Abstract
OA1 (GPR143) is a pigment cell-specific intracellular glycoprotein consisting of 404 amino acid residues that is mutated in patients with Ocular Albinism Type 1, the most common form of ocular albinism. While its cellular localization is suggested to be endolysosomal and melanosomal, the physiological function of OA1 is currently unclear. Recent reports predicted that OA1 functions as a G protein coupled receptor (GPCR) based on its weak amino acid sequence similarity to known GPCRs, and on demonstration of GPCR activity in OA1 mislocalized to the plasma membrane. Because mislocalization of proteins is often caused by or induces defects in their proper folding/assembly, the significance of these studies remains unclear. A characteristic feature of GPCRs is a seven transmembrane domain structure. We analyzed the membrane topology of OA1 properly localized to intracellular lysosomal organelles in COS-1 cells. To accomplish this analysis, we established experimental conditions that allowed selective permeabilization of the plasma membrane while leaving endolysosomal membranes intact. Domains were mapped by the insertion of a hemagglutinin (HA) tag into the predicted cytosolic/luminal regions of OA1 molecule and the accessibility of tag to HA antibody was determined by immunofluorescence. HA-tagged lysosome associated membrane protein 1 (LAMP1), a type I membrane protein, was employed as a reporter for selective permeabilization of the plasma membrane. Our results show experimentally that the C-terminus of OA1 is directed to the cytoplasm and that the protein spans the intracellular membrane 7 times. Thus, OA1, properly localized intracellularly, is a 7 transmembrane domain integral membrane protein consistent with its putative role as an intracellular GPCR.
Keywords: Membrane topology, Lysosome, Cellular localization, Ocular albinism type 1, Melanosome, G protein coupled receptor, Selective membrane permeabilization
1. Introduction
Ocular albinism type 1 (OA1) is the most common form of ocular albinism (van Dorp DB., 1987; Shen et al., 2001b). OA1 is inherited in an X-linked recessive fashion and is characterized by a severe reduction of visual acuity, refractive errors, nystagmus, iris translucency, fundus hypopigmentation, foveal hypoplasia, and loss of stereoscopic vision due to misrouting of the optic fibers at the optic chiasm (Charles et al., 1993; Creel et al., 1990; Kriss et al., 1992; King et al., 1995). Microscopic examination of eye and skin tissue from OA1 patients reveals the presence of macromelanosomes or melanin macroglobules (MMGs) (Garner et al., 1980; Wong et al., 1983).
The human OA1 gene was identified by positional cloning (Bassi et al., 1995). The OA1 gene product encodes a pigment cell-specific glycoprotein consisting of 404 amino acid residues. While its cellular localization is suggested to be endolysosomal and melanosomal, the physiological function is currently unknown (Schiaffino et al., 1996; Samaraweera et al., 2001). While early analysis suggested that the protein had 6 transmembrane domains (Newton et al., 1996), homology analysis indicates a weak similarity to the G protein coupled receptor superfamily (GPCR; type A, B, and E) (Schiaffino et al., 1999; Schiaffino and Tacchetti, 2005). When expressed in non-melanocytic cells such as COS-1, the mouse ortholog Oa1 displays a vesicular distribution and colocalizes with the late endosomal/lysosomal markers, LAMP2 and CD63 (Shen et al., 2001a).
Recent results obtained with several experimental systems suggest that OA1 can function as a GPCR (Schiaffino et al., 1999; Staleva and Orlow, 2006; Innamorati et al., 2006). A study employing Saccharomyces cerevisiae as a model system (Staleva and Orlow, 2006) revealed that 1) mouse Oa1 localizes to the prevacuolar compartment (functionally equivalent to the mammalian late endosome), 2) Oa1 can function as a GPCR in a yeast-based GPCR signaling assay, and 3) candidate ligands for Oa1 exist in melanocyte extracts. Studies involving purposeful mislocalization of human OA1 to the plasma membrane in mammalian cells (Schiaffinoet et al., 1999; Innamorati et al., 2006) indicate that the mislocalized OA1 can activate heterotrimeric G proteins and may be associated with arrestin. Both experimental systems suggest that OA1 functions as a GPCR in a similar fashion to GPCRs known to be localized at the plasma membrane.
Given that OA1 is a GPCR, it is expected to span integral membranes 7 times, with its C-terminus oriented to the cytoplasm. However, OA1 was originally predicted to have 6 transmembrane (6TM) regions (Bassi et al., 1995) based on Kyte-Doolittle plots. The SOSUI algorithm (Hirokawa et al., 1998.), which classifies integral membrane proteins, also predicts 6TM regions for OA1. However, biochemical studies regarding the mechanism of insertion of transmembrane domains into the lipid bilayer have shown that hydrophobicity is not the unique crucial factor for determining the membrane topology of integral membrane proteins (Ota et al., 1998a; Ota et al., 1998b).
Employing Saccharomyces cerevisiae as a model system, Staleva and Orlow demonstrated the existence of candidate ligands for OA1 in melanocytes (Staleva and Orlow, 2006). However a specific ligand has not yet been identified. If OA1 is truly an intracellular 7TM GPCR, then its ligand should exist with the lumen of intracellular organelles such as endolysosomes or melanosomes. As a prelude to embarking upon a major search for ligands, we deemed it critical to establish the membrane topology of intracellular OA1.
In addition, two separate dileucine motifs that purportedly target the OA1 molecule to lysosomal organelles impinge upon one of the predicted TM regions suggesting two possible orientations for OA1. To definitively establish the transmembrane orientation of OA1, we developed methodologies allowing selective permeabilization of plasma membrane without disruption of intracellular endolysosomal membranes. We adapted a technique involving cold osmotic shock to selectively permeabilize the plasma membrane (Eckhart et al., 1999) of cells and then determined the orientation of an HA-tag inserted at various points in the predicted hydrophilic domains of the OA1 molecule. Our study demonstrates that OA1 has 7TM regions and that its C-terminus is found in the cytosol, consistent with the concept that OA1 may function as an intracellular GPCR localized to endolysosomal organelles.
2. Materials and methods
Antibodies
A BD Living Colors A.v. peptide antibody, directed against GFP (rabbit polyclonal), was obtained from BD Biosciences (Palo Alto, CA). Monoclonal antibody 16B12, directed against the hemagglutinin (HA) epitope YPYDVPDYA, was obtained from COVANCE (Richmond, CA), and monoclonal antibody H5C6, directed against CD63, was obtained from Developmental Studies Hybridoma Bank (Ames, IA). HRP-conjugated goat anti-rabbit IgG was from BIO-RAD (Hercules, CA), and Texas Red conjugated goat antimouse IgG from Jackson ImmunoResearch (West Grove, CA). cDNA constructs
OA1 cDNA cloned into the EcoRI-NotI site of pCDNA 3.2/V5–DEST (invitrogen, CA), was donated by Dr. Christopher E. Touloukian (Indiana University, Indianapolis, IN). pMS402 and pMS411 are bacterial expression vectors encoding the OA1 gene. pMS420 is a mammalian expression vector carrying the OA1 gene, and pMS430 contains the OA1 gene fused to the N-terminal region of GFP on a mammalian expression vector. For the construction of pMS402, a 1.2-kilobase pair BglII-EcoRI fragment containing OA1 gene was excised from the Touloukian plasmid described above and cloned into the BamHI-EcoRI site of pBluescriptII-SK(−) (Stratagene, La Jolla, CA). The site generated by the ligation between BglII site from OA1 gene fragment and BamHI site from pBluescriptII-SK(−) (GGATCT) was mutated to a BglII site (AGATCT). For the construction of pMS420, the 1.2-kilobase pair BglII-EcoRI fragment from pMS411 containing the OA1 gene was cloned into BglII-EcoRI site of pAcGFP-N1 (BD Bioscience). Similarly, pMS421, pMS422, pMS423, pMS424, pMS425, pMS426, pMS427, and pMS428 were constructed by cloning the corresponding BglII-EcoRI fragment from pMS412, pMS413, pMS414, pMS415, pMS416, pMS417, pMS418, and pMS419, respectively, into the BglII-EcoRI site of pAcGFP-N1. pMS412, pMS413, pMS414, pMS415, pMS416, pMS417, pMS418, and pMS419 were constructed as indicated below in “Site-directed Mutagenesis”. For the construction of pMS430, the stop codon (TGA) at the C-terminus of the OA1 gene on pMS420 was mutated to AGA. As to pMS431, pMS432, pMS433, pMS434, pMS435, pMS436, pMS47, and pMS438, their stop codons (TGA) at the C-terminus of OA1 gene were mutated as well.
pMS445 carries the LAMP1 (lysosome associated membrane protein 1) gene on pBluescriptII-SK(−). For its construction, a 1.5-kilobase pair XhoI-BamHI fragment containing the LAMP1 gene was excised from pSLVhL1 (Williams and Fukuda., 1990), which was provided by Dr. Minoru Fukuda (Cancer Research Center, La Jolla CA), and cloned into the XhoI-BamHI site of pBluescriptII-SK(−). pMS448 and pMS453 carries the LAMP1-HA gene and HA-LAMP1, respectively, on pBluescriptII-SK(−). pMS448 and pMS453 were constructed as indicated below in “Site-directed Mutagenesis”. For the construction of pMS449 and pMS454, 1.5-kilobase pair XhoI-BamHI fragment were excised from pMS448 and pMS453, respectively, and cloned into the corresponding site of pAcGFP-N1.
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). HA epitope tags encoding a 9 amino acid sequence (YPYDVPDYA) were introduced into pMS411 by site directed mutagenesis. The positions of HA epitope insertions into OA1 and LAMP1, and the synthetic oligonucleotides used as mutagenic primers are summarized with the generated plasmids’ names in Table I. pMS412, pMS413, pMS414, pMS415, pMS416, pMS417, and pMS419 were constructed by Quick Change site directed mutagenesis using the primers shown in Table I and their complimentary oligomer DNA. In terms of pMS418, a 0.39-kilobase pair EcoRV-EcoRI fragment containing the C-terminal portion of OA1 coding region was amplified by the primer shown in Table I and replaced the corresponding site of pMS411. For the construction of pMS448, the 1.5-kilobase pair XhoI-BamHI fragment, amplified by the primer shown in Table. I and T7 primer, were inserted into pBluescriptII-SK(−).
Table I.
Oligonucleotides used for HA-tag insertion

|
In terms of pMS453, as an initial step, the HindIII site on the multicloning site of pMS445 (pBluescriptII-SK(−) part) was abolished by treating pMS445 with HindIII and Klenow enzyme successively, and self-ligating the linearized plasmid. In the next step, the HindIII site was introduced into the LAMP1 gene on the generated pMS450 by site-directed mutagenesis, which caused a Methionine to Serine mutation on Met 30, using the primers: 5′GCATTGTGCGTCAGCAGCAAGCTTTATGGTGAAAAATGGC3′ and its complementary oligomer DNA (codon for serine is underlined). The plasmid into which the HindIII site was introduced was named pMS452. As the final step, the 1.2-kilobase pair EcoRI-BamHI fragment, amplified by the primer shown in Table I and T3 primer, using pMS452 as a template, was used to replace the corresponding site of pMS452. This plasmid carrying HA-LAMP1 on pBluescriptII-SK(−) was named pMS453. Regarding mutagenesis other than HA tag insertion, oligonucleotides used as mutagenic primers were as follows; 5′CGCTCTAGAACTAGTAGATCTATGGCCTCCC3′ and its complimentary oligomer DNA for BglII site generation on pMS402 (BglII site is underlined), 5′CCCATGGAGACCTAAGAATTCTGCAGTCG3′ and its complementary oligomer DNA for stop codon elimination on pMS420 (stop codon is underlined). All the insertions and mutations were confirmed by sequencing.
Cell Culture and Transfections
COS-1 cells were cultured at 37°C and 5 % CO2 in DMEM supplemented with 10 % fetal bovine serum, penicillin, and streptomycin. For exogenous protein expression, a final 0.5 mg/ml of plasmids were transfected into 1 X 106 cells in 4 ml of media by using FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Mannheim, Germany).
Preparation of Membrane Fraction and Immunoblotting
Cells were washed with PBS three times 24 h after transfection. The cells were scraped with a rubber policeman and solubilized at 4°C for 1h in 0.5X Triton X-114 buffer (1 % Triton X-114, 75 mM NaCl, 5 mM Tris HCl pH 7.4) in the presence of 1 mM PMSF, 3 μg/ml Leupeptin, and 3 μg/ml pepstatin. The solubilized cell extract was layered on top of a 1.5X volume of Sucrose cushion (6 % Sucrose, 0.06 % Triton X-114, 150 mM NaCl, 10 mM Tris HCl pH 7.4), and incubated for 5 min at 30°C. After centrifugation at 300g for 3 min, the lower detergent phase was stored on ice and the phase separation above was repeated once more with the upper aqueous phase. The combined detergent phase was applied to the next step as a membrane protein-enriched fraction (Bordier, 1980). Solubilized and separated samples were treated with N-glycosidase F (PNGase F) for 20 h at 37°C after the removal of detergent by acetone extraction. Protein samples to be examined by immunoblotting were separated by 7.5 % SDS-PAGE (Laemmli, 1970) and electrophoretically blotted onto a PVDF membrane filter (PerkinElmer Life Science, Inc., Boston, MA). The filters were treated with anti-GFP and HRP-conjugated goat anti-rabbit IgG. The protein bands were detected by ECL detection kit (Amersham, Buckinghamshire, UK).
Immunofluorescence Microscopy and Selective Permeabilization of the Plasma Membrane
Cells on glass coverslips were washed twice with PBS 24 h after the transfection and fixed with methanol at −20 °C for 5 min. Fixed cells were washed with PBS three times and incubated with the primary antibody for 30 min at room temperature. After washing 4 times with PBS, cells were incubated with secondary antibody (Texas Red conjugated goat antimouse IgG) and again washed 3 times with PBS. The coverslips were mounted on glass slides and cells were visualized using a Zeiss Axiovert 2000 (Carl Zeiss, Thornwood, NY).
For selective permeabilization of the plasma membrane, we utilized cold osmotic shock in the presence of 0.005 % digitonin (Eckhart et al., 1999). Cells washed twice with PBS were fixed with 4% paraformaldehyde for 20 min at room temperature. Fixed cells were washed with PBS twice and the residual paraformaldehyde was neutralized by incubation for 5 minutes in 50 mM NH4Cl in PBS, followed by two additional PBS washes. Unless otherwise noted, all procedures involving selective permeabilization were performed at room temperature. Following the removal of PBS, ice-cold permeabilization buffer (0.005 % digitonin, 0.3 M sucrose, 0.1 M KCl, 2.5 mM MgCl2, 1 mM EDTA, and 10 mM Hepes (pH 6.9)) was added to the cells, which were then incubated at 4°C for 20 min. As a positive or a negative control, permeabilization was performed in PBS containing 0.1% Triton X-100 or in the absence of detergent, instead of 0.005 % digitonin. Partially permeabilized cells were labeled by the appropriate antibodies as described above.
3. RESULTS
Construction and Expression of HA-Tagged Mutant OA1
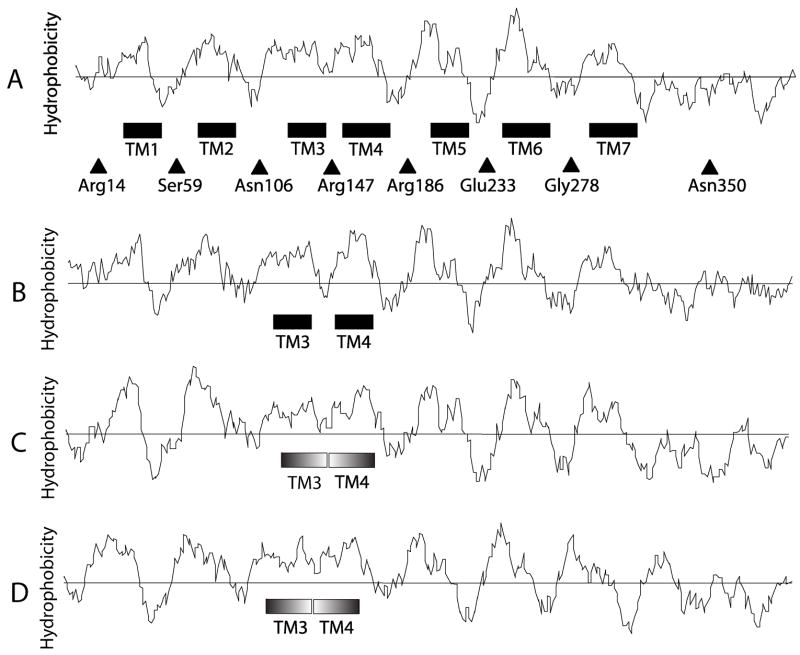
When applied to the amino acid sequence of OA1, different hydropathy algorithms predicted the presence of 6 or 7 transmembrane regions (6TM or 7TM) (Bassi et al., 1995; Schiaffino et al., 1999; Hirokawa et al., 1998; Fig. 1 A). The frequently-utilized Kyte and Doolittle hydrophobicity scale (Kyte and Doolittle., 1982), suggests that OA1 contains 7 hydrophobic regions, each of which may serve as a transmembrane domain (approximately amino acids ~30–50, TM1; ~70–90, TM2; ~120–140, TM3; ~150–180, TM4; ~200–220, TM5; ~240–260, TM6; ~290–320, TM7) (Fig. 1 A). However, the loop region between TM3 and TM4 (L3) is extremely short, and TM3 and TM4 might instead form a single TM domain. In addition, according to the SOSUI algorithm, which classifies the TM domains of integral membrane proteins based upon physicochemical features without reference to sequence similarity (Hirokawa et al., 1998), the region around the TM1 predicted in other models would in fact not be predicted as a TM domain. On the other hand, some reports indicate that hydrophobicity is not an absolute requirement for the formation of TM domains (Ota, et al., 1998a; Ota et al., 1998b).
Fig. 1. Hydropathy profile of OA1.
The amino acid sequence of Human OA1 (amino acids, 1 – 404) (A) and its homologues in other species (B for Mouse, C for Zebrafish, D for Xenopus) were analyzed by the method of Kyte and Doolittle (21). The predicted TM domains in human OA1 are boxed and numbered in the order of amino to carboxy terminus. Filled triangles with residue numbers indicate the position of HA-tag insertions.
We sought to establish the membrane topology of OA1 by assessing the accessibility of an HA-tag on different sites of an OA1 molecule with GFP fused to its carboxy terminus (Samaraweera et al., 2001; Shen et al., 2001) using an anti-HA antibody. An HA-tag (YPYDVPDYA) was inserted into the N-terminal region, C-terminal hydrophilic region or each loop region located between the predicted TMs indicated above. The amino acid positions of HA-tag insertions we constructed were the C-terminal side of Arg14, Ser59, Asn106, Arg147, Arg186, Glu233, Gly278, and Asn350. For the sake of convenience, these mutant forms of OA1-GFP are designated as summarized in Table I. For instance, OA1-GFP (L4) indicates the derivative HA-tag is inserted C-terminal to Arg147 in the predicted 4th loop.
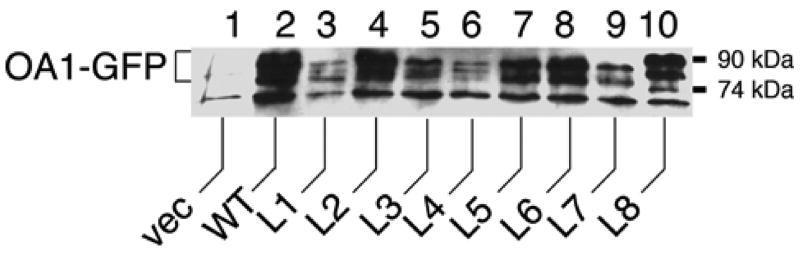
The expression level of each OA1-GFP derivative was examined by subjecting a crude membrane fraction of transfected COS-1 cells to SDS-PAGE and visualizing GFP by immunoblotting (Samaraweera et al., 2001). No OA1-GFP derivative was fractionated into the soluble fraction (data not shown). Specific immunoreactive bands were detected in cells transfected with each of the OA1-GFP derivatives (Fig. 2). Multiple bands observed in Fig. 2 represent the varied glycosylation levels for each mutant, as a result of incomplete deglycosylation by PNGase F, presumably due to OA1-GFP’s highly hydrophobic nature (M. Sone and S. J. Orlow, unpublished data.). While the difference in the cellular accumulation of the derivatives is partly caused by the difference in the rates of their synthesis, the contribution of the varied degradation level for each derivative cannot be excluded due to the long incubation for PNGase F treatment. Although protease inhibitors were added in the process of collecting and solubilizing the protein, trace amounts of resided cellular proteases might digest OA1-GFP derivatives differently due to their varied HA insertion site. However, this degradation would not affect the membrane topology of OA1, since any of the digestion would have occurred after the membrane insertion of OA1. These results indicate that all of the HA-tagged OA1 constructs were successfully expressed at detectable levels in the COS-1 cells.
Fig. 2. Expression of HA-tagged OA1-GFP derivatives in COS-1 cells.
COS-1 cells were transfected with the plasmids coding OA1-GFP derivatives as indicated below. Equal numbers of cells were collected 24 h post-transfection. Membrane fractions obtained by the phase separation described in EXPERIMENTAL PROCEDURES were analyzed by SDS-PAGE followed by immunoblotting with anti-GFP. Plasmids transfected were: lane 1, pAcGFP-N1 (vector control); lane 2, pMS430 (wild-type OA1-GFP); lane 3, pMS431 (OA1-GFP (L1)); lane 4, pMS432 (OA1-GFP (L2)); lane 5, pMS433 (OA1-GFP (L3)); lane 6, pMS434 (OA1-GFP (L4)); lane 7, pMS435 (OA1-GFP (L5)); lane 8, pMS436 (OA1-GFP (L6)); lane 9, pMS437 (OA1-GFP (L7)); lane 10, pMS438 (OA1-GFP (L8)). Multiple bands observed in each derivative represent the varied glycosylation levels for each mutant, as a result of incomplete deglycosylation by PNGase F, presumably due to the highly hydrophobic nature of OA1-GFP. Differences in the accumulation level of each derivative may partly be caused by differences in the synthesis rate of each derivative itself. However, differences in the stability and degradation level for each derivative also cannot be excluded, due to the long incubation required for PNGase F treatment.
Cellular Localization of Insertion Mutants
The wild-type and HA-tagged mutant forms of OA1-GFP were transiently expressed in COS-1 cells, and their cellular localizations were determined by immunofluorescence. All the OA1-GFP derivatives colocalized extensively in perinuclear granules with CD63, a marker for the late endosomal/lysosomal compartment,(see Fig. 3 for wild-type OA1-GFP, OA1-GFP (L3) and OA1-GFP (L6); data not shown for the other mutants). Insertion of the HA-tag did not affect the cellular localization of OA1-GFP. These results indicate that all the HA-tagged OA1-GFP derivatives undergo proper insertion into the membrane and traffic correctly after synthesis in COS-1 cells. Thus, in combination with the data above regarding the cellular expression level of each derivative, the overall membrane topology of each HA-tagged version of OA1-GFP reflects that of the wild-type.
Fig. 3. Insertion of HA-tag does not affect the subcellular localization of OA1-GFP derivatives.
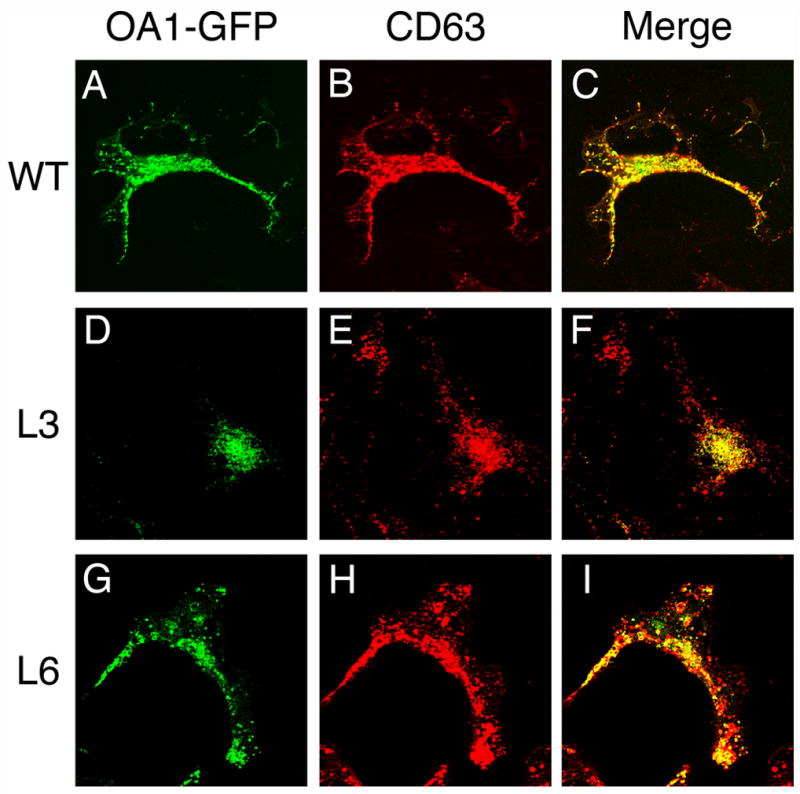
COS-1 cells transiently expressing OA1-GFP derivatives were fixed with methanol and were incubated with monoclonal anti-CD63 antibody H5C6 followed by the labeling with Texas Red conjugated goat antimouse IgG. The cells were visualized by fluorescence microscopy with the fluorescence from fused GFP (A, D, and G) and anti-CD63 (B, E, and H). The merged images (C, F, and I) indicate the colocalization of OA1-GFP derivatives and CD63, the marker for late endosome/lysosome, in perinuclear granules. OA1-GFP derivatives expressed in COS-1 cells were: wild-type OA1-GFP (A, B, and C); OA1-GFP (L3) (D, E, and F); and OA1-GFP (L6) (G, H, and I). The other derivatives that are not shown here were similarly colocalized with CD63.
Selective Permeabilization of the Plasma Membrane
Because of OA1’s intracellular localization, we needed to first establish experimental conditions that selectively permeabilized the plasma membrane of COS-1 cells while leaving lysosomal membranes intact. To do so, we adapted the conditions described by Eckhardt et al. for selective permeabilization of the plasma membrane leaving the Golgi membrane intact. (Eckhart et al., 1999). Human lysosome associated membrane protein 1 (LAMP1), a type I membrane protein, was utilized as a control. An HA-tag was inserted into the N-terminus of mature LAMP1 (oriented to the lysosomal lumen) or the C-terminus of LAMP1 (oriented to the cytoplasm) as summarized in Table I. HA-LAMP1 or LAMP1-HA was transiently expressed in COS-1 cells, and the accessibility of the HA-tag to HA antibody was assessed by immunofluorescence utilizing formaldehyde-fixed and digitonin-permeabilized cells. In the case of partial permeabilization of cells, LAMP1-HA should be labeled by HA antibody while HA-LAPM1 should not to be accessible. As shown in Fig. 4 B and E, in the cells permeabilized by low concentration of digitonin and cold osmotic shock as detailed in EXPERIMENTAL PROCEDURES, HA-LAMP1 was not labeled by anti-HA (Fig. 4 B) while LAMP1-HA was labeled as well as in the completely permeabilized cells (Fig. 4 D, E). As controls, accessibilities of both LAMP1 derivatives to anti-HA were examined using completely permeabilized cells and intact cells. HA-tags on both LAMP1 derivatives are extensively labeled by anti-HA in the completely permeabilized cells (Fig. 4 A, D) whereas not labeled at all in the intact cells (Fig. 4 C, F). Thus, the experimental condition employed here are suitable for the partial permeabilization of the plasma membrane, while leaving the lysosomal membrane intact.
Fig. 4. Detection of HA-tag fused to N- or C- terminus of LAMP1 by immunofluorescence.
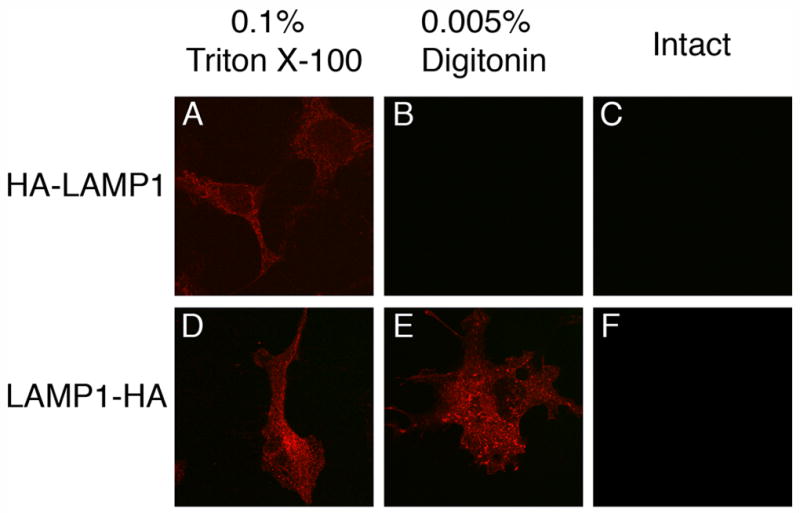
COS-1 cells transiently expressing HA-LAMP1 (A, B, C) and LAMP1-HA (D, E, F) were fixed with formaldehyde and partially permeabilized with 0.005% digitonin (B, E). As a positive or negative control regarding permeabilization, fixed cells were also incubated with 0.1% Triton X-100 (A, D) or in the absence of detergent (C, F), respectively. Cells were subjected to immunofluorescence using monoclonal anti-HA antibody 16B12 and Texas Red conjugated goat antimouse IgG. In selectively permeabilized cells (B, E), HA-tag fused to the C-terminus of LAMP1 that is oriented to the cytoplasm was labeled by anti-HA (E), while HA-tag fused to the N-terminus of LAMP1 that is oriented to the lumen of the endolysosome was not (B).
Orientation of HA-Tags
The orientations of HA-tags on OA1-GFP were determined by immunofluorescence for each HA-tagged OA1-GFP using the conditions established above for HA-tagged LAMP1s. In the completely permeabilized cells, anti-HA detected all of the OA1-GFP derivatives (Fig. 5 A, D, G, J, M, P, S, V), while as expected, none were detected in the intact cells (Fig. 5 C, F, I, L, O, R, U, X). In the partially permeabilized cells, OA1-GFP (L2), OA1-GFP (L4), OA1-GFP (L6), and OA1-GFP (L8) were labeled by anti-HA. Conversely, anti-HA did not detect OA1-GFP (L1), OA1-GFP (L3), OA1-GFP (L5), and OA1-GFP (L7) in partially permeabilized cells. 50 cells for each mutant were counted in 3 different experiments. The percentages of the cells labeled by anti-HA were more than 99 % for the former group (100% for OA1-GFP (L2), OA1-GFP (L4) and OA1-GFP (L8); 99% for OA1-GFP (L6)), and less than 3 % for the latter group (2.6% for OA1-GFP (L1); 1.3 % for OA1-GFP (L3); 2 % for OA1-GFP (L5); 0.7% for OA1-GFP (L7)). Thus, the HA-tagged positions on OA1-GFP (L2), OA1-GFP (L4), OA1-GFP (L6), and OA1-GFP (L8) are oriented to the cytoplasm while the HA-tagged positions on the other derivatives are oriented to the lysosomal lumen. These results indicate that OA1 spans intracellular membranes 7 times and its C-terminus is oriented to the cytoplasm.
Fig. 5. Orientation of HA-tag inserted in OA1-GFP derivatives as assessed by immunofluorescence of partially permeabilized cells.
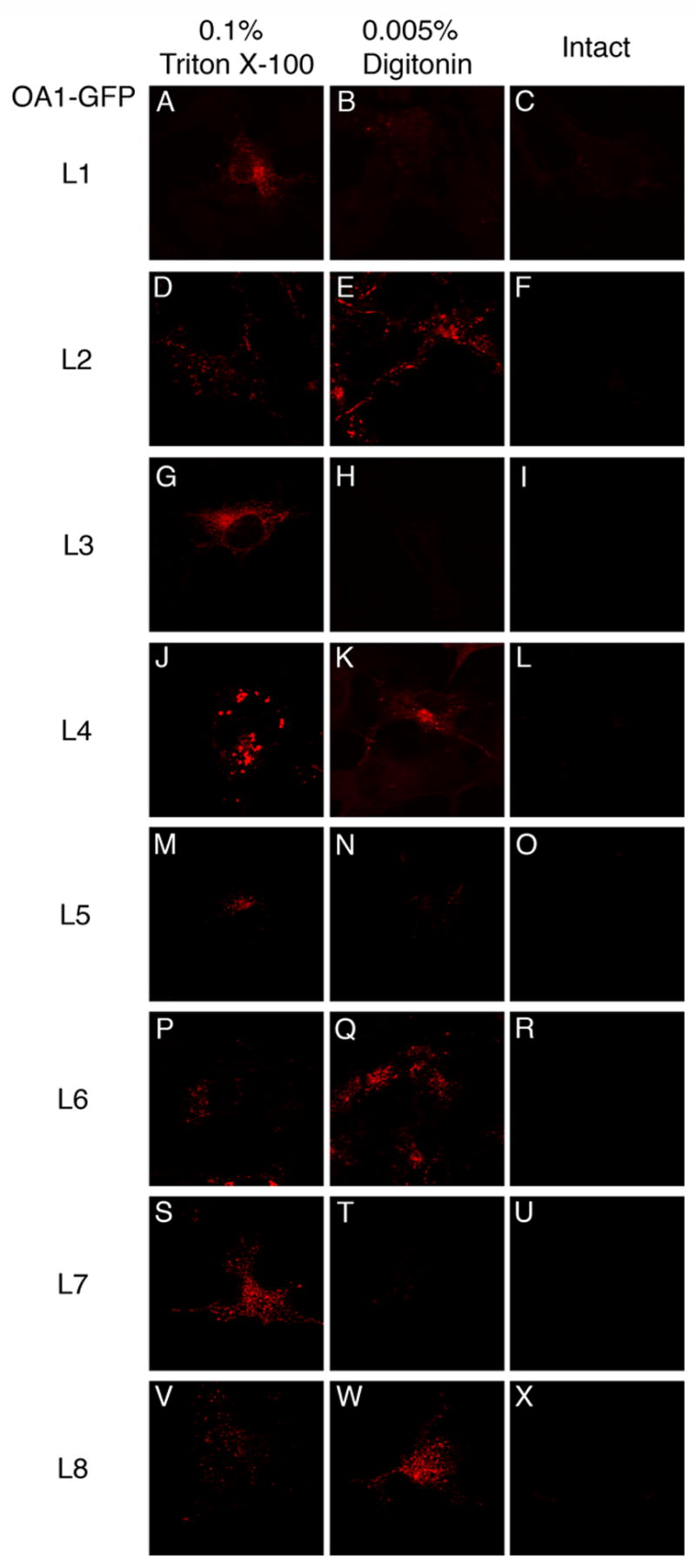
COS-1 cells transiently expressing OA1-GFP derivatives were fixed with formaldehyde and partially permeabilized with the condition as in Fig. 4. OA1-GFP derivatives expressed in COS-1 cells were: OA1-GFP (L1) (A, B, and C); OA1-GFP (L2) (D, E, and F); OA1-GFP (L3) (G, H, and I) ; OA1-GFP (L4) (J, K, and L); OA1-GFP (L5) (M, N, and O); OA1-GFP (L6) (P, Q, and R); OA1-GFP (L7) (S, T, and U); OA1-GFP (L8) (V, W, and X). HA-tag inserted to L2, L4, L6, or L8 (E, K, Q, W) was labeled by anti-HA, while that to L1, L3, L5, or L7 (B, H, N, T) was not.
4. DISCUSSION
Artifacts in the determination of membrane topology by hydropathy analysis using certain algorithms can be overcome by examining the proteins in question directly in the cell, as we have done in this study. The systematic study of integration of a multispanning membrane protein, human band 3 protein, into the ER membrane demonstrated that hydrophilic regions can be TM domains and that a TM domain is not necessarily a hydrophobic region (Ota, et al., 1998a; Ota et al., 1998b). Furthermore, SecG, a cytoplasmic membrane protein with 2TM in E.coli, was shown to invert its membrane topology when accompanied by the action of another soluble cytoplasmic protein (Nishiyama et al., 1996). In view of these and other reports regarding the membrane topology of integral membrane proteins (Skach et al., 1994; Zhang et al., 1996, Gafvelin et al., 1997; von Heijne., 2006), we sought to definitively establish the membrane topology of OA1.
OA1 was proposed to be an intracellular GPCR, due to (1) weak similarity to GPCR families (Type A, B, and E), (2) 7TM predicted by other algorithms, (3) the existence of some amino acid residues highly conserved in GPCRs (Schiaffino and Tacchetti, 2005) and (4) a yeast-based GPCR signaling assay (Staleva and Orlow, 2006). The results presented here indicate that OA1 is a 7TM integral membrane protein and that its C-terminus is oriented to the cytoplasm. Originally, OA1 was predicted to have 6TM by Kyte and Doolittle’s hydrophobicity scale (Bassi et al., 1995). As shown in Fig. 1, Kyte-Doolittle analysis fails to clearly identify two distinct hydrophobic regions (residues ~120–140, TM3 and ~150–180, TM4) around the residues 120–180. Also regarding its homologues in other species except mouse, the hydrophobicity around the corresponding portion of each molecule shows the similar profile (Fig. 1 B, C, D). In addition, the SOSUI algorithm (Hirokawa et al., 1998.) does not identify TM1 on human OA1 as a TM region. On the other hand, the presence of an E/DRY motif, which is highly conserved in type A GPCRs (rhodopsin-like family), was suggested to be present on the predicted loop between TM3 and TM4 by Schiaffino’s group (Schiaffino et al., 1999; Schiaffino and Tacchetti, 2005). However, the sequence suggested as a DRY-like motif was DAY. In a typical E/DRY motif, the arginine residue located between glutamate/asparate and tyrosine is considered to play an important role for the activation of GPCR by facilitating its proper folding (Devi, 2005). Therefore, in the absence of a critical arginine, the DAY sequence in OA1 is insufficient to be used as the basis for establishing its membrane topology. In any case, predictions based only on bioinformatics need experimental support. Moreover, the arginine is not conserved in other vertebrate OA1 homologues including Mouse, Zebrafish and Xenopus.
Definition of the number of TM domains and characterization of protein orientation has significant implications for further studies concerning the cellular function of OA1, specifically with regards to GPCR functionality, ligand screening and identification of proteins located upstream or downstream of OA1 signaling pathways. Furthermore, two dileucine motifs, “Ser222, Leu223, Leu224” proposed by Schiaffino’s group to act as a lysosomal targeting signal for OA1 (Piccirillo et al., 2006), and “Glu264, Ser265, Leu266, Leu267”, are on opposite sides of TM6. For dileucine signal activity, an acidic amino acid must be located at the N-terminal side of the two leucines, although phosphoryrated Ser can be substituted (Dietrich et al., 1997). In the report from Schiaffino’s group, a large portion of the loop containing “Ser222, Leu223, Leu224” was deleted in order to achieve high expression of OA1 on the plasma membrane. This deletion may have significantly affected the proper folding of OA1 and potentially led to alteration of OA1 properties. Due to the possibility of the inversion of membrane topology mentioned above, we wished to establish definitively the membrane topology of OA1, the number of TM regions and the protein’s orientation. There are some reports which give additional clues to the orientations of portions of the OA1 molecule; for example, glycosylation of Asn106 (Shen and Orlow, 2001c) and studies involving the C-terminal region (Schiaffino et al., 1999), however, because of the reasons mentioned above, systematical and experimental topology analysis is indispensable.
We employed an experimental system that has been widely applied to the topology analysis of plasma membrane proteins (Kast et al., 1998; van Geest et al., 2000), but here applied uniquely to a membrane protein on an intracellular organelle. Insertion of a small HA-tag was expected to have the least influence on the cellular behavior of OA1-GFP. Each HA-tagged OA1-GFP derivative was expressed in detectable levels (Fig. 2) and exhibited the same cellular localization as the wild-type (Fig. 3) suggesting that the tagged derivatives reached their final destination in the cell, the late endosome/lysosome, without any remarkable degradation or significant defects in folding/assembly. The tagged derivatives are thus expected to exhibit the same membrane topology as the wild-type and, therefore, the orientation of HA-tag on OA1-GFP derivatives reflects that of the corresponding position on wild-type OA1-GFP.
We searched for conditions suitable for the partial permeabilization of the plasma membrane. Simply controlling the concentration or type of detergent for the permeabilization did not allow us to selectively permeabilize cells (data not shown). We adapted the methodology described by Eckhardt et al. to establish suitable conditions utilizing the combination of 1) low concentration of digitonin and cold osmotic shock (Eckhardt et al., 1999), and 2) HA-tagged LAMP1s. This condition for partial permeabilization should prove useful not only for immunostaining, but for biochemical studies as well, and potential broad utility of the study of other endolysosomal membrane proteins.
For insertion of the HA-tag into the OA1 molecule, we selected positions in the middle of the loops between the hydrophobic regions predicted by Kyte and Doolittle’s hydrophobicity scale. For a helical structure which spans a lipid bilayer, at least 20 – 30 amino acid residues are needed. In view of the length requirements of TM domains and the hydropathy profile indicated in Fig. 1 A, not more than one TM domain possibly exists between the sequential two residues among Arg14, Ser59, Asn106, Arg147, Arg186, Glu233, Gly278, and Asn350. As a result of assessment by immunofluorescence, we found that HA-tags on OA1-GFP (L1) and OA1-GFP (L8), the N- and C- terminus of OA1, showed an opposite orientation (Fig. 5). This result demonstrates that the OA1 molecule contains an odd number of TM domains. The existence of an odd number of TM domains between them supports our determination of each tag’s position in combination with the possible length of TM domain and hydropathy profile described above. The membrane topology of OA1 demonstrated here is consistent with its putative role as a GPCR (15, 16) and the reports from Schiaffino’s group regarding the cytosolic motif required for lysosomal targeting (Piccirillo et al., 2006).
In several prior experimental systems (Staleva and Orlow, 2006; Innamorati et al., 2006), which suggest OA1 (and Oa1)’s ability to function as a GPCR, the protein was purposely localized to the plasma membrane. Mislocalization of protein is often caused by or can be accompanied by the abnormal overexpression/folding/assembly of the protein (Robinson et al., 1988; Rothman et al., 1986; Rothman et al., 1989). Our results presented here were all obtained with OA1 constructs whose distribution is entirely intracellular and mirrors that of endogenous OA1, and eliminate the possibility of artifacts due to plasma membrane mislocalization (Staleva and Orlow, 2006; Innamorati et al., 2006).
The possibility, supported by our results, that OA1’s physiologic function is that of an “intracellular GPCR” is highly interesting and perhaps unique. These results provide direction regarding the screening for potential ligands for OA1, since they revealed that such ligands should exist within lumen of the endolysosomal/melanosomal group of organelles in the developing retinal pigmented epithelium. Melanin polymer, tyrosinase, or melanin precursors such as tyrosine and Dopa all represent possible ligands. However, due to its membrane topology with its ligand binding domain facing the interior of the endolysosomal organelle, a simple plasma membrane based assay system for ligand screening cannot be employed. G-proteins and Arrestin have been suggested as potential interacting partners for OA1 (Schiaffino et al., 1999; Innamorati et al., 2006). Additional novel interacting partners for OA1 may also exist.
In summary, we have shown that OA1, properly localized intracellularly, has 7TM domains and its C-terminus is oriented to the cytoplasm. The results presented here provide a well-grounded basis for proceeding with future investigations of OA1 as a GPCR, including ligand screening, the identification of associated proteins, and the elucidation of OA1-related signaling cascades (Fig. 6).
Fig. 6. A model for the membrane topology of OA1 as determined by HA insertion.
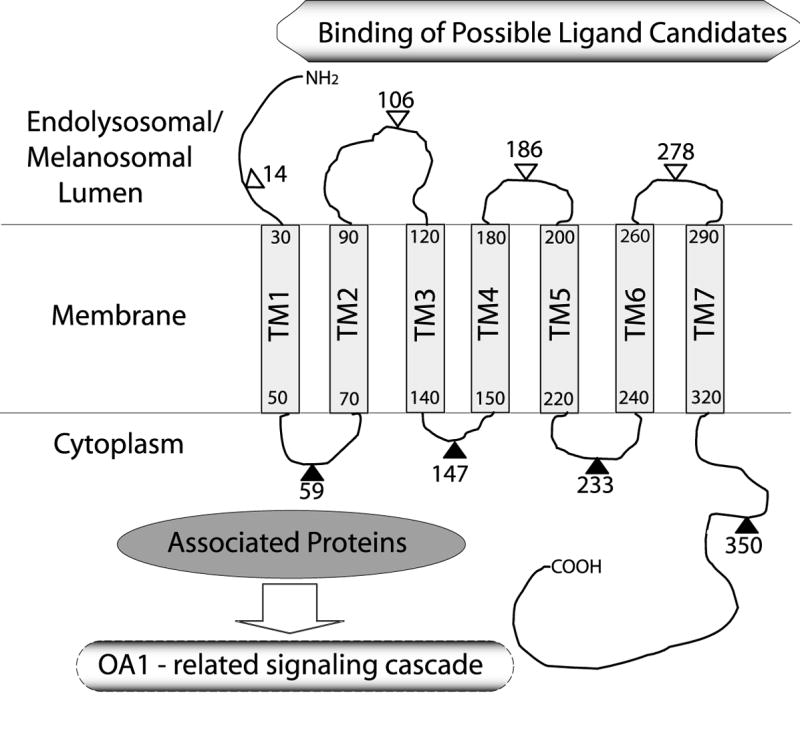
Transmembrane regions are represented by hatched boxes. Filled triangles with amino acid number indicate the sites of inserted HA tags which were accessible to HA antibody after partial permeabilization, whereas open triangles mark those which were not accessible under the same condition. Amino acid numbers at the N- and C- terminus of each hatched box indicate the approximate TM regions estimated by Kyte and Doolittle’s hydrophobicity scale (21) shown in Fig. 1 A.
Acknowledgments
We thank Dr. Christopher E. Touloukian for generously providing the OA1 clone, Dr. Minoru Fukuda for pSLVhL1, the LAMP1 clone. We thank Prashiela Manga for critical reading of the manuscript. This work was supported by grant EY10223 from the National Eye Institute.
Abbreviations
- OA1
Ocular Albinism Type I
- GPCR
G protein coupled receptor
- HA
influenza hemagglutinin epitope
- LAMP
lysosome associated membrane protein
- MMGs
melanin macroglobules
- TM
transmembrane
- DEME
Dulbecco’s modified Eagle’s Medium
- PAGE
polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
- HRP
horseradish peroxidase
- PBS
phosphate buffered saline
- GFP
green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bassi MT, Schiaffino MV, Renieri A, De Nigris F, Galli L, Bruttini M, Gebbia M, Bergen AA, Lewis RA, Ballabio A. Cloning of the gene for ocular albinism type 1 from the distal short arm of the X-chromosome. Nature Genet. 1995;10:13–19. doi: 10.1038/ng0595-13. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1980;256:1604–1607. [PubMed] [Google Scholar]
- Charles SJ, Green JS, Grant JW, Yates JR, Moore AT. Clinical features of affected males with X linked ocular albinism. Br J Ophthalmol. 1993;77:222–227. doi: 10.1136/bjo.77.4.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creel DJ, Summers CG, King RA. Visual anomalies associated with albinism. Ophthal Paediatr Genet. 1990;11:193–200. doi: 10.3109/13816819009020979. [DOI] [PubMed] [Google Scholar]
- Devi LA, editor. The G-Protein-Coupled Receptors Handbook. Humana Press Inc; New Jersey: 2005. [Google Scholar]
- Dietrich J, Kastrup J, Nielsen BL, Odum N, Geisler C. Regulation and function of the CD3gamma DxxxLL motif: a binding site for adaptor protein-1 and adaptor protein-2 in vitro. J Cell Biol. 1997;138:271–281. doi: 10.1083/jcb.138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M, Gotza B, Gerardy-Schahn G. Membrane topology of the mammalian CMP-sialic acid transporter. J Biol Chem. 1999;274:8779–8787. doi: 10.1074/jbc.274.13.8779. [DOI] [PubMed] [Google Scholar]
- Gafvelin G, Sakaguchi M, Anderson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- Garner A, Jay BS. Macromelanosomes in X-linked ocular albinism. Histopathology. 1980;4:243–254. doi: 10.1111/j.1365-2559.1980.tb02919.x. [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Innamorati G, Piccirillo R, Bagnato P, Palmisano L, Schiaffino MV. The melanosomal/lysosomal protein OA1 has properties of a G protein-coupled receptor. Pig Cell Res. 2006;19:125–135. doi: 10.1111/j.1600-0749.2006.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast C, Gros P. Epitope insertion favors a six transmembrane domain model for the carboxy-terminal portion of the multidrug resistance-associated protein. Biochemistry. 1998;37:2305–2313. doi: 10.1021/bi972332v. [DOI] [PubMed] [Google Scholar]
- King RA, Mentink MM, Oetting WS. The Metabolic Basis of Inherited Disease. New York, NY: McGraw-Hill; 1995. pp. 4353–4392. [Google Scholar]
- Kriss A, Russel-Eggitt I, Harris CM, Lloyd IC, Taylor D. Aspects of albinism. Ophthal Paediatr Genet. 1992;13:89–100. doi: 10.3109/13816819209087609. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Newton JM, Orlow SJ, Barsh GS. Isolation and characterization of a mouse homologue of the X-linked ocular albinism (OA1) gene. Genomics. 1996;37:219–25. doi: 10.1006/geno.1996.0545. [DOI] [PubMed] [Google Scholar]
- Nishiyama K, Suzuki T, Tokuda H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- Ota K, Sakaguchi M, Hamasaki N, Mihara K. Assessment of topogenic functions of anticipated transmembrane segments of human band 3. J Biol Chem. 1998a;273:28286–28291. doi: 10.1074/jbc.273.43.28286. [DOI] [PubMed] [Google Scholar]
- Ota K, Sakaguchi M, von Heijne G, Hamasaki N, Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol Cell. 1998b;2:495–503. doi: 10.1016/s1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- Piccirillo R, Palmisano L, Innamorati G, Bagnato P, Altimare D, Schiaffino MV. An unconventional dileucine-based motif and a novel cytosolic motif are required for the lysosomal and melanosomal targeting. J Cell Sci. 2006;119:2003–2014. doi: 10.1242/jcs.02930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Hunter CP, Valls LA, Stevens TH. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci USA. 1986;83:3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Yamashiro CT, Raymond CK, Kane PM, Stevens TH. Acidification of the lysosome-like vacuole and the vacuolar H+-ATPase are deficient in two yeast mutants that fail to sort vacuolar proteins. J Cell Biol. 1989;109:93–100. doi: 10.1083/jcb.109.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaraweera P, Shen B, Newton JM, Barsh JS, Orlow SJ. The mouse ocular albinism 1 gene product is an endolysosomal protein. Exp Eye Res. 2001;72:319–329. doi: 10.1006/exer.2000.0962. [DOI] [PubMed] [Google Scholar]
- Schiaffino MV, Baschirotto C, Pellegrini G, Montalti S, Tacchetti CC, DeLuca M, Ballabio A. The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc Natl Acad Sci. 1996;93:9055–9060. doi: 10.1073/pnas.93.17.9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino MV, d’Addio M, Alloni A, Baschirotto C, Valetti C, Cortese K, Puri C, Bassi MT, Colla C, DeLuca M, Tacchetti M, Ballabio A. Ocular albinism: evidence for a defect for a defect in an intracellular signal transduction system. Nature Genet. 1999;23:108–112. doi: 10.1038/12715. [DOI] [PubMed] [Google Scholar]
- Schiaffino MV, Tacchetti M. The ocular albinism type 1 (OA1) protein and the evidence for an intracellular signal transduction system involved in melanosome biogenesis. Pig Cell Res. 2005;18:227–233. doi: 10.1111/j.1600-0749.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- Shen B, Rosenberg B, Orlow SJ. Intracellular distribution and late endosomal effects of the ocular albinism type 1 gene product: Consequences of disease-causing mutations and implications for melanosome biogenesis. Traffic. 2001a;2:202–211. doi: 10.1034/j.1600-0854.2001.020306.x. [DOI] [PubMed] [Google Scholar]
- Shen B, Samaraweera P, Rosenberg B, Orlow SJ. Ocular albinism type 1: More than meet the eye. Pig Cell Res. 2001b;14:243–248. doi: 10.1034/j.1600-0749.2001.140403.x. [DOI] [PubMed] [Google Scholar]
- Shen B, Orlow SJ. Ocular albinism type 1 gene product is an N-glycoprotein but glycosylation is not required for its subcellular distribution. Pig Cell Res. 2001c;14:485–490. doi: 10.1034/j.1600-0749.2001.140609.x. [DOI] [PubMed] [Google Scholar]
- Skach WR, Lingappa VR. Transmembrane orientation and topogenesis of the third and fourth membrane-spanning regions of human P-glycoprotein (MDR1) Cancer Res. 1994;54:3202–3209. [PubMed] [Google Scholar]
- Staleva L, Orlow SJ. Ocular albinism 1 protein: trafficking and function when expressed in Saccharomyces cerevisiae. Exp Eye Res. 2006;82:311–318. doi: 10.1016/j.exer.2005.07.003. [DOI] [PubMed] [Google Scholar]
- van Dorp DB. Albinism, or the NOACH syndrome (the book of Ecnoch c.v 1–20) Clin Genet. 1987;31:228–242. doi: 10.1111/j.1399-0004.1987.tb02801.x. [DOI] [PubMed] [Google Scholar]
- van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nature Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Williams MA, Fukuda M. Accumulation of membrane glycoproteins in lysosomes requires a tyrosine residue at a particular position in the cytoplasmic tail. J Cell Biol. 1990;111:955–966. doi: 10.1083/jcb.111.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, O’Donnell LE, Green WR. Giant pigment granules in the retinal pigment epithelium of a fetus with X-linked ocular albinism. Ophthal Paediatre Genet. 1983;2:47–65. [Google Scholar]
- Zhang JT. Sequence requirements for membrane assembly of polytopic membrane proteins: molecular dissection of the membrane insertion process and topogenesis of the human MDR3 P-glycoprotein. Mol Biol Cell. 1996;7:1709–1721. doi: 10.1091/mbc.7.11.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]