Abstract
Infection with Mycobacterium avium subsp. paratuberculosis is associated with high levels of morbidity, decreased production, and early culling in dairy cattle. Clinical symptoms of Johne's disease include persistent diarrhea, inappetence, and resultant weight loss due to chronic inflammation of the small intestine. Although the presence or absence of intestinal lesions cannot be used as a definitive indicator of M. avium subsp. paratuberculosis infection, most infected cattle exhibit significant changes to intestinal mucosa, with the focus of pathology surrounding the ileal cecal junction. Typical pathology of M. avium subsp. paratuberculosis infection includes inflammation, thickening of the lumenal wall, and hyperplasia in draining lymph nodes. To further understand the pathology of Johne's disease, we compared the gene expression profiles of ileal tissues from Johne's disease-positive (n = 6), and Johne's disease-negative (n = 5) Holstein cattle. Gene expression profiles were compared with a bovine total leukocyte (BOTL-3) cDNA microarray. Genes that were expressed at significantly higher levels (>1.5-fold; P < 0.05) in tissues from Johne's disease-infected animals relative to noninfected animals included those encoding tumor necrosis factor receptor-associated protein 1 (TRAF1), interleukin-1α (IL-1α), MCP-2, N-cadherin, and β1 integrin (CD29). Dramatic upregulation of IL-1α (21.5-fold) and TRAF1 (27.5-fold) gene expression in tissues of Johne's disease-positive cows relative to tissues from control cows was confirmed by quantitative real-time PCR. Western blot analysis confirmed that IL-1α and TRAF1 mRNA levels resulted in increased protein expression in tissues of Johne's disease-positive cattle relative to tissues from control cattle. High levels of IL-1α can produce symptoms similar to those found in clinical Johne's disease. Taken together, the data presented in this report suggest that many outward symptoms of Johne's disease may be due to IL-1α toxicity. In addition, enhanced levels of TRAF1 could result in cells within the lesions of Johne's disease-positive cattle that are highly resistant to TNF-α-induced signaling.
Johne's disease is a chronic granulomatous enteritis of ruminants, with symptoms that include watery diarrhea, lowered nutrient absorption, and untreatable wasting, eventually resulting in death. The causative agent of Johne's disease is the facultative intracellular bacterium Mycobacterium avium subsp. paratuberculosis. Johne's disease is found worldwide, and the average economic loss due to the disease has been calculated to equal $227.00 for each individual cow in an infected herd per year (21). This economic loss becomes even more dramatic when coupled with an estimated 20% positive diagnosis across dairy herds in the United States alone (34).
Johne's disease is normally characterized by three stages: subclinical nonshedding, (infected carrier), subclinical shedding, and clinical shedding (35). Initial infections with M. avium subsp. paratuberculosis can occur through several pathways, including fecal-oral transfer, in utero infection, and ingestion of milk or colostrum from an infected dam (26, 29, 31). Once inside the host, it is suspected that M. avium subsp. paratuberculosis crosses epithelial linings of the gut through membranous epithelial cells (M cells), where they are phagocytosed by local macrophage cells (20, 27). Upon phagocytosis of M. avium subsp. paratuberculosis by naive macrophage cells, normal processing through the endocytic pathway may be arrested, with M. avium subsp. paratuberculosis-containing phagosomes failing to fuse with lysosomes, allowing M. avium subsp. paratuberculosis to survive and proliferate inside macrophage cells (1, 39, 40).
Following initial infection with M. avium subsp. paratuberculosis, which typically occurs before 6 months of age, a long subclinical nonshedding phase ensues for 2 to 5 years, during which M. avium subsp. paratuberculosis proliferates in the gut region, eventually spreading to the mesenteric lymph nodes and possibly throughout the body. In cattle and other ruminants, the focus of disease pathology is the gut, specifically the jejunum, ileum, ileocaecal junction, and immediately adjacent areas (2, 4). As with other mycobacteria, granuloma formation can occur at sites of M. avium subsp. paratuberculosis infection, but the more common pathology in clinical-stage cattle is diffuse granulomatous enteritis with mucosal thickening due to large numbers of infiltrating macrophages (4, 6, 17). Development of disease symptoms, including diarrhea, loss of appetite, and resultant weight loss, is gradual and varies among individuals. If not identified in subclinical stages, Johne's disease will progress to a clinical shedding phase, characterized by wasting, lowered milk production, inappetence, and shedding of M. avium subsp. paratuberculosis in feces.
Generally, peripheral immune cells from M. avium subsp. paratuberculosis-infected cattle display increased Th2-like immune response to M. avium subsp. paratuberculosis antigens over the course of infection (22, 23, 28). A strong Th2-like immune response, characterized by enhanced production of IgG1 antibodies, can suppress Th1-like cytotoxic responses, and this appears to happen in peripheral blood mononuclear cells during late-stage Johne's disease (8, 28). Because M. avium subsp. paratuberculosis is an intracellular pathogen, Th2-like immune responses are ineffective in controlling infections. In contrast toTh2-like immune responses identified in the periphery during M. avium subsp. paratuberculosis infection, inflammation of the ileum is strongly associated with Johne's infection and has been shown to persist throughout M. avium subsp. paratuberculosis infection (17, 28, 32). Local inflammation is typically associated with a Th1-like cytotoxic immune response, characterized by expression of proinflammatory cytokines, including gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), interleukin (IL)-1, IL-2, IL-6, and IL-12 (5, 17). It appears likely that the progressive inflamed state of M. avium subsp. paratuberculosis-infected ileum and adjacent digestive tract could directly contribute to decreases in nutritional status typically observed in M. avium subsp. paratuberculosis-infected cattle.
Mechanisms leading to intestinal tissue damage as a result of M. avium subsp. paratuberculosis infection are, however, largely unknown. To date few studies have focused on defining immune activity and molecular mechanisms of M. avium subsp. paratuberculosis pathogenesis at sites of infection (5, 17). In the current study we tested the hypothesis that chronic exposure to M. avium subsp. paratuberculosis and the resulting proinflammatory response within intestinal tissues of infected cattle would lead to dramatic changes in gene expression patterns relative to healthy uninfected cattle. To test this hypothesis we compared gene expression profiles of ileal tissues from Johne's disease-positive and Johne's disease-negative control cattle with a bovine-specific cDNA microarray.
Our novel results demonstrate upregulation of IL-1α mRNA and protein expression in ileal tissues of cattle infected with M. avium subsp. paratuberculosis, leading to the possibility that some symptoms of Johne's disease may result from IL-1 toxicity. In addition, enhanced levels of TNF receptor-associated factor 1 (TRAF1) mRNA and protein may result in reduced sensitivity of cells within these lesions to TNF-α-induced signaling.
MATERIALS AND METHODS
Experimental animals.
The infected and control animals used in this study were all Holstein cattle ranging in age from 12 to 48 months. The immune status of all infected cattle with regard to M. avium subsp. paratuberculosis infection had been monitored by serum enzyme-linked immunosorbent assay (ELISA) bimonthly for 24 months prior to initiation of the experiments. Periodic fecal culture testing with a U.S. Department of Agriculture-approved testing laboratory (Michigan State University Animal Health Diagnostic Laboratory, East Lansing) was conducted to confirm infection status for control and infected cattle.
Control uninfected cattle (n = 5) were serum ELISA negative, IFN-γ negative, and fecal culture negative. Infected cattle (n = 6) were positive by serum ELISA, suffered from intermittent diarrhea, and exhibited moderate weight loss at the time of necropsy. Additionally, infected cattle were confirmed as fecal culture positive but were considered low shedders (5 to 50 CFU per g of feces). Ileal tissues from infected and noninfected animals were collected at the time of slaughter over a period of 6 months. At slaughter all animals were macroscopically examined for pathological signs of Johne's disease. All infected animals exhibited thickening of the ileal mucosa, convolution of the interior of the ileum, and enlargement of adjacent draining lymph nodes. All control animals exhibited healthy ileum and mesenteric lymph nodes, with no visible pathological signs of Johne's disease.
Three individual sections (≈10g each) of ileal tissue were harvested from different regions immediately adjacent (within 20 cm) to the ileal-cecal junction from each infected and control animal. Individual tissues sections were divided in two for RNA and protein extraction or histological examination. All tissues for RNA extraction and Western blotting were immediately placed into 50-ml conical tubes containing RNAlater (Ambion Corp, Austin, Tex.) and frozen in liquid nitrogen for storage at −80°C until use. Tissues for histological examination were placed into 50 ml conical vials containing Safefix (Biochemical Science Inc., Swedesboro, N.J.), and stored at −80°C until slides were prepared. (Michigan State University Department of Pathobiology, East Lansing, Mich.).
RNA extraction.
Ileal tissues were removed from −80°C and ≈2 g of tissue from each infected and control animal was powdered with a sterile RNase-free mortar and pestel in liquid nitrogen. RNA was extracted from all powdered ileal tissues with Trizol reagent (Invitrogen Life Technologies Corp., Carlsbad, Calif.) essentially as recommended by the manufacturer (Invitrogen Life Technologies Corp., Carlsbad, Calif.). The quality and quantity of extracted total RNA was estimated by UV spectrophotometry and electrophoresis on 1.0% native agarose gels.
Experimental design.
Because tissues were collected over a period of six months a reference design was used in cDNA microarray experiments. In this design, individual tissue RNA samples from infected cattle (n = 6) were compared against the same pool of RNA from control cattle (the reference) in a flipped-fluor manner. In total, six microarray experiments were conducted, such that three randomly selected samples from infected cattle were labeled with Cy3 dye and compared directly to the reference sample labeled with Cy5 dye, each on a separate microarray slide. Each of the remaining three samples from infected cattle were labeled with Cy5 dye, and directly compared to the reference sample labeled with Cy3 dye, each on a separate microarray slide. This design, although less robust than other possibilities (16), accounts for systematic variation due to dye effects and allowed immediate analysis of multiple samples collected over an extended time period.
Preparation of labeled cDNA.
To evaluate gene expression profiles in tissues from infected cattle versus the noninfected reference sample, total RNA (5 to 10 μg) from each animal was used as template in reverse transcription reactions (BD Atlas Powerscript fluorescent labeling kit, BD Biosciences, Alameda, Calif.) with oligo(dT)15-18 as primer. First-strand cDNA synthesis, was completed by combining 5 to 10 μg of RNA, oligo(dT)15-18 primer, and 650 ng of synthetic lambda Q gene (a positive control for cDNA synthesis) in a 0.2-ml microcentrifuge tube. The cDNA reaction was placed in a thermocycler with the following steps: 5 min at 70°C, 5 min at 20°C, 65 min at 42°C, 5 min at 70°C, and 20 min at 37°C. A master mix consisting of deoxynucleoside triphosphates, Powerscript reverse transcriptase (BD Biosciences Alameda, Calif.), dithiothreitol, and a buffer supplied by the reverse transcriptase manufacturer (BD Biosciences) was added after the first 5-min 70°C denaturation step, when samples had cooled to 20°C. Once reactions reached the final step of 37°C, RNase H was added to remove any remaining template RNA. Finally, cDNAs were purified with quick-clean resin (BD Biosciences).
Resulting cDNAs from tissues of infected cattle and the reference were differentially labeled with Cy3 and Cy5 dyes (BD Atlas powerscript fluorescent labeling kit, BD Biosciences, Alameda, Calif.) as described in Materials and Methods. Labeled cDNAs were purified to remove unincorporated dyes, combined, and concentrated to 5 to 10 μl with Microcon 30 spin concentrators (Millipore Corp., Bedford, Mass.). SlideHyb solution (Ambion Inc., Austin, Tex.) was added to bring labeled cDNA reactions to a final volume of 110 μl. Hybridizations of labeled cDNA probes to the BOTL-3 microarray were conducted for 18 h in a GeneTAC Hybstation with a step down procedure (Genomic Solutions, Inc., Ann Arbor, Mich.). Following hybridization, microarray slides were washed twice in low stringency buffer and once in high stringency buffer in the Hybstation unit (Genomic Solutions, Inc.). Finally, microarrays were rinsed once in 2X SSC (saline sodium citrate, 0.15 M NaCl plus 0.015 M sodium citrate) and once in double-distilled H2O. Microarrays were dried by centrifugation in a cushioned 50-ml conical tube and immediately scanned with a GeneTAC LS IV microarray scanner. GeneTAC LS software (Genomic Solutions, Inc., Ann Arbor, Mich.) was used to process microarray images, find spots, integrate robot-spotting files with the microarray image, and finally to create reports of raw spot intensities.
Microarray data analysis.
Raw spot intensity data from each BOTL-3 microarray was organized by gene name and array address. Normalization was then performed considering a robust local regression technique with the procedure PROC LOESS of the SAS statistical software package (24, 25, 37). LOESS normalized values were imported into a Microsoft Excel spreadsheet for further analysis. Mean loge (LN) expression differences (infected versus reference) for each of the six infected cattle were combined and used to calculate an overall mean LN expression difference (infected versus reference) for each gene. Combined data were also used to calculate standard errors of the mean LN expression value, t statistic and t distribution (P value) for each gene.
Quantitative real-time PCR validation of gene expression.
Validation of selected gene expression changes observed on cDNA microarrays was performed by quantitative real-time reverse transcription-PCR (Q-RT-PCR) in an Applied Biosystems 7000 DNA Sequence detection system (Perkin Elmer Corp., Foster City, Calif.). Total RNA was extracted from individual ileal tissues of M. avium subsp. paratuberculosis-infected and noninfected cattle, quantified, and quality checked as described above for microarray analysis. RNA was converted into first-strand cDNA by adding 1 μg of total RNA to a 12 μl reaction containing 10 mM oligo(dT)15 primer. Following a 5 min incubation at 70°C, the reaction was quick-chilled on ice and adjusted by addition of 4 μl of a buffer supplied by the reverse transcriptase manufacturer (final reagent concentrations were 50 mM Tris-HCl, pH 8.3, 75 mM KCl, and 3 mM MgCl2), 1 mM deoxynucleoside triphosphates, 200 units of Superscript II RNase H reverse transcriptase (Invitrogen Life Technologies, Carlsbad, Calif.), and a final concentration of 10 mM dithiothreitol in a total reaction volume of 20 μl. The reverse transcriptase reaction was allowed to progress at 42°C for 60 min, heated to 70°C for 15 min, and cooled to 37°C prior to the addition of 2 units of DNase-free RNase H (Invitrogen Life Technologies). Incubation at 37°C continued for 30 min in the presence of RNase H to remove the original RNA templates. Heating at 70°C for 15 min subsequently inactivated RNase H. The concentration of first-strand cDNAs was measured by UV spectrophotometry and final working solutions diluted to 10 ng per μl. Diluted cDNAs were stored at −80°C until use.
Q-RT-PCR was performed with SYBR Green PCR Master Mix (Perkin Elmer Corp.) and gene-specific primers. All primers were designed with Primer Express Software (Perkin Elmer Corp.) and were synthesized by a commercial facility (Qiagen Inc, Valencia Calif.). Primer sequences for the genes analyzed in this report were: Interleukin-1 α (forward, GGCCCTGCATCACTTCAT; reverse, TTGTTGAGGACAGCGACAATG) and TRAF1 (forward, ACGGCACCTTCCTCTGGA; reverse, TGCCACAGGCCGACTCA). β-Actin was used as a control gene for Q-RT-PCR (forward, CGCCATGGATGATGATATTGC; reverse, AAGCCGGCCTTGCACAT). Prior to Q-RT-PCR, first-strand cDNA from control cattle were randomly assigned into one of two pools with two animals per pool, and cDNA from infected cattle were randomly assigned into one of three pools with two animals per pool.
Q-RT-PCR Data Analysis.
Analysis of Q-RT-PCR data was performed with the 2-ΔΔCt method as described (6a, 19). In our analysis, β-actin served as the control gene to calculate initial ΔCt values. Mean control pool ΔCt values for each gene were then used as calibrator. For each pool of cDNA from infected cattle (n = 3), a separate 2-ΔΔCt value was calculated, relative to the calibrator, with within pool Q-RT-PCR measurements. Finally these values were used to establish an overall mean 2-ΔΔCt value, standard error, t statistic and t distribution (P value) for expression of each gene in tissues from infected cattle relative to tissues from uninfected controls.
Western blot analysis of IL-1α and TRAF1.
Protein was isolated from 1.0-g samples of ileal tissue from control and infected cattle. Tissues were pulverized under liquid nitrogen within a sterile mortar and pestle. Powdered tissue was suspended in 2.5 ml radioimmunoprecipitation assay lysis buffer (1% Triton-X, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.15 M NaCl, 0.02 M Tris-HCl) containing complete protease inhibitor (Roche Inc., Basel, Switzerland). Protein lysates were clarified by centrifugation at 4000 × g for 20 min. Protein containing supernatants were transferred to new tubes and protein concentrations determined with the Bradford method (Bio-Rad Co., Hercules, Calif.). Protein aliquots were diluted with RIPA buffer to a final protein concentration of 1 μg per μl. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), 20 μl (20 μg total protein) of each sample was denatured in 20 μl of 2x sample buffer (Bio-Rad Co.) and electrophoresed for 2 h on 4% to 15% polyacrylamide gradient gels (Bio-Rad Co.). Duplicate polyacrylamide gels were run for all samples and stained with Coomassie brilliant blue to ensure equal loading of proteins in all lanes.
Separated proteins were electrophoretically transferred to ECL Hybond (Amersham, Piscataway NJ) membranes for 30 min at 80 V. Membranes were incubated in blocking buffer (ECL Western kit, Amersham, Piscataway, N.J.) overnight. Blocked membranes were washed 3 times for 15 min each in PBS containing 0.5% Tween 20 (wash buffer) with mild agitation on an orbital shaker. Membranes were subsequently incubated for 2 h in a 1:750 dilution of IL-1α (SC7929, Santa Cruz Biotechnologies Inc., Santa Cruz, Calif.) or TRAF1 (SC7186, Santa Cruz Biotechnologies Inc.) primary antibody at room temperature. After incubation with primary antibody, membranes were again subjected to three 15 min wash sessions in wash buffer. For detection of bound primary antibodies, membranes were incubated in horseradish peroxidase-conjugated anti-rabbit antibody (1:5,000 dilution in wash buffer) (ECL Western Kit, Amersham Piscataway, N.J.) for 1 h with mild agitation. Finally, membranes were washed twice for 15 min with mild agitation in wash buffer and rinsed once for 5 min in PBS only.
Secondary antibodies bound to membranes were detected with an ECL kit (Amersham, Piscataway, N.J.) essentially as described by the manufacturer. Briefly, membranes were covered in approximately 750 μl of detection reagent and sealed between two acetate sheets. Membranes were exposed to Hyperfilm ECL (Amersham, Piscataway, N.J.), and resulting images weredeveloped with a Futura 2000E Automatic X-Ray film processor (Fischer Industries Inc., Geneva, Ill.). Protein expression values were quantified with a Bio-Rad densitometer and Molecular Analyst software (Bio-Rad Co., Hercules, Calif.).
RESULTS
Gene expression analysis of Ileal tissue from Johne's disease-positive cattle.
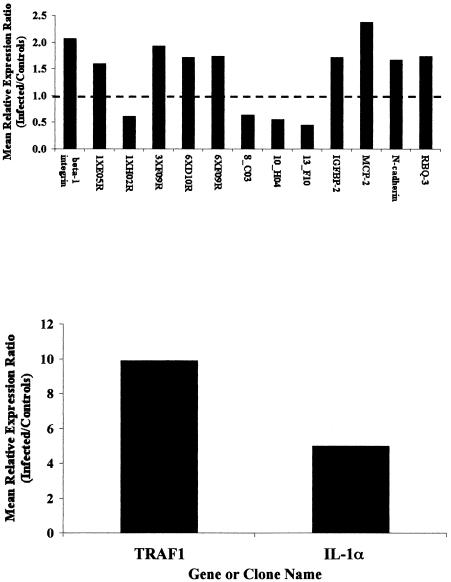
Gene expression profiles of ileal tissues from six cattle infected with M. avium subsp. paratuberculosis were directly compared to a reference consisting of pooled ileal tissues from five healthy (noninfected) cattle with a previously described cDNA microarray (6a, 7). Results from microarray experiments identified several genes that were significantly differentially expressed (>1.5-fold; P < 0.05) in tissues from M. avium subsp. paratuberculosis-infected cattle, relative to the reference sample (Fig. 1). The most profound differences in gene expression were observed for transcripts encoding IL-1α and TRAF1. Mean expression levels of transcripts encoding TRAF1 were upregulated almost 10-fold in tissues from infected cattle relative to reference, while mean expression of transcripts encoding IL-1α was upregulated almost fivefold in tissues from infected cattle, relative to reference. Other significant differences were greater than twofold upregulation of β1 integrin (CD29) and MCP-2 in ileal tissues of infected cattle, relative to reference. In addition, several genes, represented by expressed sequence tag (EST) clone inserts, were expressed at lower levels in ileal tissues from infected cattle, relative to the uninfected reference pool (Fig. 1).
FIG. 1.
Genes and EST clones found to be significantly differentially expressed (>1.5-fold and P < 0.05) in ileal tissues of Johne's disease-positive cattle (n = 6), relative to a reference comprising pooled tissues from five uninfected control cattle. Data are presented as the antilog of mean LN differences observed in six separate cDNA microarrays where gene expression profiles in tissues from infected cattle were directly and individually compared to the control reference sample. All cDNA microarrays were prepared and processed as described previously (8) and in Materials and Methods. In this representation, the antilog of mean LN differences approximates fold change relative to the reference. IL-1α and TRAF1 were plotted separately due to relatively large changes evident with these two genes. EST clone designations are abbreviations of the full clone name. Full clone names and additional information on each clone appear in a web-accessible database (URL: www.nbfgc.msu.edu). Abbreviated names shown are sufficient to access this information with the “clone name search” function within this database.
Validation of IL-1α and TRAF1 upregulation in ileal tissues from Johne's disease-positive cows by Q-RT-PCR.
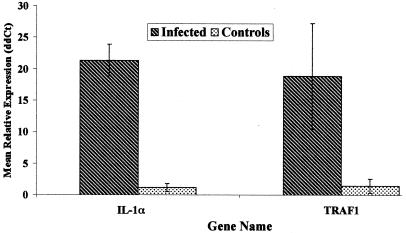
Q-RT-PCR was used to validate and extend differences in expression of IL-1α and TRAF1 transcripts observed in microarray studies. In general, the range of Q-RT-PCR tends to be much broader than cDNA microarray and thus can yield a more accurate estimate of relative mRNA abundance in multiple samples (6a, 30). As measured by Q-RT-PCR, mean expression of transcripts encoding IL-1α was 21.5-fold higher in tissues from infected cattle, relative to tissues from uninfected control cattle. Similarly, mean expression of transcripts encoding TRAF1 was 27.5-fold higher in infected tissues from infected cattle, relative to the control reference pool (Fig. 2).
FIG. 2.
Expression of IL-1α and TRAF1 transcripts in ileal tissues from M. avium subsp. paratuberculosis-infected and uninfected control cattle by Q-RT-PCR. Total RNA isolated from ileal tissues of Johne's disease-positive (Infected) and uninfected (Control) cattle was converted to first-strand cDNA and subjected to Q-RT-PCR as described previously (8, 30) and in Materials and Methods. Data presented are the mean ± standard error of the mean relative expression levels of each gene within group, calculated as 2-(ΔΔCt), as described (8, 20), with β-actin as a control gene and the mean ΔCt value for each gene in the control samples as the calibrator. Thus, data presented represent the approximate difference in expression of each gene in tissues from infected cattle relative to expression in control cattle.
Enhanced expression of IL-α and TRAF1 proteins in ileal tissues of Johne's disease-positive cattle.
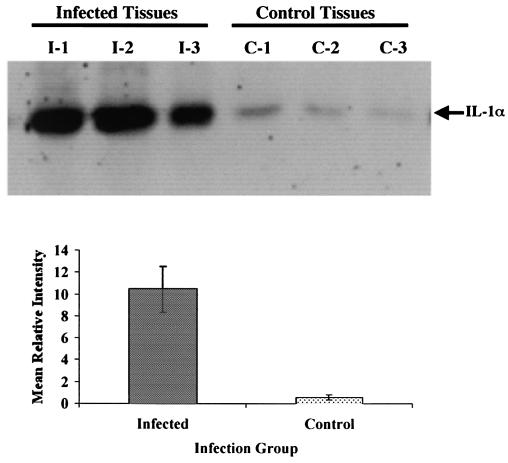
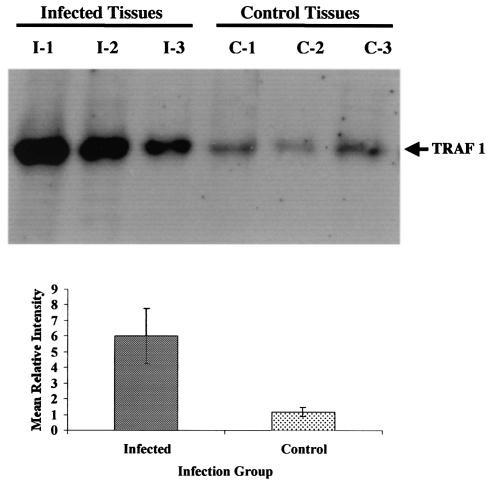
Western blot analyses were used to determine if enhanced IL-1α and TRAF1 mRNA abundance observed in cDNA microarray and Q-RT-PCR analyses resulted in an increased production of the respective proteins within tissues from infected cattle, relative to tissues from control uninfected cattle. Ileal tissues from 3 M. avium subsp. paratuberculosis-infected cattle, and 3 noninfected control cattle were processed and examined by Western blot analysis as described in Materials and Methods. The mean protein expression level of IL-1α in ileal tissues from M. avium subsp. paratuberculosis-infected cattle was ≈18-fold higher than that observed in tissues from the 3 control cattle (Fig. 3). Similarly mean protein expression of TRAF1 was 6-fold higher in tissues from infected cattle than in tissues from control cattle (Fig. 4).
FIG. 3.
Western blot analysis for expression of IL-1α protein in ileal tissues of cattle (n = 3) infected with M. avium subsp. paratuberculosis (infected tissues; I1 to I3) and uninfected control cattle (control tissues; C1 to C3). Proteins were separated by SDS-PAGE and Western blot analysis was performed as described in Materials and Methods. Final autoradiograms were analyzed by densitometry and background-subtracted intensities were obtained. Relative intensities were combined within a group and used to calculate mean relative intensities for each infection group as well as standard error of the means.
FIG. 4.
Western blot analysis for expression of TRAF1 protein in ileal tissues of cattle (n = 3) infected with M. avium subsp. paratuberculosis (infected tissues; I1 to I3) and uninfected control cattle (control tissues C1 to C3). Proteins were separated by SDS-PAGE and Western blot analysis was performed as described in Materials and Methods. Final autoradiograms were analyzed and mean relative intensity values were calculated as described in the legend to Fig. 3.
DISCUSSION
The goal of our ongoing studies is to understand pathological changes observed in intestinal tissues of M. avium subsp. paratuberculosis-infected cows and how these changes might interact with local and peripheral immune cells. It is reasonable to assume that M. avium subsp. paratuberculosis-infected tissues exhibit a chronic inflammatory pattern of gene expression, resulting in local tissue damage and influencing the peripheral immune system. Recent studies indicate that, indeed, inherent gene expression programs of peripheral blood mononuclear cells from Johne's disease-positive cattle are different from those of uninfected control cattle (6a). We propose that chronic exposure of immune cells to M. avium subsp. paratuberculosis during the long subclinical phase of Johne's disease is responsible for many of these differences.
In initial studies presented here, we detected significant (>1.5-fold; P < 0.05) differential expression of several genes in ileal tissues of M. avium subsp. paratuberculosis-infected cows, relative to a reference composed of pooled ileal tissues from control uninfected cattle. Genes expressed at higher levels in infected tissues relative to reference tissues included those encoding TRAF1, IL-1α, monocyte chemoattractant protein 2 (MCP-2), insulin-like growth factor-binding protein 2, N-cadherin, and CD29. Genes expressed at significantly lower levels in tissues from infected cattle, relative to reference tissues included those encoding an ortholog of the Notch signaling mediator protein Herp (BOTL-8_C03), the growth factor granulin (BOTL-1XH02R), and two EST clones (BOTL-6XD10R and BOTL-10_H04), representing genes with unknown function (Table 1).
TABLE 1.
Putative identification and functions of genes expressed at significantly different levels in ileal tissues of Johne's disease-afflicted cattle relative to uninfected cattle
| Gene or clone name | Description | Ontology or function | Change (fold) | P |
|---|---|---|---|---|
| β-1 integrin (CD29) | Amplicon representing bovine β-1 integrin (CD29). | Adhesion molecule | 2.06 | 0.012 |
| 1XE05R | Bovine EST clone highly similar to Homo sapiens CDC10 (cell division cycle 10) mRNA. | Cell cycle regulator | 1.59 | 0.022 |
| 1XH02R | Bovine EST clone weakly similar to Homo sapiens granulin mRNA | Growth factor | −1.64 | 0.017 |
| 3XF09R | Bovine EST clone highly similar to human mRNA for LCA homolog LAR protein (leukocyte antigen-related) mRNA | Cell cycle regulator | 1.93 | 0.044 |
| 6XD10R | Bovine EST clone not similar to any known gene | Unknown | 1.70 | 0.039 |
| 6XF09R | Bovine EST clone highly similar to human stimulatory G protein (of receptor-regulated K+ channels) alpha subunit mRNA | Potassium channel regulator | 1.73 | 0.016 |
| 8_C03 | Bovine EST clone highly similar to Homo sapiens mRNA for stress protein Herp mRNA | Mediator of Notch signaling and cell fate determination | −1.58 | 0.040 |
| 10_H04 | Bovine EST clone not similar to any known gene | Unknown | −1.82 | 0.025 |
| 13_F10 | Bovine EST clone moderately similar to Mus musculus serine dehydratase mRNA | Enzyme involved in glucose metabolism | −2.22 | 0.031 |
| TRAF1 | Amplicon representing bovine TNF-α receptor-associated factor 1 | Proapoptotic intracellular signaling molecule | 9.87 | 0.027 |
| IGFBP-2 | Amplicon representing bovine insulin-like growth factor-2 | 1.71 | 0.039 | |
| IL-1α | Amplicon representing bovine interleukin-1 | Proinflammatory cytokine | 4.98 | 0.030 |
| MCP-2 | Amplicon representing bovine monocyte chemotactic protein-2 | Chemotactic factor | 2.37 | 0.022 |
| N-cadherin | Amplicon representing bovine neural cadherin | Adhesion molecule | 1.66 | 0.0056 |
| RBQ-3 | Amplicon representing bovine retinoblastoma-binding protein RBQ-3 mRNA | Transcription coactivator | 1.73 | 0.034 |
CD29 is associated with proinflammatory Th1-like immune responses, promoting migration of T-lymphocytes to areas of inflammation, and adhesion between T cells and antigen presenting cells (36). Enhanced expression of CD29 is therefore consistent with inflammatory responses in M. avium subsp. paratuberculosis-infected ileal tissues. Likewise, MCP-2 is a crucial element of innate immunity, involved with directing migration of monocytes to regions of inflammation (33). MCP-2 has also been linked to granuloma formation and infections with gram-positive bacteria have been correlated with enhanced expression of MCP-2 (33).
The most dramatic increases in gene expression observed in samples of ileal tissue from Johne's disease-positive cattle, relative to reference tissues were for genes encoding IL-1α and TRAF1. Because of this and the potential importance of these two genes in regulating immune responses and immune cell fate, we investigated their regulation in infected ileal tissues further. Q-RT-PCR analysis confirmed dramatic overexpression of genes encoding IL-1α and TRAF1 in infected ileal tissues, indicating that actual expression levels were much higher than suggested by cDNA microarrays. This is consistent with previous data on IFN-γ expression in peripheral blood mononuclear cells from M. avium subsp. paratuberculosis-infected cattle where the lower dynamic range of cDNA microarrays grossly underestimated the actual level of enhanced expression (6a).
Enhanced gene expression does not always correlate with increased protein levels and thus it was important to determine if IL-1α and TRAF1 proteins were expressed at higher levels in ileal tissues from infected cattle, relative to similar tissues from uninfected control cattle. Western blot analysis indeed confirmed that both proteins were expressed at significantly (P < 0.05) higher levels in tissues from infected cattle, relative to controls. Although our results to date do not address pathological consequences of elevated IL-1α and TRAF1 expression in ileal tissues of cattle infected with M. avium subsp. paratuberculosis, known functions of these two proteins in model systems suggest they will be important in understanding both the pathology of Johne's disease and how intestinal lesions might influence the peripheral immune system.
TRAF1 is expressed in lymphocytes and dendritic cells, and is closely involved with other proteins regulating cell survival and a wide range of genes involved with immune response and inflammation (37). Specifically, TRAF1 associates with the cytosolic domain of CD30 as well as other TNF-α receptor family members, including CD40 and TNF-α receptor 1 (TNFR1). TRAF1 expression inhibits antigen-induced cell death of CD8+ T cells and TNF-α-induced apoptosis (27a, 33a). However, following cleavage by caspase-8 during TNF-α-induced apoptosis the TRAF1 carboxyl-terminal fragment (TRAF-c) may heterodimerize with TRAF2 (13). TRAF2 is associated with TNFR1 through the TNFR1 death domain-associated protein (TRADD), where it induces antiapoptotic signals, including activation of NF-κB (12, 18). Thus, association of caspase-8 cleaved TRAF1-c with TRAF2 sequesters TRAF2 from TNFR1 increasing sensitivity to TNF-α-induced apoptotic signals. TRAF1 is also known to play an antiapoptotic role via an amino-terminal fragment binding to specific regions of cytosolic CD30 (3). Thus, in future experiments, it will be important to determine cell types responsible for enhanced expression of TRAF1, the relative ratios of intact TRAF1, TRAF-c, and TRAF-n in those cells, as well as activity of caspase-8, and sensitivity of TRAF1 overexpressing cells from ileal tissues to TNF-α-induced apoptosis.
IL-1α is a known proinflammatory cytokine directly involved with differentiation of Th0 cells to Th1 or Th2 cells in mice (10). IL-1α is also essential for production and maintenance of granulomas in mycobacterial infections (15, 15). At low concentrations, IL-1α acts to recruit leukocytes to sites of infection by activating adhesion molecules on endothelial cells (10). In studies of inflamed intestinal tissues in mice IL-1α was implicated as having a key role in inflammatory responses, whereas in the same study IL-1β, TNF-α, and IFN-γ all did not significantly affect the inflammatory response (10). The pathogenesis of M. avium subsp. paratuberculosis infection is thus consistent with enhanced expression of IL-1α, particularly since lesions associated with Johne's disease typically have high numbers of associated macrophages.
Importantly, several of the genes upregulated in M. avium subsp. paratuberculosis-infected tissues relative to control tissues discovered in this report are linked via regulatory and transcriptional pathways. For example, IL-1α is associated with enhanced MCP-2 expression and has also been implicated in regulation of TRAF1 and insulin-like growth factor-binding protein 2 expression (3, 11, 33). In addition, enhanced expression of IL-1α induces adhesion molecule expression in endothelial cells (10) and our results suggest that CD29 and N-cadherin are upregulated in ileal tissues from Johne's disease-positive cattle.
In summary we used a bovine-specific cDNA microarray to demonstrate that several genes directly involved in inflammatory responses, immune cell adhesion, and immune cell fate are differentially expressed in ileal tissues from M. avium subsp. paratuberculosis-infected cattle relative to noninfected control cattle. Dramatic overexpression of transcripts encoding IL-1α and TRAF1 in ileal tissues of Johne's disease-positive cattle was confirmed by Q-RT-PCR, and gene expression results were extended to demonstrate enhanced levels of these two proteins in M. avium subsp. paratuberculosis-infected tissue, compared to similar tissues from noninfected controls. Our novel results suggest new avenues for further research on pathogenesis associated with M. avium subsp. paratuberculosis infection, particularly with regard to the causes of extensive intestinal tissue damage and effects on the peripheral immune system. The results presented here open the possibility that at last some of the symptoms of Johne's disease may be due to IL-1α toxicity. Furthermore, overexpression of TRAF1 may lead to decreased macrophage apoptosis, explaining, at least in part, the accumulation of these cells in lesions associated with Johne's disease.
Acknowledgments
We acknowledge the outstanding technical support of Chris Colvin and thank Jeanne Burton for many helpful discussions and critical review of the manuscript. We also acknowledge the kind assistance of Janet Ireland and Patty S. D. Weber in conducting Western blot analysis and Sue Sipkovsky for assistance with cDNA microarray analysis.
We also acknowledge the generous financial support of the College of Agriculture and Natural Science, the Michigan State University Agricultural Experiment Station, and the Office of the Vice President for Research and Graduate Studies at Michigan State University. Additional support for this project was provided by the Michigan Animal Industry Coalition and U.S. Department of Agriculture IFAFS grant number 2001-52100-11211.
Editor: B. B. Finlay
REFERENCES
- 1.Bannantine, J. P., and J. R. Stabel. 2002. Killing of Mycobacterium avium subspecies paratuberculosis within macrophages. BMC Microbiol. 2:2. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begara-McGorum, I., L. A. Wildblood, C. J. Clarke, K. M. Connor, K. Stevenson, C. J. McInnes, J. M. Sharp, and D. G. Jones. 1998. Early immunopathological events in experimental ovine paratuberculosis. Vet. Immunol. Immunopathol. 63:265-287. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, J. R., and J. S. Pober. 2001. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20:6482-6491. [DOI] [PubMed] [Google Scholar]
- 4.Buergelt, C. D., C. Hall, K. McEntee, and J. R. Duncan. 1978. Pathological evaluation of paratuberculosis in naturally infected cattle. Vet Pathol. 15(2):196-207. [DOI] [PubMed] [Google Scholar]
- 5.Burrells, C., C. J. Clarke, A. Colston, J. M. Kay, J. Porter, D. Little, and J. M. Sharp. 1999. Interferon-gamma and interleukin-2 release by lymphocytes derived from the blood, mesenteric lymph nodes and intestines of normal sheep and those affected with paratuberculosis (Johne's disease). Vet. Immunol. Immunopathol. 68:139-148. [DOI] [PubMed] [Google Scholar]
- 6.Corpa, J. M., J. Garrido, J. F. Garcia Marin, and V. Perez. 2000. Classification of lesions observed in natural cases of paratuberculosis in goats. J. Comp. Pathol. 122:255-265. [DOI] [PubMed] [Google Scholar]
- 6a.Coussens, P. M., C. J. Colvin, G. J. M. Rosa, J. Perez Lasplur, and M. D. Elftman. 2003. Evidence for a Novel Gene Expression Program in Peripheral Blood Mononuclear Cells from Mycobacterium avium subsp. paratuberculosis-Infected Cattle. Infect. Immun. 71:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coussens, P., C. Colvin, A. Abouzied, K. Wiersma, and S. Sipkovsky. 2002. Gene expression profiling of peripheral blood mononuclear cells from cattle infected with M. avium subsp. paratuberculosis. Infect. Immun. 70:5494-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussens, P. M. 2001. Interactions between Mycobacterium avium subsp. paratuberculosis and the bovine immune system. Animal Health Res. Rev. 2:141-161. [PubMed] [Google Scholar]
- 9.Dube, P. H., P. A. Revell, D. D. Chaplin, R. G. Lorenz, and V. L. Miller. 2001. A role for IL-1 alpha in inducing pathologic inflammation during bacterial infection. Proc. Natl. Acad. Sci. 98:10880-10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, J. X., and A. C. Issekutz. 1996. Expression of VCAM-1 and VLA-4 dependent T-lymphocyte adhesion to dermal fibroblasts stimulated with proinflammatory cytokines. Immunology 89:375-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoeflich, A., R. Reisinger, H. Lahm, W. Kiess, W. F. Blum, H. J. Kolb, M. M. Weber, and E. Wolf. 2001. Insulin-like growth factor-binding protein 2 in tumorigenesis: protector or promoter? Cancer Res. 61:8601-8610. [PubMed] [Google Scholar]
- 12.Hsu, H., H. B. Shu, M. G. Pan, and D. V. Goeddel. 1996. TRADD-TRAF-2 and TRAD-FADD interactions define two distinct signal transduction pathways. Cell 84:299-308. [DOI] [PubMed] [Google Scholar]
- 13.Jang, H. D., Y. M. Chung, J. H. Baik, Y. G. Choi, I. S. Park, Y. K. Jung, and S. Y. Lee. 2001. Caspase-cleaved TRAF1 negatively regulates the antiapoptotic signals of TRAF-2 during TNF-induced cell death. Biochem. Biophys. Res. Commun. 28:499-505. [DOI] [PubMed] [Google Scholar]
- 14.Juffermans, N. P., S. Florquin, L. Camoglio, A. Verbon, A. H. Kolk, P. Speelman, S. J. van Deventer, and T. van Der Poll. 2000. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J. Infect. Dis. 182:902-908. [DOI] [PubMed] [Google Scholar]
- 15.Juffermans, N. P., A. Verbon, S. J. van Deventer, H. van Deutekom, P. Speelman, and T. van der Poll. 1998. Tumor necrosis factor and interleukin-1 inhibitors as markers of disease activity of tuberculosis. Am. J. Respir. Crit. Care Med 157:1328-1331. [DOI] [PubMed] [Google Scholar]
- 16.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77:123-128. [DOI] [PubMed] [Google Scholar]
- 17.Lee, H., J. R. Stabel, and M. E. Kehrli, Jr. 2001. Cytokine gene expression in ileal tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis. Vet. Immunol. Immunopathol. 82:73-85. [DOI] [PubMed] [Google Scholar]
- 18.Liu, D. G., H. Hsu, D. V. Goeddel, and M. Karin. 1996. Dissection of TNF receptor 1 effector function; JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell 87:565-576. [DOI] [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data with real-time quantitative PCR and the 2(-Delta Delta C[t]) method. Methods 4:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Momotani, E., D. L. Whipple, A. B. Thiermann, and N. F. Cheville. 1988. Role of M cells and macrophages in the entrance of Mycobacterium avium subsp. paratuberculosis into domes of ileal Peyer's patches in calves. Vet. Pathol. 25:131-137. [DOI] [PubMed] [Google Scholar]
- 21. National Animal Health Monitoring System. 1997. Johne's disease on U.S. dairy operations. N245.1097. National Animal Health Monitoring System, U.S. Department of Agriculture, Washington, D.C.
- 22.Navarro, J. A., G. Ramis, J. Seva, F. J. Pallares, and J. Sanchez. 1998. Changes in lymphocyte subsets in the intestine and mesenteric lymph nodes in caprine paratuberculosis. J. Comp. Pathol. 118:109-121. [DOI] [PubMed] [Google Scholar]
- 23.Perez, V., J. Tellechea, J. M. Corpa, M. Gutierrez, and J. F. Garcia Marin. 1999. Relation between pathologic findings and cellular immune responses in sheep with naturally acquired paratuberculosis. Am. J. Vet. Res. 60:123-127. [PubMed] [Google Scholar]
- 24.SAS Institute. 1990. Procedures guide: version 6, 3rd ed. SAS Institute, Inc. Cary, N.C.
- 25.SAS Institute. 2000. SAS/STAT software, 8th ed. SAS Institute, Inc., Cary, N.C.
- 26.Seitz, S. E., L. E. Heider, W. D. Heuston, S. Bech-Nielsen, D. M. Rings, and L. Spangler. 1989. Bovine fetal infection with Mycobacterium avium subsp. paratuberculosis. J. Am. Vet. Med. Assoc. 194:1423-1426. [PubMed] [Google Scholar]
- 27.Sigur-Dardottir, O. G., C. M. Press, and O. Evensen. 2001. Uptake of Mycobacterium avium subsp. paratuberculosis through the distal small intestinal mucosa in goats: an ultrastructural study. Vet. Pathol. 38:184-189. [DOI] [PubMed] [Google Scholar]
- 27a.D. E. Speiser, S. Y. Lee, B. Wong, J. Arron, A. Santana, Y. Y. Kong, P. S. Ohashi, and Y. Choi. 1997. A regulatory role for TRAF1 in antigen-induced apoptosis of T cells. J. Exp. Med. 185:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stabel, J. R. 2000. Transitions in immune responses to Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 77:465-473. [DOI] [PubMed] [Google Scholar]
- 29.Streeter, R. N., G. F. Hoffsis, S. Bech-Nielsen, W. P. Shulaw, and D. M. Rings. 1995. Isolation of Mycobacterium avium subsp. paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 56:1322-1324. [PubMed] [Google Scholar]
- 30.Suchyta, S., S. Sipkovsky, R. Halgren, R. Kruska, M. Elftman, M. Weber-Nielson, M. Vandeharr, and P. Coussens. 2003. Bovine mammary gene expression profiling with a cDNA microarray enhanced for mammary specific transcripts. Physiol. Genomics, in press. [DOI] [PubMed]
- 31.Sweeney, R. W. 1996. Transmission of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 12:305-312. [DOI] [PubMed] [Google Scholar]
- 32.Sweeney, R. W., D. E. Jones, P. Habecker, and P. Scott. 1998. Interferon-gamma and interleukin 4 gene expression in cows infected with Mycobacterium. M. avium subsp. paratuberculosis. Am. J. Vet. Res. 59:842-847. [PubMed] [Google Scholar]
- 33.Tsuneyama, K., K. Harada, M. Yasoshima, K. Hiramatsu, C. R. Mackay, I. R. Mackay, M. E. Gershwin, and Y. Nakanuma. 2001. Monocyte chemotactic protein-1, -2, and -3 are distinctively expressed in portal tracts and granulomata in primary biliary cirrhosis: implications for pathogenesis. J. Pathol. 193:102-109. [DOI] [PubMed] [Google Scholar]
- 33a.Wang, C. Y., M. W. Mayo, R. G. Korneluk, D. V. Goeddel, and A. S. Baldwin, Jr. 1998.. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 nd c-IAP2 to suppress caspase-8 activation. Science 281:1680-1683. [DOI] [PubMed] [Google Scholar]
- 34.Wells, S. J., and B. A. Wagner. 2000. Herd-level risk factors for infection with MycobacteriuM. avium subsp. paratuberculosis in US dairies and association between familiarity of the herd manager with the disease or prior diagnosis of the disease in that herd and use of preventive measures. J. Am. Vet. Med. Assoc. 216:1450-1457. [DOI] [PubMed] [Google Scholar]
- 35.Whitlock, R. H., and C. Buergelt. 1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. North Am. Food Anim. Pract. 12:345-356. [DOI] [PubMed] [Google Scholar]
- 36.Woods, M. L., and Y. Shimizu. 2001. Signaling networks regulating beta1 integrin-mediated adhesion of T lymphocytes to extracellular matrix. J. Leukoc. Biol. 69:874-880. [PubMed] [Google Scholar]
- 37.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zapata, J. M., M. Krajewska, S. Krajewski, S. Kitada, K. Welsh, A. Monks, N. McCloskey, J. Gordon, T. J. Kipps, R. D. Gascoyne, A. Shabaik, and J. C. Reed. 2000. TNFR-associated factor family protein expression in normal tissues and lymphoid malignancies. J. Immunol. 16:5084-5096. [DOI] [PubMed] [Google Scholar]
- 39.Zurbrick, B. G., and C. J. Czuprynski. 1987. Ingestion and intracellular growth of Mycobacterium avium subsp. paratuberculosis within bovine blood monocytes and monocyte-derived macrophages. Infect. Immun. 55:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zurbrick, B. G., D. M. Follett, and C. J. Czuprynski. 1988. Cytokine regulation of the intracellular growth of Mycobacterium avium subsp. paratuberculosis in bovine monocytes. Infect. Immun. 56:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]