Abstract
β selection is a major checkpoint in early thymocyte differentiation, mediated by successful expression of the pre-T cell receptor (TCR) comprising the TCRβ chain, CD3 proteins, and a surrogate TCRα chain, pTα. The mechanism of action of the pre-TCR is unresolved. In humans and mice, the pTα gene encodes two RNAs, pTαa, and a substantially truncated form, pTαb. This study shows that both are biologically active in their capacity to rescue multiple thymocyte defects in pTα−/− mice. Further active alleles of pTα include one that lacks both the major ectodomain and much of the long cytoplasmic tail (which is unique among antigen receptor chains), and another in which the cytoplasmic tail is substituted with the short tail of TCR Cα. Thus, very little of the pTα chain is required for function. These data support a hypothesis that the primary role of pTα is to stabilize the pre-TCR, and that much of the conserved structure of pTα probably plays a critical regulatory role.
Keywords: pre-TCR, thymocyte development, α/β T cells, allelic exlusion, transgenic
Introduction
Most α/β thymocytes develop and mature within the thymus. Progression through this intrathymic differentiation can be defined by the sequential expression of particular cell surface markers. Thus, whereas most mature α/β T cells express either CD4 or CD8 coreceptors, their earliest immature thymic progenitors are CD4−CD8− double negative (DN). The earliest such DN progenitors express high levels of heat stable antigen (HSA) and CD44 (DN subset I), whereafter the cells acquire CD25 (DN II), then lose CD44 (DN III) and subsequently CD25 (DN IV) 1.
Although these subset classifications are purely operational and mask additional heterogeneity within each subset, the onset of TCR β gene rearrangement can be largely attributed to DN II and DN III. Only those thymocytes that succeed in generating a functional TCRβ chain selectively survive through the transition from DN III to DN IV 2 3. As a result of such “β selection,” cells survive, become activated, expand in size, and proliferate rapidly before acquiring CD4 and CD8 4. Such CD4+CD8+ double-positive (DP) cells account for the majority (∼80%) of thymocytes 5.
β selection is mediated by the pre-TCR which consists of—at minimum—the β chain, CD3 components, and a surrogate TCRα chain termed pre-Tα (pTα) 6 7 8. Thymocyte differentiation in mice deficient in components of the pre-TCR, or in signaling molecules downstream of it, are inhibited in their transition across “β selection” 9 10. Therefore, in pTα−/− mice there are very few DP cells, and the total number of thymocytes is usually only 1–10% of normal. Those DP cells that develop are largely driven by the contribution of TCRγ/δ or the precocious expression of TCRα/β 11 12. Furthermore, it has been reported that thymocytes in pTα−/− mice more commonly express two productively rearranged TCRβ chain genes, suggesting a defect in allelic exclusion 13.
By contrast, γ/δ cell differentiation does not depend on pTα, and in pTα−/− mice, γ/δ cell numbers are conspicuously increased. In particular, there occurs a subset of γ/δ cells coexpressing CD4 that is not readily detected in normal mice 14. Such observations are consistent with the proposal that αβ/γδ lineage determination in thymocytes is somewhat flexible, strongly affected by the expression of TCR chains and of the pre-TCR 15 16. In sum, the pre-TCR promotes a maturational program that includes cell survival; cell activation and growth; proliferation; differentiation to DP cells; the arrest of further β chain gene rearrangement; and possibly fate determination. Hence, the mechanism of action of the pre-TCR is of considerable interest.
The pTα genes in mice and humans each give rise to two transcripts 17 18. One, pTαa, encodes a transmembrane protein comprising a single Ig-like extracellular domain of ∼110 amino acids, a transmembrane domain containing two charged amino acids, and a COOH terminal cytoplasmic region of ∼30 amino acids 7. This COOH tail distinguishes pTα from other TCR or Ig chains that lack cytoplasmic regions of anything greater than a few amino acids. The second transcript, pTαb, splices out the second exon encoding the Ig-like ectodomain, and could therefore produce a significantly smaller isoform with very limited capacity to interact with molecules extracellularly 17. One important question is whether both naturally occurring alleles of pTα can promote thymocyte development, or whether the smaller form might be an inactive isoform that might, for example, perform a regulatory role. In this paper, a genetic complementation approach has been used to answer this question. Additionally, this paper examines which regions of the pTα protein are required for biological activity.
Because the pre-TCR is expressed in vivo at very low levels, and because there are few cell lines representative of immature thymocytes, there has been little biochemical characterization of the pre-TCR. Likewise, there has been no identification of a pre-TCR ligand. Instead, information on the active form of the pre-TCR has been gained primarily from genetic or cell biological experiments. Such experiments have provided some seemingly contradictory observations. First, a truncated pTα chain which lacks the highly conserved extracellular Ig-like loop can, together with a truncated TCRβ chain, restore the development of DP cells in mice that are deficient in recombinase activating genes (RAG) 1 or 2, and that as a result cannot rearrange their endogenous TCRβ chain genes 19. This result seems consistent with studies indicating that the pre-TCR can aggregate in the plasma membrane with lck in detergent insoluble glycolipids (DiGs), commonly termed rafts, even in the absence of any overt ligand 20. By this view, the more important components of pTα would appear to be the charged transmembrane domain that facilitates pairing with CD3; a juxtamembrane cysteine in the intracellular tail that may contribute to raft association; and any other components of the cytoplasmic tail that regulate signaling or pre-TCR stability. In particular, there is a proline-rich motif, occurring once in human pTα and repeated in murine pTα that has similarities to protein kinase C substrate sites 7, and to regions in CD2 that mediate signal transduction by binding to CD2BP2 21. Indeed, in somatic cell transfection studies, mutants lacking the proline motifs show differences in properties compared with full length versions (unpublished data). However, other experiments indicate that thymocyte development in pTα−/− mice can be rescued by a pTα allele lacking the bulk of the cytoplasmic tail 14. Combining these data, one might hypothesize that only a very small portion of pTα (∼80 amino acids), lacking much of the ectodomain and its intracellular region, is required for biological activity. This hypothesis is confirmed in this report. The implications of these findings and of the transgenic methodologies commonly used in such studies are discussed.
Materials and Methods
Generation of Transgenic Mice.
cDNAs of the different pTα forms were generated by PCR. The full length pTαa, “p600,” and the second isoform, pTαb, “p300,” have been made previously 17. Truncated forms of pTαa and pTαb, named p600ΔP and p300ΔP, respectively, in which the last cytoplasmic 16 amino acids (containing the two proline rich regions and the two potential PKC phosphorylation sites) were deleted, were generated by PCR using the following primers: for p600ΔP, 5′-AATAGATCTCTACCATCAGGCATCGCT-3′ and 5′-AATCCGCGGCTACTGGAGGTGCTGGCCCGC-3′. For p300ΔP, 5′-AATAGATCTCTACCATCAGGGGAATCT-3′ and 5′-AATCCGCGGCTACTGGAGGTGCTGGCCCGC-3′. The pTαCα construct, in which the connecting peptide, transmembrane region, and cytoplasmic tail of p600 were substituted with those of TCR-Cα, was generated in two fragments using two sets of primers. For the 5′ part of pTαCα, 5′-AATAGATCTCTACCATCAGGCATCGCT-3′ and 5′-AGCACACACCCCCTCCAGCTGTCAGACGTTCCCTGTGATGCCACGTTGACCGAG-3′. For the 3′ part of pTαCα, 5′-CTCGGTCAACGTGGCATCACAGGGAACGTCTGACAGCTGGAGGGGGTGTGTGCT-3′ and 5′-AATCCGCGGTCAACTGGACCACAGCCTCAGCGT-3′. Both PCR products were annealed for 30 min at 45°C, and subsequently amplified using the primers 5′-AATAGATCTCTACCATCAGGCATCGCT-3′ and 5′-AATCCGCGGTCAACTGGACCACAGCCTCAGCGT-3′.
In each case, products were cloned in-frame (via BglII/SacII) into the expression vector pDisplay (Invitrogen). The Igκ leader sequence, HA tag, and cDNA were then subcloned from pDisplay into the BamH1 site of the T cell lineage–specific expression vector, p1017 22 23. A Not1-Not1 fragment from p1017 was then microinjected into C57.BL6 × SJL zygotes, that were implanted into pseudopregnant females. Progeny were screened by PCR and Southern, and transgenic lines crossed to pTα2/−.
Isolation of Lymphocytes.
Intraepithelial lymphocytes were harvested as described 24. Single thymocyte suspensions were obtained by crushing whole thymi between the edges of frosted glass slides into FACS® buffer (1× PBS/2% FCS/0.1% azide). Live cells were determined by trypan blue exclusion.
Flow Cytometry.
Thymocytes or intestinal intraepithelial lymphocytes at ≤2 × 107/ml were stained with the antibodies listed below and analyzed, on either a FACStarPlus™ (Becton Dickinson) or a FACS Vantage™ (Becton Dickinson) flow cytometer. Data were analyzed with CELLQuest™ software. Monoclonal antibody reagents obtained from BD PharMingen were: αCD4-FITC (GK1.5), αCD8α-PE (53–6.7), αCD25-FITC (7D4), αCD44-APC (IM7), αCD44-cy-Chrome (IM7), αHSA-PE (M1/69), αTCR αβ-PE (H57–597), αTCR γδ-PE (GL3), αCD3ε-biotin (145–2C11). Other antibodies and reagents for flow cytometry included avidin Red670 (GIBCO BRL) and rat normal serum (GIBCO BRL).
RNA Isolation, cDNA Synthesis, and Semiquantitative PCR.
RNA isolated using Trizol reagent (GIBCO BRL) was DNase treated (GIBCO BRL) and quantitated by spectrophotometry. AMV reverse transcriptase (Roche) reactions were primed with Pd(N) (Amersham Pharmacia Biotech). Standard reverse transcription (RT)-PCRs using 200 ng thymocyte RNA from pTα−/− transgenic, and nontransgenic littermates used primers as follows: HA-For, 5′-CCA TAT GAT GTT CCA GAT TAT GCT-3′; ΔP1ΔP2-Rev, 5′-CTG GAG GTG CTG GCC CGC-3′; PTA REV, 5′-CTA TGT CCA AAT TCT GTG GGT-3′; HGH REV, 5′-GGA TAT AGG CTT CTT CAA AC-3′.
Immunoprecipitation and Immunoblotting.
For immunoprecipitations, 2 × 107 freshly isolated thymocytes or pTα transfectants 17 were washed in cold PBS buffer, lysed in 300 μl ice-cold lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCI, 1% NP-40, 0.25% Na-deoxycholate, 10 mM NaF, 10 mM Na4P2O7, 1 mM EGTA, 1 mM MgCl2, 1 mM Na3VO4, 1 mM PMSF, 20 μg/ml aprotinin, 20 μg/ml leupeptins, 10 μg/ml Pepstatin A, 20 μg/ml antipain). Lysates were incubated for 20 min on ice before centrifugation at 13,000 rpm for 15 min at 4°C. Postnuclear lysates were agitated for 1 h at 4°C with 2 μl anti-HA monoclonal antibody HA.11 (BabCO). 25 μl protein A-Sepharose beads (Amersham Pharmacia Biotech), swollen and washed in lysis buffer, were added and incubated overnight at 4°C. The beads were washed 3× in cold lysis buffer, and proteins eluted by boiling for 5 min in SDS sample buffer, separated by 15% SDS-PAGE gel, and transferred to nitrocellulose for immunoblotting. The membranes were blocked with 4% nonfat milk in TBS (10 mM Tris-HCl, pH 7.6, 150 mM NaCl) and incubated with HA.11 (1:3,000). Bound Ab was revealed with 1:8,000 diluted horseradish peroxidase (HRP)-conjugated donkey anti–mouse IgG (Jackson ImmunoResearch Laboratories) using Western blot chemiluminescence reagent (NEN Life Science Products).
Allelic Exclusion.
Single CD4+CD8+ double-positive thymocytes were sorted using a MoFlo high speed cell sorter (Cytomation) into a 96-well plate, containing 10 μl of PCR buffer containing proteinase K (250 μg/ml). Plates were incubated at 55°C for 1 h and then the proteinase K was inactivated by heating at 95°C for 15 min. Plates were then stored at −20°C. TCRβ rearrangements were amplified by a seminested two-step PCR using primers as described by Aifantis and colleagues 13 25. Briefly, 40 μl of a mixture containing dNTPs, buffer, 3 pmol of each Vβ, Dβ, and Jβ primer, and 0.1 U of Taq polymerase (QIAGEN) was added to each well. Amplification comprised five cycles in which the denaturing step was 96°C and the annealing temperature decreased from 68–60°C, followed by a further 25 cycles (30 s at 94°C, 1 min at 58°C, 1 min at 72°C) and finally 7 min at 72°C. For the second round of amplification, 1 μl of the first round product was transferred into a fresh tube containing a single 5′ primer in conjunction with the nested Jβ1 or Jβ2 primer (10 pmol of each), dNTPs, buffer, and Taq polymerase (0.1 U) in final volume of 25 μl. Amplification was for 35 cycles following the same procedure as the first round PCR (except denaturing was always at 94°C). Rearrangements were detected by migration of the PCR product on a 1.5% ethidium bromide stained agarose gel and positives purified using QIAGEN purification kits. Direct sequencing of the PCR products was performed using the BigDye Ready Reaction sequencing mix (ABI) and automated sequencing performed on a 96 lane ABI 377 sequencer.
Results
Generation of Transgenic pTα2/− Mice Expressing Different Forms of pTαb.
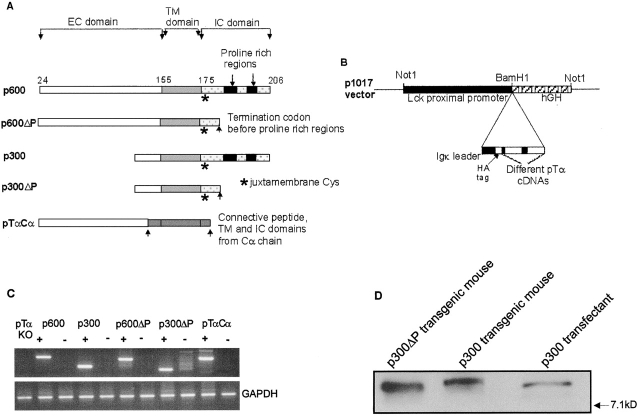
To determine whether the naturally occurring pTαb gene and derivatives thereof could functionally promote thymocyte development, numerous lines of transgenic mice were generated expressing either pTαb (p300) or a truncated form lacking the proline-rich regions and much of the remainder of the cytoplasmic tail (p300ΔP). As a positive control, mice were also generated that expressed pTαa (p600) or a corresponding cytoplasmic tail deletion mutant (p600ΔP). Transgenic mice of the latter type have previously been reported 14. Additionally, transgenic mice were generated that expressed a chimeric cDNA encoding the extracellular region of pTα joined to the connecting peptide, transmembrane, and intracellular regions of the mature Cα gene (pTαCα, Fig. 1 A). In each case, relevant cDNAs were cloned into the p1017 vector, containing the lck proximal promoter to ensure early T lineage expression (22 23; Fig. 1 B). Transgenic lines were established (genotyping not shown) and backcrossed onto the pTα−/− background, generating F2 animals that expressed the different transgenes in the absence of endogenous pTα.
Figure 1.
Schematic representation of different pTα transgenes and their expression. (A) Schematic representation of the five different pTα cDNAs constructed as described in Materials and Methods. (B) Each of the pTα cDNAs was cloned (together with a HA tag and Igκ leader sequence) into the BamH1 site of the p1017 vector. The purified Not1 fragment of p1017 recombinants was microinjected into the pronuclei of fertilized eggs. (C) Reverse transcription (RT)-PCR detection of pTα mRNA expression from thymocytes of the different pTα transgenics. Transgenics positive by genotyping (+) gave a positive mRNA expression when compared with those that were negative by genotyping (−). GAPDH was positive in all samples. (D) The expression of p300 and p300ΔP protein in thymocytes from transgenic mice compared with protein expression in a p300 transfected cell line. 7.1 kD denotes the migration of a protein size marker on the same gel.
In every case, pTα−/− mice that genotyped positively for a transgene showed transgene RNA expression in the thymus (Fig. 1 C). As the emphasis of this manuscript is on the biological activity of pTαb, the expression of wild-type and mutant forms of pTαb protein was additionally examined (Fig. 1 D). Levels detected by Western analysis of HA-tagged protein in thymocyte lysates were comparable to those in a pTαb-transfected cell line 17 26. Not surprisingly, both RNA and protein expression varied markedly among different founder transgenics, limiting the degree to which one should cross-compare phenotypes of transgenic mice generated with different pTα alleles.
pTαb Transgenes Induce DP Cell Representation.
The surface phenotypes of thymocytes in the pTα transgenic mice were analyzed by flow cytometry. The majority of pTα−/− thymocytes are blocked as DN cells (Fig. 2 A). Conversely, pTαb, as well as the smallest transgene, p300ΔP, could completely restore DP thymocyte representation (DP cells are 84% of the total thymocyte number in 300ΔP mice, compared with 7% in pTα2/− mice; Fig. 2 A). The lack of any essential functional requirement for the pTα cytoplasmic tail was confirmed by the restored phenotypes of pTα-Cα, and p600ΔP transgenic mice (Fig. 2 B). In all, substantive restoration of DP differentiation was shown by at least one line of transgenic mice generated with each of the alleles (p300, p300ΔP, p600, p600ΔP, and pTα-Cα; Fig. 2 B).
Figure 2.

Restoration of phenotype by expression of pTα transgenes. (A) CD4 versus CD8 expression on thymocytes from pTα−/− (left) and p300ΔP.pTα−/− (right). The percentage of thymocytes in each quadrant are shown. (B) The percentage of DP (i.e., CD4+CD8+) cells in the thymi of mice expressing different pTα transgenes. Individual vertical lines represent independent lines of mice; individual data points on those vertical lines represent the percentage of DP cells in the thymi of individual mice. (C and D) The percentage of DN γ/δ cells (C) or CD4+ γ/δ cells (D) in the thymi of mice expressing different pTα transgenes. Individual vertical lines represent independent lines of mice; individual data points on those vertical lines represent the percentage of γ/δ cells in the thymi of individual mice. (E) Total cellularity of the thymus (×106 cells) was established by trypan blue exclusion. Individual vertical lines represent independent lines of mice; individual data points on those vertical lines represent the cell numbers in individual mice.
pTαb Transgenes Reduce γ/δ Cell Representation in the Thymus and Gut.
Normally, thymic γ/δ cells compose ∼1% of thymocytes. These levels are increased to 5–20% of thymocytes in pTα−/− mice, and comprise both DN cells and CD4+ γ/δ cells, a subset rarely detected in wild-type thymi (Fig. 2C and Fig. D). Conversely, the conventional, low level of γ/δ cell representation (both DN and CD4+) was restored in transgenic pTα2/− mice expressing the full length or the truncated forms of pTαb (Fig. 2C and Fig. D). Indeed, the normal phenotype was restored in at least one line of transgenic mice generated with each of the five pTα alleles under study (Fig. 2C and Fig. D). In all cases the influence of pre-TCR expression over the percentages of γ/δ cells correlated reasonably well with the absolute numbers of γ/δ cells (Table ), indicating that changes in the percentages of γ/δ cells are real changes and not simply an indirect effect of changes in other cell subsets, e.g., DP thymocytes.
Table 1.
Percentage of γ/δ Cells Correlates Well with Absolute Cell Numbers
| Transgenic mouse | Mean percentage of total γ/δ cells | Mean DN γ/δ cells (×105) | Mean CD4+ γ/δ cells (×104) |
|---|---|---|---|
| Wild type | 0.64 | 1.2 | 4.8 |
| p300ΔP | 0.79 | 1.4 | 5.4 |
| p300 | 1.11 | 2.2 | 2.2 |
| p300 | 12 | 13 | 19 |
| pTα2/− | 16 | 12 | 21 |
The mean percentage of total γ/δ cells is compared with the absolute numbers of DN (×105) or CD4+ (×104) γ/δ cells in wild-type mice, pTα2/− mice, or in pTα2/− mice expressing either p300 or p300ΔP transgenes that did or did not recover normal phenotypes.
Consistent with the block in α/β T cell development in pTα−/− mice, the intestinal intraepithelial lymphocytes (IELs) are depleted of α/β T cells, and instead comprise ∼90% γ/δ cells (Table ). Because the representation of γ/δ cells in the pTα−/− thymus was significantly suppressed by the expression of the small p300ΔP construct, the question arose as to whether γ/δ cells would be similarly reduced to normal levels in the gut. This was indeed the case: IELs from pTα−/− mice expressing the p300ΔP transgene included fewer γ/δ cells than the pTα−/− gut, with a representation (∼50% of CD3+ IELs) that was comparable to normal (Table ).
Table 2.
p300ΔP Transgene Expression Recovers α/β IELs
| Wild type | p300ΔP | pTα−/− | |
|---|---|---|---|
| Recovery (×106) | 6.8 | 14 | 13.1 |
| Percent α/β cells | 29.82 | 36.4 | 1.31 |
| Percent γ/δ cells | 54.49 | 51.63 | 86.8 |
| Percent CD8α | 60 | 48.5 | 60.86 |
| Percent CD8β | 14.2 | 30.3 | 1.93 |
| Percent CD4 | 6.15 | 1.65 | 0.88 |
IELs were collected from wild-type, pTα−/−, and pTα−/− mice expressing the p300ΔP transgene. Cells were stained with the antibodies indicated and the data represent the percentage of cells (mean of two mice in each case) expressing the particular markers.
Hypocellularity of the pTα2/− Thymus Is Rescued.
The thymi of 6–8-wk-old wild-type mice contain an average of 1.2 × 108 thymocytes, with a range of ∼0.9–1.9 × 108 cells. pTα−/− mice contain 10–50-fold fewer thymocytes. Two transgenic lines, one expressing p300 (pTαb) and one expressing p300ΔP showed rescue of an approximately normal range (Fig. 2 E). In other lines, thymic cellularity was often not well rescued even where there was substantial and parallel restoration of normal DP and TCRγ/δ1 thymocyte phenotypes (Fig. 3). However, this does not reflect a failure of pTα transgenes to regulate cellularity because there is an additional effect whereby transgenes expressing either TCR chains or pre-TCR chains commonly reduce thymus cellularity, even on a wild-type background (unpublished data). As an example, two p600ΔP mice on a pTα+/+ background contain 4.8 × 107 and 6.2 × 107 thymocytes, respectively, while one p300ΔP pTα+/− strain contained 6.6 × 107 cells. The fact that these cell numbers, albeit lower than normal, are comparable in transgenic mice on a pTα−/− or a pTα+ background indicates that each of the trangenes has overcome the negative influence of pTα deficiency on thymocyte numbers (see Discussion).
Figure 3.
Schematic comparison of different transgenic lines. Two parameters of thymocyte phenotype (DP or γ/δ cell numbers) segregate together in different transgenic lines, but segregate variably with thymus size. Gradation of shading represents movement away from wild-type phenotype (see key).
Allelic Exclusion Occurs in p300ΔP Mice.
One component of normal thymocyte development is allelic exclusion. To test whether this occurs in the DP thymocyte compartment that is restored in mice expressing the smallest pTα transgene, p300ΔP, TCRβ rearrangements were examined by single cell PCR. Multiple primers were used that amplify many (but not all) Vβ segments, and that distinguish rearrangements of V or D to Jβ1 or Jβ2. Products were obtained from all cells analyzed. Conspicuously, the biased Vβ usage seen in immature thymocytes of normal mice (e.g., preferential usage of Vβ8) was also seen in the p300ΔP pTα−/− mice 27.
Many cells gave more than two PCR products but when these rearrangements were analyzed they corresponded to DNA excision loops formed alongside correct chromosomal rearrangements. 25% of cells exhibited a single in-frame VDJ rearrangement together with a DJ rearrangement. A further 1/3 of cells showed only one rearrangement which was an in-frame VDJ junction. The remaining cells showed one in-frame VDJ rearrangement together with a nonproductive VDJ rearrangement (Table ). No cell tested produced more than one in frame VDJ rearrangement, indicating that allelic exclusion operates in the p300ΔP transgenic mouse.
Table 3.
Allelic Exclusion Occurs in the p300ΔP Transgenic Line
| Allelic exlusion | Allelic inclusion | |||
|---|---|---|---|---|
| 1VDJ+ | VDJ+/VDJ− | VDJ+/DJ | VDJ+/VDJ+ | |
| 3 | 3 | 2 | 0 | |
| p300DP | Vβ8.1Jβ2.3 | Vβ8.2Jβ2.3 | Vβ8.3Jβ1.1 Vβ2Jβ1.5 | |
| Vβ8.2Jβ2.1 | Vβ12Jβ2.6 | |||
| Vβ9Jβ2.6 | Vβ9Jβ1.5 | |||
Allelic exclusion was assessed in single DP cells taken from thymi of pTα−/− mice expressing the p300ΔP transgene as described in Materials and Methods. Of the eight single cells tested, none expressed more than one correctly rearranged Vβ chain. The productive VDJ rearrangement of the Vβ chain is shown.
Discussion
This study has investigated whether the second naturally occurring pTα transcript, pTαb encodes a biologically active pTα isoform. It has also asked whether thymocyte development can be sustained by an even smaller pTαb allele that lacks both the major Ig-like ectodomain and the bulk of the cytoplasmic tail. Finally, it has asked whether thymocyte development can be functionally sustained by a pTα allele in which the cytoplasmic tail has been exchanged for the short tail of Cα. The measurement of biological activity was the functional complementation of the pTα−/− mouse. The four primary defects reported in pTα−/− mice are: the paucity of DP thymocyte differentiation (correlated with a relative increase in the percentage of DN thymocytes); increases in TCRγ/δ1 cells of both DN and CD4+ phenotypes; the failure of allelic exclusion at the TCRβ locus; and decreases in thymocyte numbers. Both the full length and truncated alleles of pTαb as well as the pTα-Cα allele showed the capacity to largely or fully restore these phenotypes, although allelic exclusion was assessed in only one of the strains (p300ΔP). Thus, highly truncated forms of pTα are biologically active, including the naturally occurring form, pTαb that is conserved in humans and mice 17 18. Interestingly, a targeted mutation of the pTα locus that would be predicted to leave intact the coding potential of pTαb showed a very mild phenotype in terms of altered thymocyte differentiation, consistent with the idea that pTαb is biologically active in vivo 10.
There is some contention over which of the four primary thymocyte defects reported in pTα−/− mice reflect direct targets of pTα. More than one of these events may be directly downstream of pTα, but some may have greater dependence on pTα function than do others. Data presented here show that all four parameters are rescued in at least one line expressing either the full length or the heavily truncated pTα alleles. Nonetheless, in several mice in which thymocytes could be classified into largely normal subset distributions, cellularity was not always normal (Fig. 3). Similar variability in thymus cellularity is evident when data from other mice transgenic for pre-TCR components are considered 14 19. At minimum, these data demonstrate that appropriate thymocyte differentiation can occur independent of extensive proliferation. A similar situation appears to characterize the IL-7−/− mouse 28.
The lack of normal cellularity seems in part due to an inhibitory effect of the expression of pTα transgenes, as a similar reduction, relative to normal, was seen in the transgenic mice on a pTα+ background. Similar observations have been made in mice expressing TCR transgenes. This may reflect the inappropriate prolonged expression of the pre-TCR that is a variable characteristic of transgenic mice. It is known that constitutively activated lck provokes loss of DP cells 29 possibly because the cells interpret continued signaling as a negative selection stimulus. Sustained expression of the pre-TCR may do likewise. Ordinarily, the pre-TCR is expressed at very low levels, and may be easily displaced by TCRα/β after TCRα gene rearrangement. In transgenic mice, the physiologic expression of pTα will not be precisely mimicked because of the heterologous promoter elements, and because of integration sites that will vary from founder to founder. Additionally, the displacement of pTα from TCRβ will likely depend on active signaling mechanisms that regulate the stability and intracellular localization of the pTα protein 30. These mechanisms may not function properly in the context of mutant pTα transgenes. For these various reasons, the transgenic mice may harbor a situation similar to that reported in lck transgenic mice. These are significant qualifications that must be applied to the interpretation of such transgenic studies.
The simplest explanation for the biological activity of very small forms of pTα is that pTα functions merely to stabilize the β chain (aiding interaction with CD3 and other downstream signaling molecules), and that a minimal peptide of pTα is sufficient to accomplish this. This is consistent with other instances of expression of truncated pTα or TCRβ alleles. For example, mice transgenic for a TCRβ chain lacking the Vβ region showed appropriate DN to DP transition 31. The biological activity of truncated versions of pTα is consistent with evidence that, independent of any ligand, the pre-TCR spontaneously clusters and associates with signaling molecules such as p56lck, CD3 molecules, and Zap-70 via sequestration in lipid rafts 20.
Presumably all active forms of pTα will elicit signaling to nuclear factor (NF)-κB implicated in promoting cell survival 26, and Vav-1 and Rac-1 32 33 implicated in modulating actin dynamics that are important in spatially orienting signaling molecules to coordinate and sustain signal transduction (for a review, see reference 34). Indeed, electrophoretic mobility shift assays indicate high levels of NF-κB activity in DN thymocytes from pTα transgenic mice (unpublished data). Nonetheless, it may be that signaling from the physiologic pre-TCR; signaling from a pre-TCR containing the pTα-Cα chimeric molecule, and signaling in the absence of the pre-TCR but via cross-linking CD3, each activate the same signaling molecules, but by different means. For example, the palmitoylation of the juxtamembranous Cys residue present in pTα that is implicated in raft association, cannot obviously occur in the pTα−Cα chimeric protein that lacks the juxtamembrane cysteine. Yet, both of the pTα−Cα transgenic lines showed comparable restoration of thymocyte phenotypes, and were not obviously less effective than the other transgenes. Possibly, the pTα−Cα protein facilitates signal transduction largely independent of raft association, because the connecting peptide of Cα (as opposed to the equivalent domain in pTα) has a much stronger association with CD3ζ 35. This would be consistent with the hypothesis that regulated activation of common downstream signaling pathways is the rate determining step to β-selection, and can be achieved by distinct, albeit related mechanisms.
If only a very small part of pTα is required for its function, the question arises as to why pTα shows conservation of its structure. One explanation is that the biological activity of the pre-TCR is so potent that many of its structural features are essential for downregulation, ensuring that the pre-TCR is expressed only at appropriate levels only during the appropriate time window. In addition to the low level and restricted time frame of pre-TCR expression, several other experiments are suggestive of this. For example, sustained expression of a transgenic TCRβ allele that lacks the V region led to thymic lymphomas 36. Although such lymphomas did not characterize any of the several pTα transgenic mice reported here, this may be because ectopically expressed pTα can be displaced, albeit inefficiently, by TCRα, whereas a mutant TCRβ chain would not be, thus sustaining ligand-independent signaling. Likewise, severe lymphomas with some characteristics of pre-T cells (including sustained pTα expression) developed with >80% penetrance in several lines of mice transgenic for an activated form of Notch 3, the regulated expression of which normally characterizes the β-selection stage (for a review by Rothenberg, see reference 37). In striking illustration of the potency of sustained pre-TCR signaling, the development of Notch 3–induced tumors is dependent on pTα expression (unpublished data).
According to this view, the pre-TCR is a potent agent of cell growth and survival that must be downregulated after β-selection. Therefore, there are likely to be active signaling components of the pre-TCR pathway, and possibly an as yet unidentified ligand that may target regions of pTα in order to negatively regulate its expression and activity. This would lead to conservation of those regions of pTα. This remains to be tested biochemically, although the recent report that the pTα tail can serve as an endoplasmic reticulum retention signal is consistent with this outlook 38. Studies of pre-B cell receptor signaling have indicated that the expression level of pre antigen receptors is a product both of the structure of pre-antigen receptor chains and the cell biological characteristics unique to the immature cells in which the pre-antigen receptors are expressed 39.
Acknowledgments
Grants from the Wellcome Trust, the Charitable Foundation of Guy's and St. Thomas' Hospitals, and the National Institutes of Health (GM37759) (A.C. Hayday), and the MSTP program [N.C. Douglas]. Support provided (D.F. Barber) by Spanish government fellowship (Ministerio de Educacion y Ciencia).
Footnotes
D. Gibbons, N.C. Douglas, and D.F. Barber contributed equally to this work.
D.F. Barber's present address is Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD 20852.
L. Geng's present address is Dana Farber Cancer Institute, Boston, MA 02115.
Q. Liu's present address is Brigham and Women's Hospital, Boston, MA 02115.
References
- Godfrey D.I., Kennedy J., Suda T., Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Mallick C., Dudley E., Viney J., Owen M., Hayday A. Rearrangement and diversity of T cell receptor beta chain genes in thymocytesa critical role for the beta chain in development. Cell. 1993;73:513–519. doi: 10.1016/0092-8674(93)90138-g. [DOI] [PubMed] [Google Scholar]
- Hoffman E., Passoni L., Crompton T., Leu T., Schatz D., Koff A., Owen M.J., Hayday A.C. Productive T cell receptor beta chain gene rearrangementclose interplay of cell cycle and cell differentiation during T cell development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., von Boehmer H. Early alpha beta T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 1997;2:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Ungewiss K., Azogui O., Palacios R., Owen M.J., Hayday A., von Boehmer H. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33kD glycoprotein. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- Saint-Ruf C., Ungewiss K., Groettrup M., Bruno L., Fehling H.J., von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science. 1994;266:1208–1212. doi: 10.1126/science.7973703. [DOI] [PubMed] [Google Scholar]
- Del Porto P., Bruno L., Mattei M.-G., von Boehmer H., Saint-Ruf C. Cloning and comparative analysis of the human pre-T-cell receptor alpha-chain gene. Immunology. 1995;92:12105–12109. doi: 10.1073/pnas.92.26.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova A., Saint-Ruf C., von Boehmer H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- Xu Y., Davidson L., Alt F.W., Baltimore D. Function of the pre-T-cell receptor alpha chain in T-cell development and allelic exclusion at the T-cell receptor beta locus. Proc. Natl. Acad. Sci. USA. 1996;93:2169–2173. doi: 10.1073/pnas.93.5.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passoni L., Hoffman E.S., Kim S., Crompton T., Pao W., Dong M., Owen M.J., Hayday A.C. Intrathymic δ selection events in γδ cell development. Immunity. 1997;7:83–95. doi: 10.1016/s1074-7613(00)80512-9. [DOI] [PubMed] [Google Scholar]
- Buer J., Aifantis I., DiSanto J.P., Fehling H.J., von Boehmer H. Role of different T cell receptors in the development of pre-T cells. J. Exp. Med. 1997;185:1541–1547. doi: 10.1084/jem.185.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aifantis I., Buer J., von Boehmer H., Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 1997;7:601–607. doi: 10.1016/s1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Iritani B.M., Krotkova A., Forbush K.A., Laplace C., Perlmutter R.M., von Boehmer H. Restoration of thymopoiesis in pT alpha−/− mice by anti-CD3ε antibody treatment or with transgenes encoding activated Lck or tailless pT alpha. Immunity. 1997;6:703–714. doi: 10.1016/s1074-7613(00)80446-x. [DOI] [PubMed] [Google Scholar]
- Dudley E.C., Girardi M., Owen M.J., Hayday A.C. Alpha/beta and gamma/delta T cells can share a late common precursor. Curr. Biol. 1995;5:659–669. doi: 10.1016/s0960-9822(95)00131-x. [DOI] [PubMed] [Google Scholar]
- Bruno L., Sheffold A., Radbruch A., Owen M.J. Threshold of pre-T-cell-receptor surface expression is associated with alpha beta T-cell lineage commitment. Curr. Biol. 1999;9:559–568. doi: 10.1016/s0960-9822(99)80259-0. [DOI] [PubMed] [Google Scholar]
- Barber D.F., Passoni L., Wen L., Geng L., Hayday A.C. The expression in vivo of a second isoform of pT alphaimplications for the mechanism of pT alpha action. J. Immunol. 1998;161:11–16. [PubMed] [Google Scholar]
- Saint-Ruf C., Lechner O., Feinberg J., von Boehmer H. Genomic structure of the human pre-T cell receptor alpha chain and expression of two mRNA isoforms. Eur. J. Immunol. 1998;28:3824–3831. doi: 10.1002/(SICI)1521-4141(199811)28:11<3824::AID-IMMU3824>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Irving B., Alt F., Killeen N. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 1998;280:905–908. doi: 10.1126/science.280.5365.905. [DOI] [PubMed] [Google Scholar]
- Saint-Ruf C., Panigada M., Azogui O., Debey P., von Boehmer H., Grassi F. Different initiation of pre-TCR and gamma-delta TCR signalling. Nature. 2000;406:521–527. doi: 10.1038/35020093. [DOI] [PubMed] [Google Scholar]
- Nishizawa K., Freund C., Li J., Wagner G., Reinherz E.L. Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc. Natl. Acad. Sci. USA. 1998;95:1497–1902. doi: 10.1073/pnas.95.25.14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin K., Beals C., Wilkie T., Forbush K., Simon M., Perlmutter R. Dissection of thymocyte signalling pathways by in vivo expression of pertussis toxin ADP-ribosyltransferase. EMBO. J. 1990;12:3821–3829. doi: 10.1002/j.1460-2075.1990.tb07600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin A., Abraham K., Forbush K., Farr A., Davison B., Perlmutter R. Disruption of thymocyte development and lymphomagenesis is induced by SV40 T-antigen. Int. Immunol. 1990;2:173–180. doi: 10.1093/intimm/2.2.173. [DOI] [PubMed] [Google Scholar]
- Davies M.D., Parrott D. Preparation and purification of lymphocytes from the epithelium and lamina propria of murine small intestine. Gut. 1981;6:481–488. doi: 10.1136/gut.22.6.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aifantis I., Pivniouk V.I., Cartner F., Feinber J., Swat W., Alt F.W., von Boehmer H., Geha R.S. Allelic exclusion of the T cell receptor beta locus requires the SH2 domain-containing leukocyte protein (SLP)-76 adaptor protein. J. Exp. Med. 1999;190:1093–1102. doi: 10.1084/jem.190.8.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll R.E., Jimi E., Phillips R.J., Barber D.F., Rincon M., Hayday A.C., Flavell R.A., Ghosh S. NF-κB activation by the pre-T cell receptor serves as a selective survival signal in T-lymphocyte development. Immunity. 2000;13:677–689. doi: 10.1016/s1074-7613(00)00067-4. [DOI] [PubMed] [Google Scholar]
- Wilson A., Marechal C., MacDonald H.R. Biased V beta usage in immature thymocytes is independent of DJ beta proximity and pT alpha pairing. J. Immunol. 2001;166:51–57. doi: 10.4049/jimmunol.166.1.51. [DOI] [PubMed] [Google Scholar]
- von-Freeden-Jeffry U., Vieira P., Lucian L.A., McNeil T., Burdach S.E., Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a non redundant cytokine. J. Exp. Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham K., Levin S., Marth J., Forbush K., Perlmutter R. Delayed thymocyte development induced by augmented expression of p56lck. J. Exp. Med. 1991;173:1421–1432. doi: 10.1084/jem.173.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trop S., Rhodes M., Wiest D.L., Hugo P., Zuniga-Pflucker J.C. Competitive displacement of pT-alpha by TCR-alpha during TCR assembly prevents surface coexpression of pre-TCR and alpha-beta TCR. J. Immunol. 2000;165:5566–5572. doi: 10.4049/jimmunol.165.10.5566. [DOI] [PubMed] [Google Scholar]
- Ossendorp F., Jacobs H., van der Horst G., de Vries E., Berns A., Borst J. T cell receptor-alpha beta lacking the beta-chain V domain can be expressed at the cell surface but prohibits T cell maturation. J. Immunol. 1992;148:3714–3722. [PubMed] [Google Scholar]
- Turner M., Mee J., Walters A.E., Quinn M.E., Mellor A.L., Zamoyska R., Tybulewicz V.L.J. A requirement for the Rho-family GTP exchange factor Vav in positive and negative selection of thymocytes. Immunity. 1997;7:451–460. doi: 10.1016/s1074-7613(00)80367-2. [DOI] [PubMed] [Google Scholar]
- Gomez M., Tybulewicz V., Cantrell D.A. Control of pre-T cell proliferation and differentiation by the GTPase Rac-1. Nat. Immunol. 2000;1:348–352. doi: 10.1038/79808. [DOI] [PubMed] [Google Scholar]
- Acuto O., Cantrell D. T cell activation and the cytoskeleton. Annu. Rev. Immunol. 2000;18:165–184. doi: 10.1146/annurev.immunol.18.1.165. [DOI] [PubMed] [Google Scholar]
- Trop S., Steff A.M., Denis F., Wiest D.L., Hugo P. The connecting peptide domain of pT alpha dictates weak association of the pre-T cell receptor with the TCR zeta subunit. Eur. J. Immunol. 1999;29:2187–2196. doi: 10.1002/(SICI)1521-4141(199907)29:07<2187::AID-IMMU2187>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Jacobs H., Ossendorp F., de Vries E., Ungewiss K., von Boehmer H., Borst J., Berns A. Oncogenic potential of a pre-T cell receptor lacking the TCR beta V domain. Oncogene. 1996;12:2089–2099. [PubMed] [Google Scholar]
- Rothenberg E.V. Notchless T cell maturation? Nat. Immunol. 2001;2:189–290. doi: 10.1038/85231. [DOI] [PubMed] [Google Scholar]
- Carrasco Y., Ramiro A., Trigueros C., de Yebenes V., Garcia-Peydro M., Toribio M. An endoplasmic reticulum retention function for the cytoplasmic tail of the human pre-T cell receptor (TCR) {alpha} chainpotential role in the regulation of cell surface pre-TCR expression levels. J. Exp. Med. 2001;193:1045–1058. doi: 10.1084/jem.193.9.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns G., de Vries E., Neefjes J., Borst J. Assembled pre-B cell receptor complexes are retained in the endoplasmic reticulum by a mechanism that is not selective for the pseudo-light chain. J. Biol. Chem. 1996;271:19272–19278. doi: 10.1074/jbc.271.32.19272. [DOI] [PubMed] [Google Scholar]