Abstract
Proteins contained on purified COPII vesicles were analyzed by matrix-assisted laser desorption ionization mass spectrometry combined with database searching. We identified four known vesicle proteins (Erv14p, Bet1p, Emp24p, and Erv25p) and an additional nine species (Yip3p, Rer1p, Erp1p, Erp2p, Erv29p, Yif1p, Erv41p, Erv46p, and Emp47p) that had not been localized to ER vesicles. Using antibodies, we demonstrate that these proteins are selectively and efficiently packaged into COPII vesicles. Three of the newly identified vesicle proteins (Erv29p, Erv41p, and Erv46p) represent uncharacterized integral membrane proteins that are conserved across species. Erv41p and Erv46p were further characterized. These proteins colocalized to ER and Golgi membranes and exist in a detergent-soluble complex that was isolated by immunoprecipitation. Yeast strains lacking Erv41p and/or Erv46p are viable but display cold sensitivity. The expression levels of Erv41p and Erv46p are interdependent such that Erv46p was reduced in an erv41Δ strain, and Erv41p was not detected in an erv46Δ strain. When the erv41Δ or ev46Δ alleles were combined with other mutations in the early secretory pathway, altered growth phenotypes were observed in some of the double mutant strains. A cell-free assay that reproduces transport between the ER and Golgi indicates that deletion of the Erv41p–Erv46p complex influences the membrane fusion stage of transport.
Keywords: ER, Golgi, vesicles, coat proteins, trafficking
Introduction
Membrane-bound transport vesicles are central intermediates for intracellular trafficking of proteins and lipids. The production of many types of transport vesicles depends on coat protein complexes that form vesicles and select the desired set of cargo (Schekman and Orci 1996). Although the importance of vesicle carriers has been appreciated, the molecular mechanisms underlying distinct stages in vesicle-mediated transport are not well understood. Several approaches have been used to identify and characterize this protein machinery, including the isolation and characterization of specific vesicle carriers involved in synaptic transmission, endocytosis, exocytosis, and intra-Golgi transport (for reviews see Söllner and Rothman 1996; Arvan and Castle 1998; Foletti et al. 1999; Smith and Pearse 1999). We have undertaken a comprehensive analysis of ER-derived transport vesicles to elucidate the molecular events associated with transport between the ER and Golgi compartments.
Transport between these early compartments of the secretory pathway is bidirectional and mediated by the COPI and COPII coat complexes. In general, it is thought that the COPII coat buds vesicles from the ER for anterograde transport, whereas the COPI coat is responsible for retrograde transport of recycled proteins from Golgi and pre-Golgi compartments back to the ER (Mellman and Warren 2000). Formation of COPII-coated vesicles may be reproduced in a cell-free reaction with purified soluble components (the Sar1p GTPase, the Sec23p complex, and the Sec13p complex) and washed ER membranes (Salama et al. 1993; Barlowe et al. 1994). A highly purified preparation of uncoated ER-derived vesicles can be obtained through a scaled up version of the cell-free budding assay. Examination of purified vesicles on protein-stained gels reveals a characteristic set of polypeptides that are solubilized by detergents but not by an elevated pH treatment (Barlowe et al. 1994; Rexach et al. 1994). Some of the abundant ER vesicle (Erv) proteins have been characterized (Barlowe et al. 1994; Schimmöller et al. 1995; Belden and Barlowe 1996; Powers and Barlowe 1998) and are found to cycle between the ER and Golgi compartments and function in the processes of vesicle formation and/or site-specific membrane fusion. In this report, we extend our studies to identify additional Erv proteins contained on ER-derived vesicles by mass spectrometry coupled to database searching. Several new Erv proteins were identified and two of these, Erv41p and Erv46p, were characterized further. Erv41p and Erv46p exist in a complex that is probably conserved across species and appears to influence the vesicle fusion stage of transport between the ER and Golgi compartments.
Materials and Methods
Yeast Strains and Media
Strains used for this study are listed in Table . Unless noted otherwise, cultures were grown at 30°C in rich medium (YPD, 1% Bacto-yeast extract, 2% Bacto-peptone, 2% dextrose) or in minimal medium (YMD, 0.67% yeast nitrogen base without amino acids, 2% dextrose) containing the appropriate amino acid supplements. Standard yeast (Sherman 1991) and cloning protocols (Ausubel et al. 1987) were used.
Table 1.
Strain List
| Strain | Genotype | Reference |
|---|---|---|
| FY833 | MATa his3200 ura3-52 leu21 lys2202 trp163 | Winston et al. 1995 |
| FY834 | MATα his3200 ura3-52 leu21 lys2202 trp163 | Winston et al. 1995 |
| RSY255 | MATα ura3-52 leu2-3,-112 | Kaiser and Schekman 1990 |
| RSY263 | MATα sec12-4 ura3-52 leu2-3,112 | Kaiser and Schekman 1990 |
| RSY265 | MATα sec13-1 ura3-52 his4-619 | Kaiser and Schekman 1990 |
| RSY267 | MATα sec16-2 ura3-52 his4-619 | Kaiser and Schekman 1990 |
| RSY277 | MATα sec21-1 ura3-52 | Kaiser and Schekman 1990 |
| RSY281 | MATα sec23-1 ura3-52 his4-619 | Kaiser and Schekman 1990 |
| RSY309 | MATα sec12-1 leu2-3,112 | Kaiser and Schekman 1990 |
| RSY962 | MATa sec35-1 lys2-801 | Wuestehube et al. 1996 |
| RSY976 | MATa ura3-52 ypt1-3 | Wuestehube et al. 1996 |
| CBY99 | FY834 with emp24::LEU2 | Belden and Barlowe 1996 |
| CBY263 | MATα trp1-1 ade2-1 ura3-1 leu2-3,112 can1-100 sed5-1 | Cao et al. 1998 |
| CBY300 | FY834 with uso1-1 | Barlowe 1997 |
| CBY356 | FY834 with erv14::HIS3 | Powers and Barlowe 1998 |
| CBY358 | FY833 with erv14::HIS3 | Powers and Barlowe 1998 |
| CBY453 | FY833 x FY834 | Powers and Barlowe 1998 |
| CBY763 | CBY453 diploid with ERV41/erv41::HIS3 | This study |
| CBY767 | CBY453 diploid with ERV46/ERV46::HIS3MX6-PGAL1-3HA | This study |
| CBY770 | FY834 with ERV46::HIS3MX6-PGAL1-3HA | This study |
| CBY771 | FY833 with ERV46::HIS3MX6-PGAL1-3HA | This study |
| CBY782 | CBY453 diploid with ERV41/ERV41::TRP1-PGAL1-3HA | This study |
| CBY783 | FY834 with ERV41::TRP1-PGAL1-3HA | This study |
| CBY784 | FY833 with ERV41::TRP1-PGAL1-3HA | This study |
| CBY794 | FY833 with erv41::HIS3 erv46::KAN | This study |
| CBY795 | FY834 with erv41::HIS3 erv46::KAN | This study |
| CBY796 | FY833 with erv41::HIS3 | This study |
| CBY797 | FY834 with erv41::HIS3 | This study |
| CBY798 | FY833 with erv46::KAN | This study |
| CBY799 | FY834 with erv46::KAN | This study |
| CBY801 | FY834 with YIF1::HIS3MX6-3HA | This study |
| CBY822 | FY833 with erv14::HIS3 erv46::KAN | This study |
| CBY823 | FY834 with erv14::HIS3 erv46::KAN | This study |
| CBY825 | FY834 with erv14::HIS3 erv41::HIS3 | This study |
| CBY826 | FY833 with erv14::HIS3 erv41::HIS3 | This study |
| CBY829 | FY834 with ypt1-3 | This study |
| CBY831 | FY833 with emp24::LEU2 erv41::HIS3 erv46::KAN | This study |
| CBY832 | FY834 with emp24::LEU2 erv41::HIS3 erv46::KAN | This study |
| CBY836 | CBY822 containing pRS426-ERV46 | This study |
| CBY840 | FY833 with erv41::HIS3 ypt1-3 | This study |
| CBY841 | FY834 with erv41::HIS3 ypt1-3 | This study |
| CBY842 | FY833 with erv46::KAN ypt1-3 | This study |
| CBY843 | FY834 with erv46::KAN ypt1-3 | This study |
| CBY844 | FY833 with erv41::HIS3 erv46::KAN ypt1-3 | This study |
| CBY845 | FY834 with erv41::HIS3 erv46::KAN ypt1-3 | This study |
| CBY847 | CBY826 containing pRS424-ERV41 | This study |
| CBY848 | FY833 with erv41::HIS3 sec21-1 | This study |
| CBY849 | FY834 with erv41::HIS3 sec21-1 | This study |
| CBY850 | FY833 with erv46::KAN sec21-1 | This study |
| CBY851 | FY834 with erv46::KAN sec21-1 | This study |
| CBY852 | FY834 with erv41::HIS3 erv46::KAN sec21-1 | This study |
| CBY853 | FY833 with erv41::HIS3 sed5-1 | This study |
| CBY854 | FY833 with ade2-1 erv46::KAN sed5-1 | This study |
| CBY855 | FY834 with erv46::KAN sed5-1 | This study |
| CBY856 | FY833 with erv41::HIS3 erv46::KAN sed5-1 | This study |
| CBY857 | FY834 with ade2-1 erv41::HIS3 erv46::KAN sed5-1 | This study |
| CBY859 | FY833 with erv41::HIS3 sec12-1 | This study |
| CBY860 | FY834 with erv41::HIS3 sec12-1 | This study |
| CBY861 | FY833 with erv46::KAN sec12-1 | This study |
| CBY862 | FY834 with erv46::KAN sec12-1 | This study |
| CBY863 | FY833 with erv41::HIS3 erv46::KAN sec12-1 | This study |
| CBY877 | FY833 with erv14::HIS3 rer1::KAN | This study |
| CBY878 | FY834 with erv14::HIS3 rer1::KAN | This study |
| CBY884 | FY833 with erv41::HIS3 nce102::KAN | This study |
| CBY885 | FY834 with erv41::HIS3 nce102::KAN | This study |
| Strain | Genotype | Reference |
| CBY893 | FY833 with erv14::HIS3 erv41::HIS3 erv46::KAN | This study |
| CBY894 | FY834 with erv14::HIS3 erv41::HIS3 erv46::KAN | This study |
| CBY909 | FY833 with erv46::KAN uso1-1 | This study |
| CBY910 | FY834 with erv46::KAN uso1-1 | This study |
| CBY911 | FY833 with erv41::HIS3 uso1-1 | This study |
| CBY912 | FY834 with erv41::HIS3 uso1-1 | This study |
| CBY913 | FY833 with erv41::HIS3 erv46::HIS3 uso1-1 | This study |
| CBY914 | FY834 with erv41::HIS3 erv46::HIS3 uso1-1 | This study |
| CBY915 | FY833 with erv46::KAN sec23-1 | This study |
| CBY916 | FY834 with erv46::KAN sec23-1 | This study |
| CBY917 | FY833 with erv41::HIS3 sec23-1 | This study |
| CBY918 | FY833 with erv41::HIS3 erv46::KAN sec23-1 | This study |
| CBY919 | FY834 with erv41::HIS3 erv46::KAN sec23-1 | This study |
| CBY920 | FY833 with erv46::KAN sec16-2 | This study |
| CBY921 | FY834 with erv46::KAN sec16-2 | This study |
| CBY922 | FY833 with erv41::HIS3 sec16-2 | This study |
| CBY923 | FY834 with erv41::HIS3 sec16-2 | This study |
| CBY924 | FY834 with erv41::HIS3 erv46::KAN sec16-2 | This study |
| CBY930 | FY834 with erv41::HIS3 erv46::KAN sec13-1 | This study |
| CBY931 | FY833 with erv41::HIS3 erv46::KAN sec13-1 | This study |
| CBY932 | FY834 with erv41::HIS3 sec13-1 | This study |
| CBY933 | FY834 with erv46::KAN sec13-1 | This study |
| CBY934 | FY833 with erv46::KAN sec13-1 | This study |
| CBY940 | FY833 with erv46::KAN sec35-1 | This study |
| CBY941 | FY834 with erv46::KAN sec35-1 | This study |
| CBY942 | FY833 with erv41::HIS3 sec35-1 | This study |
| CBY943 | FY834 with erv41::HIS3 sec35-1 | This study |
| CBY944 | FY833 with erv41::HIS3 erv46::KAN sec35-1 | This study |
| CBY945 | FY834 with erv41::HIS3 erv46::KAN sec35-1 | This study |
| CBY948 | FY833 with erv41::HIS3 rer1::KAN | This study |
| CBY949 | FY834 with erv41::HIS3 rer1::KAN | This study |
| CBY950 | CBY453 diploid with ERV29/ERV29::HIS3MX6-PGAL1-3HA | This study |
| CBY963 | FY833 with erv14::HIS3 rer1::KAN | This study |
| CBY964 | FY834 with erv14::HIS3 rer1::KAN | This study |
| CBY965 | FY833 with erv29::KAN | This study |
| CBY966 | FY834 with erv29::KAN | This study |
| CBY967 | FY833 with erv29::KAN erv41::HIS3 | This study |
| CBY968 | FY834 with erv29::KAN erv41::HIS3 | This study |
| CBY969 | FY833 with erv14::HIS3 erv29::KAN | This study |
| CBY970 | FY834 with erv14::HIS3 erv29::KAN | This study |
Plasmid Construction
Primer sequences are listed in Table . The sequences of ERV41 (YML067c) and ERV46 (YAL042w) were obtained from the Saccharomyces Genome Database. To generate plasmid pRS424-ERV41, the ERV41 sequence, and ∼300 bp of its flanking upstream and downstream, sequences were cloned from genomic DNA prepared from strain RSY255 using primers YML067c-NotI and YML067c-BamHI. The product was ligated into the NotI and BamHI restriction sites of the pRS424 vector (Christianson et al. 1992). ERV46 and its flanking regions of ∼300 bp were amplified using primers YAL042w-NotI and YAL042w-BamHI and inserted into the NotI and BamHI sites of the pRS426 vector (Christianson et al. 1992) to yield plasmid pRS426-ERV46. Correct amplification and integration were verified by DNA sequencing.
Table 2.
Primer Sequences
| Primer | Sequence |
|---|---|
| YML067c-NotI | 5′-ATAAGAATGCGGCCGCAACTCCCCTTCTCCAGACTTG-3′ |
| YML067c-BamHI | 5′-CGCGGATCCGCGAAGTATGCGGGACGCTG-3′ |
| YAL042w-NotI | 5′-ATAAGAATGCGGCCGCCTGCAGATGACATTGCGCTGC-3′ |
| YAL042w-BamHI | 5′-CGCGGATCCGCCATGATCTCTCGGGTTGG-3′ |
| YML067c-pQE-F | 5′-CGCGGATCCACGAAATGTGATTGGTTGC-3′ |
| YML067c-pQE-R | 5′-CCCAAGCTTCACCTTTAGCAGCGACATC-3′ |
| YAL042w-pQE-F | 5′-CGCGGATCCTGTGACCTGGTGAATCTCG-3′ |
| YAL042w-pQE-R | 5′-CCCAAGCTTCCTGTCCGGGAACACC-3′ |
| OCH1-pQE-F | 5′-CGCGGATCCGATCCAAGATTTGAAGAAG-3′ |
| OCH1-pQE-R | 5′-CCCAAGCTTGGGTATGATGAAAGGAGAG-3′ |
| YML067c-KO-F | 5′-TGCTTTTGCTGTAACGTACAGGCAGAGACTTCAGGAGTATATAGGCCTCCTCTAGTACACTC-3′ |
| YML067c-KO-R | 5′-GAAACGTTATCTTGGGTATACTGCATTCTCTTTTCTTTTACATATGCGCGCCTCGTTCAGAATG-3′ |
| HIS3 | 5′-GCCTCATCCAAAGGCGC-3′ |
| YAL042w-F4 | 5′-CGCCATTATAAAGGAAAGCTAGTTTTATGTCTCGTATACAGAATTCGAGCTCGTTTAAAC-3′ |
| YAL042w-R3 | 5′-TAGCGAATGCGTCCAGCGACAGCAACGTGGACCTTTTCATGCACTGAGCAGCGTAATCTG-3′ |
| YML067c-F4 | 5′-GATATTATCAATGTTCTATTCTTACTGAGAAAAGCGTTCCGAATTCGAGCTCGTTTAAAC-3′ |
| YML067c-R3 | 5′-ATAACATACGAAACGCATCAAATGTCTTCAATCCTGCCATGCACTGAGCAGCGTAATCTG-3′ |
| YGR284c-F4 | 5′-ACGAAAGATCAAAGGTGTCCTTATTTACTTACAATAGCTGGAATTCGAGCTCGTTTAAAC-3′ |
| YGR284c-R3 | 5′-GGCATACCGCCAAAATTTCCAATAGGTCCTCTGTAAGACATGCACTGAGCAGCGTAATCTG-3′ |
| MHF2 | 5′-TGGCTTCATTTGGCAAAATGTTCTAATGTGGTTAATGGGTCGGATCCCCGGGTTAATTAA-3′ |
| MHR1 | 5′-GCATGAAATTAAATCCTCTCTTTGATCTCTTCAATCAAGAGAATTCGAGCTCGTTTAAAC-3′ |
For the overexpression of fusion proteins bearing NH2-terminal 6x histidine tags, fragments of ERV41, ERV46, and OCH1 were inserted into the BamHI and HindIII restriction sites of the pQE-30 vector (QIAGEN) to yield plasmids pQE-30-ERV41, pQE-30-ERV46, and pQE-30-OCH1, respectively. Primers YML067c-pQE-F and YML067c-pQE-R for ERV41, YAL042w-pQE-F and YAL042w-pQE-R for ERV46, and primers OCH1-pQE-F and OCH1-pQE-R for OCH1 were used to amplify portions of these genes from genomic RSY255 DNA.
Strain Construction
ERV41 was targeted for disruption with the HIS3 gene (Baudin et al. 1993). An erv41::HIS3 construct was amplified using primers YML067c-KO-F and YML067c-KO-R and pHISKO as a template. The product contains the HIS3 gene flanked by 43-bp upstream of the ERV41 start codon and 45-bp downstream of the ERV41 stop codon and was used to transform CBY453 cells. Several transformants showing histidine prototrophy were screened by PCR using primers YML067c-NotI and HIS3. One (CBY763) tested positive and was sporulated. Strains carrying erv46Δ, rer1Δ, or erv29Δ null alleles disrupted with a KAN marker were generated by a deletion project (Winzeler et al. 1999; Research Genetics) and crossed several times through the FY833/FY834 background to yield an isogenic set of spores.
Erv46p, Erv41p, and Erv29p were tagged at their NH2 termini by the addition of three repeated influenza virus hemagglutinin (HA) epitopes and put under control of the GAL1 promoter (Longtine et al. 1998): plasmid pFA6a-His3MX6-PGAL1-3HA and primers YAL042w-F4 and YAL042w-R3 were used to amplify the construct targeted to the ERV46 locus. CBY453 cells were transformed with this product and selected for histidine prototrophy. Several isolates were grown in YP with 2% galactose and 0.2% glucose and screened for expression of tagged protein by Western blot of membrane fractions using an anti-HA antibody. Approximately 50% of transformants tested positive, and one (CBY767) was sporulated to yield strains CBY770 and CBY771. ERV41 was targeted with a PCR product obtained using plasmid pFA6a-TRP1-PGAL1-3HA and primers YML067c-F4 and YML067c-R3. Transformants were selected for tryptophan prototrophy and screened for expression of HA-tagged protein. One positive diploid strain (CBY782) was sporulated to obtain strains CBY783 and CBY784. Primers YGR284c-F4 and YGR284c-R3 and plasmid pFA6a-His3MX6-PGAL1-3HA were used to amplify the construct directed to the ERV29 locus. CBY453 cells were transformed and selected for growth on minimal media lacking histidine. Three transformants were screened by Western blot with the anti-HA antibody, and one tested positive (CBY950). Yif1p was tagged by the insertion of three HA epitopes at its COOH terminus (Longtine et al. 1998), using primers MHF2 and MHR1 and plasmid pFA6a-3HA-His3MX6 as template. FY834 cells were transformed with the PCR product and selected for histidine prototrophy. 7 out of 10 transformants expressed tagged protein, and one was analyzed further (CBY801).
Genetic Analyses
To generate strains carrying multiple mutations, erv14Δ, erv41Δ, and erv46Δ, strains were mated with other mutants. If possible, diploids were selected using markers of the parent strains, otherwise, zygotes were picked under the microscope. Diploid strains were sporulated, and asci were dissected on YPD plates using a micromanipulator. Plates were incubated at room temperature, and germinated spores were scored for mating types, markers, and growth on YPD plates at 16°C, room temperature, 30°C, 36°C, and 38°C. In cases where several loci had been replaced by the HIS3 gene, deletions were scored by PCR using YML067c-NotI and the HIS3 internal primer for the detection of the erv41::HIS3 allele and the ERV14 specific primer GP3 (Powers and Barlowe 1998) and the internal HIS3 primer to detect the erv14::HIS3 deletion. In cases where one of the parent strains had a genetic background different from FY833/FY834, spores were backcrossed several times to obtain isogenic strains.
Antibodies and Immunoblotting
Polyclonal antibodies were raised against 6x histidine–tagged NH2-terminal fusion proteins of fragments of Erv41p (amino acid positions 75–274), Erv46p (amino acid positions 80–296), and Och1p (amino acid positions 302–480) expressed from plasmids pQE-30-ERV41, pQE-30-ERV46, and pQE-30-OCH1, respectively. All of the recombinant proteins localized to the insoluble fraction of cells disrupted in a French Press. These fractions were solubilized with 8 M urea, and the fusion proteins were purified on Ni-NTA agarose (QIAGEN) as recommended by the manufacturer. The recombinant proteins were used to immunize rabbits according to standard procedures. For Western blotting, these antisera were diluted 1:1,000.
Antibodies directed against carboxypeptidase Y (CPY), Emp47p, Erv14p, Erv25p, Kar2p, plasma membrane ATPase, Sec12p, Sec23p, Sec61p (Powers and Barlowe 1998), Bos1p (Cao and Barlowe 2000), Rer1p (Boehm et al. 1997), and Yip1p (Yang et al. 1998) were described earlier. A monoclonal anti-HA antibody was obtained from Berkeley Antibody Co. Western blots were developed using the ECL method (Amersham Pharmacia Biotech). For densitometric analysis, films were scanned and plotted using NIH Image 1.52.
Subcellular Fractionation
Membrane fractions were prepared by the bead-beat method in lysis buffer (25 mM Hepes, pH 7.0, 50 mM potassium acetate, 2 mM EDTA, 1 mM PMSF). Pellets were resolved on 12.5% polyacrylamide gels. Microsomes (Wuestehube and Schekman 1992) and semiintact cells (Baker et al. 1988) were prepared as described. Sucrose gradient fractionation of membrane organelles was performed according to Powers and Barlowe 1998. To determine whether Erv41p and Erv46p are integral membrane proteins, semiintact FY834 cells were suspended in buffer (20 mM Hepes, pH 7.0, 150 mM potassium acetate, 2 mM EDTA), in buffer with 1% Triton X-100, or in 0.1 M sodium carbonate, pH 11.0, 2 mM EDTA, incubated on ice for 10 min and centrifuged at 60,000 rpm in a TLA100.3 rotor (Beckman Coulter) for 12 min. Equivalent amounts of the total, supernatant, and pellet fractions were resolved on a 12.5% polyacrylamide gel.
In Vitro Vesicle Budding and Transport Assays
The synthesis of COPII vesicles was performed on a preparative scale by incubating washed microsomes in the absence or presence of the proteins Sar1p, Sec13–31p complex, and Sec23–24p complex as described previously (Barlowe et al. 1994; Belden and Barlowe 1996). After collection of fractions from nycodenz density gradients, peak fractions were diluted fourfold with buffer 88 and centrifuged at 60,000 rpm for 25 min in a TLA100.3 rotor (Beckman Coulter) to collect vesicles. The membrane pellets were dissolved in 40 μl of sample buffer, and solubilized proteins were resolved on a 15% polyacrylamide gel (Novex). Gels were developed with a colloidal blue staining kit (Novex), and polypeptide staining bands were excised for analysis by mass spectrometry as described (Jensen et al. 1999). Analytical scale budding reactions were performed as described (Barlowe et al. 1994), and packaging efficiencies were determined by densitometry of scanned blots. Vesicle tethering and fusion assays following 35S-labeled glyco-pro–α factor (gp-α-F) were described by Cao et al. 1998. The data plotted in these experiments are the average of duplicate determinations, and the error bars represent the range. Pulse–chase experiments were performed according to Belden and Barlowe 1996.
Immunoprecipitation Experiments
Microsomes were solubilized on ice with buffer 88 containing 2% Triton X-100 and 1 mM PMSF for 5 min, followed by a centrifugation at 14,000 g for 5 min. Portions (25 μl) of the supernatant were mixed with 1 ml of IP buffer (15 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100), 25 μl of a 50% protein A–Sepharose slurry (Amersham Pharmacia Biotech), and a saturating amount of anti-HA antibody and were incubated at 4°C for 2 h. The precipitates were washed four times with cold IP buffer, eluted from the beads by heating at 95°C in sample buffer, and resolved on polyacrylamide gels.
Results
Identification of ERV Proteins
A protocol to generate ER-derived vesicles in vitro from washed microsomes and purified COPII components (Barlowe et al. 1994) was scaled up to prepare sufficient amounts of material to observe polypeptides on protein-stained polyacrylamide gels (Belden and Barlowe 1996). A set of polypeptides was observed (Fig. 1), and individual bands were subjected to tryptic peptide mass mapping by matrix-assisted laser desorption/ionization mass spectrometry followed by sequence database searching as described (Shevchenko et al. 1996; Jensen et al. 1999). Several proteins were identified, some that had been previously described and others that had not been characterized. Importantly, three of these proteins (Erv14p, Emp24p, and Erv25p) had been identified in our initial approaches using automated NH2-terminal sequencing (Belden and Barlowe 1996; Powers and Barlowe 1998). Therefore, the isolated ER-derived vesicles are comparable to previous preparations, which suggests that this method should allow for the identification of additional Erv proteins. The SNARE protein Bet1p (Newman and Ferro-Novick 1987) was also identified in this preparation and had been previously detected on ER-derived vesicles by immunoblot analysis and is required for transport between the ER and Golgi (Newman et al. 1992; Cao and Barlowe 2000).
Figure 1.
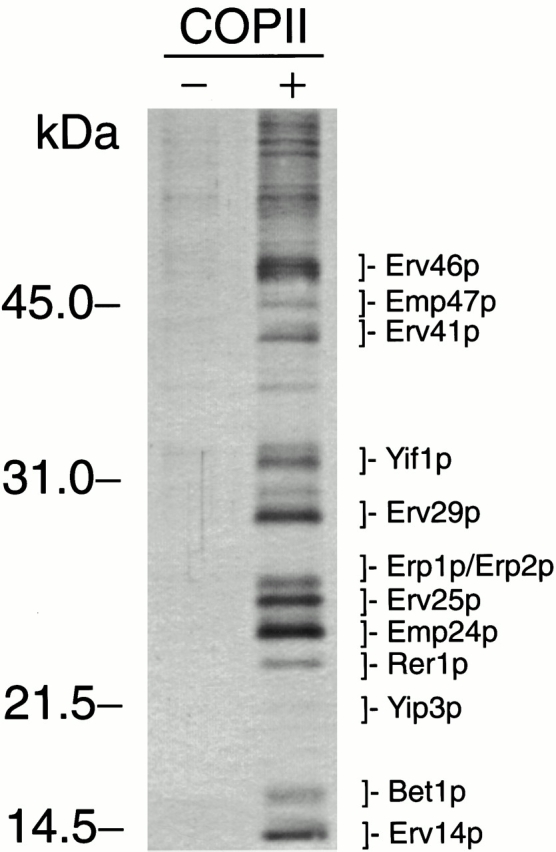
Identification of Erv proteins. COPII-coated vesicles were synthesized in vitro from ER membranes in the presence of COPII proteins (+), were solubilized in sample buffer, and were resolved on a 15% polyacrylamide gel. As a negative control, a mock reaction without COPII proteins (−) was performed. Proteins were silver stained for this figure. For mass spectroscopy, proteins were stained with colloidal blue, and individual bands were excised.
Nine other polypeptides identified in this preparation had not been localized to ER-derived vesicles before. Yip3p, Rer1p, Erp1p, Erp2p, Yif1p, and Emp47p are conserved proteins that had been characterized and are thought to operate in transport through the early secretory pathway (Sato et al. 1995; Schröder et al. 1995; Andrulis et al. 1998; Yang et al. 1998; Marzioch et al. 1999). Erv29p, Erv41p, and Erv46p represent uncharacterized proteins. Peptides derived from the Erv29p band correspond to the ORF YGR284c that encodes a nonessential 35-kD integral membrane protein terminating in the sequence KKKIY. According to its apparent molecular weight on polyacrylamide gels, we have designated this protein Erv29p. YML067c encodes an uncharacterized protein that has recently been immunoisolated from an early Golgi compartment (Cho et al. 2000). We have designated this protein Erv41p according to its molecular weight. YAL042w encodes a protein of unknown function that was named Fun9p (“function unknown”) (Coleman et al. 1986; Diehl and Pringle 1991) but not further characterized. We propose to name this protein Erv46p referring to its localization and molecular weight.
Selective Packaging of Erv Proteins In Vitro
Authentic Erv proteins should be selectively and efficiently packaged into ER-derived vesicles in the presence of COPII proteins. To test specific packaging of the uncharacterized proteins, we first generated strains that contained epitope-tagged versions of these proteins. Strain CBY801 expresses a COOH terminally HA-tagged version of Yif1p that fully complements for YIF1 function. Erv29p (CBY950), Erv41p (CBY782), and Erv46p (CBY767) were modified to contain the 3HA epitope at their NH2 termini and were placed under the control of the GAL1 promoter (Longtine et al. 1998). Galactose induction resulted in stable expression of these tagged proteins and localization to ER membranes. However, we cannot assess if these proteins are fully functional because ERV29, ERV41, and ERV46 are not essential. In vitro budding experiments were performed with microsomes from these strains (Fig. 2 A), and the relative packaging efficiencies were determined by comparing the lanes that represent 10% of the complete reactions (T) with the lanes that contain vesicles synthesized in the presence of COPII (+). Yif1p-3HA (8%), 3HA-Erv29p (13%), 3HA-Erv46p (10%), and 3HA-Erv41p (13%) were incorporated into vesicles at levels comparable to the positive controls Erv25p (14% for CBY801 and 5% for CBY950) and the v-SNARE Bos1p (11 for CBY767 and 5% for CBY782), whereas the ER resident proteins Sec12p and Sec61p were excluded. These results indicate that Yif1p, Erv29p, Erv41p, and Erv46p are selectively packaged into COPII vesicles.
Figure 2.
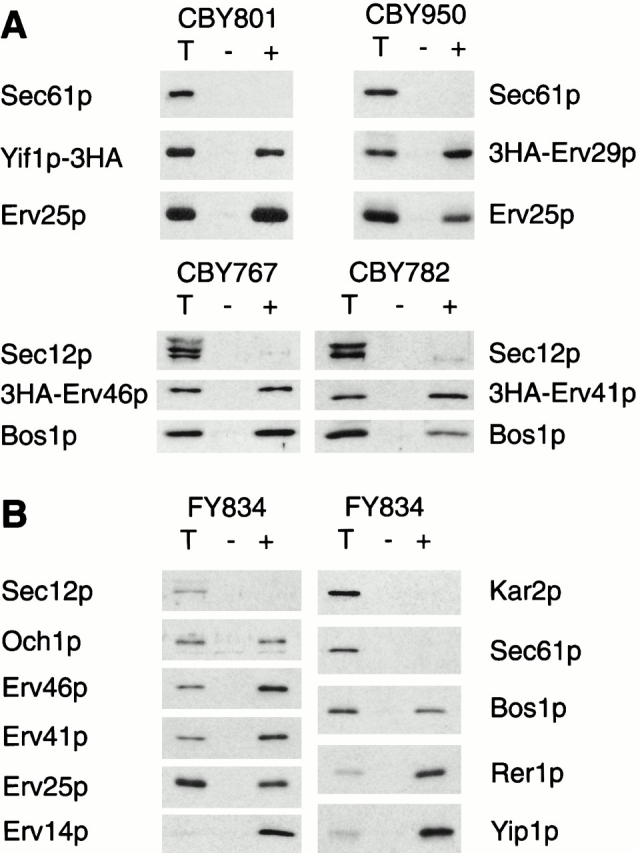
Selective packaging of Erv proteins into COPII-coated vesicles in vitro. (A) In vitro budding reactions with microsomes prepared from strains expressing tagged versions of Erv proteins: Yif1p-3HA (CBY801), 3HA-Erv29p (CBY950), 3HA-Erv46p (CBY767), and 3HA-Erv41p (CBY782). One tenth of a total reaction (T), budded vesicles isolated after incubation with COPII proteins (+), or a mock reaction without COPII proteins (–) were separated on a 12.5% polyacrylamide gel. Tagged proteins were visualized by immunoblot with an anti-HA antibody, and Sec61p or Sec12p (ER resident proteins) as negative controls and Erv25p (Erv protein) or Bos1p (v-SNARE) as positive controls were detected using polyclonal antisera. (B) In vitro budding reactions with FY834 wild-type microsomes. The same budding protocol as in A was used. Proteins were detected with polyclonal antisera against Sec12p, Kar2p, and Sec61p (ER residents), Bos1p (v-SNARE), Och1p (early Golgi marker), and the Erv proteins Erv46p, Erv41p, Erv25p, Erv14p, Rer1p, and Yip1p.
Next, we prepared or obtained specific polyclonal antibodies against several of the newly identified Erv proteins to measure their packaging efficiencies when endogenously expressed. As seen in Fig. 2 B, vesicles budded from wild-type microsomes contained Erv41p and Erv46p at levels (20% and 26%, respectively) that are higher than those of HA-tagged versions. Yip1p was packaged very efficiently (65%), as was Rer1p (51%; Sato et al. 1995). The early Golgi marker Och1p (Nakayama et al. 1992) was packaged to a significantly lesser extent (8%). The positive controls Erv14p, Erv25p, and Bos1p were efficiently packaged, whereas the ER marker proteins Sec12p, Sec61p, and Kar2p were excluded. Thus, Erv29p, Erv41p, Erv46p, Yif1p, Yip1p, and Rer1p meet the initial requirement of Erv proteins as they are selectively exported from ER membranes under reconstituted vesicle budding conditions.
Molecular Characterization of Erv41p and Erv46p
For the remainder of this report, we investigate the Erv41p and Erv46p proteins and their potential role in transport through the early secretory pathway. Erv29p will be described elsewhere. Erv41p is encoded by the ORF YML067c on chromosome XIII, and conceptual translation yields a protein of 352–amino acid residues and a predicted molecular weight of 40.6 kD. The ORF YAL042w on chromosome I encodes Erv46p, a protein predicted to be composed of 415–amino acid residues with a molecular weight of 46 kD. Database alignments showed that Erv41p and Erv46p exhibit significant sequence similarity. Both proteins also have one putative homologue each in Caenorhabditis elegans, Drosophila melanogaster, and humans, however, no known functions have been reported for these proteins. Sequence identity scores (Table ) between Erv41p and its homologues (CDA14, EG:65F1.1 and C18B12.6) are higher than those between Erv41p and Erv46p. Similarly, Erv46p and its homologues in other species (the 43.2-kD protein, CG70, and K09E9.2) show even higher identity scores. The Erv46p group is further characterized by eight conserved cysteine residues in their NH2-terminal half. Erv46p also terminates in the COPI binding motif KKXX (Jackson et al. 1990; Cosson and Letourneur 1994), a feature that is conserved across species. These results suggest that a conserved set of Erv41p–Erv46p proteins are found in other species and are likely to perform a similar function.
Table 3.
Percent Sequence Identities between Erv46p, Erv41p, and Their Homologues
| Erv46p | Erv41p | K09E9.2 | C18B12.6 | CG7011 | EG:65F1.1 | 43.2 kD | CDA14 | |
|---|---|---|---|---|---|---|---|---|
| Erv46p (S. cerevisiae) | 100 | 28 | 38 | 12 | 38 | 27 | 41 | 25 |
| Erv41p (S. cerevisiae) | 100 | 32 | 25 | 28 | 32 | 31 | 30 | |
| K09E9.2 (C. elegans) | 100 | 20 | 41 | 27 | 43 | 32 | ||
| C18B12.6 (C. elegans) | 100 | 20 | 20 | 17 | 21 | |||
| CG7011 (D. melanogaster) | 100 | 27 | 47 | 34 | ||||
| EG:65F1.1 (D. melanogaster) | 100 | 31 | 35 | |||||
| 43.2 kD protein (H. sapiens) | 100 | 38 | ||||||
| CDA14 (H. sapiens) | 100 |
Full-length sequences were aligned using the CLUSTAL V algorithm of GeneInspector v1.5 with the BLOSUM62 table and default settings (k-tuple 1, maximum gap length 5, gap penalty 3, 5 top diagonals).
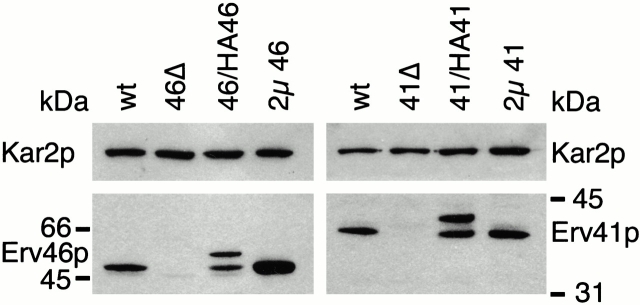
ERV41 as well as ERV46 were amplified from genomic DNA and inserted into the multicopy shuttle vectors pRS424 and pRS426, respectively, to yield plasmids pRS424-ERV41 and pRS426-ERV46. Polyclonal antisera were raised against recombinant forms of Erv41p and Erv46p and then tested on wild-type, deletion, and overproducing strains to confirm their specificity (Fig. 3). No immunoreactive species were detected in the deletion strains, whereas the heterozygous diploid strains carrying one wild-type and one tagged copy of the respective genes expressed two immunoreactive species at the expected size. This result also demonstrated that expression levels from the GAL1 promoter were comparable to wild-type expression levels at a concentration of 1.5% galactose. Strains harboring ERV41 or ERV46 on 2μ based multicopy plasmids expressed slightly higher levels of Erv41p or Erv46p than wild-type strains.
Figure 3.
Immunoblot analysis of wild-type, erv41Δ, and erv46Δ deletion and overproducing strains. FY834 wild-type (wt), erv46Δ (46Δ, CBY799), and erv41Δ (41Δ, CBY797) strains were grown in YPD medium. Heterozygous diploid strains expressing NH2 terminally 3HA-tagged Erv46p (46/HA46, CBY767) or Erv41p (41/HA41, CBY782) were grown in YP with 1.5% galactose and 0.5% glucose. erv14Δ erv46Δ deletion strains carrying pRS426-ERV46 plasmid (2μ 46, CBY836) or pRS424-ERV41 plasmid (2μ 41, CBY847) were grown in minimal media lacking uracil or tryptophan, respectively. Membrane fractions were resolved on a 12.5% polyacrylamide gel and immunoblotted with an anti-Kar2p antiserum as loading control and with polyclonal antisera raised against Erv46p or Erv41p.
Both Erv41p and Erv46p contain two segments of sufficient length and hydrophobicity to form transmembrane domains, and the short NH2- and COOH-terminal regions of both proteins are predicted to be located on the cytoplasmic side with a large lumenal domain separating the two transmembrane regions. The hydrophilicity plots of both proteins are superimposable (Fig. 4 A), and a similar topology is predicted for all of the known homologues. There is no evidence for a cleavable signal sequence. To confirm that Erv41p and Erv46p are indeed integral membrane proteins, we examined their fractionation behavior under conditions that release lumenal and peripherally bound membrane proteins (carbonate buffer, pH 11.0) or solubilize integral membrane proteins (1% Triton X-100). The fractionation profile for Erv41p and Erv46p was the same as for the integral membrane protein Bos1p, as they could only be solubilized by detergent and not by carbonate treatment (Fig. 4 B).
Figure 4.
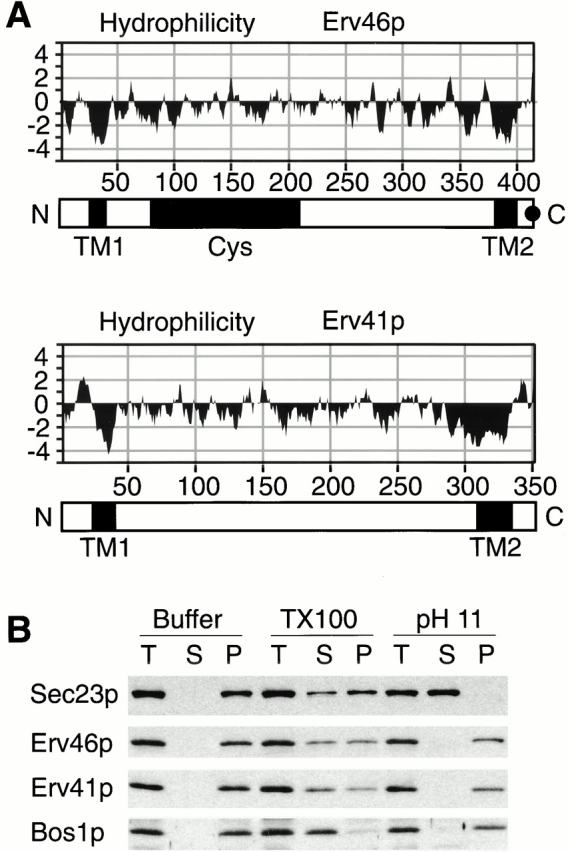
Erv46p and Erv41p are integral membrane proteins. (A) Hydrophilicity plots according to Kyte and Doolittle 1982 with a window size of 7. The schematic representations of the polypeptide chains indicate the positions of the putative transmembrane domains (TM1 and TM2) predicted by the HMMTOP program (Tusnady and Simon 1998), the region containing conserved cysteine residues (Cys) and the KKXX motif (black dot). (B) Semiintact FY834 cells were treated with buffer (Materials and Methods), buffer containing 1% Triton X-100 (TX100), or 0.1 M Na2CO3 (pH 11) and centrifuged at 100,000 g. Totals before centrifugation (T), supernatant (S), and pellet (P) fractions were analyzed on a 12.5% polyacrylamide gel and immunoblotted for Sec23p (peripheral membrane protein), Erv46p, Erv41p, and Bos1p (integral membrane protein).
The abundant Erv proteins are hypothesized to cycle between the ER and Golgi (Belden and Barlowe 1996; Powers and Barlowe 1998) because they localize to both of these membrane compartments, and, in contrast to abundant secretory proteins, their levels are not diminished by cycloheximide treatment (Yeung et al. 1995). Therefore, we examined the subcellular localization of Erv41p and Erv46p by resolution of membrane organelles on sucrose gradients (Fig. 5). Erv41p and Erv46p sedimented in two peaks, one that coincided with the Golgi marker Emp47p and the other with the ER marker Kar2p. This subcellular localization pattern was similar to that of Erv14p (Powers and Barlowe 1998) and Erv25p (Belden and Barlowe 1996). Rer1p has been shown to be predominantly localized to the Golgi (Sato et al. 1995) and to recycle between the Golgi and ER (Boehm et al. 1997). Consequently, we find that the majority of Rer1p cosedimented with Emp47p, and a second smaller peak cosedimented with Kar2p. In contrast, Och1p was localized almost exclusively to the Golgi. The localization of the epitope-tagged Erv46p was also examined by indirect immunofluorescence using the anti-HA antibody (data not shown). We observed a perinuclear staining pattern that partially overlapped with the ER marker Kar2p and was similar to that described for Erv14p (Powers and Barlowe 1998). We conclude that Erv41p and Erv46p localize to the early secretory pathway.
Figure 5.
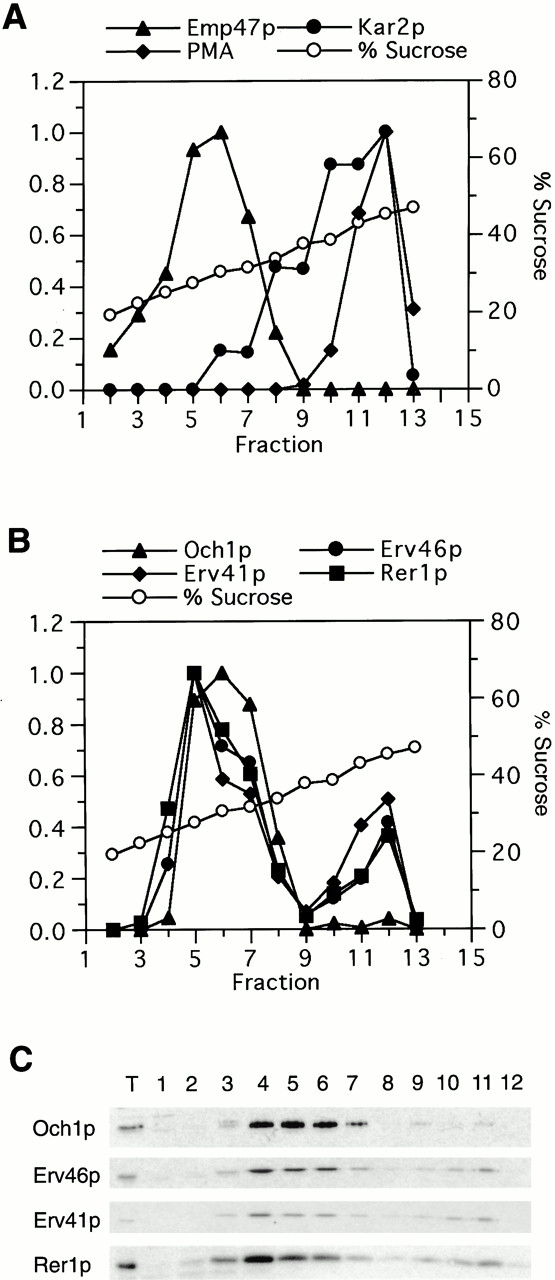
Sucrose gradient fractionation of Erv41p, Erv46p, Och1p, and Rer1p. An FY834 whole cell lysate was separated on an 18–60% sucrose density gradient, and fractions were collected, starting with fraction 1 at the top. (A) Relative levels of Emp47p (Golgi marker), Kar2p (ER marker), and plasma membrane marker (PMA) in each fraction were quantified by densitometry of immunoblots. (B) Relative levels of Och1p, Erv46p, Erv41p, and Rer1p as determined by densitometry of the immunoblots shown in C.
Analysis of the erv41Δ and erv46Δ Strains
To investigate the function of Erv41p and Erv46p, we analyzed strains bearing null alleles at the ERV41 or ERV46 loci. A PCR-based approach was used to direct the HIS3 gene to the ERV41 locus, replacing the entire ERV41 ORF. A heterozygous ERV41/erv41Δ diploid strain (CBY763) was sporulated, and dissection of asci produced four viable spores. Tetrad analysis of these spores for the HIS3 marker confirmed that ERV41 was not essential for vegetative growth, although spores carrying the deletion were slightly delayed in their germination (data not shown). A haploid strain in which the ERV46 reading frame had been replaced by a KAN marker (Winzeler et al. 1999) was crossed three times through the FY833/FY834 background to obtain the isogenic haploid erv46Δ strains CBY798 and CBY799.
The haploid erv41Δ strains (CBY796 and CBY797) grew at rates comparable to wild-type strains at 30°C and 37°C but displayed a reduced growth rate at 16°C (Fig. 6 B). The cellular morphology as determined by light microscopy as well as the mating and sporulation efficiencies of an erv41Δ strain were indistinguishable from the isogenic wild-type strains. Intracellular transport of the secretory proteins CPY and Gas1p was not detectably altered in an erv41Δ strain and the major secretory proteins contained in culture supernatants were also unchanged (data not shown). Therefore, deletion of ERV41 does not appear to interfere with general secretion. We performed a similar set of analyses on erv46Δ strains and observed phenotypes that were identical to erv41Δ strains, notably a reduced growth rate at 16°C. Furthermore, erv41Δ erv46Δ strains did not exhibit any exacerbated phenotypes compared with the single deletions strains (Fig. 6 B).
Figure 6.
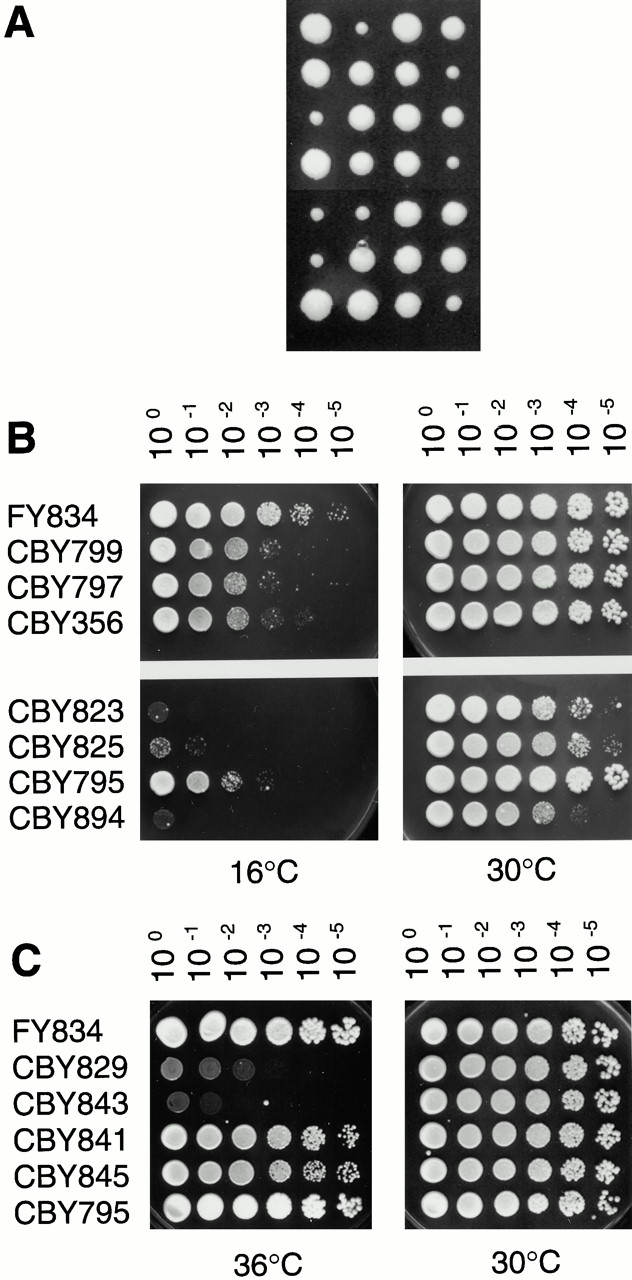
Genetic experiments with erv41Δ and erv46Δ strains. (A) An erv14Δ strain (CBY358) was mated with an erv46Δ strain (CBY799), and spores were dissected on a YPD plate. Spores that germinated and grew slower were shown to carry both deletions. (B) Cold sensitivity of erv14Δ, erv41Δ, and erv46Δ strains. Wild-type (FY834), erv46Δ (CBY799), erv41Δ (CBY797), erv14Δ (CBY356), erv14Δ erv46Δ (CBY823), erv14Δ erv41Δ (CBY825), erv41Δ erv46Δ (CBY795), and erv14Δ erv41Δ erv46Δ (CBY894) strains were grown to saturation in YPD, adjusted to an OD600 of 3.0, and 5 μl of a 10-fold dilution series were spotted onto YPD plates. (C) Effects of erv41Δ and erv46Δ mutations on the ypt1-3 mutation. Wild-type (FY834), ypt1-3 (CBY829), erv46Δ ypt1-3 (CBY843), erv41Δ ypt1-3 (CBY841), erv41Δ erv46Δ ypt1-3 (CBY845), and erv41Δ erv46Δ (CBY795) cells were spotted on YPD plates as in B.
Next, we investigated possible influences of the erv41Δ and erv46Δ deletions on other mutations that impede transport through the early secretory pathway (Table and Fig. 6). First, we examined phenotypes combined with null alleles of other known ERV genes. An erv14Δ strain (CBY356) also exhibits a delayed rate of spore germination, a slightly slower growth rate in YPD at 30°C (increase in doubling time of ∼18% over the wild type), and cold sensitivity. These phenotypes are significantly exacerbated by either erv41Δ or erv46Δ. In erv14Δ erv41Δ (CBY825) and erv14Δ erv46Δ (CBY823) strains, germination is delayed by several days (Fig. 6 A), the doubling time is increased by ∼40% compared with the wild type, and cold sensitivity is increased. The triple erv14Δ erv41Δ erv46Δ (CBY894) mutant does not show any further exacerbation of these phenotypes except that its germination is extremely delayed by ∼1 wk at room temperature. In contrast to the erv14Δ effects, combining emp24Δ with erv41Δ or erv46Δ did not alter growth phenotypes.
Table 4.
Effects of erv41 and erv46
| Strain number | Genotype | Phenotype* |
|---|---|---|
| FY834 | wild type | — |
| CBY356 | erv14Δ | Delayed germination, slightly slower growth, cold sensitive |
| CBY799 | erv46Δ | Cold sensitive |
| CBY797 | erv41Δ | Delayed germination, cold sensitive |
| CBY795 | erv41Δ erv46Δ | No effect |
| CBY823 | erv14Δ erv46Δ | Slow germination, slow growth, cold sensitive |
| CBY825 | erv14Δ erv41Δ | Very slow germination, slow growth, cold sensitive |
| CBY894 | erv14Δ erv41Δ erv46Δ | Extremely slow germination, slow growth, cold sensitive |
| CBY832 | erv41Δ erv46Δ emp24Δ | No effect |
| CBY841 | erv41Δ ypt1-3 | Significant decrease in temperature sensitivity |
| CBY843 | erv46Δ ypt1-3 | Slight increase in temperature sensitivity |
| CBY845 | erv41Δ erv46Δ ypt1-3 | Significant decrease in temperature sensitivity |
| CBY912 | erv41Δ uso1-1 | No effect |
| CBY910 | erv46Δ uso1-1 | No effect |
| CBY914 | erv41Δ erv46Δ uso1-1 | No effect |
| CBY943 | erv41Δ sec35-1 | No effect |
| CBY941 | erv46Δ sec35-1 | No effect |
| CBY945 | erv41Δ erv46Δ sec35-1 | No effect |
| CBY849 | erv41Δ sec21-1 | No effect |
| CBY851 | erv46Δ sec21-1 | No effect |
| CBY852 | erv41Δ erv46Δ sec21-1 | No effect |
| CBY853 | erv41Δ sed5-1 | No effect |
| CBY855 | erv46Δ sed5-1 | No effect |
| CBY857 | erv41Δ erv46Δ sed5-1 | No effect |
| CBY860 | erv41Δ sec12-1 | No effect |
| CBY862 | erv46Δ sec12-1 | No effect |
| CBY863 | erv41Δ erv46Δ sec12-1 | No effect |
| CBY923 | erv41Δ sec16-2 | No effect |
| CBY921 | erv46Δ sec16-2 | No effect |
| CBY924 | erv41Δ erv46Δ sec16-2 | No effect |
| CBY932 | erv41Δ sec13-1 | No effect |
| CBY933 | erv46Δ sec13-1 | No effect |
| CBY930 | erv41Δ erv46Δ sec13-1 | No effect |
| CBY917 | erv41Δ sec23-1 | No effect |
| CBY916 | erv46Δ sec23-1 | No effect |
| CBY919 | erv41Δ erv46 sec23-1 | No effect |
| CBY964 | erv14Δ rer1Δ | No effect |
| CBY949 | erv41Δ rer1Δ | No effect |
| CBY970 | erv29Δ erv14Δ | No effect |
| CBY968 | erv29Δ erv41Δ | No effect |
*Conditions tested: YPD at 16°C, 25°C, 30°C, 36°C, and 38°C.
Also, we investigated the influences of erv41Δ and erv46Δ deletions on thermosensitive mutations in known budding, tethering, and fusion genes. An erv41Δ ypt1-3 strain (CBY841) showed a significantly decreased thermosensitivity compared with ypt1-3 alone (CBY829). The same effect was observed in a ypt1-3 erv41Δ erv46Δ strain (CBY845), whereas erv46Δ alone slightly exacerbated the thermosensitivity of ypt1-3 (CBY843, Fig. 6 C). In contrast, no effect was observed with the other tethering mutations uso1-1 and sec35-1. The erv41Δ or erv46Δ null mutations did not influence any of the growth phenotypes associated with mutations in the COPI (sec21-1), COPII (sec12-4, sec13-1, sec16-1, and sec23-1), or fusion (sed5-1) proteins. Finally, we did not observe any synthetic effects when the erv41Δ or erv46Δ mutations were combined with rer1Δ or erv29Δ.
Effects of erv41Δ and erv46Δ on Transport to the Golgi
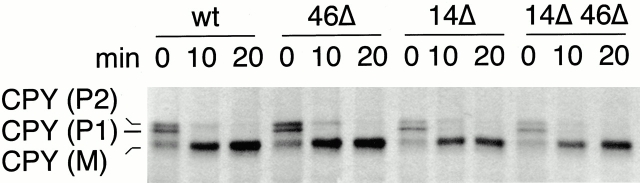
Since erv41Δ and erv46Δ strains did not exhibit major defects in secretion, but the erv14Δ erv46Δ strains showed an exacerbated growth phenotype, we examined the transport kinetics in this strain compared with wild-type and the single erv14Δ and erv46Δ strains. A pulse–chase experiment was performed in which cells were grown in minimal media and pulsed for 7 min with [35S]methionine and [35S]cysteine to label newly synthesized proteins. Excess unlabeled cysteine and methionine were added for the chase phase, and the maturation of CPY was followed by immunoprecipitation with an anti-CPY antibody. CPY first appears in the ER as the P1 precursor form of 67 kD and is then modified in the Golgi to yield its P2 form of 69 kD and finally processed in the vacuole to the mature M form of 61 kD (Stevens et al. 1984). As seen in Fig. 7, a slight delay in transport of CPY was observed in an erv14Δ strain (Powers and Barlowe 1998), and, when combined with erv46Δ, no further decrease in the transport rate was detected.
Figure 7.
Pulse–chase analysis of CPY maturation in wild-type and erv14Δ erv46Δ deletion strains. Wild-type (wt, FY833), erv46Δ (46Δ, CBY798), erv14Δ (14Δ, CBY358), and erv14Δ erv46Δ (14Δ 46Δ, CBY822) strains were pulsed for 7 min with 35S-labeled cysteine and methionine and then chased for 10 or 20 min. Labeled CPY was immunoprecipitated from cell extracts, resolved on a 10% polyacrylamide gel, and visualized by autoradiography.
To further investigate a possible defect in ER to Golgi transport, we used an in vitro transport assay that permitted us to differentiate between the budding, tethering, and fusion stages (Barlowe 1997; Cao et al. 1998). We have found this assay to be a more sensitive method to measure transport between the ER and Golgi. In some instances, mutant strains that display normal transport kinetics in pulse–chase experiments show defects in distinct stages of cell-free transport (Conchon et al. 1999). Washed semiintact cells containing 35S-labeled gp-α-F bud 35S-labeled gp-α-F–containing vesicles in the presence of COPII proteins. Packaged 35S-labeled gp-α-F in vesicles can be quantified by precipitation with concanavalin A–Sepharose, allowing us to assay budding efficiencies. The vesicle tethering stage may be monitored as the decrease in diffusible COPII vesicles upon addition of the tethering protein Uso1p. Lastly, reconstituted transport to the Golgi complex can be measured after addition of COPII, Uso1p, and LMA1 to semiintact cells by precipitation of Golgi-modified forms of 35S-labeled gp-α-F with α1,6-mannose–specific antiserum. As seen in Fig. 8 A, the budding and tethering stages of transport were not impaired in mutant strains compared with a wild-type strain. However, there was a modest but significant decrease in transport to the Golgi complex in the erv41Δ, erv46Δ, and the double erv41Δ erv46Δ membranes (Fig. 8 B). Notably, the effects of the single deletions were neither additive nor cooperative when combined in the double mutant strain. Together, these results suggest that the transport defect occurred during the fusion stage of this assay. Also, we examined transport efficiencies in the presence of a crude cytosol to determine if the erv41Δ erv46Δ membranes required additional factors not provided by purified reconstitution proteins. As shown in Fig. 8 C, a similar transport defect was observed for reactions using crude cytosol or purified proteins to drive transport.
Figure 8.
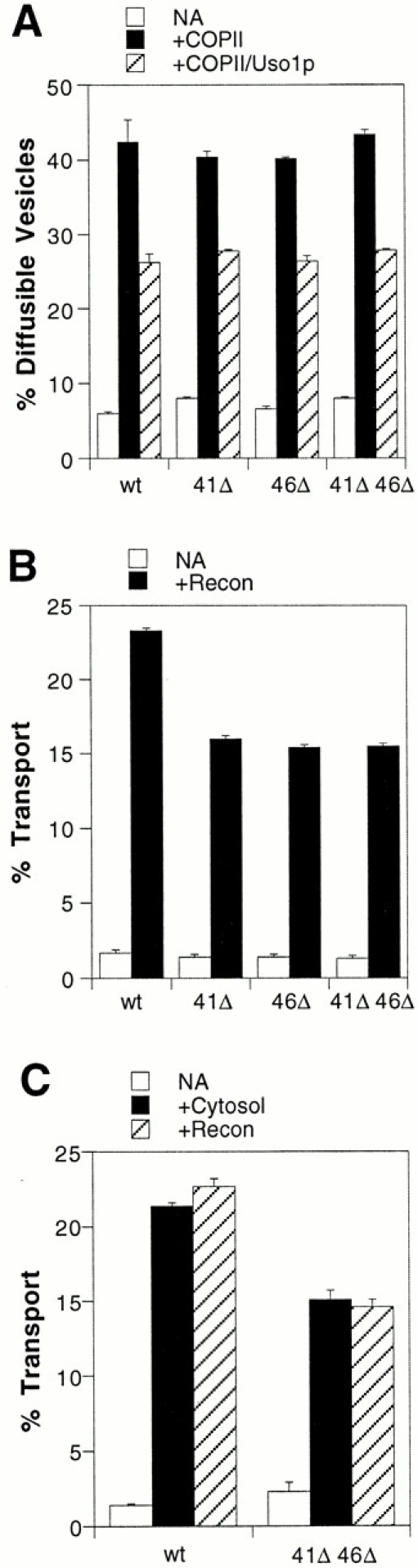
Influences of erv41Δ and erv46Δ mutations on ER–Golgi transport. (A) Vesicle budding and tethering in washed semiintact cells prepared from wild-type (wt, FY834), erv41Δ (41Δ, CBY797), erv46Δ (46Δ, CBY799), and erv41Δ erv46Δ (41Δ 46Δ, CBY795) strains. The levels of diffusible vesicles in reactions without reconstitution proteins (NA), with COPII proteins (+COPII), and with COPII proteins plus the tethering factor Uso1p (+COPII/Uso1p) are indicated. (B) Overall transport of 35S-labeled gp-α-F to the Golgi complex in the same strains. Semiintact cells were incubated alone (NA) or with the reconstitution proteins COPII, Uso1p, and LMA1 (+Recon). (C) Overall transport of 35S-labeled gp-α-F factor to the Golgi complex in wild-type (wt, FY834) and erv41Δ erv46Δ (41Δ 46Δ, CBY795) strains. Semiintact cells were incubated alone (NA), with cytosol, or with the reconstitution proteins COPII, Uso1p, and LMA1 (+Recon).
Association of Erv41p and Erv46p
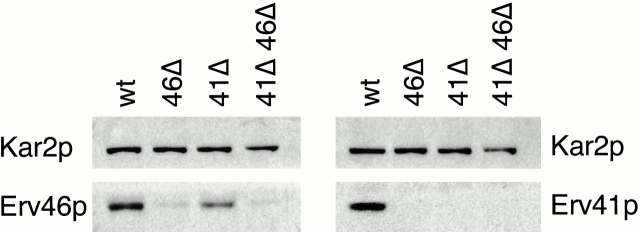
Based on the colocalization of Erv41p and Erv46p, the similarity in phenotypes displayed by erv41Δ and erv46Δ in our genetic experiments and the nonadditive effects of the erv41Δ and erv46Δ mutations on ER–Golgi transport assays, we suspected that these proteins act together and possibly form a complex. Therefore, we investigated whether the expression of Erv41p and Erv46p is interdependent by immunoblotting cells containing the erv41Δ, erv46Δ, and erv41Δ erv46Δ alleles with antisera against Erv41p and Erv46p. The erv46Δ strain did not express a detectable level of Erv41p, and the erv41Δ strain exhibited a reduced expression of Erv46p compared with the wild type (Fig. 9). Consistent with these findings, we observed that Erv46p could be expressed at higher levels from a 2μ plasmid than Erv41p (Fig. 3). A strain carrying both ERV41 and ERV46 on 2μ plasmids did not express either of them at levels higher than a strain just overexpressing Erv46p (not shown). Also, the expression level of endogenous Erv41p could be elevated (approximately twofold) if Erv46p was overproduced under control of the GAL1 promoter (compare the total [T] lanes in Fig. 10). These results indicate that the expression of Erv41p is highly dependent on the presence of Erv46p, and to a lesser extent Erv46p expression depends on the presence of Erv41p. One interpretation of these results is that Erv41p and Erv46p are associated in a multisubunit protein complex, and when one of the members of this complex is absent, the other subunit(s) is destabilized.
Figure 9.
Erv41p and Erv46p depend on each other for their expression. wt, wild-type strain FY834; 46Δ, erv46Δ deletion strain CBY799; 41Δ, erv41Δ deletion strain CBY797; and 41Δ 46Δ, erv41Δ erv46Δ double deletion strain CBY795. Semiintact cells were resolved on a 12.5% polyacrylamide gel and immunoblotted with polyclonal anti-Erv41p and anti-Erv46p antisera. Kar2p is shown as a loading control.
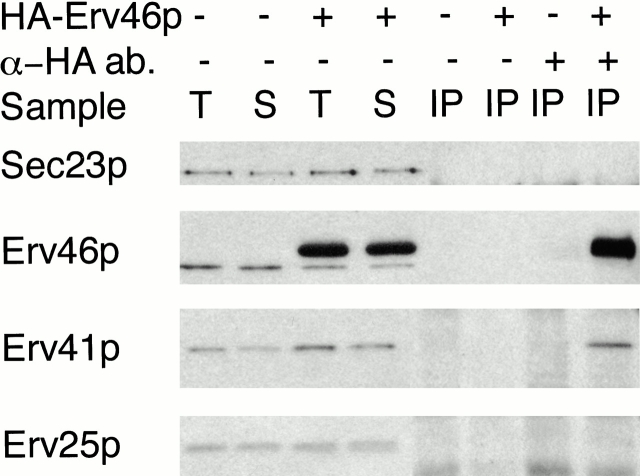
Figure 10.
Erv41p is coimmunoprecipitated with HA-tagged Erv46p. Microsomes were prepared from FY833 (− HA-Erv46p) and CBY767 (+ HA-Erv46p) cells grown in YP with 2% galactose and 0.2% glucose and solubilized with buffer containing 2% Triton X-100 (T). The soluble extracts (S) were incubated in absence (− anti-HA ab) or presence (+ anti-HA ab) of the monoclonal anti-HA antibody. Precipitates were resolved on a 12.5% polyacrylamide gel and immunoblotted for Sec23p, Erv46p, Erv41p, and Erv25p.
To determine if Erv41p and Erv46p are physically associated, we performed a native immunoprecipitation experiment from detergent-solubilized membranes. Microsomes from a wild-type strain and from a strain expressing 3HA-Erv46p were solubilized with Triton X-100, and tagged Erv46p was precipitated by the addition of anti-HA antibodies. As seen in Fig. 10, Erv41p coprecipitated with 3HA-Erv46p in the tagged strain, whereas no Erv41p was precipitated from the untagged wild-type microsomes. As controls, neither Sec23p nor Erv25p were precipitated, indicating that the association of 3HA-Erv46p with Erv41p was specific. In other experiments, we found that HA-tagged Erv41p could be coimmunoprecipitated with Erv46p when Erv46p specific polyclonal antibodies were used (not shown). Finally, we observed that endogenous Erv46p was not detected in complexes that were isolated with 3HA-Erv46p (Fig. 10; Otte, S., unpublished observation). Collectively, the expression studies and immunoprecipitation results indicate that Erv41p and Erv46p are physically associated in a heteromeric complex and that these complexes probably contain single Erv46p and Erv41p subunits.
Discussion
Using a reconstituted budding assay combined with mass spectrometry, we have identified four known transmembrane protein constituents of COPII-coated vesicles (Erv14p, Emp24p, Erv25p, and Bet1p), and have localized six other characterized proteins (Yip3p, Rer1p, Erp1p, Erp2p, Yif1p, and Emp47p) and three additional novel proteins (Erv29p, Erv41p, and Erv46p) to these carrier vesicles. We went on to characterize Erv41p and Erv46p, two related proteins that are conserved across species. Both are integral transmembrane proteins and are predicted to have large lumenal domains with shorter COOH- and NH2-terminal cytoplasmic segments. Localization studies show that both proteins reside in the Golgi and ER at steady state. erv41Δ and erv46Δ strains are cold-sensitive, and these deletions exacerbate the erv14Δ phenotype. The erv41Δ and erv46Δ mutations also influence the thermosensitivity of a ypt1-3 strain. Strains carrying the erv41Δ and erv46Δ null alleles show normal vesicle budding and tethering but a reduced overall transport efficiency between the ER and Golgi, suggesting a defect downstream of the tethering stage. Expression of Erv41p and Erv46p is interdependent, and both proteins could be coimmunoprecipitated, indicating that these proteins are physically associated.
In discussing potential functions for Erv41p and Erv46p, it may be informative to consider other characterized vesicle proteins. We have now identified four p24 proteins on isolated ER-derived vesicles. Yeast has eight members of this family, Erv25p (Belden and Barlowe 1996), Emp24p (Schimmöller et al. 1995), and Erp1p to Erp6p (Marzioch et al. 1999). These abundant, conserved, integral type I transmembrane proteins have been found in COPI and COPII vesicles and have been shown to shuttle between the ER and Golgi. Their cytosolic tails have a high affinity for coat proteins and have been proposed to act as a scaffold during the formation of the protein coat (Bremser et al. 1999), as transport receptors for secretory cargo (Schimmöller et al. 1995) or as negative regulators of vesicle budding that influence cargo sorting (Elrod-Erickson and Kaiser 1996). However, a strain that lacks all eight p24 proteins is viable and did not show defects in overall COPI- or COPII-mediated transport (Kaiser 2000; Springer et al. 2000), indicating that they do not perform an essential role in transport through the early secretory pathway in yeast. The phenotypes associated with p24 deletion strains are consistent with a role in protein and/or lipid sorting during vesicle budding either by association with cargo or through formation of specialized sorting regions or membrane compartments. The expression levels of the Erv25p, Emp24p, Erp1p, and Erp2p are interdependent, these proteins are found associated in a heterooligomeric complex, and strains lacking any one subunit of this complex exhibit similar phenotypes (Marzioch et al. 1999). Our detection of the Erp1p and Erp2p proteins on COPII vesicles is consistent with these observations, and we are testing the possibility that this heterooligomeric p24 complex is packaged en bloc during vesicle formation.
Yip3p and Yif1p, two previously identified proteins, were also encountered on ER-derived vesicles. The Yip proteins were initially discovered as Ypt1p interacting proteins (Yang et al. 1998) and Yif1p as a Yip1p interacting factor (Andrulis et al. 1998). Ypt1p is a small GTPase required for vesicle transport through the early secretory pathway (Rexach and Schekman 1991; Segev et al. 1988) and therefore the Yip and Yif proteins are thought to operate in conjunction with GTPases to catalyze vesicle fusion. Yip1p and Yif1p are integral membrane proteins that localize predominantly to Golgi membranes and possess hydrophilic NH2-terminal domains facing the cytosol. Recently, a Yip1p–Yif1p complex has been proposed to function as a receptor in recruiting GTPases to specific membranes, perhaps acting as a GDI displacement factor (Yang et al. 1998; Matern et al. 2000). We find the Yip1p–Yif1p complex is efficiently packaged into COPII vesicles, suggesting that these proteins actively cycle between the ER and Golgi compartments. It seems possible that the Yip1–Yif1p complex on vesicles could act directly to target vesicles to Ypt1p on the surface of Golgi membranes or Yip1p–Yif1p could perform a more general role in recruiting GTPases to several distinct intracellular membranes. Further studies will be needed to distinguish between these possibilities.
Rer1p is required for the ER localization of type II transmembrane proteins (e.g. Sec12p) and has been proposed to couple retrieved proteins to the COPI coat during retrograde Golgi to ER transport (Sato et al. 1995; Boehm et al. 1997; Sato et al. 1997). We have found that Rer1p is included in COPII vesicles with a high efficiency, suggesting this protein actively cycles between the ER and Golgi compartments. Our observation seems consistent with the proposed function of Rer1p in retrieval of ER residents from post-ER compartments. However, it remains to be determined if the COPII-dependent forward transport of Rer1p reflects recycling necessary to perform its function in retrieval of ER residents or whether this protein performs additional roles in anterograde transport.
We had expected that the abundant Erv proteins would represent transport machinery involved in vesicle formation, cargo selection, and membrane fusion (Belden and Barlowe 1996). To a large extent, our current study bears this out. However, we also found that the outer-chain mannosyltransferase (Och1p) was packaged into COPII vesicles as has been previously reported (Bednarek et al. 1995). Och1p was not detected in our mass spectral analysis, but antibodies against this protein were generated to allow for a comparison of packaging efficiencies with other Erv proteins. We find that Och1p is packaged into COPII vesicles, albeit at a lower efficiency than most other Erv proteins (Fig. 2), and is localized predominantly to Golgi membranes (Fig. 5). The packaged Och1p could represent newly synthesized protein in transit to the Golgi or may belong to a class of Golgi localized proteins that cycle between the ER and Golgi compartments at a significant rate as has been observed for mammalian galactosyltransferase (Zaal et al. 1999). It is interesting to note that other Golgi localized proteins such as Ypt1p and GDPase were not efficiently packaged into COPII vesicles (Cao and Barlowe 2000). Furthermore, in vivo studies indicate that some Golgi-localized proteins cycle rapidly through the ER, whereas others do not (Wooding and Pelham 1998; Barrowman et al. 2000). It remains to be determined if cycling rates necessarily reflect an important functional property. Regardless, the fact that some proteins typically thought to be Golgi residents are found in COPII vesicles suggests that uncharacterized Erv proteins could potentially belong to this group.
Erv41p and Erv46p are predicted to have large lumenal domains and short NH2- and COOH-terminal cytosolic tails that potentially interact with the COPII and/or COPI coats. Based on these and other considerations discussed above, we can envisage a few possible roles for the Erv41p–Erv46p complex that are consistent with our experimental observations. First, this complex could perform a role similar to the p24 complex in sorting during vesicle formation. The complex could fulfill this role through direct interaction with cargo molecules or by establishing domains on the surface of budding membranes. In some respects, the heteromeric arrangement, the membrane topology, and the nonessential phenotypes exhibited by the Erv41p–Erv46p complex are reminiscent of the p24 deletion strains. We have not detected any specific secretory cargo that accumulates in the erv41Δ and/or erv46Δ deletion strains, however, we currently have a very limited capability of monitoring individual secretory cargo. If the Erv41p–Erv46p complex is required for the efficient transport of a small subset of nonessential cargo proteins, we may not easily detect an accumulation. Second, the Erv41p–Erv46p complex could operate in the retention and/or retrieval of transport machinery to the early secretory pathway. For example, as Rer1p acts in the retrieval of Sec12p and other ER residents, the Erv41p–Erv46p complex could operate in localizing proteins to Golgi membranes. In keeping with this idea, the erv41Δ erv46Δ strain displayed a modest defect in the fusion stage of in vitro transport between the ER and Golgi, suggesting the Erv41p–Erv46p complex could interact with or serve to correctly localize the membrane fusion machinery (e.g., SNAREs and SNARE regulatory proteins). Third, the Erv41p–Erv46p complex may not act in movement or localization of proteins, but could be involved in lipid transport. A majority of cellular phospholipid, glycolipid, and sterol synthesis occurs in the ER (for review see Daum et al. 1998), and it seems probable that specific proteins act to sort and transport these species to their proper location. Although nonsecretory routes exist for lipid transport, much of this lipid transport occurs through the classical secretory pathway and presumably through COPII vesicles. Therefore, the observed in vitro defect during the vesicle fusion stage could be caused by a suboptimal lipid composition of vesicle or acceptor membrane bilayers, leading to a reduction in membrane fusion efficiency. Finally, the Erv41p–Erv46p complex could perform a role in the posttranslational maturation of secretory proteins such as protein folding or glycosylation in the early secretory pathway. If this last possibility were the case, we again would speculate that the effect is on a subset of secretory cargo because we do not detect any general defects in folding, no extracellular secretion of kar2p (Otte, S., unpublished observation), and no obvious alterations in N- or O-linked oligosaccharide modifications.
Several recent reports suggest a role for COPII-dependent transport in regulation of cellular homeostasis (Niwa et al. 1999; Nohturfft et al. 2000; Travers et al. 2000). Although the underlying mechanisms of these regulatory pathways remain to be elucidated, our investigation of Erv proteins may shed light on these questions. Notably, several of the proteins we have identified on COPII vesicles, including the novel proteins Erv29p, Erv41p, and Erv46p, are induced upon activation of the unfolded protein response pathway (Travers et al. 2000). With respect to the Erv41p–Erv46p complex, it will be important to determine binding partners that potentially include additional members of the heteromeric Erv41p–Erv46p complex, vesicle coat proteins, and/or specific cargo molecules. We will combine these biochemical approaches with cell-free transport assays and molecular genetic analyses of Erv41p–Erv46p to elucidate their function.
Acknowledgments
We thank Hans Dieter Schmitt and Dieter Gallwitz for gifts of Rer1p and Yip1p antisera, and for comments on this manuscript.
S. Otte was supported through a fellowship from Deutsche Forschungsgemeinschaft. W. Belden was supported by a pre-doctoral fellowship from the National Institutes of Health. This work was supported by grants from the National Institute of General Medical Sciences and the Pew Scholars Program in the Biomedical Sciences.
Footnotes
Abbreviations used in this paper: CPY, carboxypeptidase Y; Erv, ER vesicle; gp-α-F, glyco-pro–α factor; HA, hemagglutinin.
References
- Andrulis E.D., Neimann A.M., Zappula C.D., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Arvan P., Castle D. Sorting and storage during secretory granule biogenesislooking backward and looking forward. Biochem. J. 1998;332:593–610. doi: 10.1042/bj3320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, R.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York.3.0.1–3.14.3.
- Baker D., Hicke L., Rexach M., Schleyer M., Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J. Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M., Ravazzola M., Amherdt M., Schekman R. COPIIa membrane coat formed by Sec proteins that drive vesicle budding from the ER. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barrowman J., Sacher M., Ferro-Novick S. TRAPP stably associates with the Golgi and is required for vesicle docking. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:862–869. doi: 10.1093/emboj/19.5.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C.A. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek S.Y., Ravazzola M., Hosobuchi M., Amherdt M., Perrelet A., Schekman R., Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Belden W.J., Barlowe C. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- Boehm J., Letourneur F., Ballensiefen W., Ossipov D., Démollière C., Schmitt H.D. Sec12p requires Rer1p for sorting to coatomer (COPI)-coated vesicles and retrieval to the ER. J. Cell Sci. 1997;110:991–1003. doi: 10.1242/jcs.110.8.991. [DOI] [PubMed] [Google Scholar]
- Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C.A., Söllner T., Rothman J.E., Wieland F.T. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Cao X., Barlowe C. Asymmetric requirements for a Rab GTPase and SNARE proteins in fusion of COPII vesicles with acceptor membranes. J. Cell Biol. 2000;149:55–65. doi: 10.1083/jcb.149.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Ballew N., Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cho J.-H., Noda Y., Yoda K. Proteins in the early Golgi compartment of Saccharomyces cerevisiae immunoisolated by Sed5p. FEBS Lett. 2000;469:151–154. doi: 10.1016/s0014-5793(00)01268-0. [DOI] [PubMed] [Google Scholar]
- Coleman K.G., Steensma H.Y., Kaback D.B., Pringle J.R. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation and characterization of the CDC24 gene and adjacent regions of the chromosome. Mol. Cell. Biol. 1986;6:4516–4525. doi: 10.1128/mcb.6.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchon S., Cao X., Barlowe C., Pelham H.R.B. Got1p and Sft2pmembrane proteins involved in traffic to the Golgi complex. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3934–3946. doi: 10.1093/emboj/18.14.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P., Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Daum G., Lees N.D., Bard M., Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae . Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Diehl B.E., Pringle J.R. Molecular analysis of Saccharomyces cerevisiae chromosome Iidentification of additional transcribed regions and demonstration that some encode essential functions. Genetics. 1991;127:287–298. doi: 10.1093/genetics/127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M.J., Kaiser C.A. Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol. Biol. Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletti D.L., Prekeris R., Scheller R.H. Generation and maintenance of neuronal polaritymechanisms of transport and targeting. Neuron. 1999;23:641–644. doi: 10.1016/s0896-6273(01)80022-2. [DOI] [PubMed] [Google Scholar]
- Jackson M.R., Nilsson T., Peterson P.A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N., Wilm M., Shevchenko A., Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- Kaiser C. Thinking about p24 proteins and how transport vesicles select their cargo. Proc. Natl. Acad. Sci. USA. 2000;97:3783–3785. doi: 10.1073/pnas.97.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydrophathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marzioch M., Henthorn D.C., Herrmann J.M., Wilson R., Thomas D.Y., Bergeron J.J.M., Slari R.C.E., Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a Yeast p24 complex. Mol. Biol. Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern H., Yang X., Andrulis E., Sternglanz R., Trepte H.-H., Gallwitz D. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO (Eur. Mol. Biol. Org.) J. 2000;19:4485–4492. doi: 10.1093/emboj/19.17.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Warren G. The road takenpast and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferaseouter chain elongation of asparagine-linked oligosaccharides. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.P., Ferro-Novick S. Characterization of new mutants in the early part of the yeast secretory pathway isolated by a [3H] mannose suicide selection. J. Cell Biol. 1987;105:1587–1594. doi: 10.1083/jcb.105.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.P., Groesch M.E., Ferro-Novick S. Bos1p, a membrane protein required for ER to Golgi transport in yeast, co-purifies with the carrier vesicles and with Bet1p and the ER membrane. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:3609–3617. doi: 10.1002/j.1460-2075.1992.tb05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M., Sidrauski C., Kaufman R.J., Walter P. A role for presenilin-1 in nuclear accumulation of Ire1 fragments and induction of the mammalian unfolded protein response. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- Nohturfft A., Yabe D., Goldstein J.L., Brown M.S., Espenshade P.J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Powers J., Barlowe C. Transport of Axl2p depends on Erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J. Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M.F., Schekman R.W. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J. Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M.F., Latterich M., Schekman R.W. Characteristics of endoplasmic reticulum-derived transport vesicles. J. Cell Biol. 1994;126:1133–1148. doi: 10.1083/jcb.126.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N.R., Yeung T., Schekman R. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nishikawa S., Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER)characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol. Biol. Cell. 1995;6:1459–1477. doi: 10.1091/mbc.6.11.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M., Nakano A. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc. Natl. Acad. Sci. USA. 1997;94:9693–9698. doi: 10.1073/pnas.94.18.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schimmöller F., Singer-Krüger B., Schröder S., Krüger U., Barlowe C., Riezman H. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S., Schimmöller F., Singer-Krüger B., Riezman H. The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in α-COP. J. Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., Mulholland J., Botstein D. The yeast GTP-binding Ypt1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Smith C.J., Pearse B.M. Clathrinanatomy of a coat protein. Trends Cell Biol. 1999;9:335–338. doi: 10.1016/s0962-8924(99)01631-1. [DOI] [PubMed] [Google Scholar]
- Söllner T.H., Rothman J.E. Molecular machinery mediating vesicle budding, docking and fusion. Experientia. 1996;52:1021–1025. doi: 10.1007/BF01952097. [DOI] [PubMed] [Google Scholar]
- Springer S., Chen E., Duden R., Marzioch M., Rowley A., Hamamoto S., Merchant S., Schekman R. The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA. 2000;97:4034–4039. doi: 10.1073/pnas.070044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxy peptidase Y to the vacuole. Cell. 1984;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Tusnady G.E., Simon I. Principles governing amino acid composition of integral membrane proteinsapplication to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse L.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wooding S., Pelham H.R. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestehube L.J., Schekman R. Reconstitution of transport from the endoplasmic reticulum to the Golgi complex using an ER-enriched membrane fraction from yeast. Methods Enzymol. 1992;219:124–136. doi: 10.1016/0076-6879(92)19015-x. [DOI] [PubMed] [Google Scholar]
- Wuestehube L.J., Duden R., Eun A., Hamamoto S., Korn P., Ram R., Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Matern H.T., Gallwitz D. Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4954–4963. doi: 10.1093/emboj/17.17.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T., Barlowe C., Schekman R. Uncoupled packaging of targeting and cargo molecules during transport vesicle budding from the endoplasmic reticulum. J. Biol. Chem. 1995;270:30567–30570. doi: 10.1074/jbc.270.51.30567. [DOI] [PubMed] [Google Scholar]
- Zaal K.J., Smith C.L., Polishchuk R.S., Altan N., Cole N.B., Ellenberg J., Hirschberg K., Presley J.F., Roberts T.H., Siggia E., Phir R.D., Lippincott-Schwartz J. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]