Abstract
Anchorage of microtubule minus ends at spindle poles has been proposed to bear the load of poleward forces exerted by kinetochore-associated motors so that chromosomes move toward the poles rather than the poles toward the chromosomes. To test this hypothesis, we monitored chromosome movement during mitosis after perturbation of nuclear mitotic apparatus protein (NuMA) and the human homologue of the KIN C motor family (HSET), two noncentrosomal proteins involved in spindle pole organization in animal cells. Perturbation of NuMA alone disrupts spindle pole organization and delays anaphase onset, but does not alter the velocity of oscillatory chromosome movement in prometaphase. Perturbation of HSET alone increases the duration of prometaphase, but does not alter the velocity of chromosome movement in prometaphase or anaphase. In contrast, simultaneous perturbation of both HSET and NuMA severely suppresses directed chromosome movement in prometaphase. Chromosomes coalesce near the center of these cells on bi-oriented spindles that lack organized poles. Immunofluorescence and electron microscopy verify microtubule attachment to sister kinetochores, but this attachment fails to generate proper tension across sister kinetochores. These results demonstrate that anchorage of microtubule minus ends at spindle poles mediated by overlapping mechanisms involving both NuMA and HSET is essential for chromosome movement during mitosis.
Keywords: chromosome, kinetochore, spindle pole, NuMA, HSET
Introduction
The spindle is a complex microtubule-based superstructure responsible for chromosome movement and segregation during mitosis and meiosis (McIntosh and Koonce 1989; Mitchison 1989a; Rieder 1991; Hyman and Karsenti 1996; Compton 2000). Chromosome movement on spindles during mitosis in cultured cells has been well documented (Gorbsky 1992; Rieder and Salmon 1994; Inoué and Salmon 1995; Rieder and Salmon 1998; Khodjakov et al. 1999) and is driven by three different force-generating mechanisms (Mitchison 1989b; Gorbsky 1992; Rieder and Salmon 1994; Khodjakov and Rieder 1996; Khodjakov et al. 1999). Poleward chromosome movement is driven by forces derived from kinetochore-associated microtubule motor proteins and the continuous poleward flux of tubulin subunits within the spindle lattice. Kinetic analyses indicate that a majority (60–70%) of poleward chromosome movement in cultured somatic cells is driven by forces generated by kinetochore-associated motors (Mitchison and Salmon 1992), and candidates for these motors are the kinesin-related proteins CENP-E and MCAK/XKCM1 as well as cytoplasmic dynein (Rieder and Alexander 1990; Pfarr et al. 1991; Steuer et al. 1991, Yen et al. 1991; Walczak et al. 1996; Schaar et al. 1997; Wood et al. 1997; Maney et al. 1998). Chromosome movement away from spindle poles during congression is driven by polar ejection forces (Rieder et al. 1986) that may be generated by chromosome-associated kinesin-related proteins (Antonio et al. 2000; Funabiki and Murray 2000).
An implicit assumption in how these force-generating mechanisms cause chromosome movement is that microtubule minus ends are firmly anchored at spindle poles. It has been suggested that this anchorage is necessary in anaphase to “bear the load when chromosomes move, so that the chromosomes move toward the poles rather than the poles toward the chromosome” (Nicklas 1989). In its extreme form, this idea posits that if the microtubule minus ends were not appropriately anchored at spindle poles, then the poleward forces generated by kinetochore-associated motors (coupled to microtubule depolymerization) would pull the microtubules in toward the chromosome rather than move the chromosome toward the pole. Indeed, direct observation of microtubule-chromosome interactions under defined in vitro conditions has demonstrated that microtubules are reeled in toward the chromosomes in the absence of anchorage at minus ends (Koshland et al. 1988). Likewise, the polar ejection forces would extrude the microtubules past the region of the spindle pole toward the cell cortex instead of pushing the chromosome toward the spindle equator.
Our understanding of the mechanisms for microtubule minus end anchorage at spindle poles has grown substantially in the past few years (Merdes and Cleveland 1997; Compton 1998). Centrosomes are the dominant site for microtubule nucleation and, when present, are located at spindle poles as a consequence of their function in microtubule nucleation. However, a variety of evidence demonstrates that centrosomes are neither necessary nor sufficient to act as functional spindle poles, and chromosome movement occurs normally in cells lacking centrosomes and in cells where centrosomes have been experimentally removed (Szollosi et al. 1972; Nicklas 1989; Heald et al. 1996; Gaglio et al. 1997; Khodjakov et al. 2000). These observations demonstrate that centrosomes cannot be the primary anchorage sites for microtubule minus ends at spindle poles in vertebrate cells. Thus, we hypothesize that noncentrosomal proteins provide the primary anchorage site for microtubule minus ends at poles to counterbalance the forces involved in chromosome movement. To test this hypothesis, we monitored chromosome movement in cultured cells after perturbation of nuclear mitotic apparatus protein (NuMA) and human homologue of the KIN C motor family (HSET), two noncentrosomal proteins involved in focusing microtubule minus ends at spindle poles (McDonald et al. 1990; Kallajoki et al. 1991; Hatsumi and Endow 1992; Yang and Snyder 1992; Endow et al. 1994; Gaglio et al. 1995; Kuriyama et al. 1995; Matthies et al. 1996; Merdes et al. 1996, Merdes et al. 2000; Walczak et al. 1997; Matuiene et al. 1999; Mountain et al. 1999). The results show that simultaneous perturbation of both NuMA and HSET virtually abolishes chromosome movement in cultured cells despite bi-oriented spindle assembly and microtubule attachment to kinetochores. These results indicate that NuMA and HSET act through overlapping mechanisms to hold microtubule minus ends at spindle poles and permit chromosome movement to be driven by kinetochore- and polar ejection–derived forces.
Materials and Methods
Cell Culture
CFPAC-1 cells were maintained at 37°C in a 5%-CO2 atmosphere in Iscove's Modified Dulbecco's Medium containing 10% fetal bovine serum, 50 IU/ml penicillin, and 50 μg/ml streptomycin.
Antibodies
Rabbit polyclonal antibodies against NuMA were generated by immunization with the full-length recombinant NuMA protein (Gaglio et al. 1995). Rabbit polyclonal antibodies against HSET were generated by immunization with the recombinant protein representing the COOH-terminal 377 amino acids (Mountain et al. 1999). Tubulin was detected with the DM1α mouse monoclonal antibody (Sigma-Aldrich). Kinetochores were detected with the human anticentromere antibody ACA-m (provided by Kevin Sullivan, Scripps Research Institute, San Diego, CA), and CENP-E was detected using the mouse monoclonal antibody mAb177 (Yen et al. 1991; provided by Tim Yen, Fox Chase Cancer Center, Philadelphia, PA).
Antibody Microinjection
CFPAC-1 cells growing on photo-etched alphanumeric glass cover slips (Bellco Glass Co.) were microinjected following the procedures of Compton and Cleveland 1993 and Capecchi 1980. IgG was purified from whole serum for microinjection by affinity chromatography using protein A-conjugated agarose (Roche Molecular Biochemicals). PD-10 Sephadex G-25 columns (Amersham Pharmacia Biotech) were used for buffer exchange to microinjection buffer (100 mM KCl, 10 mM KPO4, pH 7.0), followed by concentration using Centricon spin columns (Millipore). The following antibody concentrations refer to their concentrations in the microinjection needle: 20 mg/ml α-NuMA, 10 mg/ml α-HSET, and 19 mg/ml nonimmune IgG.
Time-Lapse Microscopy
Cells were injected in the cytoplasm during interphase and monitored by phase-contrast microscopy until they entered prophase (as judged by chromosome condensation). The coverslip was then mounted on a custom-built stainless steel modified Rose chamber containing growth medium and sealed with VALAP (Vaseline, lanolin, and paraffin wax in a 1:1:1 mass ratio). The chamber was placed on a heated stage, and differential interference contrast (DIC) images were acquired at 30-s intervals with a Hamamatsu Orca II cooled CCD camera using the Openlab software (Improvision Inc.) on an Axioplan 2 microscope using a Plan-Apochromat 63× (NA 1.4) objective (Carl Zeiss, Inc.). To minimize file sizes, the supplemental video sequences accompanying this article only contain images acquired every minute.
Chromosome velocities were obtained from the digital time-lapse record of each cell. The microscopy system used for time-lapse recordings was calibrated using a stage micrometer under the same conditions used for image acquisition. Individual chromosome movement was tracked by frame-by-frame analysis of digital images using Openlab software (Improvision, Inc.). The straight line calibration tool in the Openlab software package was used to determine the distance traveled by an individual chromosome between different time points. Velocities were then calculated by dividing the total distance traveled (microns) by the time interval in which the measurements were made (minutes). The spindle equator was used as a frame of reference in many images due to the lack of organized poles, and was assigned as the position where a bulk of the chromosomes were aligned. Chromosome movement was judged to be directed when the chromosome was displaced by >3 μm in a linear fashion. Displacement of this magnitude is easily distinguishable from Brownian motion (see Alexander and Rieder 1991).
Indirect Immunofluorescence Microscopy
Cells to be processed for immunofluorescence were first immersed in microtubule stabilizing buffer (MTSB: 4 M glycerol, 100 mM PIPES, pH 6.8, 1 mM EGTA, 5 mM MgCl2) for 1 min. Cells were then extracted with MTSB/0.5% Triton X-100 for 2 min, followed by a 2-min rinse in MTSB. Cells were then fixed in 1% glutaraldehyde in PBS for 10 min, followed by two 10-min treatments with 0.5 mg/ml NaBH4, and a 5-min wash in TBS (10 mM Tris, pH 7.5, 150 mM NaCl) containing 1% BSA (TBS-BSA). Primary antibody was diluted in TBS-BSA and added to the cells for 30 min in a humidified chamber. The cells were washed for 5 min in TBS-BSA and stained with the appropriate fluorochrome-conjugated, species-specific secondary antibodies in TBS-BSA along with 1 μg/ml DAPI (Sigma-Aldrich). Coverslips were washed for 5 min in TBS-BSA, and then mounted in Vectashield (Vector Laboratories).
Cells to be stained for kinetochore components were fixed in 3.5% paraformaldehyde in PBS for 5 min at room temperature, followed by extraction with TBS-BSA/0.5% Triton X-100 for 5 min and a 5-min wash in TBS-BSA. All subsequent antibody treatments were as described above.
Fluorescent images were captured with a Hamamatsu Orca II cooled CCD camera mounted on an Axioplan 2 microscope equipped for epifluorescence. A series of 0.5-μm optical sections were collected in the z plane for each channel (DAPI, fluorescein, and/or Texas red) and deconvolution using the Openlab software (Improvision Inc.) was used to eliminate extraneous fluorescence background. Selected planes from the z series were then overlayed to generate the final image.
Electron Microscopy
The position of injected CFPAC-1 cells on the alphanumeric coverslips was noted for subsequent selection for examination by electron microscopy. Cells were rinsed in MTSB for 1 min, extracted with MTSB/2% Triton X-100 for 5 min, and washed with MTSB for 2 min at room temperature. Cells were then fixed in 1% glutaraldehyde in 0.1 M Na-cacodylate buffer for at least 30 min. After fixation, the cells were rinsed twice with 0.1 M Na-cacodylate buffer, and then postfixed with 1% OsO4 in Na-cacodylate buffer for 30 min at room temperature, and en bloc stained in 2% uranyl acetate for 30 min at room temperature. Cells were dehydrated through a graded series of ethanols and propylene oxide, and flat embedded in epon (LX112)/araldite(502). The glass coverslip was removed by etching in cold concentrated hydrofluoric acid, as described by Moore 1975 and Rieder and Bowser 1987. The area containing the injected cells was identified under a dissecting microscope, cut out of the flat embedded rectangle, and remounted onto epoxy blanks. Sections (90–200 nm) were stained with 2% aqueous uranyl acetate for 45 min at 50°C and with Reynold's lead citrate for 20 min at room temperature. Electron micrographs were taken at 80 or 100 kV on a JEOL 100CX.
Online Supplemental Material
Quicktime videos to accompany Fig. 1 Fig. 2 Fig. 3 Fig. 4 are available at http//:www.jcb.org/cgi/content/full/152/3/425/DC1.
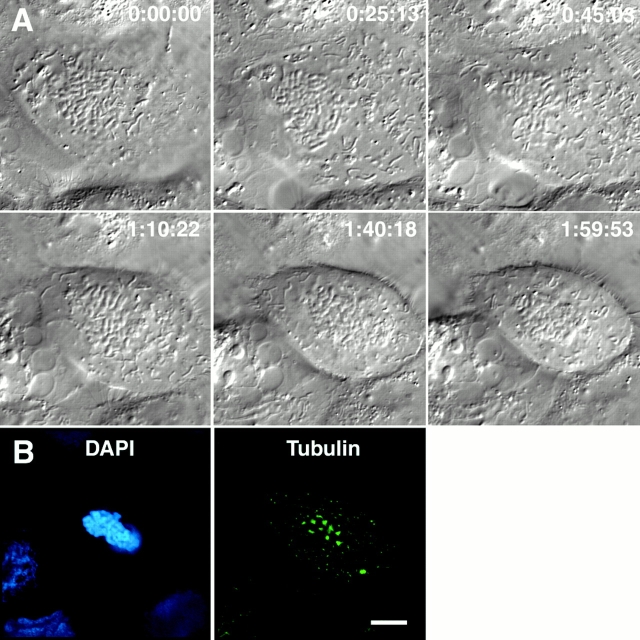
Figure 1.

Chromosome movement in a CFPAC-1 cell microinjected with antibodies to NuMA. (A) Selected DIC images from the video record of a mitotic cell that has been microinjected with an antibody to NuMA. Times are indicated in hours:minutes:seconds. The arrow highlights a chromosome that makes a pronounced movement toward the cell equator. (B and C) The cell featured in A (B) and a second, unrelated injected mitotic CFPAC-1 cell (C) were fixed and processed for immunofluorescence microscopy using antibodies specific for tubulin, the injected NuMA antibody, and the DNA-specific dye DAPI, as indicated. Arrows indicate centrosomes and arrowheads indicate sites of microtubule convergence (see Video 1 in online supplement). Bar: 20 μm.
Figure 2.
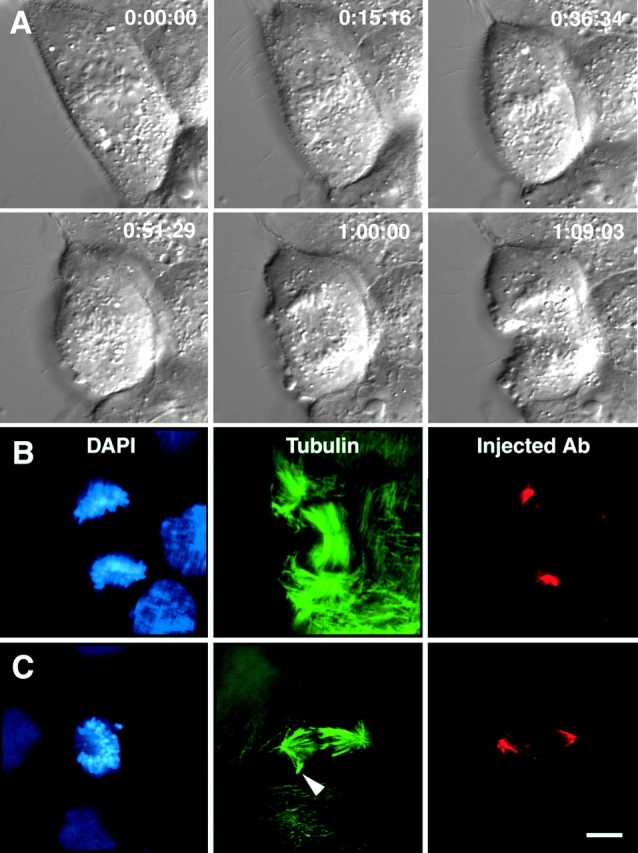
Chromosome movement in a cell microinjected with antibodies to HSET. (A) Selected DIC images from the video record of a mitotic cell that has been microinjected with an antibody to HSET. Times are indicated in hours:minutes:seconds. (B and C) The cell featured in A (B) and a second, unrelated injected mitotic CFPAC-1 cell (C) were fixed and processed for immunofluorescence microscopy using antibodies specific for tubulin, the injected HSET antibody, and the DNA-specific dye DAPI, as indicated. The arrowhead highlights a microtubule bundle protruding from the main body of the spindle (see Video 2 in online supplement). Bar: 20 μm.
Figure 3.
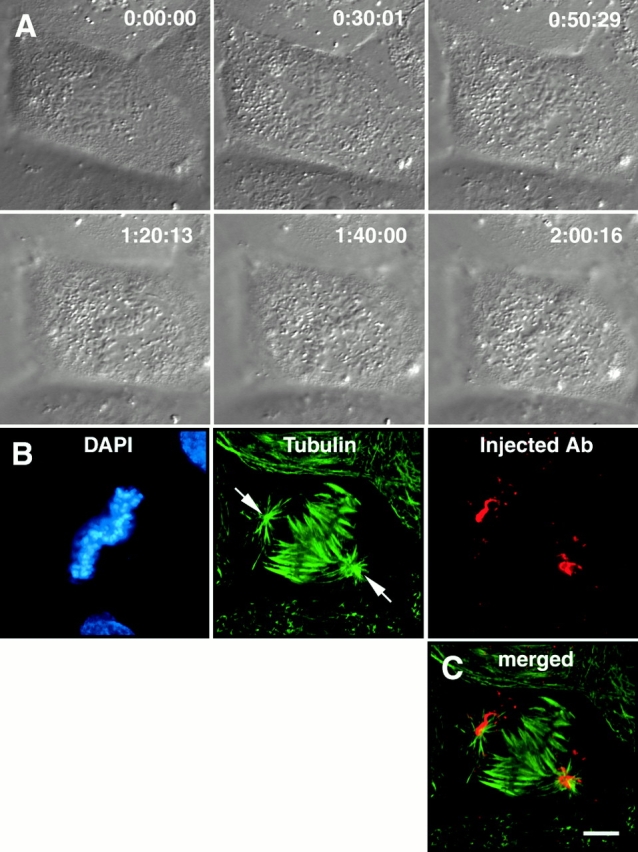
Chromosome movement in a cell microinjected with antibodies to both NuMA and HSET. (A) Selected DIC images from the video record of a mitotic cell that has been microinjected with antibodies to both NuMA and HSET. Times are indicated in hours:minutes:seconds. (B) The cell featured in A was fixed and processed for immunofluorescence microscopy using antibodies specific for tubulin, the injected antibodies (since both the NuMA- and HSET-specific antibodies were generated in rabbits, this panel shows the location of both), and the DNA specific dye DAPI, as indicated. (C) Merged image of the tubulin and injected antibody signals (see Video 3 in online supplement). Bar: 20 μm.
Figure 4.
Chromosome movement in a cell treated with nocodazole. (A) Selected DIC images from the video record of a mitotic cell cultured in the presence of 200 nM nocodazole. Times are indicated in hours:minutes:seconds. (B) The cell featured in A was fixed and processed for immunofluorescence microscopy using antibodies specific for tubulin and the DNA-specific dye DAPI, as indicated (see Video 4 in online supplement). Bar: 20 μm.
Results
Chromosome Movement in Cells Lacking Organized Poles
To determine whether spindle pole organization is required for chromosome movement in mitosis, we monitored chromosome dynamics in cultured human CFPAC-1 cells by time-lapse DIC microscopy. Uninjected control cells and cells injected with a nonimmune IgG progressed from the prophase/prometaphase transition (as judged by nuclear envelope breakdown) to anaphase onset in 62.4 ± 14.6 min (data not shown). Control cells displayed characteristic chromosome movements, including initial poleward motion in prometaphase, congression to the metaphase plate, oscillations during prometaphase and metaphase, and poleward movement in anaphase A. The rates for chromosome movements in control cells are shown in Table .
Table 1.
Chromosome Velocities
| Treatment | Poleward | Away from pole | Anaphase A | |||
|---|---|---|---|---|---|---|
| μm/min | n | μm/min | n | μm/min | n | |
| Uninjected | 3.55 ± 0.85 | 30 | 3.55 ± 0.8 | 55 | 2.05 ± 0.37 | 19 |
| Control IgG injected | 3.52 ± 0.69 | 25 | 3.66 ± 0.47 | 38 | 2.09 ± 0.07 | 5 |
| α-NuMA IgG injected | 3.24 ± 1.39 | 18 | 3.67 ± 1.28 | 20 | N/A | |
| α-HSET IgG injected | 3.51 ± 0.69 | 25 | 3.53 ± 0.77 | 34 | 2.16 ± 0.07 | 5 |
| α-HSET/NuMA IgG injected | 1.74 ± 0.35 | 16 | 1.79 ± 0.30 | 16 | N/A | |
To determine whether chromosome movement in mitosis is affected by disruption of NuMA function, we microinjected NuMA-specific antibodies into the cytoplasm of interphase cells and then monitored chromosome movements by time-lapse DIC microscopy of those injected cells that subsequently entered mitosis. Perturbation of NuMA function by either antibody microinjection into somatic cells or immunodepletion from frog egg extracts causes microtubule minus ends to splay at spindle poles and centrosomes to dislocate from the spindle (Gaglio et al. 1995; Merdes et al. 1996). Time-lapse DIC microscopy of a mitotic cell that had been injected with NuMA-specific antibodies showed that chromosome movement resembles control cells (Fig. 1 A). Chromosomes were observed moving poleward, congressing toward the cell equator (Fig. 1 A, arrow), and undergoing frequent oscillations at the metaphase plate. The rates of poleward and away from the pole chromosome movements in cells injected with NuMA-specific antibodies were not significantly different from uninjected control cells (t test, P = 0.26 and 0.33 for poleward and away from the pole motion, respectively; Table ) despite the fact that the spindle lacks well-organized poles (Fig. 1 B). The injected antibody concentrated in discrete aggregates in the cytoplasm (Fig. 1 B), and we have previously shown that the endogenous NuMA protein is trapped in these aggregates and is prevented from interacting properly with microtubules (Gaglio et al. 1995). This distribution is different from the typical localization of NuMA at the polar ends of spindles (Gaglio et al. 1995; Merdes et al. 1996, Merdes et al. 2000; Kallajoki et al. 1991; Yang and Snyder 1992). Only two differences were detectable in α-NuMA–injected cells relative to control cells. In approximately half of the injected cells, we observed that one or two chromosomes (in a given focal plane) failed to undergo detectable directed movement for extended periods. Also, these cells never entered anaphase during the time of observation (up to 3 h after nuclear envelope break down). These data indicate that disruption of NuMA function does not have a major impact on chromosome movement in prometaphase despite the disorganization of spindle poles.
In many of the α-NuMA–injected cells, we noticed that microtubule minus ends were loosely focused into pole-like regions (Fig. 1 B, arrowheads). In some cases, nearly bipolar spindles formed with two focused poles, although the centrosomes were not associated with those pole-like regions (Fig. 1 C, see also Figure 9 F in Gaglio et al. 1995). This suggests that other factors promote microtubule focusing at poles in the absence of NuMA activity. A strong candidate for this activity is the minus end–directed KIN C motor, which has been shown to play a role in spindle pole organization in numerous different systems (McDonald et al. 1990; Hatsumi and Endow 1992; Endow et al. 1994; Kuriyama et al. 1995; Matthies et al. 1996; Walczak et al. 1997; Matuiene et al. 1999; Mountain et al. 1999). To determine whether perturbation of HSET affects chromosome movement, we microinjected interphase cells in the cytoplasm with antibodies against HSET and monitored chromosome dynamics in those cells that subsequently entered mitosis (Fig. 2). Time-lapse DIC microscopy of a cell injected with HSET-specific antibodies showed that chromosome movement resembles control cells (Fig. 2 A) with the rates of poleward, away from the pole, and anaphase movements being not significantly different from uninjected control cells (t test, P = 0.40, 0.46, and 0.27 for poleward, away from the pole, and anaphase motion, respectively; Table ). We are confident that these antibodies block HSET function for several reasons. First, these antibodies have previously been shown to block microtubule organization into poles under acentrosomal conditions in mitotic extracts and in mouse oocytes (Mountain et al. 1999). Second, the injected antibody is concentrated near the spindle poles, suggesting it interacts with HSET and displaces it from its typical localization throughout the spindle (Fig. 2B and Fig. C). Third, the duration of prometaphase in α-HSET–injected cells increased to 77.5 ± 30.0 min compared with control cells that complete prometaphase, on average, in 38.5 ± 10.3 min, consistent with previous results showing that perturbation of KIN C motor proteins causes a decrease in spindle assembly efficiency and increases the duration of prometaphase (Matthies et al. 1996; Walczak et al. 1997; Matuiene et al. 1999). Finally, examination of spindle structure in injected cells during metaphase frequently showed microtubule bundles protruding from the main body of the spindle (Fig. 2 C, arrowhead), a hallmark of the loss of KIN C motor function (Endow et al. 1994; Hatsumi and Endow 1992; Matthies et al. 1996; Walczak et al. 1997; Matuiene et al. 1999; Mountain et al. 1999). Thus, the perturbation of the KIN C motor HSET perturbs spindle structure prolonging prometaphase, but there is no detectable effect on the rates of chromosome movement.
That microtubule minus ends were loosely organized at poles after perturbation of NuMA and tightly focused at poles after perturbation of HSET raised the possibility that these proteins play redundant roles in spindle pole function. To test this idea, we injected cells with antibodies to both NuMA and HSET and monitored chromosome dynamics by time-lapse DIC microscopy (Fig. 3). Chromosomes in cells injected with antibodies to both NuMA and HSET appeared to experience Brownian motion, but failed to undergo detectable directed movement (Fig. 3 A). Instead, the chromosomes remained loosely arranged near the cell center. In some cases (e.g., Fig. 3), chromosomes coalesced into a metaphase-like arrangement, but in other cases the chromosomes formed a loose group in the cell center and did not align efficiently (e.g., see Fig. 6 B). Spindles in cells injected with both antibodies were bi-oriented but lacked organized poles, and centrosomes were dissociated from the bulk of the microtubules connected to the chromosomes (Fig. 3 B). The injected antibody was concentrated around the centrosomes as well as in aggregates in the cytoplasm, consistent with the sequestration of NuMA and HSET away from their normal sites of localization (Fig. 3 C). Many of the chromosomes appeared to have K fibers associated with each sister kinetochore, and the K fibers extend normally toward opposite sides of the cell, but K fibers of different chromosomes did not focus at poles. These cells did not enter anaphase, similar to when cells were injected with NuMA-specific antibodies alone.
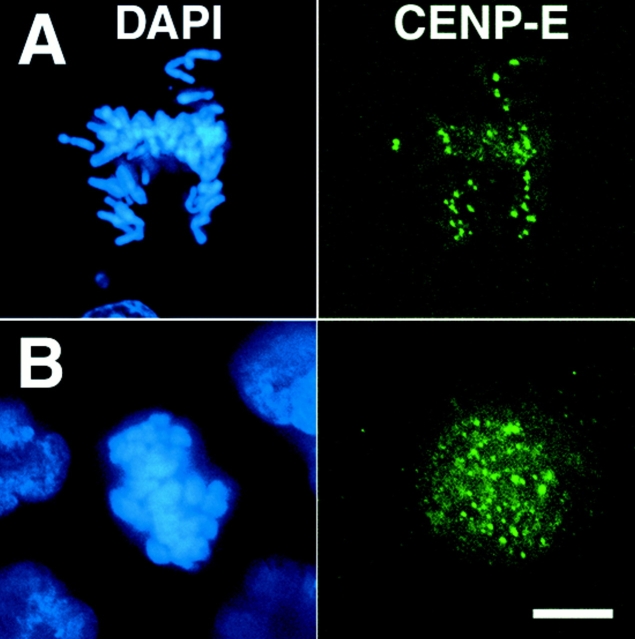
Figure 6.
CENP-E localizes normally to kinetochores in cells lacking organized spindle poles. CFPAC-1 cells that were uninjected (A) or microinjected with antibodies to both NuMA and HSET (B) were fixed and processed for immunofluorescence microscopy using antibodies to the kinetochore-associated motor protein CENP-E and the DNA-specific dye DAPI, as indicated. Bar: 20 μm.
Chromosome velocities were difficult to measure in cells like that shown in Fig. 3. However, a few of these double-injected cells showed residual chromosome movement, from which velocities could be determined. These movements were rare as the number of measurable events in double-injected cells was reduced 85% compared with control cells. Table shows the average velocity of these rare chromosome movements that was significantly slower than in uninjected control cells (t test, P < 0.0001 for both poleward and away from the pole motion; Table ). Importantly, the value reported in Table for the average velocity of chromosome movement in double-injected cells was derived only from those chromosomes that exhibited directed movement and does not include the values for chromosomes that failed to undergo directed movement. Thus, this value is a gross over-estimate of the total rate of chromosome movement in these cells. In conclusion, simultaneous perturbation of NuMA and HSET function suppressed directed chromosome movement to such a degree that no observable directed motion was detected in many cells. In the rare instances where chromosome movement was detected, the rate was significantly slower than in control cells. Furthermore, given that perturbation of either NuMA or HSET alone showed no detectable affect on the velocity of moving chromosomes, these data also indicate that the activities of HSET and NuMA overlap and are functionally redundant with respect to chromosome movement.
We also monitored chromosome movement in mitosis in cells treated with nocodazole (Fig. 4). Nocodazole prevents the assembly of spindle microtubules, thereby eliminating any microtubule-dependent chromosome movement. Upon nuclear envelope breakdown in the presence of 200 nM nocodazole, chromosomes displayed Brownian motion and coalesced near the center of the cell, but did not show detectable directed movement (Fig. 4 A). Immunofluorescence microscopy verified the lack of a spindle in this cell and the presence of only small clumps of short residual microtubules (Fig. 4 B). Thus, chromosome behavior in nocodazole-treated cells is similar to cells lacking both HSET and NuMA activities despite the fact that nocodazole-treated cells lack spindle microtubules and cells injected with both NuMA- and HSET-specific antibodies have bi-oriented spindle-like structures. Chromosomes may coalesce near the cell center under both these conditions because that is the thickest part of the cell.
Microtubule Attachment to Kinetochores in Cells Lacking Organized Poles
To verify that microtubules were attached to kinetochores in cells injected with antibodies to both NuMA and HSET, we examined injected cells by transmission electron microscopy (Fig. 5). Uninjected control cells showed organized spindles with tightly focused spindle poles (Fig. 5 A). In contrast, cells injected with antibodies to both NuMA and HSET lacked organized spindle poles (Fig. 5 B), consistent with images obtained by fluorescence microscopy (Fig. 3 B). At high magnification, many of the chromosomes had identifiable kinetochores and multiple microtubules were observed attached to both kinetochores, consistent with the formation of bi-oriented K fibers (Fig. 5 C). These data demonstrate that the lack of directed chromosome movement in the absence of organized spindle poles does not arise from a lack of microtubule binding to kinetochores or formation of K fibers, although this limited analysis does not permit us to determine whether each kinetochore has obtained a full complement of microtubules.
Figure 5.
Microtubule attachment to kinetochores in microinjected cells. CFPAC-1 cells that were uninjected (A) or microinjected with antibodies to both NuMA and HSET (B) were processed for transmission electron microscopy. Arrows in A and B point to centrosomes. The electron-dense material detected in B near the centrosomes is most likely the antibody-induced aggregates of NuMA and HSET, which are prominently detected by immunofluorescence microscopy. (C) High magnification views to highlight kinetochore-microtubule interactions in the chromosomes identified as 1–4 in B. The kinetochore regions of chromosome 2 were located in the adjacent serial section from that shown in B. Bars: (A and B) 3 μm; (C) 100 nm.
As an additional control, we stained these double-injected cells with antibodies to CENP-E, a kinetochore-specific kinesin-related protein involved in chromosome positioning on the spindle (Fig. 6; Yen et al. 1991; Schaar et al. 1997; Wood et al. 1997). Control mitotic cells showed intense kinetochore staining for CENP-E, particularly at prometaphase (Fig. 6 A). Cells injected with antibodies to the spindle pole components NuMA and HSET also showed intense kinetochore staining for CENP-E (Fig. 6 B). This demonstrates that the lack of chromosome movement in cells lacking NuMA and HSET activity is not due to the inappropriate displacement of a microtubule motor protein that has been shown to be essential for chromosome positioning and movement on the spindle. Together, these data demonstrate that properties of the kinetochore that are important to chromosome movement (microtubule capture and the presence of motor proteins) are not detectably perturbed in cells lacking organized spindle poles.
Tension Across Sister Kinetochores Is Reduced upon Disruption of Spindle Poles
Forces acting to move chromosomes during mitosis also generate tension between sister kinetochores during prometaphase and metaphase (Nicklas et al. 1995; Waters et al. 1998). To test whether proper tension was generated across sister kinetochores in cells lacking organized spindle poles, we fixed and processed control and injected cells for immunofluorescence analysis using a human autoimmune sera that specifically recognizes centromeric antigens. We then measured the distances between sister kinetochores (Fig. 7 and Table ). Uninjected cells, control injected cells, and cells injected with antibodies to either NuMA or HSET each showed an average interkinetochore distance slightly >3 μm. Cells that entered mitosis in the presence of 10 μM nocodazole lack spindle microtubules and showed an average interkinetochore distance of only 1.6 μm, consistent with previously published results showing that microtubule-dependent forces are involved in generating tension between sister kinetochores. The relaxed interkinetochore distance observed in the presence of nocodazole was similar to that observed in untreated early prometaphase cells before chromosome–microtubule interactions (data not shown). Mitotic cells injected with both NuMA- and HSET-specific antibodies displayed interkinetochore distances of ∼2.5 μm. This 20% reduction in interkinetochore distance is significant (t test, P < 0.0001), and demonstrates that full tension is not generated across sister kinetochores in the absence of organized spindle poles despite microtubule attachment to kinetochores.
Figure 7.
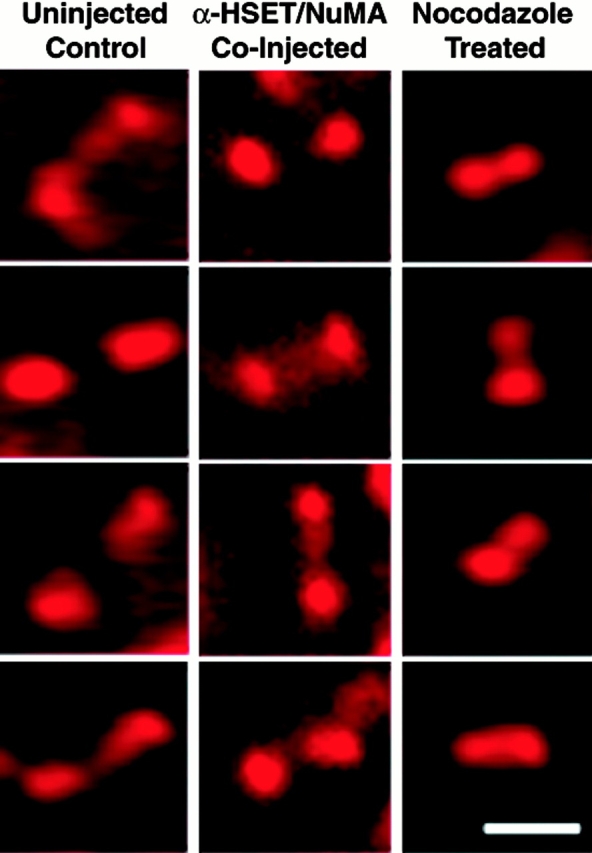
Interkinetochore distances in cells with disorganized spindle poles. Four representative pairs of kinetochores are shown for an uninjected control mitotic cell, a mitotic cell injected with antibodies to both NuMA and HSET, and a cell treated with 10 μM nocodazole, as indicated. Centromere regions were visualized by staining with an anticentromere-specific autoimmune serum. Bar: 2.5 μm.
Table 2.
Sister Kinetochore Distances
| Treatment | Interkinetochore distance |
|---|---|
| μm | |
| Nocodazole | 1.625 ± 0.186 |
| Uninjected control | 3.128 ± 0.415 |
| Control IgG injected | 3.083 ± 0.325 |
| α-NuMA IgG injected | 3.016 ± 0.425 |
| α -HSET IgG injected | 3.114 ± 0.298 |
| α -HSET/NuMA IgG injected | 2.551 ± 0.303 |
Discussion
The experiments presented here show that perturbation of both NuMA and HSET disrupts spindle pole organization, and as a consequence chromosome movement is severely suppressed. We specifically targeted NuMA and HSET because both proteins have essential roles in spindle pole organization and neither protein has been shown to participate in any aspect of spindle assembly other than spindle pole organization. Specifically, neither protein has been implicated in acting at kinetochores and we did not detect any differences in microtubule attachment to kinetochores or the association of the essential kinetochore-associated motor CENP-E with kinetochores in cells lacking organized spindle poles. Thus, we interpret the defects in chromosome movement to result from the lack of spindle pole organization. This interpretation is consistent with our hypothesis that noncentrosomal proteins provide the primary anchorage site for microtubule minus ends at spindle poles, and this anchorage is necessary to counterbalance the forces needed for chromosome movement. In the absence of adequate microtubule anchorage at spindle poles, the forces that would normally move chromosomes toward and away from the poles are dissipated in an unproductive manner that fails to generate chromosome movement.
A prediction from these data is that chromosome movement during anaphase A should be inhibited in the absence of HSET and NuMA function. Unfortunately, we were unable to examine that possibility due to the activity of the checkpoint that prevents anaphase onset. The checkpoint may remain active in these cells for a variety of reasons including the reduced tension across sister kinetochores, the potential role of spindle poles in regulating Mad2 activity (Howell et al. 2000), or the possibility that spindle pole integrity is necessary for proper cyclin B proteolysis (Clute and Pines 1999; Wakefield et al. 2000). We are currently investigating how specific components of the checkpoint (i.e., Mad2) are affected by spindle pole disruption and whether inactivation of the checkpoint permits us to follow chromosome movement during anaphase in cells lacking organized poles.
A striking feature of cells lacking both NuMA and HSET function is that bi-oriented spindle-like structures formed with well organized K fibers. This suggests that many of the constituent parts of the spindle function normally despite the absence of pole organization. For example, the paired sister kinetochores may dictate bi-orientation, and cross-linking proteins such as Eg5 may bundle microtubules into K fibers despite the fact that those K-fibers do not focus tightly at poles. Moreover, steric interactions between K fibers may inhibit the diffusion of chromosomes contributing to a metaphase plate-like arrangement of chromosomes in some of these cells. It is also striking that interkinetochore distances were only reduced by ∼20% under conditions where chromosome motion was suppressed. The most likely explanation for this is that the force needed to stretch kinetochores is smaller than the force needed for chromosome movement, and our experiments caused a reduction in poleward forces sufficient to suppress chromosome movement while only minimally reducing sister kinetochore stretching. Support for this interpretation comes from the work of Waters et al. 1996, who demonstrated that the force generated from poleward microtubule flux alone was sufficient to maintain kinetochores in a fully stretched configuration in the absence of forces generated by kinetochore-associated motors. Thus, the fact that many aspects of spindle assembly appear normal in cells injected with antibodies to both NuMA and HSET lends support to the conclusion that the only aspect of spindle function perturbed in these cells is the organization of microtubule ends at spindle poles.
Models for Microtubule Anchorage at Spindle Poles
Two models are most plausible to explain how microtubule anchorage at spindle poles contributes to chromosome movement, and both of these ideas have been proposed previously in alternate forms. One model involves the anchorage of microtubule minus ends in a matrix (McIntosh 1980; Pickett-Heaps et al. 1982; Rebhun and Palazzo 1988; Nicklas 1989). This matrix would bind to microtubules and hold the minus ends so that poleward forces generated at kinetochores would cause chromosomes to move relative to the poles rather than the poles relative to the chromosomes (Fig. 8 A). If the interaction between this matrix and microtubules were disrupted, then the drag the matrix imposes on microtubules would be relieved and chromosome movement would stall due to the lack of resistance on kinetochore microtubules.
Figure 8.

Models for the NuMA- and HSET-dependent anchorage of microtubules at spindle poles. (A) The matrix model posits that NuMA and an unidentified matrix element associated with HSET anchor microtubule ends at spindle poles. This anchorage creates drag on microtubules that provides the necessary resistance for forces generated by kinetochore-associated motors (large solid arrows) to create chromosome movement. (B) The microtubule cross-linking model posits that the cross-linking and minus end–directed motility (small arrows) of HSET and NuMA (in association with dynein) involved in focusing microtubules at spindle poles also creates physical linkages between kinetochore and nonkinetochore microtubules. This connects microtubules involved in poleward forces (large solid arrows) with microtubules involved in polar ejection forces (large open arrows), as well as interdigitating microtubules emanating from opposite poles, which constrains the positions of the spindle poles and permits chromosomes to move relative to the poles.
Support for this model comes from data showing that poleward chromosome movement in anaphase A continues if kinetochore fibers linking the chromosomes to the pole are physically severed (Nicklas 1989; Spurck et al. 1997). Those experiments indicate that microtubules do not need to extend to the pole to support chromosome movement and have been interpreted to indicate that a matrix surrounds the microtubules and holds them in place to support the forces for chromosome movement. Indeed, NuMA possesses characteristics consistent with such a hypothetical matrix. It is capable of self-association into extensive three-dimensional structures (Saredi et al. 1996, Saredi et al. 1997; Gueth-Hallonet et al. 1998), forms an insoluble matrix at spindle poles (Dionne et al. 1999), binds to microtubules (Merdes et al. 1996), and is required for spindle pole organization (Kallajoki et al. 1991; Yang and Snyder 1992; Gaglio et al. 1995; Merdes et al. 1996, Merdes et al. 2000). While NuMA may participate in microtubule anchorage at spindle poles in this manner, it cannot be the sole component of such a matrix as chromosome movement occurs at normal rates after NuMA disruption (Fig. 1). This suggests that there is a NuMA-independent matrix coupled to HSET or that the matrix is responsive to the functions of both NuMA and HSET.
An alternative model involves the interaction of kinetochore and nonkinetochore (interpolar) microtubules at spindle poles (Fig. 8 B). In this model, NuMA and HSET cross link kinetochore microtubules to interpolar microtubules. This creates direct physical connections between microtubules experiencing compressive forces (kinetochore microtubules) with microtubules that could either passively resist those compressive forces (antiparallel interacting interpolar microtubules) or directly oppose those compressive forces through repulsive forces (polar ejection forces). In the absence of both NuMA and HSET activities, the linkages between interpolar and kinetochore microtubules are broken and chromosome movement stalls due to a lack of resistance on the kinetochore microtubules. We note, however, that while the specific subset of interpolar microtubules forming antiparallel interactions between the poles probably participate in this model, they cannot provide the primary resistance to poleward forces acting on kinetochore microtubules. This is evident from the fact that chromosome movement occurs normally on monopolar spindles and during anaphase-like prometaphase, two circumstances where antiparallel interactions between microtubules emanating from opposite poles are completely absent (Bajer 1982; Rieder et al. 1986; Rieder and Alexander 1990).
This model is supported by a variety of observations in the literature. First, micromanipulation experiments in grasshopper spermatocytes have demonstrated that physical associations exist among kinetochore and nonkinetochore spindle microtubules (Nicklas et al. 1982). These physical associations were shown to be tightest near the spindle poles, although some nonkinetochore microtubules were observed linked to kinetochore microtubules a substantial distance from the poles. Moreover, examination of spindles in PtK1 cells by electron microscopy has shown that a large percentage of interpolar microtubules have their minus ends associated with K fibers, and that these linkages occur an appreciable distance away from the centrosome (Mastronarde et al. 1993). Next, interactions between kinetochore and nonkinetochore microtubules has been argued to generate forces driving chromosome movement in several systems, including plant cells, as well as crane fly, grasshopper, and flatworm spermatocytes (Molè-Bajer 1969; Salmon and Begg 1980; Steffen and Fuge 1982; Bajer and Molè-Bajer 1986; Steffen 1986; Bajer 1990; Fuge and Falke 1991; Smirnova and Bajer 1992). In some of these cell types, the velocity of chromosome movement was positively correlated with the density of nonkinetochore microtubules interacting with the kinetochore microtubules. In other cases, it was shown that treatments that selectively disassemble nonkinetochore microtubules halt chromosome movement, which was only restored upon reestablishment of nonkinetochore microtubules after removal of the offending treatment. Finally, both HSET and NuMA (in association with cytoplasmic dynein) have microtubule cross-linking and minus end–directed motor activities (Merdes et al. 1996; Mountain et al. 1999). These two minus end–directed motors act redundantly in organizing microtubule minus ends at spindle poles (Walczak et al. 1998; Mountain et al. 1999), and that redundancy may be related to the functional redundancy we show here for HSET and NuMA during chromosome movement. This raises the possibility that the functional role that HSET and NuMA play in organizing microtubules at spindle poles may directly relate to how they create connections between interpolar and kinetochore microtubules to support chromosome movement as proposed in Fig. 8 B.
In summary, we provide evidence that NuMA and HSET cooperate to anchor microtubule minus ends at spindle poles to support chromosome movement. These data begin to define the specific proteins involved in the mechanics of chromosome movement and indicates that a common mechanism provides the microtubule anchorage necessary for chromosome movement in all vertebrate cells regardless of whether centrosomes are present or not. The models proposed to explain this mechanism are not mutually exclusive and it is possible that some combination of the spindle matrix and microtubule cross-linking models participate in this essential process. Further, these models imply that spindle poles are not only the sites to which microtubule minus ends converge physically, but they also act as sites for integration of different forces generated within the spindle.
Supplemental Material
Acknowledgments
The authors thank Kevin Sullivan and Tim Yen for providing antibodies. We also thank Roger Sloboda and Gary Mack for providing comments on the manuscript and Conly Rieder for helpful discussions.
This work was supported by a grant from the National Institutes of Health (GM51542). The time-lapse microscopy equipment was purchased using a grant from the Fannie E. Rippel Foundation.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: DIC, differential interference contrast; HSET, human homologue of the KIN C motor family; MTSB, microtubule stabilizing buffer; NuMA, nuclear mitotic apparatus protein.
References
- Alexander S.P., Rieder C.L. Chromosome motion during attachment to the vertebrate spindleinitial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J. Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Nebreda A.R., Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Bajer A. Functional autonomy of monopolar spindle and evidence for oscillatory movement in mitosis. J. Cell Biol. 1982;93:33–48. doi: 10.1083/jcb.93.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajer A.S. The elusive organization of the spindle and the kinetochore fibera conceptual retrospective. Adv. Cell Biol. 1990;3:65–93. [Google Scholar]
- Bajer A.S., Molè-Bajer J. Drugs with colchicine-like effects that specifically disassemble plant but not animal microtubules. Ann. NY Acad. Sci. 1986;466:767–784. doi: 10.1111/j.1749-6632.1986.tb38458.x. [DOI] [PubMed] [Google Scholar]
- Capecchi M.R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- Clute P., Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Compton D.A. Focusing on spindle poles. J. Cell Sci. 1998;111:1477–1481. doi: 10.1242/jcs.111.11.1477. [DOI] [PubMed] [Google Scholar]
- Compton D.A. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Compton D.A., Cleveland D.W. NuMA is required for the proper completion of mitosis. J. Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne M.A., Howard L., Compton D.A. NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil. Cytoskelet. 1999;42:189–203. doi: 10.1002/(SICI)1097-0169(1999)42:3<189::AID-CM3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Endow S.A., Chandra R., Komma D.J., Yamamoto A.H., Salmon E.D. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle defects in mitosis. J. Cell Sci. 1994;107:859–867. doi: 10.1242/jcs.107.4.859. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Murray A.W. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Fuge H., Falke D. Morphological aspects of chromosome spindle fibres in Mesostaoma“microtubule fir tree” structures and microtubule association with kinetochores and chromatin. Protoplasma. 1991;160:39–48. [Google Scholar]
- Gaglio T., Dionne M.A., Compton D.A. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Compton D.A. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueth-Hallonet C., Wang J., Harborth J., Weber K., Osborn M. Induction of a regular nuclear lattice by overexpression of NuMA. Exp. Cell Res. 1998;243:434–452. doi: 10.1006/excr.1998.4178. [DOI] [PubMed] [Google Scholar]
- Gorbsky G.J. Chromosome motion in mitosis. Bioessays. 1992;14:73–80. doi: 10.1002/bies.950140202. [DOI] [PubMed] [Google Scholar]
- Hatsumi M., Endow S.A. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci. 1992;101:547–559. doi: 10.1242/jcs.101.3.547. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Beker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Howell B.J., Hoffman D.B., Fang G., Murray A.W., Salmon E.D. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 2000;150:1233–1249. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A.A., Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Inoué S., Salmon E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallajoki M., Weber K., Osborn M. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle; a microinjection study using SPN monoclonal antibodies. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3351–3362. doi: 10.1002/j.1460-2075.1991.tb04899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Gabashvili I.S., Rieder C.L. “Dumb” versus “smart” kinetochore models for chromosome congression during mitosis in vertebrate somatic cells. Cell Motil. Cytoskelet. 1999;43:179–185. doi: 10.1002/(SICI)1097-0169(1999)43:3<179::AID-CM1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol. 1996;135:315–327. doi: 10.1083/jcb.135.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D.E., Mitchison T.J., Kirschner M.W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Kuriyama R., Kofron M., Essner R., Kato T., Dragas-Granoic S., Omoto C.K., Khodjakov A. Characterization of a minus end-directed kinesin-like motor protein from cultured mammalian cells. J. Cell Biol. 1995;129:1049–1059. doi: 10.1083/jcb.129.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Hunter A.W., Wagenbach M., Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N., McDonald K.L., Ding R., McIntosh J.R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H.J.G., McDonald H.B., Goldstein L.S.B., Theurkauf W.E. Anastral meiotic spindle morphogenesisrole of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuiene J., Essner R., Ryu J.-H., Hamaguchi Y., Baas P.W., Haraguchi T., Kuriyama R. Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J. Cell Sci. 1999;112:4041–4050. doi: 10.1242/jcs.112.22.4041. [DOI] [PubMed] [Google Scholar]
- McDonald H.B., Stewart R.J., Goldstein L.S.B. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- McIntosh J.R. Microtubule polarity and interaction in mitotic spindle function. In: Schweiger H.G., editor. International Cell Biology. Springer Verlag; Berlin: 1980. pp. 359–368. [Google Scholar]
- McIntosh J.R., Koonce M.P. The mitotic spindle. Science. 1989;246:622–628. doi: 10.1126/science.2683078. [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Merdes A., Cleveland D.W. Pathways of spindle pole formationdifferent mechanisms; conserved components. J. Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W.C., Cleveland D.W. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 2000;149:851–861. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J. Mitosisbasic concepts Curr. Opin. Cell Biol. 1 1989. 67 74a [DOI] [PubMed] [Google Scholar]
- Mitchison T.J. Polewards microtubule flux in the mitotic spindleevidence from photoactivatioin of fluorescence J. Cell Biol. 109 1989. 637 652b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J., Salmon E.D. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molè-Bajer J. Fine structure studies of apolar mitosis. Chromosoma. 1969;26:427–448. doi: 10.1007/BF00326354. [DOI] [PubMed] [Google Scholar]
- Moore M.J. Removal of glass coverslips from cultures flat embedded in epoxy resins using HF. Microscopy. 1975;104:205. doi: 10.1111/j.1365-2818.1975.tb04018.x. [DOI] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D.A. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–365. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. The motor for poleward chromosome movement in anaphase is in or near the kinetochore. J. Cell Biol. 1989;109:2245–2255. doi: 10.1083/jcb.109.5.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B., Kubai D.F., Hays T.S. Spindle microtubules and their mechanical associations after micromanipulation in anaphase. J. Cell Biol. 1982;95:91–104. doi: 10.1083/jcb.95.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B., Ward S.C., Gorbsky G.J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., Tippit D.H., Porter K.R. Rethinking mitosis. Cell. 1982;29:729–744. doi: 10.1016/0092-8674(82)90435-4. [DOI] [PubMed] [Google Scholar]
- Pfarr C.M., Copve M., Grissom P.M., Hayes T.S., Porter M.E., McIntosh J.R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1991;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Rebhun L.I., Palazzo R.E. In vitro reactivation of anaphase B in isolated spindles of the sea urchin egg. Cell Motil. Cytskelet. 1988;10:197–209. doi: 10.1002/cm.970100124. [DOI] [PubMed] [Google Scholar]
- Rieder C.L. Mitosistowards a molecular understanding of chromosome behavior. Curr. Opin. Cell Biol. 1991;3:59–66. doi: 10.1016/0955-0674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Alexander S.P. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L, Bowser S. Correlative LM and EM on the same epoxy section. In: Hyat M.A., editor. Correlative Microscopy in Biology. Academic Press; New York, NY: 1987. pp. 249–277. [Google Scholar]
- Rieder C.L., Davidson E.A., Jensen L.C.W., Cassimeris L., Salmon E.D. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and the half-spindle. J. Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E.D., Begg D.A. Functional implications of cold-stable microtubules in kinetochore fibers of insect spermatocytes during anaphase. J. Cell Biol. 1980;85:853–865. doi: 10.1083/jcb.85.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saredi A., Howard L., Compton D.A. NuMA assembles into an extensive filamentous structure when expressed in the cell cytoplasm. J. Cell Sci. 1996;109:619–630. doi: 10.1242/jcs.109.3.619. [DOI] [PubMed] [Google Scholar]
- Saredi A., Howard L., Compton D.A. Phosphorylation regulates the assembly of NuMA in a mammalian mitotic extract. J. Cell Sci. 1997;110:1287–1297. doi: 10.1242/jcs.110.11.1287. [DOI] [PubMed] [Google Scholar]
- Schaar B.T., Chan G.K.T., Maddox P., Salmon E.D., Yen T.J. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E.A., Bajer A.S. Spindle poles in higher plant mitosis. Cell Motil. Cytoskelet. 1992;23:1–7. doi: 10.1002/cm.970230102. [DOI] [PubMed] [Google Scholar]
- Spurck T., Forer A., Pickett-Heaps J. Ultraviolet microbeam irradiations of epithelial and spermatocyte spindles suggest that the forces act on the kinetochore fibre and are not generated by its disassembly. Cell Motil. Cytoskelet. 1997;36:136–148. doi: 10.1002/(SICI)1097-0169(1997)36:2<136::AID-CM4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Steffen W. Relationship between the arrangement of microtubules and chromosome behavior of syntelic autosomal univalents during prometaphase in crane fly spermatocytes. Chromosoma. 1986;94:412–418. doi: 10.1007/BF00328642. [DOI] [PubMed] [Google Scholar]
- Steffen W., Fuge H. Dynamic changes in autosomal spindle fibers during prometaphase in crane fly spermatocytes. Chromosoma. 1982;87:363–371. [Google Scholar]
- Steuer E.R., Schroer T.A., Wordeman L., Sheetz M.P. Cytoplasmic dynein localizes to mitotic spindles and kinetochores. Nature. 1991;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., Donahue R.P. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- Wakefield J.G., Huang H., Raff J.W. Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr. Biol. 2000;10:1367–1370. doi: 10.1016/s0960-9822(00)00776-4. [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Mitchison T.J., Desai A. XKCM1a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Verma S., Mitchison T.J. XCTK2a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Waters J.C., Chen R.-H., Murray A.W., Salmon E.D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Mitchison T.J., Rieder C.L., Salmon E.D. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S.B., Cleveland D.W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yang C.H., Snyder M. The nuclear-mitotic apparatus protein (NuMA) is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T.J., Compton D.A., Wise D., Zinkowski R.P., Brinkley B.R., Earnshaw W.C., Cleveland D.W. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.