Abstract
Ticks serve as both the vector and the reservoir for members of the spotted fever group rickettsiae. The molecular interaction(s) that results from this close relationship is largely unknown. To identify genetic factors associated with the tick response to rickettsial infection, we utilized differential-display PCR. The majority of upregulation appeared in the infected tissue. We cloned and sequenced 54 differentially expressed transcripts and compared the sequences to those in the GenBank database. Nine of the 54 clones were assigned putative identities and included a clathrin-coated vesicle ATPase, peroxisomal farnesylated protein, Ena/vasodilator-stimulated phosphoprotein-like protein, α-catenin, tubulin α-chain, copper-transporting ATPase, salivary gland protein SGS-3 precursor, glycine-rich protein, and Dreg-2 protein. Confirmation of the rickettsial influence on the differential expression in the ovaries for a number of these clones was demonstrated by semiquantitative reverse transcription-PCR and Northern blot analyses, resulting in confirmation of six out of nine and three out of four assessed clones, respectively. Further characterization of the clones identified tissue-dependent expression in the midguts and salivary glands. The potential roles of these molecules in the maintenance and transmission of rickettsiae are discussed.
Among tick-borne human pathogens, Rickettsia rickettsii, the causative agent of Rocky Mountain spotted fever, is a serious health concern in the United States. The parameters of transmission make Rocky Mountain spotted fever exceptionally difficult in terms of management and control. Small mammals, and occasionally humans, become infected when infected ticks feed on them during their life cycles. However, spotted fever group (SFG) rickettsiae are primarily maintained in nature through transovarial transmission via the infected germinal tissues of ixodid ticks. Under both laboratory and natural conditions, it has been reported that ticks infected with one species of Rickettsia are unable to pass a second species to their progeny (1, 20). While the transovarial transmission-blocking phenomenon has been reported only for SFG rickettsiae within ticks, de la Fuente et al. (3) recently reported that experimental competition results in a selection of only one genotype of Anaplasma marginale in the tick.
The mechanism(s) allowing for SFG rickettsial interference remains elusive. We hypothesize that, in response to a primary rickettsial infection, differentially regulated tick-derived molecules prevent a secondary rickettsial infection. The category (i.e., receptor or defense) and mode of action of these tick-derived molecules are unknown, and in order to test the hypothesis, differentially expressed candidate molecules must first be identified. We have recently utilized subtractive hybridization as a tool to identify some candidate molecules (21), and characterization of these molecules is the objective of our ongoing project. The objective of this study was to assess the differential expression of cDNAs in uninfected and Rickettsia-infected tick ovaries by PCR using random primers. We used differential-display PCR to determine if rickettsial infection of ticks results in the modulation of gene expression. The results of this study indicate that several transcripts were differentially expressed in response to infection. Thus, differential-display PCR is a viable means to assess events derived from tick infection with SFG rickettsiae.
MATERIALS AND METHODS
Ticks.
Uninfected and Rickettsia montanensis-infected Dermacentor variabilis females used in this study were maintained at the tick-rearing facility at Old Dominion University, as previously described (19). For this study, virgin females that had been fed for 4 days were forcibly detached from the rabbit host, and select tissues (salivary glands, midguts, and ovaries) were dissected out of the ticks. Tissues were immediately stored in RNAlater (Ambion, Austin, Tex.) at −80°C until they were used for RNA extraction. Rickettsial infection was verified by single-step reverse transcription (RT)-PCR (Invitrogen, Carlsbad, Calif.) using the primers Rr190.70n (27) and 190.701 (28) to amplify a portion of rOmpA, as previously described (19).
RNA extraction.
Frozen aliquots of tick tissues (salivary glands, midguts, and ovaries) stored in RNAlater were centrifuged at high speed (16,000 × g) for 10 min and rinsed with RNase- and DNase-free water to remove the buffer. RNA was extracted using Trizol (Life Technologies, Rockville, Md.) according to the manufacturer's protocol. Total RNA was treated with DNase I (Promega, Madison, Wis.) at 37°C for 1 h.
Differential display.
According the manufacturer's protocol for the Delta differential-display kit (Clontech, Palo Alto, Calif.), first-strand cDNA was synthesized for either uninfected or Ricketsia-infected tick ovaries by using Moloney murine leukemia virus reverse transcriptase (200 U), diluted (1:40), and stored at −20°C until it was used in the differential-display PCR. cDNA was PCR amplified using 10 different random forward primers (P1 to P10), which were separately paired with an oligo(dT) reverse primer (T1) provided with the kit. The 20-μl reaction mixture consisted of 1 μl of cDNA, 1 μl (20 μM) of each of the forward and reverse primers, 10× PCR buffer, a mixture of the deoxynucleoside triphosphates (5 mM each), [α-33P]dATP (2 μCi), Advantage KlenTaq polymerase, and sterile water. The PCR conditions were as follows: 1 cycle of 94, 40, and 68°C for 5 min each; 2 cycles of 94°C for 2 min and 40 and 68°C for 5 min each; 25 cycles of 94°C for 1 min, 60°C for 1 min, and 68°C for 2 min; and a final incubation at 68°C for 7 min.
The PCR product (5 μl) was combined with an equal volume of Tris-borate-EDTA loading buffer (Invitrogen), incubated at 95°C for 5 min, and placed on ice for 5 min. The products were subsequently electrophoresed on a 6% polyacrylamide gel in Tris-borate-EDTA buffer for 3 h at 75 W. The gel was dried and exposed to film overnight at −80°C.
Differentially expressed bands were gel purified, reamplified by using the appropriate forward primer and the oligo(dT) reverse primer, and cloned into the TA cloning vector (Invitrogen). Sequencing was performed by the dye terminator method on a model 373 automated fluorescence sequencing system (Applied Biosystems, Foster City, Calif.). Sequence analyses were carried out using MacVector software (Accelrys, San Diego, Calif.), and similarity comparisons were assessed using the GenBank database. Putative identification was assigned based on an E (expect) value of ≤0.05 using a nonredundant or Swiss-Prot tBlastX search. For presentation purposes in this report, clones with putative identities are presented as the clone designation (e.g., OiXXX, where XXX is a number) followed by the putative protein name.
Expression analysis by RT-PCR.
DNase-treated total RNA (4.5 μg) from Rickettsia-infected and uninfected tick tissues was used for first-strand cDNA synthesis using a Superscript first-strand synthesis kit (Invitrogen) according to the manufacturer's protocol. In each 25-μl reaction mixture, 1 μl of diluted (1:10) cDNA was used as a template with clone-specific primers (Table 1). The reaction mixtures were subjected to 1 cycle at 94°C for 3 min; 25 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min; and a final incubation at 72°C for 5 min. Tick mitochondrial 16S ribosomal primers (5′-GAATGCTAAGAGAATGGAAT-3′ and 5′-GTCTGAACTCAGATCAAGT-3′) (described in reference 2) were used as a positive load control for the reactions. PCR-amplified products were visualized on an ethidium bromide-stained 1% agarose gel, and intensities of bands were quantified using Alpha Innotech (San Leandro, Calif) imaging software. Density values for bands were adjusted against the 16S band intensity for uninfected and Rickettsia-infected samples for each tissue. Two separate reactions were carried out for each molecule in each tissue.
TABLE 1.
Clone-specific primers utilized for semiquantitative RT-PCR analyses
| Clone | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| Oi619-VASP | CACTAAATGCTGGGTGACACG | TTTTGGTGGTAGCGACTTGTAG |
| Oi6113-clathrin-coated V-ATPase | CCTCACTAAATGCTGGGTGTGTC | GGTCCACTTGGCAAAGATGATG |
| Oi812-α-catenin | TCAACAACACACGGGATGCTTAC | CTGGTGACAATAAAGTGCTATGCG |
| Oi1013-tubulin α-chain | CTGGTGTCCCACAGGATTCAAG | CGCACTTTTTGGTTCCCTTAGG |
| Oi212-Cu2+ ATPase | CCTCTTGAAGCGACCAGGTGTG | TGGAACCCCCACTACAACATTC |
| Oi411-PfX | GCACACTGCTCGGGCATTC | TGGTAGGCTCACTGGGTCTCG |
| Oi312-SGS-3 | CCCTCACTAAATGCTGGTGGTGGTC | GGTGTGCCATCCGCCATCAAAC |
| Oi814-GRP | TGAGGTGGTAACGGCTACGAAG | AGAAAGGTGTCACAAAGTGCTCG |
RNA blot analysis.
For uninfected and Rickettsia-infected tick ovary samples, 4 μg of total RNA was loaded per lane on a 1% formaldehyde agarose gel, electrophoresed for 1.5 h at 80 V, and transferred to a nylon membrane. Probes were generated by linear amplification of the antisense-strand primer according to the Strip EZ PCR protocol (Ambion). Probe hybridization and subsequent analysis were carried out as described for the Northern Max protocol (Ambion). Membrane exposure to film was for 10 min, 30 min, and 12 h, depending on the intensities of the bands. Ethidium bromide-stained tick 16S ribosomal bands served as a positive load control.
Nucleotide sequence accession numbers.
Sequences of PCR-amplified fragments have been deposited in the GenBank express sequence tag database under the accession numbers 16981594 to 16981647.
RESULTS
Differential-display PCR.
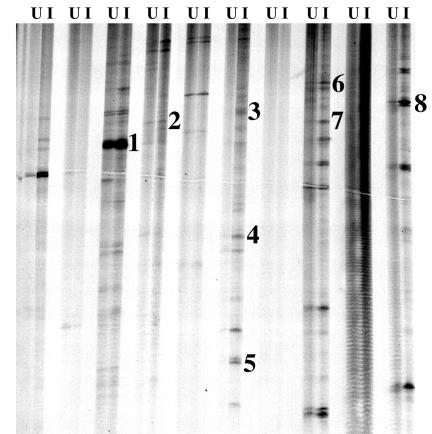
To verify rickettsial infection, RNA samples from ticks were utilized as a template in a single-step RT-PCR with primers designed to amplify a portion of the gene encoding the rOmpA protein. Amplification occurred in Rickettsia-infected ticks but not in the uninfected ticks (data not shown). For differential display, DNase-treated RNA from both uninfected and Rickettsia-infected D. variabilis ovaries was used to generate cDNAs, which were then amplified and compared. Using 10 random primers (forward) and one oligo(dT) (reverse), the results of separate reactions for each tissue were compared side by side on a polyacrylamide gel (Fig. 1). In both uninfected and Rickettsia-infected lanes, bands were differentiated by the presence or absence or by changes in intensity. A total of 54 differentially expressed bands were identified, and the DNA was recovered and reamplified by PCR. Amplified fragments ranged from 202 to 868 bp in size, with an average size of 417 bp. All PCR-amplified fragments were cloned and sequenced.
FIG. 1.
Differential-display PCR analysis of uninfected and Rickettsia-infected ovary cDNA. RNA was isolated from uninfected and Rickettsia-infected ovaries and used for first-strand cDNA synthesis, and the cDNA was used as a template for differential-display PCR. Samples were electrophoresed on a 6% denaturing polyacrylamide gel, and the dried gel was exposed to film overnight at −80°C. U and I represent uninfected and Rickettsia-infected samples, respectively. Numbers identify clones with putative identifications as follows: 1, Oi312-SGS-3; 2, Oi411-PfX; 3, Oi616-Dreg-2; 4, Oi619-VASP; 5, Oi6113-clathrin-coated V-ATPase; 6, Oi812-α-catenin; 7, Oi814-GRP; 8, Oi1013-tubulin α-chain.
Sequence analysis.
Database comparative analysis with tBlastX was carried out on each clone sequenced. Of the 54 clones, a total of 12 (∼23%) clones had significant homology (E value, ≤0.05) to characterized submissions in the database (Table 2). Three clones had significant homology to hypothetical proteins with no described or proposed function (not listed). The putative identification of the other nine clones included vascular-proton-translocating ATPase A isoform 1/clathrin-coated vesicle (Oi6113-clathrin-coated V-ATPase), peroxisomal farnesylated protein (Oi411-PfX), Ena/vasodilator-stimulated phosphoprotein-like protein (Oi619-VASP), α-catenin, cadherin (Oi812-α-catenin), tubulin α-chain (Oi1013-tubulin α-chain), copper-transporting ATPase (Oi212-Cu2+ ATPase), salivary gland protein SGS-3 precursor (Oi312-SGS-3), glycine-rich protein (Oi814-GRP), and Dreg-2 protein (Oi616-Dreg-2).
TABLE 2.
Putative identification and tissue-specific expression patterns of tick molecules identified from D. variabilis ovaries in response to rickettsial infection, as assessed by semiquantitative RT-PCR
| Code | Express sequence tag accession no. | Length of sequence (bp) | Size of mRNA (kb) | Differential tissue-specific expression in response to rickettsial infection (%)b
|
Homology | % Amino acid identity | ||
|---|---|---|---|---|---|---|---|---|
| Ovary | Midgut | Salivary gland | ||||||
| Oi619 | 16981620 | 310 | ∼5.3 | +17.6 | −7.0 | +19.9 | Ena/vasodilator-stimulated protein | 52 |
| Oi6113 | 16981624 | 230 | ∼3.7 | +21.3 | −5.4 | −13.3 | V ATPase (clathrin-coated) | 49 |
| Oi812 | 16981629 | 515 | ∼4.4, 3.9a | +12.8 | −5.7 | −18.7 | α-catenin (cadherin) | 76 |
| Oi1013 | 16981642 | 458 | ∼2.4 | +1.6 | −7.9 | +12.7 | Tubulin α-chain | 97 |
| Oi212 | 16981599 | 438 | +2.5 | −12.4 | +18.0 | Cu2+-transporting ATPase | 47 | |
| Oi411 | 16981605 | 666 | +19.7 | −2.4 | −0.1 | Peroxisomal farneslyated protein | 48 | |
| Oi312 | 16981603 | 410 | −14.8 | −79.4 | −105.5 | Salivary glue protein (SGS-3) precursor | 33 | |
| Oi814 | 16981631 | 413 | −17.6 | −18.9 | −52.0 | GRP | 44 | |
| Oi616 | 16981617 | 457 | Dreg-2 | 42 | ||||
Two isoforms were detected by Northern blot analysis, and two approximate sizes are given.
Values are calculated from one reaction that is representative of two RT-PCRs per tissue per molecule.
Validation of differential expression and tissue distribution.
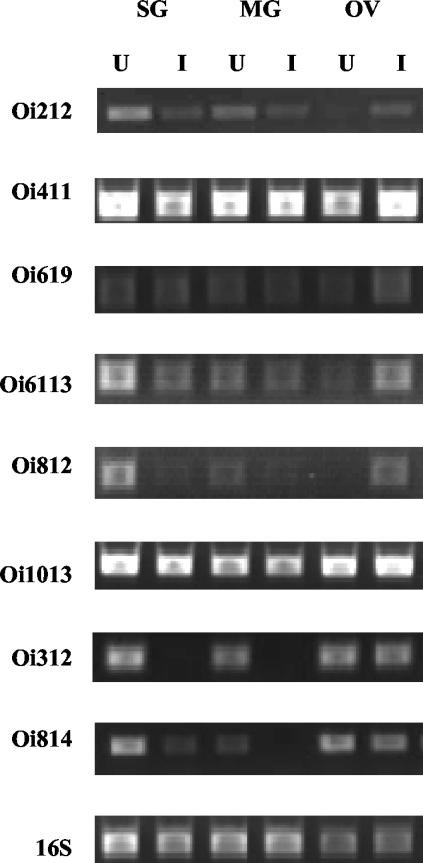
Total RNA from uninfected and rickettsia-infected ovaries, midguts, and salivary glands was used to create first-strand cDNA for each tissue. To confirm differential expression observed in the differential-display gel analysis, clone-specific primers were designed to amplify portions of the clones that were assigned a putative identification. cDNA template and specific primers were used in two separate PCRs as described above, and the intensities of amplified bands were measured with a densitogram and appropriate software. Similar patterns of expression were observed from the two reactions. Consistent with the differential-display PCR, six (Oi6113-clathrin-coated V-ATPase, Oi411-PfX, Oi619-VASP, Oi812-α-catenin, Oi1013-tubulin α-chain, and Oi212-Cu2+ ATPase) of the nine clones were also upregulated in infected ovaries when they were assessed by RT-PCR (Fig. 2). Transcription of two other putatively identified clones, Oi312-SGS-3 and Oi814-GRP, was found to be decreased in infected ovaries, despite originally having been identified as upregulated. Primers designed to amplify a portion of the Dreg-2 clone were able to amplify the plasmid preparation but were unable to amplify transcripts in tick tissues.
FIG. 2.
Semiquantitative RT-PCR analysis of D. variabilis cDNAs that were identified as differentially regulated in response to rickettsial infection. RNA from uninfected and Rickettsia-infected salivary glands (SG), midguts (MG), and ovaries (OV) was used to generate first-strand cDNA. By using clone-specific primers (Table 1) and cDNA as the template, comparative PCR analysis of expression was carried out. Above each lane, U and I represent uninfected and Rickettsia-infected samples, respectively. D. variabilis 16S primers served as a positive load control.
In order to initially characterize the expression of clones in other tick tissues, cDNA was generated from uninfected and rickettsia-infected tick salivary glands and midgut total RNA. Expression patterns varied, depending on the clone and target tissue (Table 2). Decreased transcription in the midguts of rickettsia-infected ticks was identified for each clone assessed. In fact, Oi312-SGS-3 transcriptional levels were greatly reduced in salivary glands and midguts in response to rickettsial infection (Fig. 2). In the salivary gland tissue, most transcripts were found to be downregulated except for Oi1013-tubulin α-chain and Oi619-VASP, which were upregulated in the salivary glands during rickettsial infection. Also, Oi411-PfX transcription was unchanged when uninfected and infected salivary glands were compared.
RNA blot analysis.
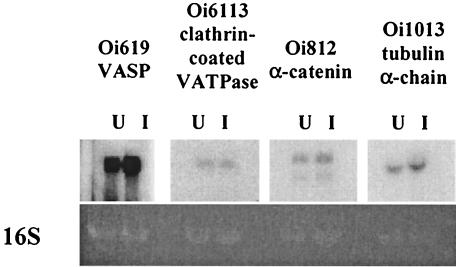
Because of our interest in four of the molecules (Oi619-VASP, Oi6113-clathrin-coated V-ATPase, Oi812-α-catenin, and Oi1013-tubulin α-chain) for their potential role in rickettsial maintenance in tick ovaries, their transcription was further assessed by Northern blot analysis. As shown in Fig. 3, the four probes, Oi619-VASP, Oi6113-clathrin-coated V-ATPase, Oi812-α-catenin, and Oi1013-tubulin α-chain, hybridized transcripts of ∼5.3, ∼3.7, ∼4.4 or 3.9, and 2.4 kb, respectively. Also, the probe for Oi812-α-2 catenin bound two transcripts, suggesting that two isoforms were present. Consistent with RT-PCR results for Oi619-VASP, Oi812-α-catenin, and Oi1013-tubulin α-chain, differential expression was observed in the ovaries (the only tissues tested). However, a difference in Oi6113-clathrin-coated V-ATPase between uninfected and infected samples was not evident.
FIG. 3.
Northern blot analysis of D. variabilis transcripts in uninfected (U) and R. montanensis-infected (I) tissues. RNA blots were prepared and probed with α-33P-labeled probes, according to the Northern Max protocol (Ambion). Blots were exposed to film for 10 min to 12 h at −80°C. The 16S band visualized on the ethidium bromide-stained gel served as a load control.
DISCUSSION
In the present study, we identified tick molecules that were differentially regulated in response to rickettsial infection by using differential-display PCR. Side-by-side comparison of randomly amplified cDNAs from uninfected and Rickettsia-infected D. variabilis ovaries allowed for the identification of 54 clones. While a few clones were identified as upregulated in the uninfected samples, the majority of the clones were identified as upregulated from the Rickettsia-infected preparations. Nine of the 54 isolated clones were found to have significant homology to known proteins in the GenBank database. Clones assigned a putative identification were originally identified as upregulated in Rickettsia-infected ovaries, and RT-PCR analysis verified this for 66.6% of the clones. Furthermore, upregulation of specific transcripts was confirmed by Northern blot analysis, indicating that in this model, differential-display analysis is a practical technique for identifying differentially expressed transcripts.
Both differential-display and subtractive-hybridization PCR allow for identification of unique cDNAs with similar success rates (6, 7, 8, 25). In this study, we found 16.6% (9 out of 54) of identified clones to have homology to known proteins. Conversely, 43.3% (13 out of 30) of the tick clones identified by subtractive hybridization were found to have significant homology to proteins in the database (21). Differences between identification rates are likely due to a high number of clones being isolated during differential-display PCR. Inherent to this technique, cDNAs are screened by their presence or absence as well as on the appearance of different levels of expression (intensities) of bands. Interestingly, while no individual transcript was identified by both techniques, similar types of molecules (i.e., adhesions and receptors) were identified. Therefore, these findings support the use of a multitechnique molecular approach to examine vector-pathogen relationships.
While the majority of clones isolated in this study were novel, the nine clones that were assigned putative identities proved to be very interesting. Several of the clones have roles in cell structure, movement, and cell-to-cell interactions, typical of what might be expected in a mitotically active tissue (9), such as the ovaries of partially fed ticks. Both VASP (a focal adhesion protein) and V-ATPase have been associated with actin assembly (26, 30); tubulin is a cytoskeleton component (11), and α-catenin is a regulator of cadherin function (15). Because the effect of rickettsial infection resulting in increased transcription of these molecules was not addressed in this study and is not known, we can only speculate on a role for these molecules during infection. For example, SFG rickettsiae utilize an intracellular-actin-based motility system to facilitate cell-to-cell spread (32) and VASP has been shown to be associated with rickettsial actin tails (14). Recent studies (12, 14) suggest that rickettsiae utilize a unique actin tail assembly mechanism comparable to those of other bacteria (e.g., Listeria and Shigella spp.) and the vaccinia virus (10). The role of this VASP-like molecule described here may be a requirement for rickettsial-actin-based motility in ticks. In addition to being associated with the cytoskeleton (30), V-ATPase is associated with clathrin-coated vesicles that facilitate protein sorting and receptor-mediated endocytosis by the cell (24), the process by which rickettsiae enter cells. Interestingly, a clathrin-coated adapter protein was identified as being upregulated in tick ovaries during rickettsial infection by subtractive hybridization (21).
The peroxisomal farnsylated protein is reported to be involved in peroxisome biogenesis, which is important for the regulation of excess fatty acids, amino acids, and H2O2 (16). The tick stress response to rickettsial infection and propagation may require increased metabolism within the cells, which therefore requires an abundance of peroxisomes. The increased transcription of a PfX-like molecule indicates increased peroxisome biogenesis, fulfilling the requirement for increased cellular metabolism. Likewise, the copper-transporting ATPase, a P-type ATPase which plays a role in excess copper removal by hepatocytes (29), may be a survival requirement of infected cells in ticks.
The upregulation of a number of these molecules in the Rickettsia-infected tissues may be correlated to the reactivation and massive replication of rickettsiae within the ovaries (13). It is known that tick-pathogen interactions are closely associated with the organ-specific function within the tick (23). Because this study utilized a chronically infected line of ticks, it is not surprising that we observed Rickettsia-mediated differential expression of most of these molecules in specific tissues. It is noteworthy that in tissues involved in either horizontal (salivary glands) or vertical (ovaries) transmission of rickettsiae, many of these molecules were noticeably upregulated or downregulated. However, downregulation of all these molecules was observed in the midgut, an organ not directly associated with vertical transmission. It is more difficult to speculate on the downregulation of specific molecules observed in this study, Oi312-SGS-3 and Oi814-GRP. The gene encoding the SGS-3 protein has been described for several species of Diptera as having an ecdysone-responsive element. Levels of ecdysone control the level of transcription of Sgs-3 in Drosophila melanogaster in an inversely proportional manner (17). Because ecdysone has been described for ticks, it is expected that hormone response element genes are present in ticks (4), specifically in the reproductive tissues. The Oi312-SGS-3 molecule was decreased in the ovaries and nearly abolished in the midguts and salivary glands of the Rickettsia-infected ticks. Although this transcript is likely under hormonal regulation, as are its paralogs in Drosophila (17), the significance of Rickettsia-mediated modulation is not clear. It is an exciting possibility that an unknown rickettsial receptor is in fact regulated either by this molecule or by the hormone. GRP is a cellular-matrix protein that has been proposed to be a component of the attachment cement secreted by the tick Haemaphysalis longicornis (22). This molecule may prove interesting in that its homologs have been associated with successful tick feeding (22), as well as control of the spread of the tobacco mosaic virus in plants (31). Although currently undetermined, either of these molecules may play a role in rickettsial establishment and maintenance in ticks.
While differential display has been used in insect systems, its utility in tick-borne disease models is only now being addressed. Since its inception, differential-display PCR has proven to be a useful tool for identification of differentially expressed mRNAs by allowing for a visual side-by-side comparison of levels of transcription (18). Technical developments have allowed for differential analysis using limited amounts of starting RNA, and the method has been adapted for use in arthropod systems (reviewed in reference 5). Identification of candidate molecules reported here will enable us to further characterize the mechanisms of maintenance and transmission of rickettsiae by ticks. Functional characterization of the molecules, including identifying their role in Rickettsia-tick interactions, is under way.
Acknowledgments
We thank D. E. Sonenshine, Old Dominion University, for technical assistance.
This work is supported by NIH grants AI-43006 (A.F.A.) and AI-051857 (F32 postdoctoral fellowship awarded to K.R.M.)
Editor: J. T. Barbieri
REFERENCES
- 1.Burgdorfer, W., S. F. Hayes, and A. J. Mavros. 1981. Nonpathogenic rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsii, p. 585-594. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, Inc., New York, N.Y.
- 2.de la Fuente, J., R. A. Van Den Bussche, and K. M. Kocan. 2001. Molecular phylogeny and biogeography of North American isolates of Anaplasma marginale (Rickettsiaceae: Ehrlichieae). Vet. Parasitol. 97: 65-76. [DOI] [PubMed] [Google Scholar]
- 3.de la Fuente, J., E. F. Blouin, and K. M. Kocan. 2003. Infection exclusion of the rickettsial pathogen Anaplasma marginale in the tick vector Dermacentor variabilis. Clin. Diagn. Lab. Immunol. 10:182-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delbecque, J. P., P. A. Diehl, and J. D. O'Connor. 1978. Presence of ecdysone and ecdysterone in the Amblyomma hebraeum Koch. Experientia 34:1379-1381. [Google Scholar]
- 5.Dimopoulos, G., and C. Louis. 1997. Differential display of mRNA, p. 261-267. In J. M. Crampton, C. B. Beard, and C. Louis (ed.), The molecular biology of insect disease vectors: a methods manual. Chapman and Hall, New York, N.Y.
- 6.Dimopoulos, G., A. Richman, A. della Torre, F. C. Kafatos, and C. Louis. 1996. Identification and characterization of differentially expressed cDNAs of the vector mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. USA 93:13066-13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimopoulos, G., A. Richman, H. M. Muller, and F. C. Kafatos. 1997. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. USA 94:11508-11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimopoulos, G., T. L. Casavant, S. Chang, T. Scheetz, C. Roberts, M. Donohue, J. Schultz, V. Benes, P. Bork, W. Ansorge, M. B. Soares, and F. C. Kafatos. 2000. Anopheles gambiae pilot gene discovery project: identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc. Natl. Acad. Sci. USA 97:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fristrom, D., M. Wilcox, and J. Fristrom. 1993. The distribution of PS integrins, laminin A and F-actin during key stages in Drosophila wing development. Development 117:509-523. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg, M. B. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hachouf-Gheras, S., M. T. Besson, and G. Bosquet. 1998. Identification and developmental expression of a Bombyx mori alpha-tubulin gene. Gene 208:89-94. [DOI] [PubMed] [Google Scholar]
- 12.Harlander, R. S., M. Way, Q. Ren, D. Howe, S. S. Grieshaber, and R. A. Heinzen. 2003. Effects of ectopically expressed neuronal Wiskott-Aldrich syndrome protein domains on Rickettsia rickettsii actin-based motility. Infect. Immun. 71:1551-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes, S. F., and W. Burgdorfer. 1982. Reactivation of Rickettsia rickettsii in Dermacentor andersoni ticks: an ultrastructural analysis. Infect. Immun. 37:779-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinzen, R. A., S. S. Grieshaber, L. S. Van Kirk, and C. J. Devin. 1999. Dynamics of actin-based movement by Rickettsia rickettsii in Vero cells. Infect. Immun. 67:4201-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano, S., N. Kimoto, Y. Shimoyama, S. Hirohashi, and M. Takeichi. 1992. Identification of a neural alpha-catenin as a key regulator of cadherin function and multicellular organization. Cell 70:293-301. [DOI] [PubMed] [Google Scholar]
- 16.James, G. L., J. L. Goldstein, R. K. Pathak, R. G. Anderson, and M. S. Brown. 1994. PxF, a prenylated protein of peroxisomes. J. Biol. Chem. 269:14182-14190. [PubMed] [Google Scholar]
- 17.Lehmann, M., F. Wattler, and G. Korge. 1997. Two new regulatory elements controlling the Drosophila Sgs-3 gene are potential ecdysone receptor and fork head binding sites. Mech. Dev. 62:15-27. [DOI] [PubMed] [Google Scholar]
- 18.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 19.Macaluso, K. R., D. E. Sonenshine, S. M. Ceraul, and A. F. Azad. 2001. Infection and transovarial transmission of rickettsiae in Dermacentor variabilis ticks acquired by artificial feeding. Vector Borne Zoonotic Dis. 1:45-53. [DOI] [PubMed] [Google Scholar]
- 20.Macaluso, K. R., D. E. Sonenshine, S. M. Ceraul, and A. F. Azad. 2002. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J. Med. Entomol. 39:809-813. [DOI] [PubMed] [Google Scholar]
- 21.Mulenga, A., K. R. Macaluso, J. A. Simser, and A. F. Azad. 2003. Dynamics of Rickettsia-tick interactions: identification and characterization of differentially expressed mRNAs in uninfected and infected Dermacentor variabilis. Insect Mol. Biol. 12:185-193. [DOI] [PubMed] [Google Scholar]
- 22.Mulenga, A., C. Sugimoto, Y. Sako, K. Ohashi, A. Musoke, M. Shubash, and M. Onuma. 1999. Molecular characterization of a Haemaphysalis longicornis tick salivary gland-associated 29-kilodalton protein and its effect as a vaccine against tick infestation in rabbits. Infect. Immun. 67:1652-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munderloh, U. G., and T. J. Kurtti. 1995. Cellular and molecular interrelationships between ticks and prokaryotic tick-borne pathogens. Annu. Rev. Entomol. 40:221-243. [DOI] [PubMed] [Google Scholar]
- 24.Nishi, T., and M. Forgac. 2000. Molecular cloning and expression of three isoforms of the 100-kDa a subunit of the mouse vacuolar proton-translocating ATPase. J. Biol. Chem. 275:6824-6830. [DOI] [PubMed] [Google Scholar]
- 25.Oduol, F., J. Xu, O. Niare, R. Natarajan, and K. D. Vernick. 2000. Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc. Natl. Acad. Sci. USA 97:11397-11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohta, S., T. Mineta, M. Kimoto, and K. Tabuchi. 1997. Differential display cloning of a novel rat cDNA (RNB6) that shows high expression in the neonatal brain revealed a member of Ena/VASP family. Biochem. Biophys. Res. Commun. 237:307-312. [DOI] [PubMed] [Google Scholar]
- 27.Regnery, R. L., C. L. Spruill, and B. D. Plikaytis. 1991. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux, V., P. E. Fournier, and D. Raoult. 1996. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J. Clin. Microbiol. 34:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theophilos, M. B., D. W. Cox, and J. F. Mercer. 1996. The toxic milk mouse is a murine model of Wilson disease. Hum. Mol. Genet. 5:1619-1624. [DOI] [PubMed] [Google Scholar]
- 30.Toyomura, T., T. Oka, C. Yamaguchi, Y. Wada, and M. Futai. 2000. Three subunit a isoforms of mouse vacuolar H(+)-ATPase. Preferential expression of the α-3 isoform during osteoclast differentiation. J. Biol. Chem. 275:8760-8765. [DOI] [PubMed] [Google Scholar]
- 31.Ueki, S., and V. Citovsky. 2002. The systemic movement of a tobamovirus is inhibited by a cadmium-ion-induced glycine-rich protein. Nat. Cell Biol. 7:478-486. [DOI] [PubMed] [Google Scholar]
- 32.Van Kirk, L. S., S. F. Hayes, and R. A. Heinzen. 2000. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect. Immun. 68:4706-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]