem.187.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet D.H., Held W., Correa I., Dorfman J., Wu M.-F., Corral L. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol. Rev. 1997;155:41–52. doi: 10.1111/j.1600-065x.1997.tb00938.x. [DOI] [PubMed] [Google Scholar]
- Kubota A., Kubota S., Lohwasser S., Mager D.L., Takei F. Diversity of NK cell receptor repertoire in adult and neonatal mice. J. Immunol. 1999;163:212–216. [PubMed] [Google Scholar]
- Roth C., Carlyle J.R., Takizawa H., Raulet D.H. Clonal acquisition of inhibitory Ly49 receptors on differentiating NK cell precursors is successively restricted and regulated by stromal cell class I MHC. Immunity. 2000;13:143–153. doi: 10.1016/s1074-7613(00)00015-7. [DOI] [PubMed] [Google Scholar]
- Held W., Raulet D.H. Expression of the Ly49A gene in murine natural killer cell clones is predominantly but not exclusively mono-allelic. Eur. J. Immunol. 1997;27:2876–2884. doi: 10.1002/eji.1830271120. [DOI] [PubMed] [Google Scholar]
- Held W., Kunz B. An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire. Eur. J. Immunol. 1998;28:2407–2416. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Chess A., Simon I., Cedar H., Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Hollander G.A., Zuklys S., Morel C., Mizoguchi E., Mobisson K., Smpson S., Terhorst C., Wishart W., Golan D.E., Ghan A., Burakoff S. Monoallelic expression of the interleukin-2 locus. Science. 1998;279:2118–2121. doi: 10.1126/science.279.5359.2118. [DOI] [PubMed] [Google Scholar]
- Bix M., Locksley R.M. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- Rhoades K.L., Singh N., Simon I., Glidden B., Cedar H., Chess A. Allele-specific expression patterns of interleukin-2 and pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr. Biol. 2000;10:789–792. doi: 10.1016/s0960-9822(00)00565-0. [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Vambrie S., Steinlein P., Kozmik Z., Rolink A., Weith A., Busslinger M. Independent regulation of the two Pax5 alleles during B-cell development. Nat. Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- Held W., Dorfman J.R., Wu M.-F., Raulet D.H. Major histocompatibility complex class I-dependent skewing of the natural killer cell Ly49 receptor repertoire. Eur. J. Immunol. 1996;26:2286–2292. doi: 10.1002/eji.1830261003. [DOI] [PubMed] [Google Scholar]
- Held W., Raulet D.H. Ly49A transgenic mice provide evidence for a major histocompatibility complex–dependent education process in NK cell development. J. Exp. Med. 1997;185:2079–2088. doi: 10.1084/jem.185.12.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.Y., George T., Dorfman J., Roland J., Kumar V., Bennett M. The role of Ly49A and 5E6 (Ly49C) molecules in hybrid resistance mediated by murine natural killer cells against normal T cell blasts. Immunity. 1996;4:67–76. doi: 10.1016/s1074-7613(00)80299-x. [DOI] [PubMed] [Google Scholar]
- Hanke T., Raulet D.H. Cumulative inhibition of lymphocytes resulting from engagement of multiple inhibitory Ly49 receptors. J. Immunol. 2001;In press doi: 10.4049/jimmunol.166.5.3002. [DOI] [PubMed] [Google Scholar]
- Zijlstra M., Bix M., Simister N.E., Loring J.M., Raulet D.H., Jaenisch R. β2-Microglobulin deficient mice lack CD4−8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- Coligan J., Kruisbeek A., Margulies D., Shevach E., Strober W. Current Protocols in Immunology. Greene Publishing Associates and John Wiley & Sons, Inc; New York: 1991. [Google Scholar]
- Smith H.R.C., Karlhofer F.M., Yokoyama W.M. Ly-49 multigene family expressed by IL-2-activated NK cells. J. Immunol. 1994;153:1068–1079. [PubMed] [Google Scholar]
- Nagasawa R., Gross J., Kanagawa O., Townsend K., Lanier L.L., Chiller J., Allison J.P. Identification of a novel T cell surface disulfide-bonded dimer distinct from the α/β antigen receptor. J. Immunol. 1987;138:815–824. [PubMed] [Google Scholar]
- Roland J., Cazenave P.A. Ly-49 antigen defines an alpha beta TCR population in i-IEL with an extrathymic maturation. Int. Immunol. 1992;4:699–706. doi: 10.1093/intimm/4.6.699. [DOI] [PubMed] [Google Scholar]
- Sentman C.L., Hackett J.J., Moore T.A., Tutt M.M., Bennett M., Kumar V. Pan natural killer cell monoclonal antibodies and their relationship to the NK1.1 antigen. Hybridoma. 1989;8:605–614. doi: 10.1089/hyb.1989.8.605. [DOI] [PubMed] [Google Scholar]
- Coles M.C., McMahon C.W., Takizawa H., Raulet D.H. Memory CD8 T lymphocytes express inhibitory MHC-specific Ly49 receptors. Eur. J. Immunol. 2000;30:236–244. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Unkeless J.C. Characterization of a monoclonal antibody directed against mouse macrophage and lymphocyte Fc receptors. J. Exp. Med. 1979;150:580–596. doi: 10.1084/jem.150.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason L., Giardina S., Hecht T., Ortaldo J., Mathieson B. LGL-1a non polymorphic antigen expressed on a major population of mouse natural killer cells. J. Immunol. 1988;140:4403–4412. [PubMed] [Google Scholar]
- Corral L., Takizawa H., Hanke T., Jamieson A.M., Raulet D.H. A new monoclonal antibody reactive with several Ly49 NK cell receptors mediates redirected lysis of target cells. Hybridoma. 1999;18:359–366. doi: 10.1089/hyb.1999.18.359. [DOI] [PubMed] [Google Scholar]
- Lanier L.L., Corliss B.C., Wu J., Leong C., Phillips J.H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- Vance R.E., Raulet D.H. Toward a quantitative analysis of the repertoire of class I MHC-specific inhibitory receptors on natural killer cells. Curr. Top. Microbiol. Immunol. 1998;230:135–160. doi: 10.1007/978-3-642-46859-9_10. [DOI] [PubMed] [Google Scholar]
- Held W., Kunz B., Lowin-Kropf B., van de Wetering M., Clevers H. Clonal acquisition of the Ly49A NK cell receptor is dependent on the trans-acting factor TCF-1. Immunity. 1999;11:433–442. doi: 10.1016/s1074-7613(00)80118-1. [DOI] [PubMed] [Google Scholar]
- Salcedo M., Diehl A.D., Olsson-Alheim M.Y., Sundbäck J., Van Kaer L., Karre K., Ljunggren H.-G. Altered expression of Ly49 inhibitory receptors on natural killer cells from MHC class I deficient mice. J. Immunol. 1997;158:3174–3180. [PubMed] [Google Scholar]