Abstract
Differentiation of naive CD4+ T cells into helper T (Th) cells is controlled by a combination of several transcriptional factors. In this study, we examined the functional role of the Runx1 transcription factor in Th cell differentiation. Naive T cells from transgenic mice expressing a dominant interfering form of Runx1 exhibited enhanced interleukin 4 production and efficient Th2 differentiation. In contrast, transduction of Runx1 into wild-type T cells caused a complete attenuation of Th2 differentiation and was accompanied by the cessation of GATA3 expression. Furthermore, endogenous expression of Runx1 in naive T cells declined after T cell receptor stimulation, at the same time that expression of GATA3 increased. We conclude that Runx1 plays a novel role as a negative regulator of GATA3 expression, thereby inhibiting the Th2 cell differentiation.
Keywords: GATA3, Runx1, Th2, T lymphocytes, transcription factor
Introduction
When encountering antigens, naive CD4+ T cells in peripheral lymphoid tissues differentiate into mature Th cells and secrete a large amount of effector cytokines. Th cells are categorized into two functionally distinct subsets based on their cytokine secretion characteristics (1). Th1 cells produce IL-2, IFN-γ, and tumor necrosis factor α. These cytokines act on macrophages and mediate their effector functions, such as eradication of intracellular organisms and organ-specific autoreactive immune responses. Th2 cells produce IL-4, IL-5, IL-6, IL-10, and IL-13, which act on B cells and regulate their humoral immune response against extracellular pathogens (2–4).
Expression of cytokine genes in a Th1- versus Th2-lineage-specific fashion is strictly regulated by the coordination of both lineage-specific and nonspecific transcription factors. Naive cells require signaling via TCRs and CD28 molecules in conjunction with signaling via the IL-4 receptor (IL-4R) to differentiate into Th2 cell (5, 6). TCR stimulation initiates the expression of a Th2-specific transcription factor, c-Maf (7, 8), and induces the nuclear translocation of NFAT (9). IL-4R–mediated signaling promotes the phosphorylation and subsequent nuclear translocation of STAT6, which in turn triggers the expression of GATA3 (10). GATA3 is a key transcription factor that regulates the expression of the entire set of Th2 cytokine genes (11). For example, ectopic expression of GATA3 can, even in the absence of IL-4–mediated STAT6 activation, induce chromatin remodeling at the IL-4 locus as well as expression of other Th2 cytokine genes (12–14). Furthermore, the expressed GATA3 protein auto-activates GATA3 transcription (13, 14). These effects of GATA3 ensure full commitment of cells to the Th2 lineage.
The Runx1 transcription factor, also known as acute myelogenous leukemia protein 1 (AML1) and core binding factor α2 (CBFα2), harbors a 128-amino-acid region which is homologous to the Drosophila gene products Runt and Lozenge. This region is termed the Runt domain and is responsible for both DNA binding and hetero-dimerization with its partner protein, PEBP2β/CBFβ (15, 16). Of clinical significance is that several chromosomal rearrangements involving Runx1 are associated with the occurrence of human acute myelogenous leukemia (17). Furthermore, gene targeting studies have revealed that the homozygous deletion of Runx1 severely impairs the development of a definitive type of hematopoiesis, thereby causing the embryonic lethality (18–20). Thus, Runx1 is involved in leukemogenesis and hematopoietic development (21–23).
We previously established several lines of transgenic mice in which we artificially modulated Runx1 activity, and used them to show that Runx1 is also involved in various aspects of T cell differentiation in the thymus (24, 25). In the current study, we examined the functional relevance of Runx1 in helper–T cell differentiation. We have found that Runx1 by itself can repress GATA3 expression and Th2 differentiation of CD4+ T cells.
Materials and Methods
Mice.
Transgenic mouse lines expressing the Runt domain of the murine Runx1 protein were established as described previously (24). Litters possessing the transgene were backcrossed with C57BL/6J mice for seven generations. Mice targeted for IL-4R were provided by F. Brombacher, Univ. Cape Town, Cape Town, Republic of South Africa (26). Establishment of mouse lines expressing GATA3 as a transgene will be described elsewhere (unpublished data). C57BL/6J and BALB/c mice were purchased from Kumagai Co. Ltd. and Sankyo Inc., respectively.
Cell Culture.
Cells were liberated from the spleens of mice and suspended in PBS. The single cell suspension was overlaid onto Lymphosepar II (IBL) and centrifuged at 400 g for 20 min at room temperature. The lymphocyte fraction was collected and incubated with DynaBeads Mouse CD4 (L3T4; Dynal). The CD4+ T cells were then dissociated from the beads by the use of DETACHaBEAD Mouse CD4 (Dynal). The CD4+ fraction obtained was more than 95% pure as judged by flow cytometrical analysis. The cells were suspended in RPMI 1640 medium containing 10% (vol/vol) FCS, 10 mM HEPES-KOH, pH 7.4, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol, and subjected to culture in a 48-well plate at a density of 2 × 105 cells/500 μl medium in each well.
For TCR stimulation, the cells were cultured in a plate that was coated beforehand with 10 μg/ml of an anti-CD3 antibody (145–2C11) and 10 μg/ml of anti-CD28 antibody (37.51; BD Biosciences) according to the previously described procedure (27). In some cases, the TCR-stimulated cells were cultured in the presence of 30 U/ml of IL-2 for 5 d.
To induce cells to differentiate toward the Th2 lineage, the cells were cultured in the presence of 4 μg/ml of anti–IFN-γ antibody (UBI) and 100 U/ml of murine IL-4 (PeproTech), together with the precoated anti-CD3/anti-CD28 antibodies. For Th1 differentiation, the cells were cultured in the presence of 4 μg/ml of anti-IL-4 antibody (UBI) and 5 ng/ml of murine IL-12 (PeproTech), together with the precoated anti-CD3/anti-CD28 antibodies. After 4 d of culture, the cells were washed with fresh media, replated at a cell density of 2 × 105 in a well precoated with anti-CD3/anti-CD28 antibodies and incubated for a further 24 h.
To assess the degree of cell proliferation, the cells were incubated with 1 μCi/well 3H-thymidine for 6 h. The incorporation of 3H-thymidine into a 5% (wt/vol) trichloroacetic acid-insoluble fraction of cells was counted with a liquid scintillation counter.
Retrovirus Infection.
The cDNAs of murine Runx1 (28), Runx2 (29), and Runx3 (AF155880, GenBank/EMBL/DDBJ) were inserted into a pMX-green fluorescent protein (GFP)* vector (30). To ensure that Runx1 (or its related protein) and GFP were translated bi-cistronically, an internal ribosomal entry site was ligated upstream of the GFP. In case, Runx1 was inserted into a pMX-rat CD2 vector and GATA3 into pMX-GFP. Each of the resulting plasmids was transfected into a packaging cell line, PLAT-T, using FuGENE6 (Roche) and, after incubation for 24 h, the culture supernatant was harvested and condensed as a viral stock.
The CD4+ T cells were stimulated by anti-CD3/anti-CD28 antibodies for 24 h. The cells were then infected with retrovirus in the presence of 0.5 μg/ml of polybrene for 24 h and cultured further in the presence of 30 U/ml of IL-2 for 5 d. In some cases, 100 U/ml of IL-4 was added to the medium together with the IL-2. The cells were washed with fresh media and restimulated by anti-CD3/anti-CD28 antibodies for 6 h for intracellular cytokine staining and for 24 h for the cytokine production assay.
Flow Cytometrical Analysis.
The single cell suspensions were first incubated with an anti-Fc receptor antibody (24G2) and then stained with appropriately diluted monoclonal antibodies. The fluorescein-conjugated antibodies used were as follows: anti-CD4 PE (H129.19), anti-CD4 FITC (RM4–5), anti-CD8a FITC (53–6.7), anti-CD8a PE (53–6.7), anti-TCRβ FITC (H57–597), anti-CD25 FITC (7D4), and anti-CD69 FITC (H1.2F3). The anti-CD4 PE was purchased from Sigma-Aldrich and all the other antibodies were purchased from BD Biosciences. Anti-IL-4Rα (M1) was provided by Immunex.
To detect intracellular IL-4 and IFN-γ, the cells were restimulated by anti-CD3/anti-CD28 antibodies and incubated in the presence of 2 μM monensin (Sigma-Aldrich) for 6 h. The cells were then fixed with 4% (wt/vol) paraformaldehyde in PBS and permeabilized in a solution containing 50 mM NaCl, 5 mM EDTA, 0.02% (wt/vol) NaN3, pH 7.5, and 0.5% (wt/vol) Triton X-100. After blocking with PBS containing 3% (wt/vol) BSA, the cells were stained with anti–IFN-γ FITC (XMG1.2) and anti–IL-4 PE (11B11). Flow cytometry was performed using a FACSCalibur™ and the data were analyzed using a CELLQuest™ software (Becton Dickinson). In some cases, the GFP+ or rat CD2+ population was sorted using a FACSVantage™. Anti-rat CD2 PE (LFA-2) was purchased from Cedarlane.
ELISA.
The amount of cytokines secreted in the tissue culture supernatant was assayed by ELISA. Kits supplied from Biosource International were used to detect IL-2, IL-4, IL-5, IL-10, and IFN-γ.
Immunoblot Analysis and EMSA.
All the procedures necessary for immunoblot analysis were performed as described previously (6). Anti-Jak1, anti-STAT6, anti-phosphorylated STAT6, and anti-phosphotyrosine antibodies were purchased from Upstate Biotechnology, Sigma-Aldrich, New England Biolabs, Inc., and Covance Research Products, Inc., respectively. Anti-Ikaros (M-20) and anti-GATA3 (HG3–31) antibodies were obtained from Santa Cruz Biotechnology, Inc. and anti-Runx1 antibody from Geneka. The protocol of EMSA was as described previously (31).
Northern Blot Analysis.
Total cytoplasmic RNA was isolated from cells using a TRIzol reagent (GIBCO BRL). 2 μg of RNA was separated on a 1% (wt/vol) agarose gel containing 2.2 M formaldehyde. Transfer of RNA onto a Hybond N membrane (Amersham Biosciences), hybridization, and washing were performed according to the procedure supplied by the manufacturer (Roche). The probes used were digoxigenin-labeled antisense-riboprobes transcribed from the cDNA template of T-bet, GATA3, and G3PDH. Signals were visualized using an alkaline phosphatase-conjugated, anti-digoxigenin antibody (Roche).
Results
A Dominant Interfering Form of Runx1 Promotes Th2 Cell Differentiation.
We previously established mouse lines that express a DNA binding domain of Runx1 from a transgene in a T-lineage-specific way (24). This Runt domain is known to function in a dominant interfering fashion against the Runx1 protein that is expressed endogenously in T lymphocytes. In the present study, we investigated the role of the Runx1 transcription factor in Th cell differentiation using these Runt-transgenic mice. Naive CD4+ T cells were isolated from the spleens of wild-type and Runt-transgenic mice and stimulated with anti-CD3/anti-CD28 antibodies. The culture supernatants were collected and the amount of cytokines secreted was measured by ELISA (Fig. 1 A). The Runt-transgenic cells produced five times as much IL-4 as the wild-type cells at 72 h after incubation. Similarly, the Runt-transgenic cells produced 1.7 and 3.4 times as much IL-5 and IL-10, respectively, as the wild-type cells. The Runt-transgenic cells thus exhibited enhanced production of Th2–type cytokines during an early phase of TCR activation. In contrast, secretion of IFN-γ and IL-2 from the Runt-transgenic cells was decreased slightly and markedly, respectively, compared with the wild-type cells. A similar result as that shown in Fig. 1 A was obtained when the TCR-stimulated cells were incubated in the presence of IL-2 for 5 d, washed with fresh media, and restimulated via TCR (Fig. 1 B). Under this neutral culture condition where neither IL-4 nor IL-12 was added to media, the Runt-transgenic cells secreted several fold more amount of Th2-type cytokines and less amount of Th1-type cytokines than the wild-type cells.
Figure 1.
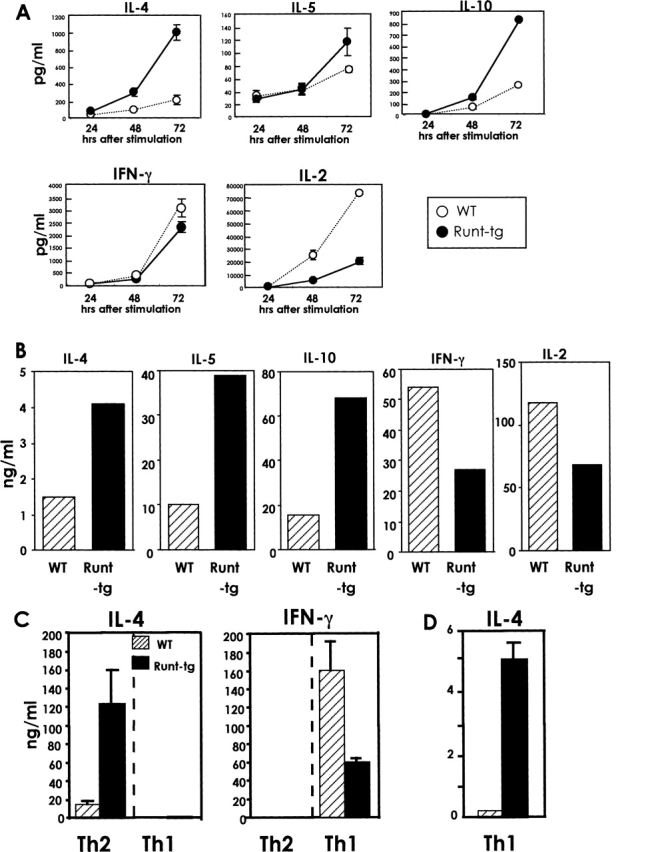
Production of Th2- and Th1-type cytokines from wild-type and Runt-transgenic CD4+ T cells. (A) Naive CD4+ T cells were isolated from the spleens of wild-type and Runt–transgenic mice and stimulated with anti-CD3/anti-CD28 antibodies, and the culture supernatants were collected at the indicated times after stimulation. (B) The TCR-stimulated cells were cultured in the presence of IL-2 for 5 d, washed with fresh media, and restimulated via TCR. The culture supernatants were collected after 24 h. (C) The cells were cultured for four days in the Th2- or Th1-inducing condition. The cells were washed with fresh media and restimulated via TCR. The culture supernatants were collected after 24 h. (D) Same as C, but note the difference of scale used in C and here. The amounts of cytokines secreted into the supernatants were measured by ELISA and their averages and standard deviations are shown. Data are representative of four independent experiments.
We next examined the cytokine production from the Runt-transgenic cells under culture conditions that favor either Th1 or Th2 differentiation. CD4+ T cells were stimulated with anti-CD3/anti-CD28 antibodies and incubated in the presence of either IL-4 and an anti-IFN-γ antibody (Th2 condition) or IL-12 and an anti-IL-4 antibody (Th1 condition) for 4 d. Cells were washed with fresh media and restimulated via TCR, and cytokine secretion was measured (Fig. 1 C). In the Th2 condition, the Runt-transgenic cells produced eight times as much IL-4 as the wild-type cells. In contrast, in the Th1 condition, although IFN-γ production from the Runt-transgenic cells was reduced compared with the wild-type cells, IL-4 production from the Runt-transgenic cells did not reach the level of 10 to 100 ng/ml concentration. Thus, the Runt-expressing cells effectively differentiate into the Th2 lineage in a Th2- but not a Th1-favorable environment.
It must be pointed here that, even in the Th1 condition, the Runt-transgenic cells actually produced 25 times as much IL-4 as the wild-type cells (Fig. 1 D). The absolute amount of IL-4 secreted from the Runt-transgenic cells was, however, 5 ng/ml in the Th1 condition (Fig. 1 D) instead of 120 ng/ml in the Th2 condition (Fig. 1 C). Therefore, the Runt-transgenic cells, though showing a Th2 tendency to some degree, could not have differentiated effectively into the Th2 lineage under the Th1 condition.
Overexpression of Runx1 Prevents Naive Cells from Differentiating Into the Th2 Lineage.
If reducing the effect of Runx1 with the Runt transgene induces Th2 differentiation, as shown in Fig. 1, we wondered whether the artificial overexpression of Runx1 protein in naive T cells would cause a decrease in Th2 differentiation. CD4+ T cells were stimulated with anti-CD3/anti-CD28 antibodies and infected by retroviruses that harbor either pMX or pMX-Runx1. The former vector encodes GFP only, and the latter encodes both GFP and Runx1. The cells were incubated in the presence of IL-2 and restimulated via TCR, and the profile of cytokine production was assessed by intracellular staining of cytokines followed by flow cytometry (Fig. 2 A). In pMX-infected cells, 71% of GFP+ cells were IL-4 positive. In contrast, the percentage of IL-4–producing cells in the pMX-Runx1-infected population was drastically decreased to 5% of GFP+ cells. However, the proportion of IFN-γ producing cells markedly increased with pMX-Runx1 infection compared with pMX infection. We also examined the effect of Runx1 expression on the secretion of other Th2-type cytokines. The GFP+ cells were sorted after retrovirus infection and the amount of IL-5 and IL-10 secreted was measured (Fig. 2 B). Overexpression of Runx1 strikingly reduced the amount of IL-5 and IL-10. The overexpression of Runx1 thus inhibits the production of Th2-type cytokines in TCR-stimulated cells in a neutral cytokine environment. Immunoblot analysis detects the Runx1 protein in the cDNA-transduced, GFP+ fraction but scarcely in the nontransduced, GFP− fraction out of pMX-Runx1-infected cells (Fig. 2 C). The effects seen in Fig. 2, A and B, are thus considered to be due to the exogenously expressed Runx1 protein.
Figure 2.
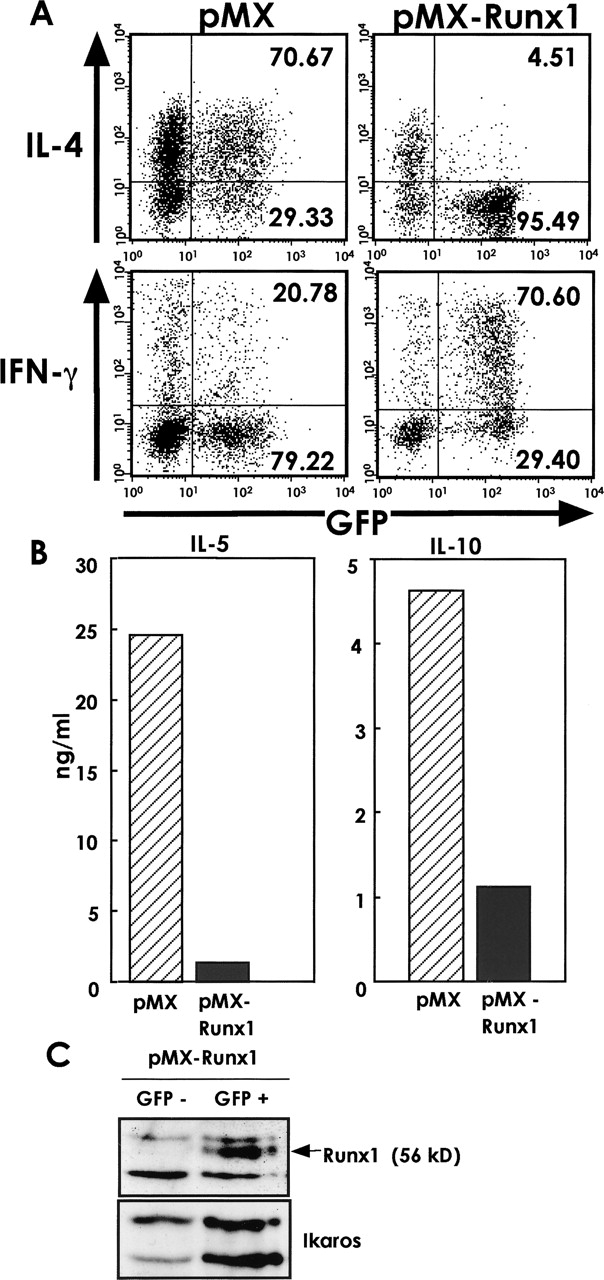
Effect of Runx1 overexpression on the production of various cytokines. (A) Naive CD4+ T cells were isolated from the spleens of BALB/c mice, stimulated with anti-CD3/anti-CD28 antibodies, and infected by retroviruses carrying pMX or pMX-Runx1. After culture in the presence of IL-2, the cells were washed and reactivated via TCR, and processed for flow cytometrical analysis of intracellular IL-4, IFN-γ, and GFP. The numbers represent the percentages of cells in each quadrant. (B) The GFP+ population was sorted after retrovirus infection and reactivated via TCR. The amount of cytokines secreted into the culture supernatant was measured by ELISA. Data are representative of four independent experiments. (C) Immunoblot analysis of Runx1 protein. The GFP− and GFP+ populations were sorted from the pMX-Runx1–infected cells. The nuclear fractions were prepared from each population and their protein extracts were processed for immunoblot analysis using anti-Runx1 and anti-Ikaros antibodies, respectively.
We next examined whether overexpression of Runx1 is inhibitory for Th2-cell differentiation even in a Th2-favorable cytokine environment. CD4+ T cells were stimulated with anti-CD3/anti-CD28 antibodies, infected by retroviruses, and cultured in the presence or absence of an excessive amount of exogenously added IL-4. The cells were restimulated via TCR and analyzed for cytokine production using flow cytometry (Fig. 3 A). In the pMX-infected cells, addition of IL-4 increased the ratio of IL-4+ cells among the GFP+ population as expected. IL-4 production was much lower in the Runx1-overexpressing cells than in the pMX-infected cells, and IL-4 supplementation did not restore IL-4 production in these cells. The amount of cytokines secreted from the GFP+ population was also measured by ELISA (Fig. 3 B). Addition of IL-4 enhanced the secretion of both IL-4 and IL-5 from the pMX-infected cells. In the Runx1-overexpressing cells, however, production of IL-4 and IL-5 did not recover even after addition of IL-4. Thus, the overexpression of Runx1 can block TCR-stimulated CD4+ cells from differentiating into the Th2 lineage even under a Th2-favorable culture condition and this inhibitory effect of Runx1 on Th2 differentiation is not due to the paucity of IL-4 itself.
Figure 3.
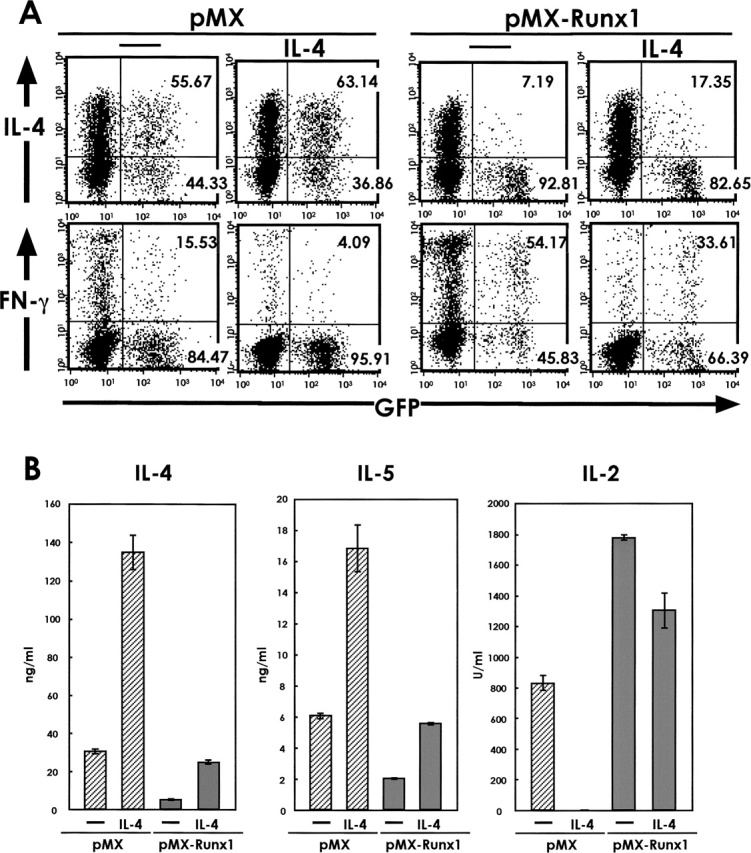
Effect of IL-4 supplementation on the production of various cytokines from Runx1-transduced cells. (A) Naive CD4+ T cells were isolated from the spleens of BALB/c mice, stimulated with anti-CD3/anti-CD28 antibodies and infected by retroviruses carrying pMX or pMX-Runx1. After culture in the presence of IL-2 alone or of both IL-2 and IL-4, the cells were washed, reactivated via TCR, and processed for flow cytometrical analysis of intracellular IL-4, IFN-γ, and GFP. The numbers represent the percentages of cells in each quadrant. (B) The GFP+ population was sorted from the retrovirus-infected cells and reactivated via TCR. The amount of cytokines secreted into the culture supernatant was measured by ELISA. Data are representative of two independent experiments.
It must be noted that the IL-4-containing medium supplies cells with a condition unfavorable to Th1 differentiation. In both the pMX- and pMX-Runx1–infected cells, the percentage of IFN-γ-positive cells as well as the amount of secreted IL-2 decreased in response to the addition of IL-4.
Overexpression of Runx1 Does Not Impair TCR and IL-4R Signaling During Th Differentiation.
As overexpression of Runx1 prevented cells from differentiating into Th2 cells, we examined whether Runx1 affects the TCR and IL-4R signaling pathways. We used flow cytometry to analyze cell surface molecules such as the TCRβ chain, CD4, CD25, CD69, and IL-4Rα (Fig. 4 A). The expression profiles of these molecules in the GFP+ fractions were comparable in the pMX- and pMX-Runx1-infected cells. To analyze signaling molecules further, protein was extracted from the GFP+ population and processed for immunoblot analysis (Fig. 4 B). The amounts of Jak1 and STAT6 were not affected by overexpression of Runx1 nor by supplementation with IL-4. Phosphorylation of a 125-kD protein (indicated in Fig. 4 B by the bottom arrow) was induced by the addition of IL-4 and likely represents Jak1 and Jak3. The degree of phosphorylation of this 125-kD protein was comparable in pMX- and pMX-Runx1–infected cells. Phosphorylation of STAT6 was also induced by IL-4 to a similar degree in both the pMX- and pMX-Runx1–infected cells. Thus, overexpression of Runx1 is not likely to affect the IL-4R signaling pathway, at least not through the phosphorylation of STAT6. It must be noted also that overexpression of Runx1 was not inhibitory for IL-4–dependent cell proliferation. Different concentrations of IL-4 were added to the medium and incorporation of 3H-thymidine into an acid-insoluble fraction of the cells was measured (Fig. 4 C). The pMX-Runx1-infected cells proliferated rather better than the pMX-infected cells.
Figure 4.
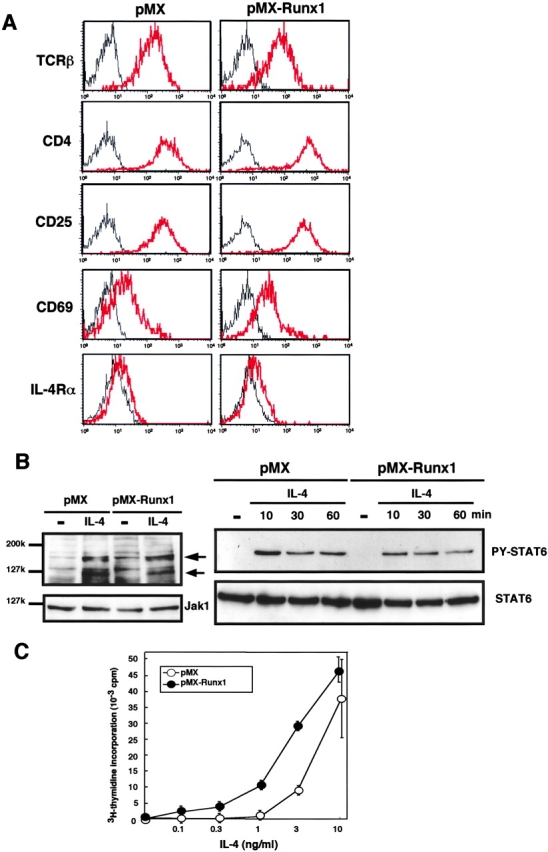
Effect of Runx1 overexpression on TCR- and IL-4R- signaling. (A) Naive CD4+ T cells were isolated from the spleens of BALB/c mice, stimulated with anti-CD3/anti-CD28 antibodies, and infected by retroviruses carrying pMX or pMX-Runx1. After culture in the presence of IL-2, the cells were washed, reactivated via TCR, and processed for flow cytometrical analysis. The red lines represent fluorescence emitted from antibody-stained cells, whereas the gray lines represent fluorescence from cells stained by control IgG. (B) The GFP+ population was sorted from the retrovirus-infected cells and cultured in the absence or presence of IL-4. In the left panel, protein was extracted at 10 min after the addition of IL-4, whereas, in the right panel, protein was extracted at 10, 30, and 60 min after the IL-4 addition. The extract was processed for immunoblot analysis using anti-Jak1, anti-STAT6, anti-phosphotyrosine, and anti-phosphorylated STAT6-specific antibodies. The bands indicated by the bottom arrow probably correspond to phosphorylated Jak1/Jak3, whereas the bands indicated by the top arrow probably represent phosphorylated IRS1/IRS2. (C) Naive CD4+ T cells were TCR-stimulated, infected by retroviruses, and cultured in the presence of IL-2 alone or of both IL-2 and the indicated concentration of IL-4. The GFP+ population was sorted and incubated in the presence of 3H-thymidine. The incorporation of radioactivity into an acid-insoluble fraction of the cells was measured. Data are representative of two independent experiments.
Runx1 Negatively Regulates Th2 Differentiation of Cells by Inhibiting GATA3 Expression.
The GATA3 transcription factor is located downstream of IL-4R signaling pathway and regulates the commitment of cells toward the Th2 lineage (11). We therefore examined whether overexpression of Runx1 affects GATA3 expression. The RNA was extracted from the GFP+ fractions and processed for Northern blot analysis (Fig. 5 A). In the pMX-infected cells, some amount of GATA3 transcript was detected (lane 1), whereas almost no GATA3 transcript was detected in the pMX-Runx1–infected cells (lane 3). Furthermore, addition of an excess amount of IL-4 into the medium induced higher expression of GATA3 in the pMX-infected cells (lane 2), but could not restore GATA3 expression in the Runx1-overexpressing cells (lane 4). The amount of T-bet, a Th1-specific transcription factor (32), decreased in response to the addition of IL-4 in both the Runx1-overexpressing and pMX-infected cells (lanes 2 and 4). Thus, Runx1 can strongly repress the induction of GATA3 expression and the inability of Runx1-overexpressing cells to differentiate into the Th2 lineage is likely due to this paucity of GATA3 induction.
Figure 5.

Effect of Runx1 overexpression and Runt-transgene on GATA3 expression. (A) Naive CD4+ T cells were isolated from the spleens of BALB/c mice, stimulated with anti-CD3/anti-CD28 antibodies, and infected by retroviruses carrying pMX or pMX-Runx1. After culture in the presence of IL-2 alone or of both IL-2 and IL-4, the cells were washed and reactivated via TCR. The GFP+ population was sorted as shown in the top panel, and RNA was extracted and processed for Northern blot analysis as shown in the lower panel. (B) Naive CD4+ T cells were isolated from the spleens of wild-type and Runt-transgenic mice, and stimulated with anti-CD3/anti-CD28 antibodies. In case, either anti–IL-4 antibody or IL-4 or IL-12 was added to the culture medium as indicated. RNA was extracted and processed for Northern blot analysis. Relative intensity of GATA3 and T-bet transcripts that were normalized by that of G3PDH are shown below the lanes. Data are representative of two independent experiments.
We further examined the degree of GATA3 expression in the Runt-transgenic cells. CD4+ T cells were stimulated by anti-CD3/anti-CD28 antibodies and the extracted RNA was processed for Northern blot analysis (Fig. 5 B). The GATA3 transcript was detected in the Runt-transgenic cells at a significantly higher level (1.5 to 2.0 fold increase) than in the wild-type cells. The result thus suggests that the effective differentiation of Runt-transgenic cells into the Th2 lineage under the neutral as well as Th2 conditions (Fig. 1, A–C) is likely attributable to the up-regulation of GATA3 expression.
It must be noted that expression of GATA3 was blocked in both the wild-type and Runt-transgenic cells by adding an anti-IL-4 antibody (Fig. 5 B), indicating that GATA3 expression is dependent on IL-4R signaling. A difference between the two types of cells is, however, clearly visible when the TCR-stimulated cells were incubated in the presence of IL-12 (a lower portion of Fig. 5 B). The Runt-transgenic cells expressed twice as much GATA3 transcript as the wild-type cells, though to a lesser degree compared with the absence of IL-12. Thus, in the case of Runt-transgenic cells, a feature of GATA3 expression in the presence of IL-12 (Fig. 5 B) appears to be in accordance with the profile of IL-4 production in the Th1 condition (Fig. 1 D).
Forced Expression of GATA3 Relieves the Inhibitory Effect of Runx1 on Th2 Differentiation.
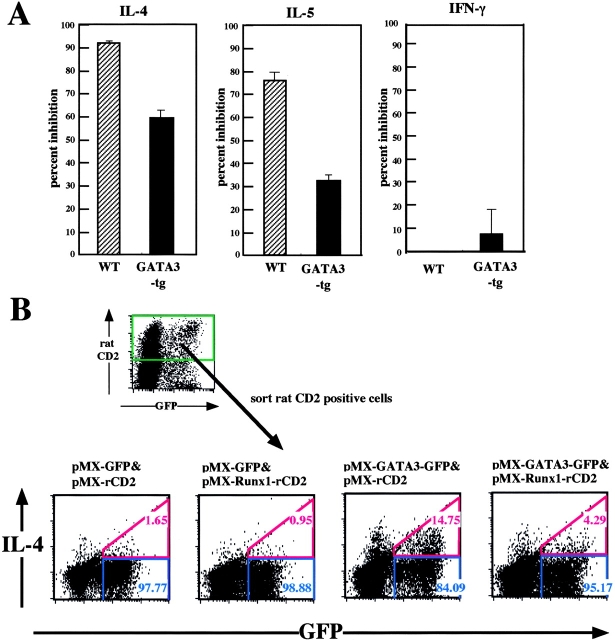
We next examined whether GATA3 transduction could recover Th2 cytokine production in the pMX-Runx1–infected cells, using GATA3-transgenic mice. CD4+ T cells were prepared from wild-type and transgenic mice, stimulated by anti-CD3/anti-CD28 antibodies, and infected by pMX or pMX-Runx1 retroviruses. The GFP+ cells were sorted and restimulated via TCR, and the amounts of secreted cytokines were assayed. In Fig. 6 A, the values obtained for the pMX-Runx1-infected cells are presented as percent inhibition of secretion, taking the values obtained for the pMX-infected cells to be 100%. The percent inhibition of IL-4 and IL-5 secretion in the GATA3-transgenic cells was roughly two thirds and one half of that in the wild-type cells, respectively. GATA3 did not cause a significant difference in the IFN-γ production of the wild-type and transgenic cells.
Figure 6.
Effect of cotransduction of GATA3 and Runx1 on Th2 cell differentiation. (A) Naive CD4+ T cells were isolated from the spleens of wild-type and GATA3-transgenic mice, TCR-stimulated, infected by retroviruses carrying pMX or pMX-Runx1, and cultured in the presence of IL-2. The GFP+ population was sorted and reactivated via TCR. The cytokines secreted into the culture supernatant were measured by ELISA. The values obtained for the pMX-Runx1–infected cells are presented as percent inhibition of secretion, taking the values observed for the pMX-infected cells to be 100%. (B) Naive CD4+ T cells were isolated from the spleens of IL-4R (−/−) mice, TCR-stimulated, coinfected by pMX-GFP and pMX-rat CD2 retroviruses as indicated, and cultured in the presence of IL-2. The rat CD2+ population was sorted, TCR-stimulated, and processed for flow cytometrical analysis of intracellular IL-4. Data are representative of two independent experiments.
We also tried cotransduction of GATA3 and Runx1 into CD4+ T cells. In this particular case, the cells were prepared from the IL-4R (−/−) mice so that it becomes possible to evaluate the effects of introduced genes independently from IL-4R signaling. The TCR-stimulated cells were doubly infected by pMX-GFP and pMX-rat CD2 retroviruses each of which harbors (or not) GATA3 or Runx1 as indicated (Fig. 6 B). The rat CD2 positive cells were sorted, restimulated via TCR and processed for intracellular staining of IL-4. Both the GATA3-solely-expressing and GATA3/Runx1-coexpressing cells showed a significant increase in the IL-4 positive fraction, compared with the cells not introduced by GATA3. The results in Fig. 6, A and B, thus indicate that forced expression of GATA3 can, at least partially, compensate for the inhibitory effect of Runx1 on Th2 differentiation.
Expression of Runx1 in Naive CD4+ T Cells Is Down-regulated after TCR Stimulation.
Runx1, when overexpressed, can negatively regulate GATA3 expression. To explore a relationship between Runx1 and GATA3 under a physiological condition, we first examined the expression pattern of endogenous proteins in naive CD4+, Th2, and Th1 cells by immunoblot analysis (Fig. 7 A). The Runx1 protein was evident as a 56-kD band in naive CD4+ cells, but not in fully differentiated Th1 cells for 7 d. In Th2 cells, a subtle amount of Runx1 protein might be present. Therefore, Runx1 appears to play its role, if any, in naive but not in differentiated Th1 nor Th2 cells.
Figure 7.
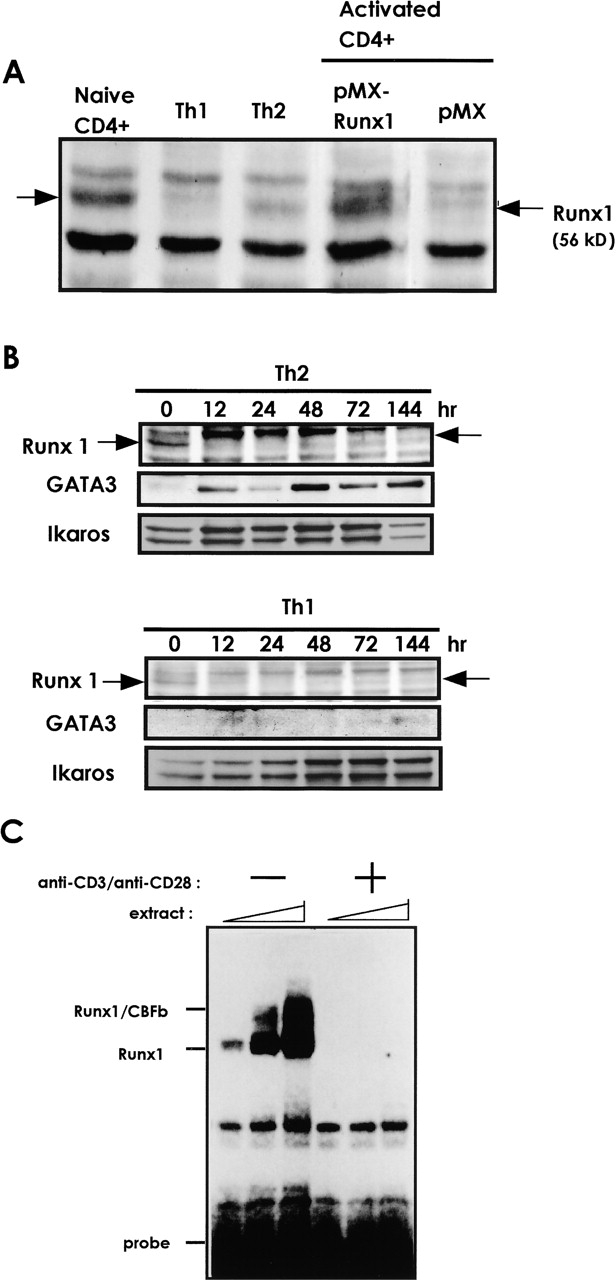
Expression of endogenous Runx1 and GATA3 proteins in CD4+ T cells. (A) Naive CD4+ T cells were isolated from the spleens of mice and cultured under the Th1- or Th2-condition for 7 d. Protein extracts were processed for immunoblot analysis, using anti-Runx1 antibody. Protein extracted from the pMX-Runx1-infected or pMX-infected, TCR-stimulated CD4+ T cells was also probed with the same antibody. (B) Protein was extracted at indicated hour after culturing in the Th2- or Th1-condition. (C) Naive CD4+ T cells were treated or not by anti-CD3/anti-CD28 antibodies for 24 h. Extracts containing 1, 3, and 5 μg protein, respectively, were processed for EMSA. Data are representative of two independent experiments.
We then examined the profile of protein expression during an early phase of TCR activation (Fig. 7 B). At 12 and 24 h after TCR stimulation, the level of Runx1 was markedly decreased in both the Th2- and Th1-culturing conditions. Concomitantly, expression of GATA3 was induced in a Th2-condition. The extract was also processed for EMSA using a Runx-binding oligonucleotide as a probe (Fig. 7 C). DNA/protein complexes containing Runx1 or a Runx1/CBFβ heterodimer (33) were detected in the naive CD4+ cells but not in the cells treated by anti-CD3/anti-CD28 antibodies for 24 h. Thus, the results in Fig. 7, B and C, indicate that the TCR stimulation of naive CD4+ T cells causes down-regulation of Runx1 protein and simultaneous induction of GATA3 protein during the early phase of activation.
We finally evaluated the degree of Runx1 overexpression in the retrovirus-infection experiments described above. The intensity of 56-kD band in the pMX-Runx1–infected cells was roughly twice as dense as that in naive CD4+ cells (Fig. 7 A). In addition, the endogenous and exogenous 56-kD bands turned out to comigrate in parallel, indicating the band to represent the authentic Runx1 protein. Thus, the amount of exogenously introduced Runx1 protein is roughly similar with that expressed endogenously in naive CD4+ T cells.
Discussion
In this study, we showed that transduction of a dominant interfering form of Runx1 in peripheral CD4+ T cells resulted in a marked and coordinate production of IL-4, IL-5, and IL-10 following TCR stimulation. Overexpression of Runx1, on the other hand, attenuated Th2-cell differentiation and led to the cessation of GATA3 expression. This inhibitory effect of Runx1 on Th2 differentiation could not be reversed by supplementation with an excess amount of IL-4, but was, at least partially, relieved by the forced expression of GATA3 itself. Thus, the Runx1 transcription factor functions as a negative regulator for Th2-cell differentiation by inhibiting GATA3 expression.
Various transcription factors have been reported to be involved in the regulation of Th2 differentiation (34). Among them, GATA3 is considered to be a crucial factor, as it directly controls chromatin remodeling of the IL-4 locus (13, 14). GATA3 expression occurs as two distinct phases: a transient initial phase that is dependent on IL-4–mediated STAT6 function (10), and a GATA3-dependent auto-activation phase (13, 14). Recently, repressor of GATA (ROG) and friend of GATA1 (FOG1) were shown to bind to GATA3 and were thereby characterized as GATA3-repressing proteins (35, 36). Expression of ROG and FOG1 is transiently detected within 24 to 48 h after primary T cell activation and is detected regardless of whether the conditions are skewed toward Th1 or Th2. Thus, these two factors conceivably participate in the regulation of GATA3-dependent auto-activation in order to prevent the excessive production of Th2 cytokines. It must be noted that the expression pattern of Runx1 in the early activation phase is distinct from that of ROG and FOG1. Runx1 is significantly expressed in the wild-type, CD4+ T cells but rapidly decreases after TCR activation. Given that the inhibitory effect of Runx1 on GATA3 expression could not be reversed by adding an excess amount of IL-4, it is conceivable that Runx1 negatively regulates the GATA3 expression in an IL-4R–independent fashion. Thus, in the case of naive T cells, TCR-mediated down-regulation of Runx1 may be a first step or a prerequisite for initiating expression of GATA3. In contrast to naive cells, Runx1 was not detected or detected only in a subtle amount in differentiated Th1 or Th2 cells. Therefore, Runx1 appears to play its role mainly in naive CD4+ cells and during the early phase of TCR activation but not in differentiation-completed Th cells.
On the other hand, as seen in transgenic mice, reduction of endogenous Runx1 activity by Runt leads to the excessive production of Th2-type cytokines under the neutral and Th2-favorable conditions. This suggests that Runx1 may also play an additional role so as to prevent cells from overwhelmingly differentiating into the Th2 lineage. It must be noted that the Runt-transgenic cells did not fully differentiate into the Th2 lineage under the Th1 culturing condition, as seen by their magnitude of IL-4 production and GATA3 expression. Thus, IL-4 signaling appears to be necessary for the Runt-transgenic cells to fully commit to the Th2 lineage. Considering how Runx1 affects GATA3 expression, the above situation may not be unreasonable. Runx1 can be a repressor of GATA3 expression and functions during an initial phase of TCR-stimulation of naive cells. The Runt domain, which acts as a dominant negative factor against the endogenous Runx1 protein, therefore could cancel only the repressive activity of Runx1 on GATA3, but not positively enhance GATA3 expression.
The function of Runx1 as a transcription factor differs depending on its interaction with different types of cofactors. On one hand, Runx1 functions as a transcriptional activator of hematopoiesis-related genes such as GM-CSF (37), IL-3 (38), and the TCRβ chain (39, 40). In these cases, Runx1 interacts with a coactivator, p300/CBP (41), which has histone acetyltransferase activity. On the other hand, Runx1 can also behave as a transcriptional repressor through interaction with a corepressor (42), and one possibility is that Runx1 represses GATA3 expression by directly binding to its regulatory region. The conserved COOH-terminal pentapeptide, VWRPY, in the Runx1 protein is a binding site for a corepressor, Groucho/TLE (43, 44). The transducion of a series of deletion mutants demonstrated that the COOH-terminal region including this VWRPY motif is responsible for the inhibitory effect of Runx1 on Th2 differentiation (unpublished data). However, as the VWRPY motif is conserved among all three Runx proteins, and the inhibition of IL-4 production is a characteristic feature of Runx1 but not of Runx2 nor of Runx3 (unpublished data), it is unlikely that the GATA3 repression by Runx1 is simply mediated by recruiting Groucho/TLE. Moreover, an alternative possibility that cannot be excluded is that Runx1 is indirectly involved in repressing GATA3 expression by regulating other factor(s) such as Mel-18 (45) and NF-κB (46). Further analysis is required to elucidate the precise mechanism by which Runx1 represses GATA3 expression. A possibility is not excluded either that Runx1 is also involved in the regulation of IL-4 transcription itself.
Acknowledgments
We express our sincere thanks to the following scientists for providing us with their valuable experimental tools: T. Kitamura for the pMX-GFP vector, F. Brombacher for IL-4R targeted mice, and D. Levanon and Y. Groner for the murine Runx3 cDNA. We also thank M. Kuji for her secretarial assistance.
This work was supported by research grants from the Ministry of Education, Science, Sports, Culture and Technology, Japan.
O. Komine and K. Hayashi contributed equally to this work.
Footnotes
Abbreviation used in this paper: GFP, green fluorescent protein.
References
- 1.Mosmann, T.R., H. Cherwinski, M.W. Bond, M.A. Giedlin, and R.L. Coffman. 1986. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136:2348–2357. [PubMed] [Google Scholar]
- 2.Paul, W.E., and R.A. Seder. 1994. Lymphocyte responses and cytokines. Cell. 76:241–251. [DOI] [PubMed] [Google Scholar]
- 3.O'Garra, A. 1998. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity. 8:275–283. [DOI] [PubMed] [Google Scholar]
- 4.Glimcher, L.H., and K.M. Murphy. 2000. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 14:1693–1711. [PubMed] [Google Scholar]
- 5.Thompson, C.B. 1995. Distinct roles for the costimulatory ligands B7-1 and B7-2 in T helper cell differentiation? Cell. 81:979–982. [DOI] [PubMed] [Google Scholar]
- 6.Kubo, M., M. Yamashita, R. Abe, T. Tada, K. Okumura, J.T. Ransom, and T. Nakayama. 1999. CD28 costimulation accelerates IL-4 receptor sensitivity and IL-4-mediated Th2 differentiation. J. Immunol. 163:2432–2442. [PubMed] [Google Scholar]
- 7.Ho, I.C., M.R. Hodge, J.W. Rooney, and L.H. Glimcher. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 85:973–983. [DOI] [PubMed] [Google Scholar]
- 8.Ho, I.C., D. Lo, and L.H. Glimcher. 1998. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and -independent mechanisms. J. Exp. Med. 188:1859–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan, W.M., B. Corthesy, R.J. Bram, and G.R. Crabtree. 1991. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 352:803–807. [DOI] [PubMed] [Google Scholar]
- 10.Kurata, H., H.J. Lee, A. O'Garra, and N. Arai. 1999. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 11:677–688. [DOI] [PubMed] [Google Scholar]
- 11.Zheng, W., and R.A. Flavell. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 89:587–596. [DOI] [PubMed] [Google Scholar]
- 12.Ferber, I.A., H.J. Lee, F. Zonin, V. Heath, A. Mui, N. Arai, and A. O'Garra. 1999. GATA-3 significantly downregulates IFN-γ production from developing Th1 cells in addition to inducing IL-4 and IL-5 levels. Clin. Immunol. 91:134–144. [DOI] [PubMed] [Google Scholar]
- 13.Lee, H.J., N. Takemoto, H. Kurata, Y. Kamogawa, S. Miyatake, A. O'Garra, and N. Arai. 2000. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 192:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang, W., M. Lohning, Z. Gao, M. Assenmacher, S. Ranganath, A. Radbruch, and K.M. Murphy. 2000. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 12:27–37. [DOI] [PubMed] [Google Scholar]
- 15.Kagoshima, H., K. Shigesada, M. Satake, Y. Ito, H. Miyoshi, M. Ohki, M. Pepling, and P. Gergen. 1993. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 9:338–341. [DOI] [PubMed] [Google Scholar]
- 16.Meyers, S., J.R. Downing, and S.W. Hiebert. 1993. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol. Cell. Biol. 13:6336–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Look, A.T. 1997. Oncogenic transcription factors in the human acute leukemias. Science. 278:1059–1064. [DOI] [PubMed] [Google Scholar]
- 18.Okuda, T., J. van Deursen, S.W. Hiebert, G. Grosveld, and J.R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 84:321–330. [DOI] [PubMed] [Google Scholar]
- 19.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A.H. Sharpe, and N.A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA. 93:3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada, H., T. Watanabe, M. Niki, H. Takano, N. Chiba, N. Yanai, K. Tani, H. Hibino, S. Asano, M.L. Mucenski, et al. 1998. AML1(−/−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene. 17:2287–2293. [DOI] [PubMed] [Google Scholar]
- 21.Ito, Y. 1999. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells. 4:685–696. [DOI] [PubMed] [Google Scholar]
- 22.Speck, N.A., T. Stacy, Q. Wang, T. North, T.L. Gu, J. Miller, M. Binder, and M. Marin-Padilla. 1999. Core-binding factor: a central player in hematopoiesis and leukemia. Cancer Res. 59:1789s–1793s. [PubMed] [Google Scholar]
- 23.Downing, J.R., M. Higuchi, N. Lenny, and A.E. Yeoh. 2000. Alterations of the AML1 transcription factor in human leukemia. Semin. Cell Dev. Biol. 11:347–360. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, K., W. Natsume, T. Watanabe, N. Abe, N. Iwai, H. Okada, Y. Ito, M. Asano, Y. Iwakura, S. Habu, et al. 2000. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J. Immunol. 165:6816–6824. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi, K., N. Abe, T. Watanabe, M. Obinata, M. Ito, T. Sato, S. Habu, and M. Satake. 2001. Overexpression of AML1 transcription factor drives thymocytes into the CD8 single-positive lineage. J. Immunol. 167:4957–4965. [DOI] [PubMed] [Google Scholar]
- 26.Barner, M., M. Mohrs, F. Brombacher, and M. Kopf. 1998. Difference between IL-4R alpha-deficient and IL-4-deficient mice reveal a role for IL-13 in the regulation of Th2 responses. Curr. Biol. 8:669–672. [DOI] [PubMed] [Google Scholar]
- 27.Fujii, M., K. Hayashi, M. Niki, N. Chiba, K. Meguro, K. Endo, J. Kameoka, S. Ito, K. Abe, T. Watanabe, and M. Satake. 1998. Overexpression of AML1 renders a T hybridoma resistant to T cell receptor-mediated apoptosis. Oncogene. 17:1813–1820. [DOI] [PubMed] [Google Scholar]
- 28.Bae, S.C., Y. Yamaguchi-Iwai, E. Ogawa, M. Maruyama, M. Inuzuka, H. Kagoshima, K. Shigesada, M. Satake, and Y. Ito. 1993. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 8:809–814. [PubMed] [Google Scholar]
- 29.Ogawa, E., M. Maruyama, H. Kagoshima, M. Inuzuka, J. Lu, M. Satake, K. Shigesada, and Y. Ito. 1993. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA. 90:6859–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misawa, K., T. Nosaka, S. Morita, A. Kaneko, T. Nakahata, S. Asano, and T. Kitamura. 2000. A method to identify cDNAs based on localization of green fluorescent protein fusion products. Proc. Natl. Acad. Sci. USA. 97:3062–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, Y., T. Watanabe, N. Chiba, M. Niki, and Y. Kuroiwa, T. Nishihira, S. Satomi, Y. Ito, and M. Satake. 1997. The protooncogene product, PEBP2β/CBFβ, is mainly located in the cytoplasm and has an affinity with cytoskeletal structures. Oncogene. 15:677–683. [DOI] [PubMed] [Google Scholar]
- 32.Szabo, S.J., S.T. Kim, G.L. Costa, X. Zhang, C.G. Fathman, and L.H. Glimcher. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. [DOI] [PubMed] [Google Scholar]
- 33.Kanno, T., Y. Kanno, L.F. Chen, E. Ogawa, W.Y. Kim, and Y. Ito. 1998. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol. 18:2444–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, K.M., W. Ouyang, J.D. Farrar, J. Yang, S. Ranganath, H. Asnagli, M. Afkarian, and T.L. Murphy. 2000. Signaling and transcription in T helper development. Annu. Rev. Immunol. 18:451–494. [DOI] [PubMed] [Google Scholar]
- 35.Miaw, S.C., A. Choi, E. Yu, H. Kishikawa, and I.C. Ho. 2000. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 12:323–333. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, M., W. Ouyang, Q. Gong, S.G. Katz, J.M. White, S.H. Orkin, and K.M. Murphy. 2001. Friend of GATA-1 represses GATA-3–dependent activity in CD4+ T cells. J. Exp. Med. 194:1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, A., M. Satake, Y. Yamaguchi-Iwai, S.C. Bae, J. Lu, M. Maruyama, Y.W. Zhang, H. Oka, N. Arai, K. Arai, et al. 1995. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 86:607–616. [PubMed] [Google Scholar]
- 38.Uchida, H., J. Zhang, and S.D. Nimer. 1997. AML1A and AML1B can transactivate the human IL-3 promoter. J. Immunol. 158:2251–2258. [PubMed] [Google Scholar]
- 39.Bae, S.C., E. Ogawa, M. Maruyama, H. Oka, M. Satake, K. Shigesada, N.A. Jenkins, D.J. Gilbert, N.G. Copeland, and Y. Ito. 1994. PEBP2αB/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol. Cell. Biol. 14:3242–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halle, J.P., P. Haus-Seuffert, C. Woltering, G. Stelzer, and M. Meisterernst. 1997. A conserved tissue-specific structure at a human T-cell receptor β-chain core promoter. Mol. Cell. Biol. 17:4220–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitabayashi, I., A. Yokoyama, K. Shimizu, and M. Ohki. 1998. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 17:2994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutterbach, B., J.J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S.W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275:651–656. [DOI] [PubMed] [Google Scholar]
- 43.Aronson, B.D., A.L. Fisher, K. Blechman, M. Caudy, and J.P. Gergen. 1997. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol. 17:5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levanon, D., R.E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA. 95:11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura, M., Y. Koseki, M. Yamashita, N. Watanabe, C. Shimizu, T. Katsumoto, T. Kitamura, M. Taniguchi, H. Koseki, and T. Nakayama. 2001. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity. 15:275–287. [DOI] [PubMed] [Google Scholar]
- 46.Das, J., C.H. Chen, L. Yang, L. Cohn, P. Ray, and A. Ray. 2001. A critical role for NF-κB in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat. Immunol. 2:45–50. [DOI] [PubMed] [Google Scholar]