Abstract
The Rieger syndrome is an autosomal dominant disease characterized by ocular, craniofacial, and umbilical defects. Patients have mutations in PITX2, a paired-bicoid homeobox gene, also involved in left/right polarity determination. In this study we have identified a family of genes for enzymes responsible for hydroxylizing lysines in collagens as one group of likely cognate targets of PITX2 transcriptional regulation. The mouse procollagen lysyl hydroxylase (Plod)-2 gene was enriched for by chromatin precipitation using a PITX2/Pitx2-specific antibody. Plod-2, as well as the human PLOD-1 promoters, contains multiple bicoid (PITX2) binding elements. We show these elements to bind PITX2 specifically in vitro. The PLOD-1 promoter induces the expression of a luciferase reporter gene in the presence of PITX2 in cotransfection experiments. The Rieger syndrome causing PITX2 mutant T68P fails to induce PLOD-1–luciferase. Mutations and rearrangements in PLOD-1 are known to be prevalent in patients with Ehlers-Danlos syndrome, kyphoscoliosis type (type VI [EDVI]). Several of the same organ systems are involved in Rieger syndrome and EDVI.
Keywords: PITX2, PLOD, Rieger, Ehlers-Danlos, promoter
Introduction
Studies of the autosomal-dominant Rieger syndrome first suggested that a single gene could be involved in the development of eye, tooth, and abdominal organs (Rieger 1935). Mutations in PITX2 were detected in patients with Rieger syndrome (Semina et al. 1996). The ocular abnormalities include an anterior and prominent Schwalbe's line (posterior embryotoxon), iris adhesions to the trabecular meshwork, iris hypoplasia, ectopic pupils, displaced pupils, full-thickness iris tears (polycoria), iridocorneal adhesions, and glaucoma. Patients with Rieger (or Axenfeld-Rieger) syndrome display the ocular features in combination with dental and umbilical abnormalities. The dental phenotypes are hypoplasia, missing upper incisors, and sometimes total lack of teeth. The umbilicus is usually a protruding stump, but severe cases have omphalocele, or an unclosed abdomen. Mice disrupted in the Pitx2 gene display phenotypes reminiscent of the corresponding human defects as well as symmetry defects (Campione et al. 1999; Gage et al. 1999; Lin et al. 1999; Lu et al. 1999). Pitx2 is also involved in the Nodal/Sonic hedgehog pathway, which determines left/right polarity of mesoderm-derived organs such as heart, gut, and stomach (for reviews see Blum et al. 1999; Tamura et al. 1999).
The PITX genes are members of the bicoid class of the homeodomain proteins. These have a lysine residue at position nine of the third helix and are especially noteworthy for a role in both DNA and RNA binding (Dubnau and Struhl 1996; Amendt et al. 1998). In Drosophila, seven different target genes have been identified epistatically and biochemically for bicoid (Driever and Nüsslein-Volhard 1989; Jackle and Sauer 1993). Target genes for Pitx2 have only been described for the pituitary; the prolactin gene is synergistically upregulated by Pit-1 and Pitx2 (Simmons et al. 1990; Szeto et al. 1996; Ryan and Rosenfeld 1997; Amendt et al. 1998. Other pituitary-specific Pitx2 target genes have also been described (Tremblay et al. 2000). Recently, we presented the first study on Pitx2 protein expression in the eye, as well as directly demonstrating asymmetric protein expression in the early development of mouse gut, heart, and lung (Hjalt and Murray 2000).
PITX2 was first identified by positional cloning of the 4q25 locus, but only 40% of patients diagnosed with classical Rieger syndrome have PITX2 mutations (Semina et al. 1996). Other loci for Rieger syndrome include RIEG2 on 13q14 and FKHL7/FOXE7 on 6p25 (for reviews see Craig and Mackey 1999; Amendt et al. 2000). Still undescribed genes/loci for Rieger syndrome may be downstream of PITX2. It is also still unknown which genes are the cognate targets for PITX2 in left/right asymmetry regulation in the development of the heart, gut, and lung.
Chromatin precipitation is a direct method for in vivo detection of target genes. It has been used successfully to identify: ultrabithorax target genes (Gould et al. 1990), polycomb-repressed domains in the bithorax complex (Orlando and Paro 1993), Pax-2 in vivo binding sites (Phelps and Dressler 1996), and a Hox-C8 target gene (Tomotsune et al. 1993).
Here we identify some members of the procollagen lysyl hydroxylase (PLOD) family, genes for enzymes that hydroxylate lysines in collagens, as cognate targets for Pitx2/PITX2, by chromatin precipitation. The hydroxylysine residues have two important functions: as attachment sites for carbohydrate units and to provide stability to intermolecular cross-links. Cross-links involving hydroxylysine-derived aldehydes are more stable than those involving lysine-derived aldehydes (Prockop and Kivirikko 1984; Kivirikko and Myllylä 1985; Kivirikko et al. 1992).
Point mutations and rearrangements in the human PLOD1 are causative for the Ehlers-Danlos syndrome, kyphoscoliosis type, type VI (EDVI), characterized by ocular, muscular, and skin defects (Hyland et al. 1992; Heikkinen et al. 1997; Valtavaara et al. 1997). The proximal promoter regions of both PLOD-1 and Plod-2 harbor multiple bicoid elements. We show these elements to bind the PITX2 protein in vitro, as well as activate a reporter gene in transient transfection assays. A Rieger syndrome causing PITX2 mutant, T68P, cannot induce the PLOD-1 promoter.
Materials and Methods
Chromatin Precipitation
We have previously characterized the Pitx2-specific antibody P2R10 (Hjalt and Murray 2000). It was used to immunoprecipitate chromatin bound in vivo to Pitx2. About 30 mouse E14 heads were fixed in 50 ml 4% paraformaldehyde in PBS on ice for 30 min. The heads were washed gently with 30 ml PBS twice, then once with 30 ml 50 mM glycine-HCl, pH 7.5, and once again with 30 ml PBS. The heads were slurried in 15 ml PBS supplemented with 1 ml 10× concentrated general protease inhibitor cocktail (Sigma-Aldrich), 200 μl saturated PMSF in isopropanol, and 20 mg powdered Calpain Inhibitor I/ALLN (Calbiochem-Novabiochem). The heads were homogenized and then sonicated on ice four times for 30 s with a 1/8 inch microtip at 30% (sonic dismembrator model 300; Fisher Scientific). The sonicate was centrifuged for 2 min at 3,000 rpm in a swinging bucket rotor. The supernatant extract was precleared for 15 min at 22°C with a 50% slurry of protein A–Sepharose CL4B in PBS (Amersham Pharmacia Biotech). The supernatant of the preclearing step was incubated with a P2R10-protein A–Sepharose CL4B affinity gel (prepared according to the recommendations of the manufacturer) for 2 h at 4°C, with a fresh 1 ml of general protease inhibitors. The gel was batch washed three times for 10 min at 22°C with 15 ml 50 mM Tris-HCl, pH 7.5/500 mM NaCl, then 3 times for 10 min with 15 ml 50 mM Tris-HCl, pH 7.5/100 mM NaCl. The gel was suspended in a fresh 1 ml of the last wash solution and treated with 100 μl proteinase K (Roche) at 55°C for 1 h (without SDS), then stopped at 70°C for 3 min. The sample was treated with phenol/chloroform, chloroform, and precipitated overnight at −20°C with sodium acetate and ethanol. The immunoenriched DNA was digested over night with EcoRI, cloned into a λZAPExpress vector (Stratagene), and packaged into phage particles (GigaPack III Gold; Stratagene), producing a primary library of 1.7 × 105 independent clones. Clones were chosen at random, excised using the ExAssist helper phage, prepared as plasmids, and the inserts were cleaved out and labeled with [32P]dCTP. The labeled inserts were used as probes for genomic Southern analyses and to probe a portion of the unamplified library in a plaque hybridization assay. Sometimes vector backbone DNA was difficult to separate from insert on a gel (same size). In these cases, vector T3 and T7 primers were used to PCR amplify the insert for probe production.
Bacterial Artificial Chromosome Screening and Sequencing
We screened bacterial artificial chromosome (BAC) libraries by PCR and BAC clones in pBeloBac11 were obtained from Research Genetics (mouse Plod-2, 249B6; human PLOD-1, 33A19). BAC DNA was prepared and sequenced directly as described previously (Hjalt and Murray 1999).
Electrophoretic Mobility Shift Assay
Oligodeoxyribonucleotides (see Table and Fig. 2) were synthesized (Integrated DNA Technologies) in complementary pairs, annealed to form double stranded DNA, and labeled by kinasing with [γ-32P]ATP. The probes were purified on Sephacryl S-200 Microspin columns (Amersham Pharmacia Biotech) and the activity was measured by scintillation counting. Bacterially overexpressed PITX2 was prepared as described previously (Hjalt and Murray 2000). Electrophoretic mobility shift assays (EMSAs) were performed essentially as described in Amendt et al. 1998. The Plod-2 G probe was modified from the sequence in the promoter. The wild-type Plod-2 G probe would form concatemers during annealing (not shown). The GC-rich 3′ sequence (GCGCCGCGGG) was substituted for an arbitrary AT-rich sequence (AAAATCATGA).
Table 1.
Primers Used
| Gene | Forward (name) | Reverse (name) | Annealing temperature | Size |
|---|---|---|---|---|
| bp | ||||
| Primer pairs for BAC library screening | ||||
| Plod-2 | AGAGGCGGTGATGGAATGAACA (b18-s2) | CTACAAAACACTCGGTAAACAAGAT (b18-15) | 63°C | 121 |
| PLOD-1 | CCTGGGGCTGCTCTAAGTGC (p1-21) | CCCTGCCGTCTCCCTCCCTTCTCT (p1-8) | 68°C | 232 |
| Primer pairs for RT-PCR | ||||
| Pitx2 | GCCAGCAAGGAAAGAATGAGGAT (Pitx2-1) | CGTAGACACTTGGGGACATTCCTT (Pitx2-6) | 67°C | 950 |
| PITX2 | CCGAGGACCCGTCTAAGAAGAAGC (PITX2-5) | GCATACTGGCAAGCACTCAGGT (PITX2-2) | 67°C | 709 |
| Plod-1 | GGGAGGACTGGAGTGTGGAT (m1-10) | CAGGTTCTGGAAGATTCGGCAGCGAT (m1-11) | 67°C | 451 |
| Plod-2 | CAATTACACTGTGAAGGTTCTTGGTC (b18-7) | GCATAGCCAATAAAGCCTCCAGAAT (b18-rt) | 67°C | 333 |
| PLOD-1 | CAGATGGCTACTATGCCCGTTC (h1x59) | CAAAGTGCTCCATCTCCTCCAC (h1x63) | 67°C | 437 |
| PLOD-2 | GACCAGAAGAAAATCTAAGTCAAGC (h2x29) | AGTTCCTTTCATTCATCTCTGAT (h2x25) | 67°C | 359 |
| Oligodeoxyribonucleotides for EMSAs (only sense strand shown) | ||||
| Bicoid | ACGGCCCATCTAATCCCGTG | |||
| PLOD-1 C | AAATATGAAATAATCCCACA | |||
| PLOD-1 H | CTCACACCTGTAATCCCAGC | |||
| PLOD-1 J | CACATGCCTGTAATCCCAGC | |||
| PLOD-2 C | ATTTTTGTTTTCATCCCTAAACACAAA | |||
| PLOD-2 E | TTTACACTTTTAGTCCCAGGATTTAAA | |||
| PLOD-2 F | CATAGACATACTAATCAAAACCCAAAG | |||
| PLOD-2 G | TGGCTGCTCTTAAGCCCAAAATCATGA | |||
| Primer pairs for quantitative PCR | ||||
| 18S-rDNA | AGCCTATTCTTTTTACTGGCTTGG (18S1F) | GGAAGCGTGGCTCGGG (18S1R) | ||
| Prolactin | CCTGCTGTTCTGCCAAAATGT (TMPL-1) | CGGAGAGAAGTCTGGCAGTCA (TMPL-2) | ||
| Plod-2 | CAGAAGGAACAGCTGGGAGTG (TMP1F) | GTGGTGACTGCGAGGGCTT (TMP1RN) | ||
| TacMan probe | ACAAGCGCCTACTCAGCCAAGCAGAC (TMm2-1) | |||
Figure 2.


The proximal promoter sequences of PLOD-1 and Plod-2. The proximal promoter DNA sequences of the human PLOD-1 (A) and mouse Plod-2 (B) are shown. PITX2 binding elements are in bold. The corresponding binding site names (A–J and A–H) are listed on the right side. The transcriptional start site of Plod-2 is not mapped, for reference purposes the 5′-most nucleotide of the cDNA (available from GenBank/EMBL/DDBJ under accession no. NM_011961) is used as +1. Bicoid-like and bicoid elements are in bold and boxed. In A, ApaI restriction sites are underlined. CAAT and TAATAA sequences are underlined with thick lines. These sequence data are available from GenBank/EMBL/DDBJ under accession nos. AF081786 (PLOD-1) and AF283255 (Plod-2).
Reverse Transcription PCR
Mouse mRNA was prepared from freshly dissected mouse tissues using a Micro Fast Track kit (Invitrogen). Human cDNAs were purchased from CLONTECH Laboratories, Inc. 20-μl reverse transcription (RT) reactions were performed using a semianchored oligo-dT primer and Superscript II reverse transcriptase (GIBCO BRL) according to the recommendations of the manufacturer. The cDNAs were diluted four or six times in 10 mM Tris-HCl, pH 8.5, and 1 μl was used per PCR reaction. The primers used for PCR are listed in Table . Standard 10- or 20-μl PCR reactions were started hot. Cycling conditions: 35 cycles of 30 s at 94°C, X°C for 1 min, 72°C for 1 min, and one cycle at 72°C for 1 min 30 s. Annealing temperatures (“X”) are listed in Table . The primer pairs chosen for PITX2/Pitx2 amplify most known cDNA isoforms. All primer pairs were designed with one to two intervening intron boundaries, and in some cases also with one primer straddling another intron–exon boundary.
Real-Time PCR
An ABI Prism Sequence Detector 7700, with the program Sequence Detector v1.7, was used for real-time PCR experiments, performed according to the recommendations of the manufacturer. Primers (Table ) were designed using PrimerExpress 1.5. Primers for 45S/18S rDNA amplify the segment between nucleotides 251 and 315 (5′ of transcription initiation) in GenBank/EMBL/DDBJ accession no. V00850. Primers for prolactin amplifies the segment between nucleotides 112–180 (exon 2) in GenBank/EMBL/DDBJ accession no. X02892. The 18S and prolactin products were detected using CyberGreen Master Mix (PerkinElmer). Primers for the Plod-2 promoter amplifies the segment between −535 and −466 in Fig. 2 B. The Plod-2 product was detected using a TaqMan probe (Table ) and TaqMan Master Mix (Applied Biosystems/Roche). 80 ng was used per reaction for both the control and the immunoenriched mouse DNA in four independent experiments. All PCR profiles were 50°C for 2 min, 95°C for 10 min, and 40 cycles at 94°C for 15 s, 60°C for 1 min. The differences in threshold cycle value, ΔCt, were obtained by comparing the Ct values for normal total mouse genomic DNA and immunoenriched mouse DNA. The results were normalized to the corresponding ΔCt values for the control primer set, 18S ribosomal DNA, such that fold enrichment was calculated as: 2[ΔCt(18S) + ΔCt(assayed gene)].
Reporter and Expression Constructs
Parts of the PLOD-1 promoter sequence were cloned into the luciferase gene expression vector pGL3 (Promega). Construct A, a 3,140-bp Apa1 fragment containing 10 bicoid elements and ending 37 bp upstream of exon 1 (positions 265–3404, in GenBank/EMBL/DDBJ accession no. AF081786; see Fig. 2 A), was excised from the human BAC 33A19 (Research Genetics) and cloned into the Apa1 site of pBluescriptIISK−. The reporter constructs were PCR amplified from this clone. The Plod-1 2561 luciferase reporter contains Plod-1 sequences from −60 to −880 and has the TAATAA and CAAT boxes (see Fig. 2). The 5′ sense primer contains a unique BamH1 site (underlined) linked to the Plod-1 sequence, 5′- CGGGATCCCAAGAGTGAAACTCTGTCTC-3′, and the 3′ antisense primer contains a unique HindIII site (underlined) 5′-CCCCAAGCTTCCCCGCCCCTCCACGCCC-3′. To make the Plod-1 261 luciferase reporter, the sequence from −60 to −3180 was PCR-amplified. The 5′ sense primer contains BamH1 site (underlined) linked to the Plod-1 sequence, 5′-CGGGATCCAAGCAGTCCTCCTGCCTT-3′, and the above 3′ antisense primer. The PCR products were digested with BamH1 and HindIII isolated and ligated into thymidine kinase (TK)-luc (Amendt et al. 1999), which was digested with BamH1 and HindIII to remove the TK promoter. The Plod-1 sequences were cloned upstream of the luciferase gene. The PCR profile consisted of 94°C for 1.5 min, 60°C for 2 min, and 72°C for 3 min for 30 cycles with Pfu DNA polymerase (Stratagene). A cytomegalovirus (CMV)–β-galactosidase reporter plasmid (CLONTECH Laboratories, Inc.) was cotransfected in all experiments as a control for transfection efficiency. Expression plasmids containing the CMV promoter linked to the PITX2 DNA were constructed in pcDNA 3.1 MycHisC (Invitrogen) as described previously (Amendt et al. 1998).
Cell Culture, Transient Transfections, Luciferase, and β-Galactosidase Assays
CHO cells were cultured in DME supplemented with 5% FBS and penicillin/streptomycin in 60-mm dishes and transfected by electroporation. CHO cells were mixed with 2.5 μg of expression plasmids, 5 μg of reporter plasmid, and 0.5 μg of CMV–β-galactosidase plasmid plated in 60-mm culture dishes and then fed with 5% FBS and DME. Electroporation of CHO cells was at 360 V and 950 μF; cells were fed 24 h before transfection. HeLa cells were mixed with 2.5 μg of expression plasmids, 5 μg of reporter plasmid, and 0.5 μg of CMV–β-galactosidase plasmid. HeLa cells were electroporated at 220 V and 960 μF (Bio-Rad Laboratories), plated in 60-mm culture dishes, and fed with 5% FBS and DME as described previously (Amendt et al. 1998). Cells were then lysed and assayed for reporter activities and protein content by Bradford assay (Bio-Rad Laboratories). Luciferase was measured using reagents from Promega. β-Galactosidase was measured using the Galacto-Light Plus reagents (Tropix, Inc.). All luciferase activities were normalized to β-galactosidase activity. Expression of transiently expressed PITX2 proteins has been demonstrated previously (Amendt et al. 1998).
Results
Plod-2 Is Immunoenriched by P2R10
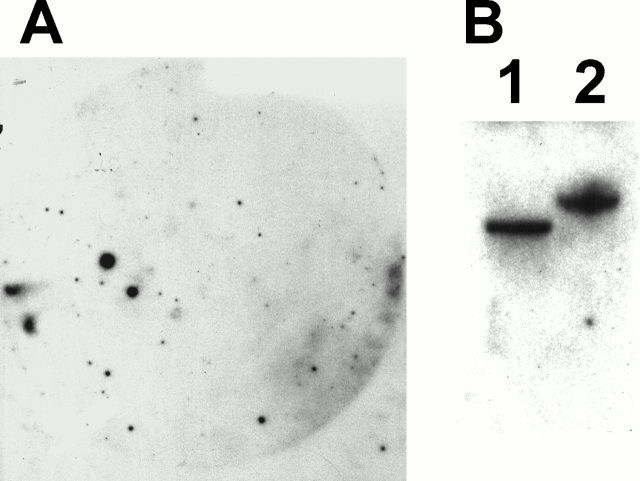
The nucleoprotein complexes of E14 mouse heads were immunoenriched by using a Pitx2-specific antibody (Hjalt and Murray 2000). The DNA was cloned into a phage library by plaque PCR using T7 and T3 flanking vector primers from 80 isolates; 90% contained inserts ranging in size from 100 to 8,000 bp, with an average size of 3,000 bp (not shown). 20 clones were excised at random one by one and inserts were labeled and used as probes for genomic Southern analyses and a plaque enrichment assay. One of the enriched clones, B18, that also represented a unique gene by Southern analysis (Fig. 1), displayed DNA sequence homology to mouse procollagen Plod-2. Normally, a 3,000-bp fragment of the mouse genome should appear 0.03 times on a 30,000 plaque filter (3E4 × 3E3/3E9). Five hits on the filter represent 165-fold enrichment. Control hybridization to a normal mouse genomic library filter of equal density was blank (not shown). The 2,701-bp B18 clone insert matched Plod-2 and PLOD-2 cDNA sequences in BLAST searches. Later we could confirm that B18 matched exon 7, intron 7, and exon 8 of PLOD-2. Since enrichment of the Plod-2 gene occurred before digestion with EcoRI and library cloning, we knew it would be possible to clone parts of the gene other than the promoter. We also confirmed that prolactin, a known Pitx2 binder (Amendt et al. 1998), was immunoenriched by probing a set of filters with a probe for mouse prolactin, exon 2 (not shown). Control hybridization to a normal mouse genomic library filter of equal density was blank (not shown). We also performed real-time PCR on normal total mouse DNA and immunoenriched DNA of the same concentrations. We used a primer set for 18S ribosomal DNA as a control and compared the results to those for the Plod-2 promoter, or exon 2 for prolactin. The normalized ΔCt values are presented in Table . The Plod-2 promoter was enriched 32-fold and the prolactin exon 2 was enriched 12-fold in this assay.
Figure 1.
Clone B18 is immunoenriched by a Pitx2-specific antibody and hybridizes to single bands in a genomic Southern analysis. (A) The insert of clone B18 from the chromatin precipitation library was labeled and used to probe a filter with 30,000 plaques of nonamplified immunoenriched library. At least five strong signals are apparent and indicate an enrichment factor of 165. (B) Southern blot of total mouse DNA digested with PstI (lane 1) and EcoRI (lane 2).
Table 2.
Enrichment Test by Real-Time PCR
| Ct control | Ct enriched | ΔCt | Enrichment (fold) | |
|---|---|---|---|---|
| 18S | 16.33 ± 0.04 | 18.22 ± 0.07 | Down 1.89 | NA |
| Plod-2 | 32.88 ± 0.30 | 29.76 ± 0.09 | Up 3.12 | 32 |
| Prolactin | 21.45 ± 0.03 | 19.73 ± 0.04 | Up 1.72 | 12 |
n = 4.
The Plod-2 Promoter Contains Multiple Bicoid Elements
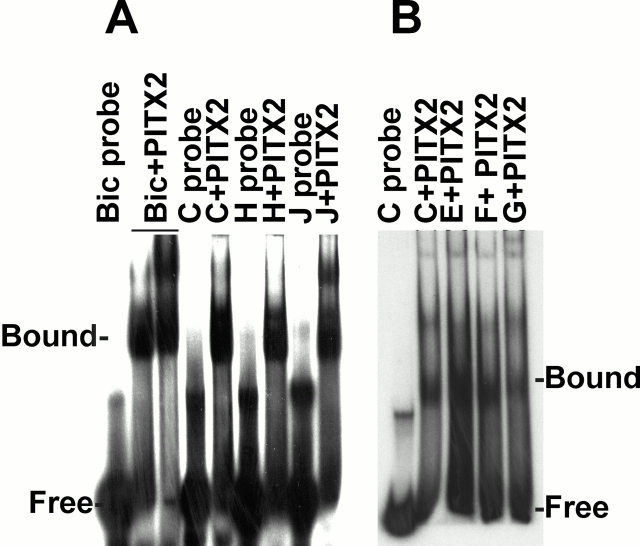
We screened a mouse genomic library and retrieved a BAC clone containing the Plod-2 gene. We sequenced 2,551 bp upstream of where the 5′ end of the published cDNA sequence starts. Here we discovered several sites shown previously to bind Drosophila bicoid specifically by DNaseI footprinting (Fig. 2). Sites A and C resemble the “X3” site, and site D resembles the “X1/X2” sites (Ma et al. 1996). Site F resembles the “B1” site (Driever and Nüsslein-Volhard 1989). Other similar sites have been shown to bind bicoid specifically (Rivera-Pomar et al. 1995; Dave et al. 2000). We confirmed that the C, E, F, and G sites also bound PITX2 by EMSA (Fig. 3 B). Negative controls were performed for all probes (probe only), but in Fig. 3 B only the control for the C probe is shown.
Figure 3.
PLOD-1 and Plod-2 promoter elements bind PITX2 in vitro. (A) EMSA of PITX2 protein incubated with radioactively labeled double stranded oligodeoxyribonucleotide probes designed from the PLOD-1 promoter. Bic, Drosophila bicoid element. For sequences of Bic and of elements C, H, and J see Fig. 2 and Table . (B) EMSA of PITX2 protein incubated with probes designed from the Plod-2 promoter. For sequences of elements C, E, F, and G see Fig. 2 and Table .
The PLOD-1 Promoter Contains Multiple Bicoid Elements
We also studied the human PLOD-1 promoter, since the sequence was available (GenBank/EMBL/DDBJ accession no. AF081786). Approximately 3 kb upstream of the PLOD-1 5′ untranslated region we detected 10 bicoid elements (TAATCCC). We confirmed the in vitro binding properties of some of these elements (C, E, and G) by EMSA, using human PITX2 protein (Fig. 3 A) and synthetic double stranded DNA.
PITX2 Regulates PLOD-1–luciferase Fusions in Cell Culture
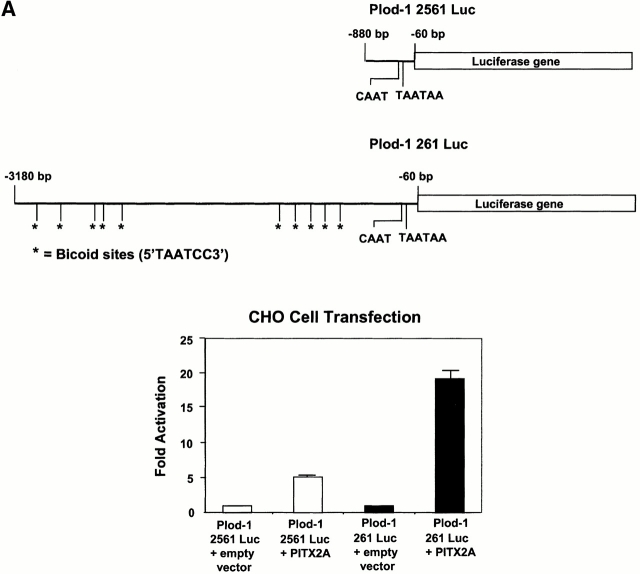
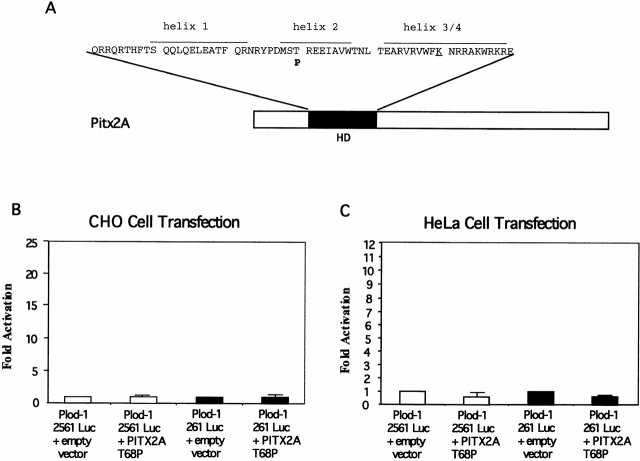
We proceeded to clone the PLOD-1 promoter in a luciferase reporter gene vector. We characterized the response to PITX2 by cotransfecting the PLOD-1–luciferase construct with a PITX2 expression vector. In CHO cells, the PLOD-1 261 construct containing 10 bicoid elements was activated 20-fold by PITX2A (Fig. 4 A). The minimal promoter constructs, PLOD-1 2561, were modestly activated (approximately fivefold) by PITX2. We attribute the modest activation to the fact that two bicoid-like sites remain, one TAACCC and one TAAGCC, in the minimal construct. Such elements are known to bind PITX2 in vitro (Amendt, B., unpublished data). In HeLa cells, PLOD-1 was activated 6.5-fold by PITX2A (Fig. 4 B). Furthermore, we found that a PITX2 gene carrying the Rieger syndrome causing mutation T68P failed to upregulate PLOD-1 in cell culture (Fig. 5). This protein can be expressed in cell culture (Amendt et al. 1998). The responses were similar to the negative controls both in CHO cells (Fig. 5 B) and in HeLa cells (Fig. 5 C).
Figure 4.
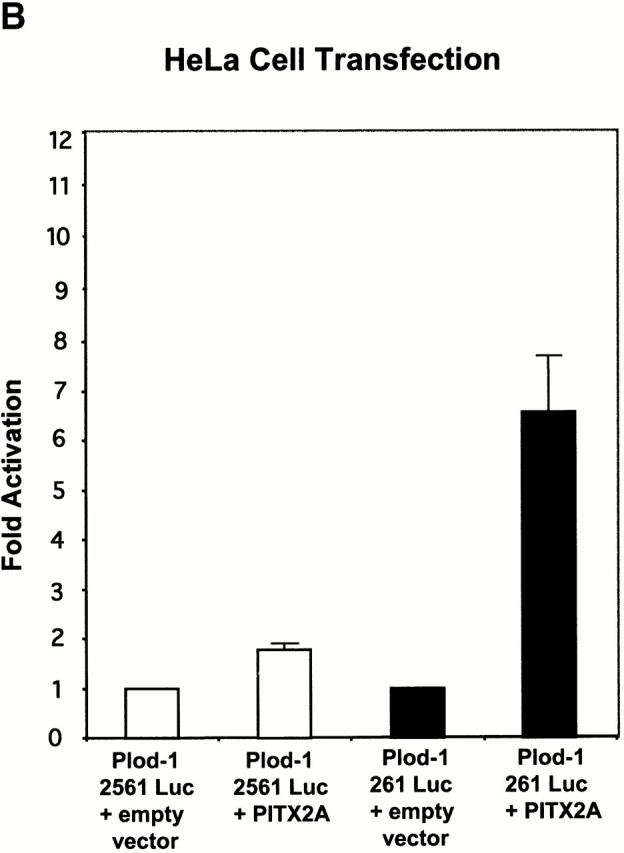
PITX2 upregulates PLOD-1–luciferase fusion constructs. The PLOD-1 promoter was fused to a luciferase reporter gene and assayed for activation by PITX2A. (A) CHO cells. (B) HeLa cells.
Figure 5.
Rieger syndrome mutant PITX2 T68P fails to upregulate PLOD-1–luciferase fusion constructs. The PLOD-1 promoter was fused to a luciferase reporter gene and assayed for activation by PITX2A-T68P. (A) Location of the mutation T68P in helix 2 of PITX2. A lysine residue in helix 3, important for binding, is underlined. (B) CHO cells. (C) HeLa cells.
Coexpression in Several Mouse Tissues of Pitx2 with Plod-1 and Plod-2
We wanted to study which tissues Pitx2 was coexpressed in with Plod-1 and Plod-2. We assayed this by RT-PCR of mRNA isolated from various mouse tissue (Fig. 6).
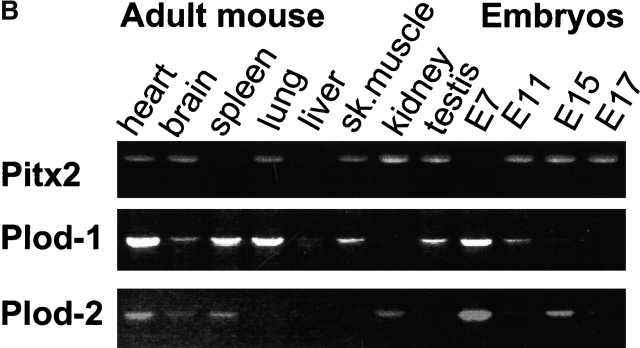
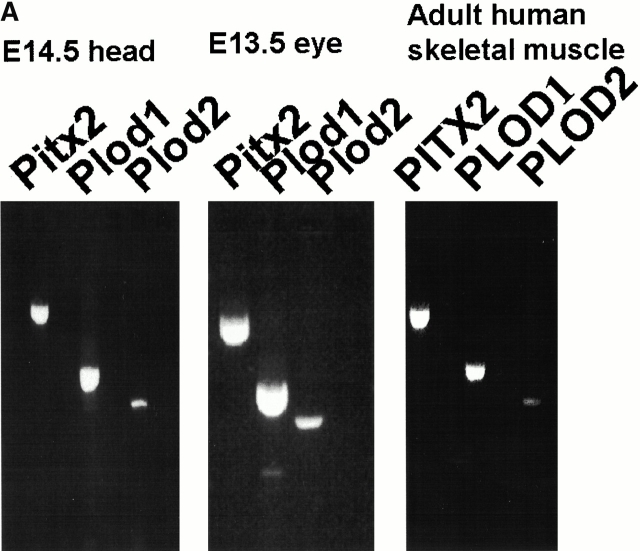
Figure 6.
Coexpression of Pitx2 and Plod-1/Plod-2. (A) RT-PCR with Pitx2, Plod-1, and Plod-2 primers on mRNA prepared from E14.5 mouse heads, E13.5 mouse eyes, and adult human skeletal muscle. (B) RT-PCR with various embryonic and adult mouse tissues.
The PCR primers for Pitx2 were designed to detect most known isoforms (see Materials and Methods). First, we could confirm that Pitx2, Plod-1, and Plod-2 are coexpressed in the tissue used to create the chromatin precipitation library, mouse E14.5 heads (Fig. 6 A). Moreover, we could confirm the same coexpression in E13.5 eyes (Fig. 6 A). We also detected coexpression of PITX2, PLOD-1, and PLOD-2 in adult human skeletal muscle (Fig. 6 A). We also detected coexpression of Pitx2, Plod-1, and Plod-2 in adult mouse heart and brain; of Pitx2 and Plod-1 in adult mouse lung, skeletal muscle, and testis; and Pitx2 and Plod-2 in mouse adult kidney (Fig. 6 B). In mRNA from whole mouse embryos, we detected coexpression of Pitx2, Plod-1, and Plod-2 at E11 and E15 (Fig. 6 B). At E17, Pitx2 and Plod-2 were coexpressed. At E7, Plod-1 and Plod-2 were expressed without Pitx2. This was also the case in adult spleen. No expression was seen of either gene in adult liver.
Clinical Similarities between Rieger and EDVI
We compared the reported clinical manifestations of Rieger syndrome and EDVI. We used the database Online Mendelian Inheritance in Man (available at http://www.ncbi.nlm.nih.gov/omim/) textbooks (e.g., Traboulsi 1998), as well as published and unpublished clinical observations (May and Beauchamp 1987; Wenstrup et al. 1989; Heikkinen et al. 1997; Murray, J.C., unpublished). Several similarities of organ system involvement are listed in Table . Most notable are the comparable ocular and abdominal abnormalities.
Table 3.
Clinical Features of Rieger and Ehlers-Danlos Syndromes
| Clinical manifestations | Rieger syndrome | EDVI |
|---|---|---|
| Ocular | ||
| Glaucoma | Yes | Yes |
| Microcornea | Yes | Yes |
| Cornea plana | Yes | Yes |
| Blue sclera | No | Yes |
| Fragile eyes/corneas | No | Yes |
| Iris hypoplasia | Yes | No |
| Iridocorneal adhesions | Yes | No |
| Myopia | No | Yes |
| Dental | ||
| Tooth abnormalities | Yes | Yes |
| Abdominal | ||
| Inguinal hernia | Yes | Yes |
| Umbilical hernia | Yes | Yes |
| Omphalocele | Yes | No |
| Skin hyperflexibility | No | Yes |
| Colon rupture | No | Yes |
Discussion
Several direct and indirect lines of evidence now support the theory that the Plod genes belong to the cognate targets for Pitx2. (a) We were able to enrich for the Plod-2 gene ∼170-fold in a chromatin precipitation assay using a Pitx2/PITX2-specific antibody. This antibody does not cross-react with PITX1 or PITX3 (Hjalt and Murray 2000). However, we do not exclude the possibility that Pitx1/Pitx3/PITX1/PITX3 might also be involved in the regulation of the Plod/PLOD genes. Indeed, it has been shown that PITX1 and PITX2 share DNA binding and transactivating properties of target genes in the pituitary (Tremblay et al. 2000). (b) The PLOD-1 and Plod-2 genes contain clusters of bicoid elements in their proximal promoter regions and we have shown that these elements bind PITX2 in vitro. (c) We have shown that the proximal PLOD-1 promoter can be induced to express a reporter gene in the presence of PITX2. (d) The Rieger syndrome causing PITX2 mutant T68P fails to induce the PLOD-1 reporter gene construct. (e) The EDVI patients share several characteristics with the Rieger syndrome patients (Table ). It is normally easy to distinguish these two disorders from each other. However, the similarities of organ systems involved pointed out here are indicative of the role that lysine hydroxylation of collagens may play in Rieger syndrome. (f) The EDVI patients also have defects in tissues corresponding to where Pitx2 is expressed in the mouse, such as cornea, skeletal muscle, aorta smooth muscle, and skin (May and Beauchamp 1987; Wenstrup et al. 1989; Heikkinen et al. 1997; Hjalt and Murray 2000). (g) Plod-1 ESTs have been isolated from rabbit corneal endothelial cells (Fujimaki et al. 1999) and strong Plod-like enzymatic activity can be isolated from chick embryonic corneas (Kao et al. 1983). These structures also strongly express the Pitx2 protein in the mouse (Hjalt and Murray 2000). We were also able to show coexpression of Pitx2, Plod-1, and Plod-2 mRNA in mouse E13.5 eye by RT-PCR.
It is also interesting to point out that Pitx2 is expressed in mouse aorta smooth muscle and some EDVI patients have aortic defects (Tsai and Grajewski 1994; Cunningham et al. 1998; Mammi et al. 1998). Rieger and ED syndromes normally have different modes of inheritance, autosomal dominant and recessive, respectively. However, it is still unclear exactly what the link is between the pathologies of each syndrome.
Our finding that PITX2 regulates PLOD-1 differently in different cellular backgrounds is indicative of the dependence of cofactors for correct function of a homeobox gene. The most appealing model for in vivo specificity of homeobox genes is probably the binding site selection model, in which cofactors define appropriate DNA binding site and later fine-tune expression levels (Mann and Chan 1996; Mann and Affolter 1998). This model has recently been very convincingly supported for Fushi tarazu, which also can either activate or repress its cognate target genes depending on cell type (Nasiadka et al. 2000). Furthermore, the estimated amount of available Fushi tarazu protein per cell is several orders of magnitude smaller than the theoretically available cognate DNA binding sites (Nasiadka et al. 2000). The support for the binding site selection model also lends encouragement to methods such as chromatin precipitation.
Some mouse tissues express Plod-1 and Plod-2 in the absence of Pitx2 (adult spleen and E7 whole embryo). It may be that another Pitx family member regulates the Plod genes in these tissues. Perhaps more likely is that Pitx2 serves as a modifier of gene expression, as is known to be the case in the regulation of prolactin by PIT-1 and PITX2 (Simmons et al. 1990; Szeto et al. 1996; Ryan and Rosenfeld 1997; Amendt et al. 1998). Either protein, by itself, accomplishes only modest transactivation of the prolactin promoter, but together a dramatic synergistic effect on gene expression is seen (Simmons et al. 1990; Szeto et al. 1996; Ryan and Rosenfeld 1997; Amendt et al. 1998). It will be interesting and important to find the cognate cofactors of Pitx2/PITX2 in the regulation of the Plod-1 and 2/PLOD-1 and 2 genes. Our discovery validates the chromatin precipitation method as a viable means of finding additional target genes of PITX2.
Acknowledgments
This study was supported by grants from the National Institutes of Health (DE09170 and EY12384 to J.C. Murray); the National Institute of Dental and Craniofacial Research (DE13941-01 to B.A. Amendt); and the American Heart Association (9960299Z to B.A. Amendt); and a postdoctoral research fellowship was supported by the Fight for Sight research division of Prevent Blindness America (PD99018 to T.A. Hjalt).
Footnotes
Abbreviations used in this paper: BAC, bacterial artificial chromosome; CMV, cytomegalovirus; EDVI, Ehlers-Danlos syndrome type VI; EMSA, electrophoretic mobility shift assay; PLOD, procollagen lysyl hydroxylase; RT, reverse transcription; TK, thymidine kinase.
References
- Amendt B.A., Sutherland L.B., Semina E.V., Russo A.F. The molecular basis of Rieger syndrome. J. Biol. Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- Amendt B.A., Sutherland L.B., Russo A.F. Multifunctional role of PITX2 homeodomain protein C-terminal tail. Mol. Cell. Biol. 1999;19:7001–7010. doi: 10.1128/mcb.19.10.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendt B.A., Semina E.V., Alward W.L.M. Rieger syndromea clinical, molecular, and biochemical analysis. Cell. Mol. Life Sci. 2000;57:1652–1666. doi: 10.1007/PL00000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M., Steinbeisser H., Campione M., Schweichert A. Vertebrate left-right asymmetryold studies and new insights. Cell. Mol. Biol. 1999;45:505–516. [PubMed] [Google Scholar]
- Campione M., Steinbesser H., Schweickert A., Deissler K., van Bebber F., Lowe L.A., Nowotschin S., Viebahn C., Haffter P., Kuehn M.R., Blum M. The homeobox gene Pitx2mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- Craig J.E., Mackey D.A. Glaucoma geneticswhere are we? Where will we go? Curr. Opin. Ophthalmol. 1999;10:126–134. doi: 10.1097/00055735-199904000-00009. [DOI] [PubMed] [Google Scholar]
- Cunningham E.T., Jr., Eliott D., Miller N.R., Maumenee I.H., Green W.R. Familial Axenfeld-Rieger anomaly, atrial septal defect, and sensorineural hearing lossa possible new genetic syndrome. Arch. Ophthalmol. 1998;116:78–82. doi: 10.1001/archopht.116.1.78. [DOI] [PubMed] [Google Scholar]
- Dave V., Zhen C., Yang F., Tung C.-S., Ma J. Reprogrammable recognition codes in Bicoid homeodomain-DNA interaction. Mol. Cell. Biol. 2000;20:7673–7684. doi: 10.1128/mcb.20.20.7673-7684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Dubnau J., Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- Fujimaki T., Hotta Y., Sakuma H., Fujiki K., Kanai A. Large-scale sequencing of the rabbit corneal endothelial cDNA library. Cornea. 1999;18:109–114. doi: 10.1097/00003226-199901000-00017. [DOI] [PubMed] [Google Scholar]
- Gage P.J., Suh H., Camper S.A. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Gould A.P., Brookman J.J., Strutt D.I., White R.A.H. Targets of homeotic gene control in Drosophila . Nature. 1990;348:308–312. doi: 10.1038/348308a0. [DOI] [PubMed] [Google Scholar]
- Heikkinen J., Toppinen T., Yeowell H., Krieg T., Steinmann B., Kivirikko K.I., Myllylä R. Duplication of seven exons in the lysyl hydroxylase gene is associated with longer forms of a repetitive sequence within the gene and is a common cause for the type VI variant of Ehlers-Danlos syndrome. Am. J. Hum. Genet. 1997;60:48–56. [PMC free article] [PubMed] [Google Scholar]
- Hjalt T.A., Murray J.C. The human BARX2 genegenomic structure, chromosomal localization, and single nucleotide polymorphisms. Genomics. 1999;62:456–459. doi: 10.1006/geno.1999.6037. [DOI] [PubMed] [Google Scholar]
- Hjalt T.A., Murray J.C. The Pitx2 protein in mouse development. Dev. Dyn. 2000;218:195–200. doi: 10.1002/(SICI)1097-0177(200005)218:1<195::AID-DVDY17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Hyland J., Ala-Kokko L., Royce P., Steinmann B., Kivirikko K.I., Myllylä R. A homozygous stop codon in the lysyl hydroxylase gene in two siblings with Ehlers-Danlos syndrome type VI. Nat. Genet. 1992;2:228–231. doi: 10.1038/ng1192-228. [DOI] [PubMed] [Google Scholar]
- Jackle H., Sauer F. Transcriptional cascades in Drosophila . Curr. Opin. Cell. Biol. 1993;5:505–512. doi: 10.1016/0955-0674(93)90017-k. [DOI] [PubMed] [Google Scholar]
- Kao W.W., Mai S.H., Chou K.L., Ebert J. Mechanism for the regulation of post-translational modifications of procollagens synthesized by matrix-free cells from chick embryos. J. Biol. Chem. 1983;258:7779–7787. [PubMed] [Google Scholar]
- Kivirikko K.I., Myllylä R. Post-translational processing of procollagens. Ann. NY Acad. Sci. 1985;460:187–201. doi: 10.1111/j.1749-6632.1985.tb51167.x. [DOI] [PubMed] [Google Scholar]
- Kivirikko K.I., Myllylä R., Pihlajaniemi T. Hydroxylation of proline and lysine residues in collagens and other animal and plant proteins. In: Harding J.J., Crabba M.J.C., editors. Focus on Post-translational Modifications of Proteins. CRC Press; Boca Raton, FL: 1992. pp. 1–51. [Google Scholar]
- Lin C.R., Kiossi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisua-Belmonte J.-C., Rosenfeld M.G. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Lu M.-F., Pressman C., Dyer R., Johnson R.L., Martin J.F. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Ma X., Yuan D., Diepold K., Scarborough T., Ma J. The Drosophila morphogenic protein Bicoid binds DNA cooperatively. Development. 1996;122:1195–1206. doi: 10.1242/dev.122.4.1195. [DOI] [PubMed] [Google Scholar]
- Mann R.S., Affolter M. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mann R.S., Chan S.K. Extra specificity from extradenticlethe partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- Mammi I., DeGiorgio P., Clementi M., Tenconi R. Cardiovascular anomaly in Rieger Syndromeheterogeneity or contiguity? Acta Ophthalmol. Scand. 1998;76:509–512. doi: 10.1034/j.1600-0420.1998.760424.x. [DOI] [PubMed] [Google Scholar]
- May M.A., Beauchamp G.R. Collagen maturation defects in Ehlers-Danlos keratopathy. J. Pediatr. Ophthalmol. 1987;Strabismus. 24:78–82. doi: 10.3928/0191-3913-19870301-07. [DOI] [PubMed] [Google Scholar]
- Nasiadka A., Grill A., Krause H.M. Mechanisms regulating target gene selection by the homeodomain-containing protein Fushi tarazu . Development. 2000;127:2965–2976. doi: 10.1242/dev.127.13.2965. [DOI] [PubMed] [Google Scholar]
- Orlando V., Paro R. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Phelps D.E., Dressler G.R. Identification of novel Pax-2 binding sites by chromatin precipitation. J. Biol. Chem. 1996;271:7978–7985. doi: 10.1074/jbc.271.14.7978. [DOI] [PubMed] [Google Scholar]
- Prockop D.J., Kivirikko K.I. Heritable diseases of collagen. New Engl. J. Med. 1984;311:376–386. doi: 10.1056/NEJM198408093110606. [DOI] [PubMed] [Google Scholar]
- Rieger H. Dysgenesis mesodermalis corneal et iridis. Z. Augenhelik. 1935;86:333. [Google Scholar]
- Rivera-Pomar R., Lu X., Taubert H., Perrimon N., Jäckle H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376:253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- Ryan A.K., Rosenfeld M.G. POU domain family valuesflexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Semina E.V., Reiter R., Leysens N.J., Alward W.L.M., Small K.W., Datson N.A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B.U., Carey J.C., Murray J.C. Cloning and characterization of a novel bicoid-related homeobox gene, RIEG, involved in Rieger syndrome. Nat. Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Simmons D.M., Voss J.W., Ingraham H.A., Holloway J.M., Broide R.S., Rosenfeld M.G., Swanson L.W. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711. doi: 10.1101/gad.4.5.695. [DOI] [PubMed] [Google Scholar]
- Szeto D.P., Ryan A.K., O'Connell S.M., Rosenfeld M.G. P-OTXa PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc. Natl. Acad. Sci. USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Yonei-Tamura S., Belmonte J.C. Molecular basis of left-right asymmetry. Dev. Growth Differ. 1999;41:645–656. doi: 10.1046/j.1440-169x.1999.00465.x. [DOI] [PubMed] [Google Scholar]
- Tomotsune D., Shoji H., Wakamatsu Y., Kondoh H., Takahashi N. A mouse homologue of the Drosophila tumour-suppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature. 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- Traboulsi E.I. Malformations of the anterior segment of the eye. In: Traboulsi E.I., editor. Genetic Diseases of the Eye. Oxford University Press; New York: 1998. pp. 81–98. [Google Scholar]
- Tremblay J.J., Goodyer C.G., Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- Tsai J.C., Grajewski A.L. Cardiac valvular disease and Axenfeld-Rieger syndrome Am . J. Ophthalmol. 1994;118:255–256. doi: 10.1016/s0002-9394(14)72910-1. [DOI] [PubMed] [Google Scholar]
- Valtavaara M., Papponen H., Pirttilä A.-M., Hiltunen K., Helander H., Myllylä R. Cloning and characterization of a novel human lysyl hydroxylase isoform highly expressed in pancreas and muscle J . Biol. Chem. 1997;272:6831–6834. doi: 10.1074/jbc.272.11.6831. [DOI] [PubMed] [Google Scholar]
- Wenstrup R.J., Murad S., Pinnell S. Ehlers-Danlos syndrome type VIclinical manifestations of collagen lysyl hydroxylase deficiency. J. Pediatr. 1989;115:405–409. doi: 10.1016/s0022-3476(89)80839-x. [DOI] [PubMed] [Google Scholar]