Abstract
The role of T cell receptor α enhancer (Eα) cis-acting elements in the developmental regulation of VDJ recombination at the TCR α/δ locus was examined in transgenic mice containing variants of a minilocus VDJ recombination substrate. We demonstrate that the 116-bp Tα1,2 core enhancer fragment of the 1.4-kb Eα is sufficient to activate the enhancer-dependent step of minilocus rearrangement, and that within Tα1,2, intact binding sites for TCF/LEF and Ets family transcription factors are essential. Although minilocus rearrangement under the control of the 1.4-kb Eα initiates at fetal day 16.5 and is strictly limited to αβ T cells, we find that rearrangement under the control of Tα1,2 initiates slightly earlier during ontogeny and occurs in both γδ and αβ T cells. We conclude that the core fragment of Eα can establish accessibility to the recombinase in developing thymocytes in vivo in a fashion that is dependent on the binding of TCF/LEF and Ets family transcription factors, but that these and other factors that bind to the Eα core cannot account for the precise developmental onset of accessibility that is provided by the intact Eα. Rather, our data suggests a critical role for factors that bind Eα outside of the core Tα1,2 region in establishing the precise developmental onset of TCR α rearrangement in vivo.
Ordered recombination of TCR variable (V), diversity (D), and joining ( J ) gene segments is a process that is crucial for the generation of the diverse antigen recognition repertoires that characterize mature αβ and γδ T cells (1–3). The most immature thymic population has a CD4low CD8−CD3−CD25−HSA+ phenotype and has all TCR genes in unrearranged configuration (4). These cells subsequently lose expression of CD4 to become CD4−CD8− double negative (DN)1 cells, which can be further subdivided into four distinct populations on the basis of CD44 and CD25 expression. DN thymocytes mature from CD44hi CD25− to CD44hiCD25+ to CD44lowCD25+ to CD44low CD25− (5, 6). The CD44lowCD25+ DN subset expresses high levels of RAG-1 and RAG-2 and undergoes extensive VDJ recombination at the TCR-β, -γ, and -δ loci (7, 8). In-frame TCR-β rearrangement directs the synthesis of a TCR-β protein which, in conjunction with pTα, forms a pre-TCR (9, 10) that functions to inhibit RAG-1 and RAG-2 expression, to drive thymocyte proliferation, and to drive thymocyte maturation through the CD44lowCD25− DN and immature single positive (ISP) stages to the CD4+CD8+ double positive (DP) stage (7, 8, 11–13). TCR-α rearrangement is activated as thymocytes transit into the DP stage (5, 7, 14).
The relationship between TCR gene rearrangement events and thymocyte commitment to the αβ or γδ lineage has been an area of intense interest. As TCR-β and TCR-γ rearrangements are found in both αβ and γδ T cells, the initiation of rearrangement events at these loci is not associated with lineage commitment. TCR-δ gene segments lie within Vα and Jα gene segments in the complex TCR-α/δ locus and are therefore deleted by Vα to Jα rearrangement (15–17). Recent reports have identified rearranged TCR-δ genes in ISP precursors of αβ T cells before the onset of TCR-α rearrangement (14) and on Vα-Jα excision products in αβ thymocytes and peripheral T cells (18–20), arguing that TCR-δ gene rearrangement is initiated in a common precursor of αβ and γδ T cells as well. Because excised TCR-δ gene VDJ recombination products are relatively depleted of in-frame rearrangements (18, 19), it appears likely that functional TCR-δ and TCR-γ rearrangement can commit thymocytes towards the γδ pathway and away from the αβ pathway. However, because at least some γδ cells show evidence of selection on the basis of functional TCR-β gene rearrangement (11), and late stage CD44lowCD25− DN thymocytes have been shown to include precursors of both αβ and γδ T cells (21), at least some thymocytes may remain uncommitted until very late in the DN population. The activation of TCR-α rearrangement as thymocytes transit into the DP stage, with concomitant deletion of TCR-δ, must irrevocably assign all remaining uncommitted thymocytes to the αβ pathway. TCR-α is therefore the only TCR gene whose rearrangement is activated in a lineage-specific fashion. The mechanisms that establish the developmental onset of TCR-α rearrangement are therefore of particular interest.
Numerous studies have demonstrated that transcriptional promoters and enhancers play an important role in the developmental regulation of VDJ recombination at TCR and Ig loci, probably by modulating accessibility of chromosomal recombination substrates to the recombinase machinery (for recent reviews, see references 22, 23). We have examined the role of transcriptional enhancers in the temporal and lineage-specific control of VDJ recombination at the TCR-α/δ locus by evaluating VDJ recombination in transgenic mice carrying variants of a human TCR-δ gene minilocus rearrangement substrate that included either the 1.4-kb TCR-δ enhancer (Eδ) or the 1.4-kb TCR-α enhancer (Eα) (24, 25). We found that the developmental regulation of minilocus rearrangement under the control of Eδ or Eα paralleled that found at the endogenous TCR-α/δ locus. Specifically, in Eδ-bearing transgenic lines, the enhancer-dependent VD to J step of minilocus rearrangement began on fetal day 14.5 and was equivalent in αβ and γδ T cells, much like endogenous VδDδJδ rearrangement. In Eαbearing transgenic lines, VD to J rearrangement was delayed until fetal day 16.5 and was limited to αβ cells, much like endogenous VαJα rearrangement (25). These results imply that Eδ and Eα play important roles in the developmental regulation of VδDδJδ and VαJα rearrangement, respectively, at the endogenous TCR-α/δ locus.
An important goal of ours has been to identify the cisacting elements of Eα that are critical in establishing the precise developmental regulation of VαJα rearrangement at the TCR-α/δ locus. The core or minimal enhancer fragment of Eα has been defined as a 116-bp fragment (Tα1,2) that contains binding sites for several transcription factors, including ATF/CREB, TCF-1/LEF-1, CBF/PEBP2, and Ets proteins (26–28). This definition as a core or minimal enhancer is based on the ability of Tα1,2 to potently activate plasmid reporter gene expression in transient transfection experiments and the critical role played by each of the defined factor binding sites for significant enhancer activity. Further supporting the role of this enhancer fragment as a discrete functional unit is its ability to support the cooperative assembly of a stable nucleoprotein complex in vitro in the presence of the various transcription factors noted above (28). These transcription factors and their cognate binding sites in Tα1,2 are therefore attractive candidates for contributing to the developmental regulation of VDJ recombination by Eα in vivo. Two of these binding sites were selected for analysis in the present study. The first binds the related TCF-1 and LEF-1 members of the high mobility group (HMG) −1 box family of DNA binding proteins (29–31). These proteins bind to the minor groove of DNA and induce a sharp bend in the DNA helix that, in the case of LEF-1, has been shown to facilitate interactions between Ets-1 and ATF/CREB proteins bound at nonadjacent sites in Tα1,2 to generate a stable and active nucleoprotein complex (28, 32). Transient transfection experiments have shown that an intact TCF-1/LEF-1 binding site is essential for Tα1,2 enhancer activity, and that LEF-1 and TCF-1 can both transactivate reporter gene expression by binding to Tα1,2 (28, 29, 31). The second cis-acting element studied is the binding site for members of the Ets family of transcription factors. Transient transfection experiments have shown that an intact Ets binding site is also required for Tα1,2 enhancer activity, and that the Ets-1 protein can bind to Tα1,2 and transactivate a Tα1,2-driven reporter gene as well (28, 33).
In the present study, we examine VDJ recombination of the TCR δ gene minilocus under the control of wild-type Tα1,2 sequences and compare it with VDJ recombination mediated by Tα1,2 fragments carrying mutations in either the TCF/LEF- or Ets binding sites. Our results indicate the Tα1,2 core enhancer can activate VDJ recombination in a fashion that is dependent on TCF/LEF and Ets family transcription factors, but that additional Eα sequences that lie outside of the enhancer core are required for precise developmental control.
Materials and Methods
Transgenic Mice.
The Ets binding site mutation (Tα1,2mEts) was generated by PCR using the 700-bp BstXI fragment of the human Eα cloned into the BamHI site of pUC13 (Eα0.7) as a template (26) (provided by J. Leiden, University of Chicago, Chicago, IL). PCR was performed using the mutagenic oligonucleotide Ets ( TATTTTAAACTCTTCTTTCCAGAACTTGTGGCTTCT ) and the −40 primer. The PCR product was digested with DraI and EcoRI to generate a 135-bp fragment that was ligated into SmaI and EcoRI cut pBluescript KS+. Dideoxynucleotide sequence analysis of the resultant plasmid confirmed that the insert carried a 3-bp change in the Tα1,2 Ets binding site. The 125-bp Tα1,2mEts was excised from this plasmid by digestion with BamHI (plus PvuI to further cleave the plasmid and prevent subsequent religation), and the ends were blunted by treatment with the Klenow fragment of Escherichia coli DNA polymerase I. The Tα1,2mEts fragment was then ligated into XbaI-digested, blunted, and phosphatase-treated pBluescript carrying the previously described enhancerless minilocus (24).
Tα1,2 with a 2-bp mutation in the TCF/LEF binding site (Tα1,2mTCF) was generated by PCR overlap extension (34) using Eα0.7 template DNA, mutagenic oligonucleotides TCF-1A (GGGAGAGCTTCTATGGGTGCCCTAC) and TCF-1B (GTAGGGCACCCATAGAAGCTCTCCC), along with the −40 and reverse primers. The final PCR product was digested with DraI and EcoRI, cloned into SmaI and EcoRI cut pBluescript KS+, and sequenced. The 125-bp Tα1,2mTCF fragment was then excised from the plasmid with BamHI, blunt ended with Klenow, and ligated into XbaI-digested, blunted, and phosphatase-treated pBluescript carrying the enhancerless minilocus.
The Tα1,2 fragment of Eα had been previously excised from the Eα0.7 plasmid using BamHI and DraI digestion, blunt ended with Klenow, and subcloned into EcoRV cut pBluescript KS+. To generate a minilocus containing wild-type Tα1,2, the insert was excised from this plasmid by digestion with HindIII and SmaI, blunt ended with Klenow, and cloned into XbaI-digested, blunted, and phosphatase-treated pBluescript carrying the enhancerless minilocus.
After confirmation of minilocus construct structures by dideoxynucleotide sequence analysis, minilocus DNA was purified as previously described (24) and microinjected into fertilized (C57BL/6 × SJL/J)F2 eggs by the Duke University Comprehensive Cancer Center Shared Transgenic Mouse Facility. Progeny tail DNA was initially characterized on Southern blots probed with radiolabeled Cδ and Vδ1 fragments. Transgene germline copy number was determined by analysis of Tα1,2, Tα1,2mEts, and Tα1,2mTCF tail DNAs, along with tail DNAs of previously identified single copy integrants, on slot blots (Schleicher & Schuell, Keene, NH). Blots were probed with a radiolabeled Cδ fragment and the resultant hybridization signals quantified using a Betascope (Betagen, Waltham, MA). Transgenes were maintained on a mixed C57BL/6 × SJL/J background.
PCR.
With the exception of experiments using sorted αβ and γδ cells depicted in Fig. 6, genomic DNA PCR templates were prepared from thymi of 4-wk-old animals by standard techniques (35). For single copy transgenic lines, 12 ng of genomic DNA was used as a template for PCR reactions. For multicopy integrants, the quantity of genomic DNA used in PCR was reduced to account for copy number and to insure that all PCR signals were in the linear range. PCR templates from sorted αβ and γδ thymocytes (as well as unsorted thymocytes from the same animals) were prepared by incubation of <1 × 106 pelleted cells in 200 μl lysis buffer (10 mM Tris, pH 8.4, 2.5 mM MgCl2, 50 mM KCl, 200 μg/ml gelatin, 0.45% NP40, 0.45% Tween-20, and 60 μg/ml proteinase K) for 1 h at 56°C and then 15 min at 95°C (36). Sample aliquots containing 5 × 103 cell equivalents were immediately used as templates in PCR reactions. All PCR reactions were performed identically using previously described reaction conditions and primers (24).
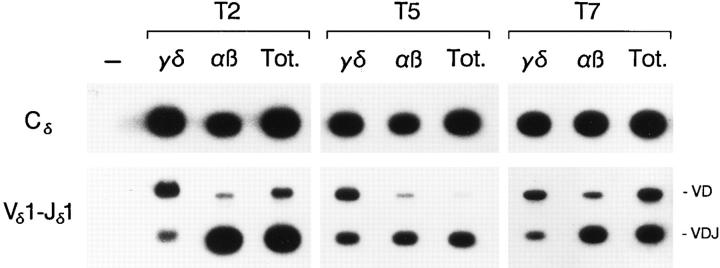
Figure 6.
Tα1,2 minilocus rearrangement in αβ and γδ thymocytes. Genomic DNA templates from sorted αβ and γδ thymocytes and total unfractionated thymocytes (Tot.) of Tα1,2 line T2, T5, and T7 mice (4 wk old), and a no DNA control (−) were amplified by PCR and probed as in Fig. 2.
Blot Hybridization of Genomic DNA and PCR Products.
Gel electrophoresis, blotting, and hybridization with 32P-labeled probes were performed as previously described (24). Hybridization signals were quantified using a Betascope and reported values for VD and VDJ rearrangement signals were normalized to the Cδ signal for each template.
Antibodies.
Biotinylated H57-597 anti-TCR-β, phycoerythrin-conjugated GL3 anti-TCR-γδ, FITC-streptavidin, and unlabeled 2.4G2 anti-FcγRII/III mAbs for flow cytometry were obtained from PharMingen (San Diego, CA). GK1.5 anti-CD4 (37) and 41-3.48 anti-Lyt-2.2 (38) mAbs were used for cell depletions as culture supernatants.
Flow Cytometric Analysis and Cell Sorting.
Enriched CD4−CD8− cells were prepared from single cell suspensions of thymocytes from 4-wk-old animals by treatment with saturating amounts of GK1.5 and 41-3.48 mAbs and rabbit complement (Cedarlane Laboratories, Ltd., Hornby, ON, Canada) for 60 min at 37°C. Viable cells were collected after centrifugation over Lympholyte-M (Cedarlane Labs. Ltd.). The enriched CD4−CD8− cells and unfractionated thymocytes from the same animal were incubated with saturating concentrations of unlabeled 2.4G2, biotinylated H57597, and phycoerythrin-GL3 for 40 min at 4°C in PBS with 0.1% BSA and 0.1% NaN3, washed twice, and then stained with FITC-Streptavidin for 20 min at 4°C. Cells were subsequently washed and sorted using a FACStar Plus® (Becton Dickinson, Mountain View, CA). H57-597+ αβ cells were sorted using stained unfractionated thymocytes as a starting population while GL3+ γδ cells were sorted using identically stained, enriched CD4−CD8− cells as a starting population. Immediate reanalysis of sorted populations by two-color flow cytometry revealed contamination with <1% of cells expressing the inappropriate cell surface TCR in all cases.
Fetal Thymus Samples.
Fetal mice were obtained from timed matings of homozygous transgenic males and 6–8-wk-old (C57BL/6 × SJL/J)F1 females (Jackson Laboratory, Bar Harbor, ME). The day of detecting a vaginal plug was designated as day 0.5 of embryonic development. Genomic DNA prepared by standard techniques from pooled fetal thymi of individual litters was analyzed for minilocus rearrangement by PCR.
Results
The 116-bp Core Eα Fragment Tα1,2 Is Sufficient to Activate Minilocus Rearrangement In Vivo.
The rearrangement substrate used in the present study has been previously described as a 22.5-kb human TCR-δ gene minilocus consisting of germline Vδ1, Vδ2, Dδ3, Jδ1, Jδ3, and Cδ gene segments (24). Frameshift mutations within the Vδ1 and Vδ2 coding segments prevent the rearranged transgene from encoding a functional TCR polypeptide that could alter normal T cell development in transgenic mice. The initial step of transgene rearrangement, V to D, is enhancer independent (24). The second step of transgene rearrangement, VD to J, is dependent on the presence of a functional enhancer in the Jδ3-Cδ intron. Thus, we infer that the enhancer is required to promote J segment accessibility to the recombinase (24). The 1.4-kb Eα has been shown to efficiently activate VD to J rearrangement in this system (25). To determine whether Tα1,2, the 116-bp core fragment of Eα, was also sufficient to activate minilocus VDJ recombination in vivo, we constructed a new TCR-δ gene minilocus containing this fragment in place of Eα (Fig. 1 A). Four independent transgenic lines denoted T2, T3, T5, and T7 were established and determined by slot blot analysis to carry 28, 1, 3, and 4 transgene copies, respectively.
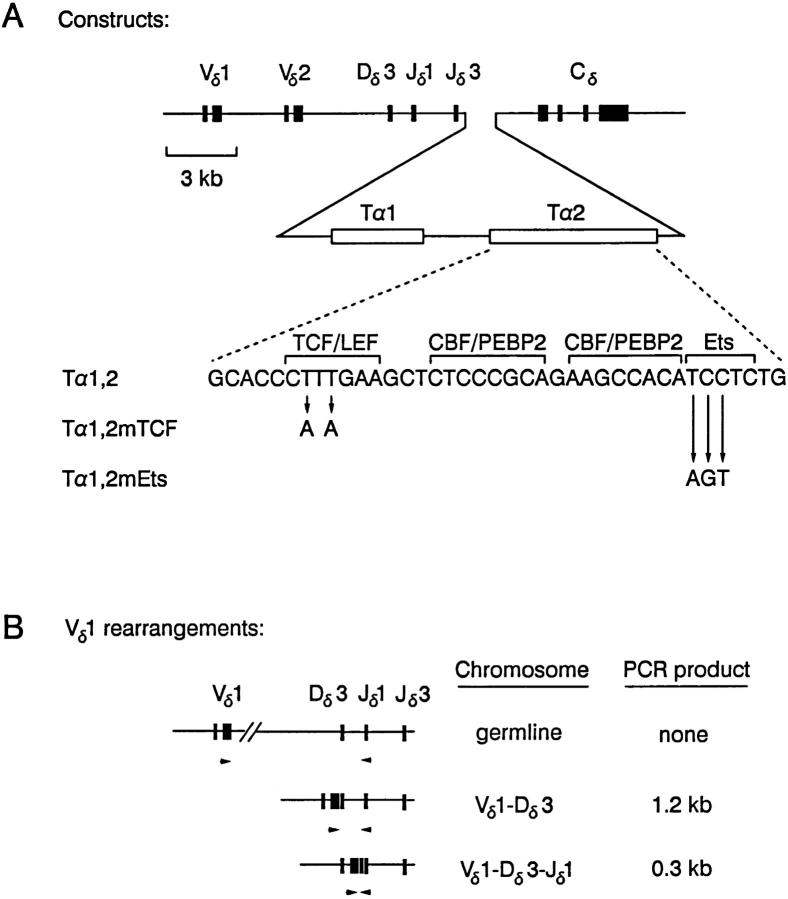
Figure 1.
Human TCR-δ gene minilocus. (A) Diagram of the three Tα1,2-containing minilocus constructs. Solid boxes, exons, open boxes, protein binding sites. Wild-type and mutant Tα2 sequences are shown. (B) PCR products generated from Vδ1 rearrangements are depicted along with the Vδ1 and Jδ1 primers (arrows) used. Similar products are generated using Vδ2 and Jδ1 primers. Specific PCR products are not generated from unrearranged templates because of the large distances between primers. The primers do not amplify products from the endogenous murine TCR-δ locus.
VDJ recombination in the four Tα1,2 transgenic lines was assessed by quantitative PCR of thymic genomic DNA templates that were amplified using primers specific for minilocus Vδ1, Vδ2, and Jδ1 gene segments (24). PCR using primer combinations Vδ1-Jδ1 or Vδ2-Jδ1 yields a 0.3-kb product resulting from transgene VDJ rearrangement and a 1.2-kb fragment resulting from VD rearrangement (Fig. 1 B), both of which can be detected on Southern blots probed with radiolabeled Vδ1- or Vδ2-specific DNA fragments. Amplification of a 0.3-kb rearrangement-independent product with a pair of Cδ primers serves as an internal control for PCR efficiency and allows quantitative comparison of rearrangement patterns between different templates. As seen in Fig. 2 and Table 1, low levels of Vδ1-Dδ3 and high levels of Vδ1-Dδ3-Jδ1 rearrangement were observed in three of four Tα1,2 lines (T2, T5, and T7). Levels of Vδ1Dδ3-Jδ1 rearrangement in thymocytes from lines T2 and T5 were 60 and 38%, respectively, of that found in thymocytes from Eα line J, which includes the intact 1.4-kb Eα (Fig. 3, Table 2). However, VD and VDJ rearranged products were barely detectable in Tα1,2 line T3 (Fig. 2, Table 1). Similar variability in rearrangement phenotype among different lines of animals bearing an identical construct has been noted in our previous studies (24, 39) and is likely due to inherent differences in transgene integration sites. We suggest that the minilocus is integrated into a relatively inactive region of chromatin in line T3; in support of this notion, preliminary in vivo footprinting studies have demonstrated markedly diminished protection of protein binding sites within Tα1,2 in thymus DNA of T3 mice as compared with thymus DNA of T2, T5, or T7 mice (HernandezMunain, C., personal communication). Taken as a whole, our results demonstrate that the 116-bp Tα1,2 fragment of Eα is, in most contexts, sufficient to mediate activation of the enhancer-dependent VD to J step of minilocus rearrangement. The apparently less efficient conversion of Vδ2Dδ3 to Vδ2-Dδ3-Jδ1 in these lines is probably related to our previous observation that Vδ2 rearrangement is only ∼10% as efficient as Vδ1 rearrangement in this system (24).
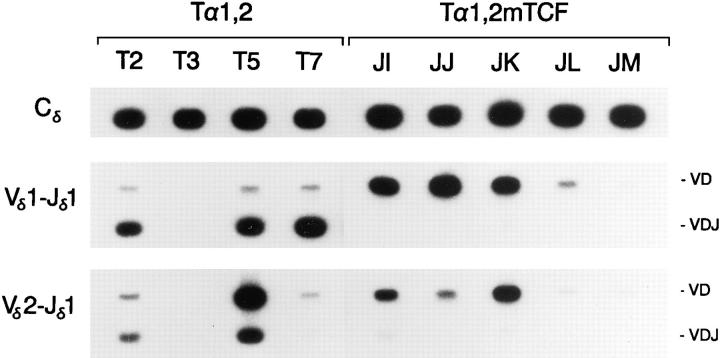
Figure 2.
PCR analysis of Tα1,2 and Tα1,2mTCF minilocus rearrangement. Genomic DNA templates from unfractionated thymocytes of Tα1,2 mice from lines T2, T3, T5, and T7, and of Tα1,2mTCF mice from lines JI, JJ, JK, JL, and JM (all 4 wk old) were amplifed by PCR using the indicated primers. Southern blots were probed with radiolabeled Cδ, Vδ1, or Vδ2 DNA fragments. The positions of 1.2-kb VD and 0.3-kb VDJ rearrangement products are indicated.
Table 1.
Minilocus Rearrangement in Thymocytes of Tα1,2 and Tα1,2mTCF Mice
| Tα1,2 | Tα1,2mTCF | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2 | T3 | T5 | T7 | JI | JJ | JK | JL | JM | ||||||||||
| Vδ1-Dδ3 | 0.16 | 0.01 | 0.15 | 0.20 | 0.97 | 2.77 | 1.17 | 0.15 | 0.03 | |||||||||
| Vδ1-Dδ3-Jδ1 | 1.00 | 0.03 | 0.88 | 1.88 | 0.03 | 0.07 | 0.05 | nd | nd | |||||||||
Blot hybridization signals from the experiment shown in Fig. 2 were quantified using a Betascope. Reported values are normalized to the Cδ signal for each sample. nd, not detected.
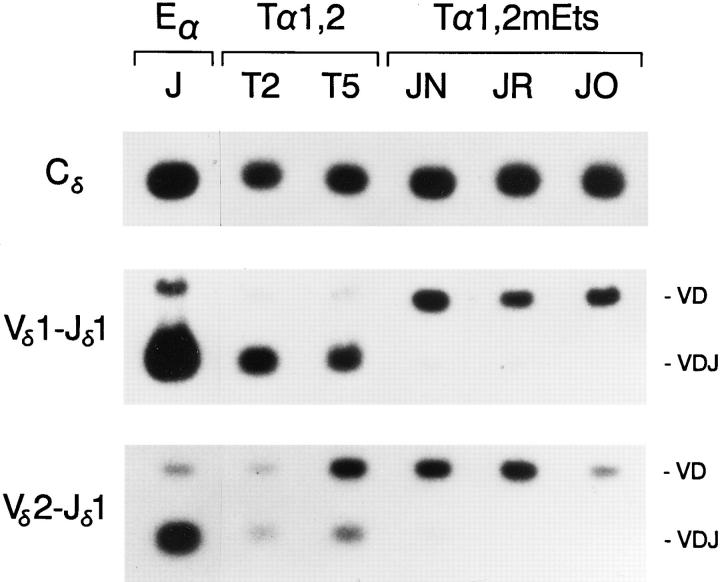
Figure 3.
PCR analysis of Eα, Tα1,2, and Tα1,2mEts minilocus rearrangement. Genomic DNA templates from unfractionated thymocytes of an Eα mouse from line J, of Tα1,2 mice from lines T2 and T5, and of Tα1,2mEts mice from lines JN, JR, and JO (all 4 wk old) were amplified by PCR and probed as in Fig. 2.
Table 2.
Minilocus Rearrangement in Thymocytes of Eα, Tα1,2, and Tα1,2mEts Mice
| Eα | Tα1,2 | Tα1,2mEts | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| J | T2 | T5 | JN | JR | JO | |||||||
| Vδ1-Dδ3 | 0.30 | 0.15 | 0.19 | 1.09 | 0.77 | 0.99 | ||||||
| Vδ1-Dδ3-Jδ1 | 4.52 | 2.67 | 1.72 | 0.14 | 0.19 | 0.11 | ||||||
Blot hybridization signals from the experiment shown in Fig. 3 were quantified using a Betascope. Reported values are normalized to the Cδ signal for each sample.
Tα1,2 and Eα minilocus rearrangement was also analyzed directly by genomic Southern blot (Fig. 4) as an independent means of corroborating results obtained by quantitative PCR. Analysis of Vδ1 rearrangements in PstI plus EcoRI-digested thymus DNA from Eα line L revealed low levels of 1.0-kb germline Vδ1 and 0.9-kb Vδ1-Dδ3 rearranged fragments, and higher levels of a 1.7-kb species resulting from Vδ1-Dδ3-Jδ1 rearrangement (Fig. 4). Similar results were obtained with Eα line J (data not shown). Analysis of similarly digested thymocyte DNA from Tα1,2 line T2 also revealed levels of the fully rearranged Vδ1Dδ3-Jδ1 fragment that were more prevalent than the 0.9-kb partially rearranged (Vδ1-Dδ3) species (Fig. 4). Hence, these results are consistent with those obtained by PCR and provide confirmation that Tα1,2 alone can efficiently activate minilocus VDJ recombination.
Figure 4.
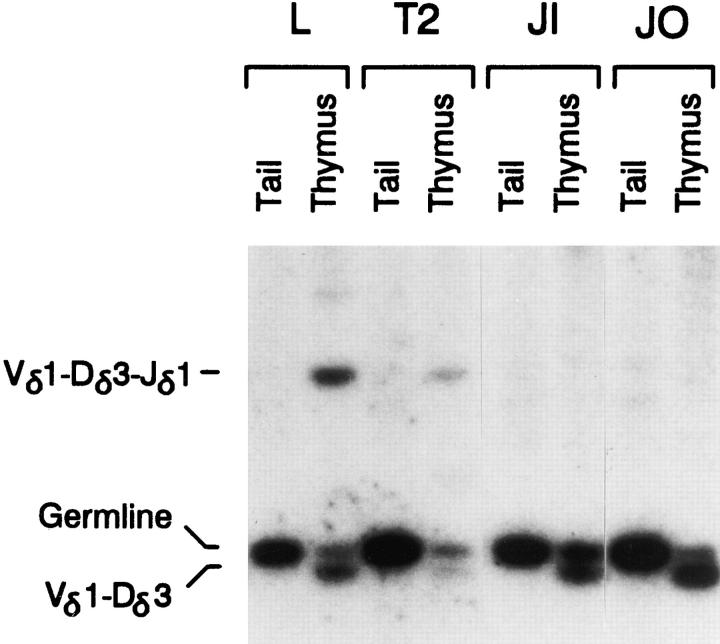
Analysis of minilocus rearrangement by genomic Southern blot. PstI plus EcoRI-digested tail and thymus genomic DNA samples from Eα line L, Tα1,2 line T2, Tα1,2mTCF line JI, and Tα1,2mEts line JO mice (all 4 wk old) were analyzed by Southern blot using a radiolabeled 1.0-kb Vδ1 genomic PstI fragment. Positions of the expected 1.0-kb germline, 0.9-kb Vδ1-Dδ3, and 3.2-kb Vδ1-Dδ3-Jδ1 rearranged fragments are indicated.
The conspicuous variations in total Vδ1 hybridization signal intensities seen in this experiment (Fig. 4) result in part from differences in germline transgene copy number between the various transgenic lines (T2 = 28, L = 13). A diminution of total Vδ1 signal intensity in thymus DNA relative to tail DNA isolated from the same animal is also apparent, particularly in line T2 (Fig. 4). Slot blot quantification revealed Cδ copy number to be 13 in line L tail and 8 in L thymus, and although 28 in T2 tail, only 4 in T2 thymus (data not shown). Such copy number loss has been noted in some other multicopy lines as well, and is likely due to thymocyte rearrangement events between V, D, and J gene segments within different, presumably concatameric, copies of the minilocus, with resultant loss of intervening sequences. These differences in thymus transgene copy number are not apparent in PCR experiments where thymus template amounts were adjusted for Cδ copy number.
An Intact TCF/LEF Binding Site Is Required for Efficient Activation of Minilocus VD to J Rearrangement by Tα1,2.
To determine whether the activation of minilocus VDJ recombination by Tα1,2 in vivo is dependent on TCF/LEF family transcription factors, a variant of the Tα1,2 minilocus containing a 2-bp mutation in the TCF-1/LEF-1 binding site (Tα1,2 mTCF) was constructed (Fig. 1 A). Five independent lines of Tα1,2mTCF transgenic mice were generated. Slot blot analysis revealed that Tα1,2mTCF lines JL and JM each carry one copy of the minilocus, whereas lines JI, JJ, and JK carry 5, 12, and 9 copies, respectively (data not shown). Analysis of Tα1,2mTCF minilocus VDJ recombination was performed by quantitative PCR, as described above. Notably, as assessed using both Vδ1-Jδ1 and Vδ2-Jδ1 primer combinations, all five Tα1,2mTCF lines displayed dramatically reduced levels of VDJ rearranged products as compared with wild-type Tα1,2 lines T2, T5, and T7 (Fig. 2). Quantification revealed levels of Vδ1-Dδ3Jδ1 rearrangement in Tα1,2mTCF lines JI, JJ, and JK that were only 3, 7, and 5%, respectively, of that observed in wild-type Tα1,2 line T2 (Table 1). In lines JL and JM, Vδ1-Dδ3-Jδ1 rearrangement was undetectable. Because V to D rearrangement was readily detectable in most transgenic lines, these results argue that mutation of the TCF/ LEF site significantly and specifically impairs the VD to J step of minilocus VDJ recombination. Verification of these results was obtained by genomic Southern blot analysis which revealed an abundance of germline Vδ1 and Vδ1Dδ3 rearranged fragments, but no detectable Vδ1-Dδ3-Jδ1 rearrangement in PstI plus EcoRI digested thymus DNA from Tα1,2mTCF line JI (Fig. 4).
The relatively low level of V to D rearrangement in Tα1,2mTCF lines JL and JM may, in part, be a reflection of unique properties of the transgene integration sites. However, it is interesting that all three transgenic lines examined in this study displaying very low levels of total rearrangement (Tα1,2 line T3, and Tα1,2mTCF lines JL and JM) are all single copy. This suggests an effect of copy number as well. Nevertheless, multicopy integrants are not required for efficient transgene VDJ recombination, as evidenced by the previously characterized Eδ-containing lines A, B, and C (24), and the Tα1,2mEts line JN (see below). It may be that while a single copy transgene experiences the full negative impact of integration into a relatively inactive region of chromatin, internal copies of a concatameric multicopy integrant might be relatively protected from such effects.
An Intact Ets Binding Site Is also Necessary for Efficient Activation of Minilocus VD to J Rearrangement By Tα1,2.
To determine whether the activation of minilocus VDJ recombination by Tα1,2 in vivo is dependent on Ets family transcription factors, a second variant of the Tα1,2 minilocus containing a 3-bp mutation in the Tα2 Ets binding site (Tα1,2mEts) was constructed. The three independent lines of transgenic mice generated, JN, JO, and JR, were found to carry 1, 13, and 21 copies of the minilocus, respectively, as assessed by slot blot analysis. VDJ rearrangement in thymocytes from Tα1,2mEts animals was compared with that of wild-type Tα1,2 and Eα mice by quantitative PCR. This analysis revealed that the VD to J step of transgene rearrangement was dramatically curtailed in all three lines of Tα1,2mEts transgenic animals (Fig. 3). Specifically, the levels of Vδ1- Dδ3-Jδ1 rearrangement in Tα1,2mEts lines JN, JR, and JO were only 5, 7, and 4% of that of wild-type Tα1,2 line T2 (Table 2); Vδ2-Dδ3-Jδ1 rearrangement was only barely detectable (Fig. 3). Nevertheless, VD rearrangement was high in all three lines (Fig. 3). These results were also corroborated by genomic Southern blot of digested thymus DNA from Tα1,2mEts line JO, which failed to reveal a discernible Vδ1-Dδ3-Jδ1 fragment despite readily detectable Vδ1Dδ3 rearrangement (Fig. 4). From these data we conclude that the presence of an intact Ets binding site within Tα1,2 is a prerequisite for efficient activation of the VD to J step of minilocus rearrangement in vivo.
Temporal and Lineage-specific Control of Minilocus Rearrangement By Tα1,2 Is Distinct from that of Eα.
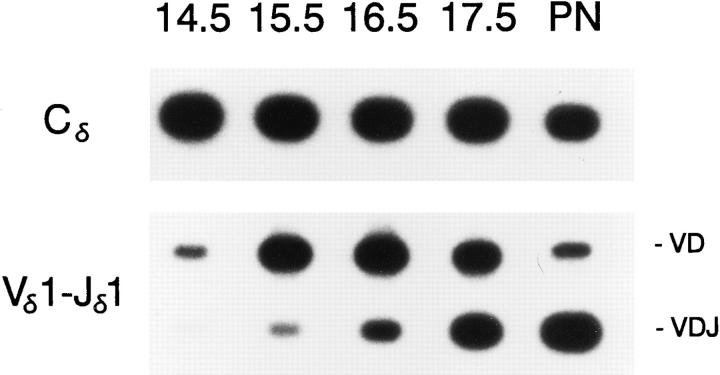
The 1.4-kb Eα has been previously shown to confer physiologically appropriate developmental control to the enhancer-dependent VD to J step of minilocus rearrangement. Because the 116-bp Tα1,2 core enhancer fragment proved sufficient to activate minilocus rearrangement, we asked whether the core enhancer fragment is sufficient to impart precise developmental control as well. Accordingly, we examined the timing of minilocus VDJ rearrangement during ontogeny in Tα1,2 transgenic animals. PCR analysis of fetal thymus genomic DNA templates from line T2 timed pregnancies revealed that the VD to J step of minilocus rearrangement began on fetal day 15.5 (Fig. 5), one day earlier than that previously noted in Eα line J (25). Identical results were obtained from analysis of Tα1,2 line T5 (data not shown). Thus, Tα1,2 appeared to be activated slightly earlier during fetal thymic ontogeny than the intact Eα.
Figure 5.
Time course of Tα1,2 minilocus rearrangement during fetal ontogeny. Genomic DNA samples from Tα1,2 line T2 thymi isolated on days 14.5–17.5 of gestation, as well as from a postnatal (PN) T2 mouse (4 wk old), were amplified by PCR and probed as in Fig. 2.
We then compared minilocus VDJ recombination in sorted αβ and γδ T cell populations from adult Tα1,2 animals. As expected, PCR analyses revealed abundant VD and VDJ rearrangement in lines T2, T5, and T7 αβ thymocytes (Fig. 6). Strikingly, however, γδ thymocytes from these animals also exhibited substantial VDJ rearrangement (Fig. 6). Quantification of these data revealed that the level of Vδ1-Dδ3-Jδ1 rearrangement in γδ cells relative to αβ cells was 8% in line T2, 65% in line T5, and 19% in line T7 (Table 3). These results cannot be explained by contamination of cell populations, since flow cytometric reanalysis demonstrated that <1% of the cells in the sorted populations bore the inappropriate TCR. The results from all three lines contrast dramatically with previous observations in Eα mice; the level of Vδ1-Dδ3-Jδ1 rearrangement in γδ cells was negligible (2.5, 0.0, and 1.3% of the signal in αβ thymocytes in Eα lines J, L, and M, respectively, levels that are probably within the limits of purity of the sorted γδ populations [25]). From these results, we conclude that truncation of Eα results in partial dysregulation of VDJ recombination during development. Specifically, the slightly premature activation of VDJ recombination with relaxed lineage control that is directed by Tα1,2 suggests that Eα elements that lie outside of Tα1,2 are critical for the tightly regulated and physiologically appropriate activation of TCR-α gene rearrangement in vivo.
Table 3.
Minilocus Rearrangement in αβ and γδ Thymocytes of Tα1,2 Animals
| T2 | T5 | T7 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| γδ | αβ | γδ | αβ | γδ | αβ | |||||||
| Vδ1-Dδ3 | 1.67 | 0.34 | 2.04 | 0.30 | 1.21 | 0.45 | ||||||
| Vδ1-Dδ3-Jδ1 | 0.63 | 7.77 | 1.32 | 2.07 | 0.54 | 2.90 | ||||||
Blot hybridization signals from the experiment shown in Fig. 6 were quantified using a Betascope. Reported values are normalized to the Cδ signal for each sample. Because quantification of T2, T5, and T7 samples was performed on separate blots probed at different times, the values for VD/C and VDJ/C are useful for comparison of the γδ and αβ samples within a line, but are not useful for comparisons between different lines.
Discussion
In this study, we examined the roles of cis-acting elements of Eα in the developmental regulation of VDJ recombination at the TCR-α/δ locus. We found that the 116-bp Tα1,2 core enhancer fragment of the 1.4-kb Eα is sufficient to activate the enhancer-dependent VD to J step of transgenic minilocus rearrangement, and that intact TCF/LEF and Ets binding sites within Tα1,2 are required. Investigation of the temporal and lineage-specific control of VDJ recombination afforded by Tα1,2 revealed that thymocyte VD to J rearrangement begins on fetal day 15.5 and occurs in both αβ and γδ cells. This contrasts with previous results obtained in transgenic lines carrying the 1.4-kb Eα, in which VD to J rearrangement was found to begin on fetal day 16.5 and to be limited to αβ cells (25). Taken together, these data indicate that the core fragment of Eα can establish accessibility to the recombinase in developing thymocytes in vivo in a fashion that is dependent on the binding of TCF/LEF and Ets family transcription factors, but that these and other factors that bind to the Eα core cannot account for the precise developmental onset of accessibility that is provided by the intact Eα. Rather, our data suggests a critical role for factors that bind Eα outside of the core Tα1,2 region in establishing the precise developmental onset of TCR-α rearrangement in vivo.
Previous studies identified the 116-bp Tα1,2 as the core fragment of Eα on the basis of its ability to potently activate plasmid reporter gene expression in transient transfection experiments, and its ability, as naked DNA, to support the assembly of a stable multiprotein complex consisting of ATF/CREB, LEF-1, Ets-1, and CBF/PEBP2 (26–28). Our experiments are the first to test the core fragment of Eα in a chromosomally integrated context. Our data indicates that this fragment is capable of modifying chromatin structure in vivo over a distance of at least 2 kb, as measured by its ability to modify the accessibility of the Jδ1 gene segment to the VDJ recombinase. Furthermore, our data are the first to provide evidence that TCF/LEF and Ets family transcription factors are important regulators of Eα in a chromosomal environment, suggesting that the multiprotein complex that was previously documented to assemble in vitro (28) may have an important role in regulating chromatin structure in vivo.
TCF-1 and LEF-1 are members of the HMG box family of transcription factors that bind at the same site within Tα1,2 (Fig. 1 A) (29–31). The roles of both of these factors have been analyzed in vivo by targeted gene disruption. Analysis of LEF-1−/− mice has revealed that, despite early postnatal lethality due to impaired development of multiple organs, TCR-α rearrangement and T cell development proceed normally (40). TCF-1−/− animals, on the other hand, are healthy and appear morphologically normal, but exhibit reduced numbers of apparently normal peripheral T cells and a significant impairment in thymocyte differentiation at the transition from the ISP to DP stages of development (41). TCR-β rearrangement appears to be unaffected in these animals (41). However, since thymocyte TCR rearrangement and expression normally begins around the ISP to DP transition that is inhibited by TCF-1 gene disruption, it is difficult to judge whether TCR-α rearrangement is inhibited, albeit incompletely, as a primary consequence of the mutation.
Our experiments, which tested the effects of a disrupted TCF-1/LEF-1 binding site within Tα1,2 in a phenotypically neutral recombination reporter construct, clearly implicate TCF-1, LEF-1, or related factors in the developmental activation of minilocus VD to J rearrangement, and by implication, in the developmental activation of Vα to Jα rearrangement at the endogenous TCR-α/δ locus. There are several reasons why our results may appear to contrast with those obtained by targeted gene disruption. First, our results may be easier to interpret unambiguously because there is no confounding effect on T cell development clouding the interpretation of perturbed transgene rearrangement. Second, our experimental approach, in which the binding site is mutated, accounts for the possibility that TCF-1 and LEF-1 might play important but redundant roles in regulating TCR-α gene rearrangement. Such redundancy might allow apparently normal TCR-α rearrangement and expression in gene targeted animals that lack either TCF-1 or LEF-1, but not both. Third, our experimental approach would detect an effect even if a related HMG family member, rather than TCF-1 or LEF-1, were the critical regulator of TCR-α gene rearrangement in vivo. Finally, our test of the TCF/LEF binding site mutation in the context of the Tα1,2 core enhancer, rather than the 1.4-kb Eα, may represent a more sensitive measure of the effect of protein binding to this site, as it eliminates the contribution of potentially redundant cis-acting Eα elements that lie outside of Tα1,2. Such elements might mask an effect in a TCF-1 or LEF-1 knockout, or in mutated versions of an otherwise intact Eα. Indeed, in transient transfection experiments, the DraI-ApaI Eα fragment containing the Tα3 and Tα4 nuclear protein–binding sites can partially compensate for deleterious mutations in Tα1,2 (27). The nature of our experiment does not allow us to conclude that TCF/LEF family members are strictly required for the activation of TCR-α gene rearrangement within the endogenous TCR-α/δ locus in vivo. We can reasonably conclude, however, that these factors are likely to contribute to the developmental activation of TCR-α gene rearrangement in vivo.
Ets transcription factors constitute a large family of DNA binding proteins, among which, Ets-1, Ets-2, GABPα, Elf-1, Fli-1, and Spi-B are all expressed in T cells (42, 43). Ets-1 in particular has been thought to be a regulator of Tα1,2 on the basis of its ability to transactivate gene expression in transient transfection experiments, and its ability to interact with ATF/CREB and CBF/PEBP2 to form a stable multiprotein complex on Tα1,2 in vitro (28, 33). Ets-1 is preferentially expressed in resting lymphocytes of adult mice, and its expression in fetal and postnatal thymocytes roughly parallels that of TCR-α (44, 45). Nevertheless, although gene disruption experiments have revealed diminished numbers of mature thymocytes and peripheral T cells with impaired activation and survival characteristics in Ets-1−/− animals, TCR expression appears to be normal (46, 47).
Our results, which clearly establish that an intact binding site for Ets family members is required for efficient activation of minilocus VD to J rearrangement by Tα1,2 in vivo, might imply that another Ets family member is the crucial regulator of Eα or compensates for the loss of Ets-1 in Ets-1−/− mice. The only other Ets-related transcription factor that is expressed in the T lineage and whose role has been examined genetically is Fli-1. Mice expressing an altered Fli-1 allele display reduced numbers of all thymocyte subsets; however, TCR-α rearrangement and expression was not specifically examined (48). Fli-1 has been shown to bind to and transactivate gene expression via Tα1,2 in transient transfection experiments. However, the magnitude of transactivation was substantially less than that observed using Ets-1, and unlike Ets-1, Fli-1 did not interact with ATF/ CREB proteins (28). Elf-1, on the other hand, has a distinct binding specificity and does not stably interact with Tα1,2 (49). Thus, the physiologically relevant factor that interacts with the Tα1,2 Ets site in vivo remains uncertain. Finally, as noted above for the TCF/LEF binding site mutation, our test of the Ets site mutation in the context of the Tα1,2 rather than the 1.4-kb Eα might, due to redundancy of cis-acting elements, detect an effect that would not be readily detected in the Ets-1 knockout or in mutated versions of the 1.4-kb Eα. While we cannot conclude that the binding of Ets family members is absolutely required for the activation of TCR-α gene rearrangement within the endogenous TCR-α/δ locus, our data does implicate these factors as potentially important contributors to the developmental activation of TCR-α gene rearrangement in vivo.
Although our data demonstrates quite clearly that the accessibility required for the activation of VDJ recombination can be established by the binding of TCF/LEF family, Ets family, and presumbably other transcription factors to the core of Eα, our data also indicates that the binding of these factors cannot account for the precise developmental onset of accessibility that is provided by the intact Eα. Rather, we detect both a partial loss of lineage specificity and a partial loss of temporal or developmental stage specificity in Tα1,2 transgenic lines. We propose that the loss of lineage specificity is a direct consequence of the loss of temporal or developmental stage specificity, as follows. Our observation that an intact Eα activates the VD to J step of minilocus rearrangement exclusively in developing αβ T cells implies that the transgenic Eα is developmentally activated either coordinately with, or subsequent to, activation of the endogenous Eα, which, by inducing endogenous Vα to Jα rearrangement, commits developing thymocytes to become αβ cells. On the other hand, the relaxed lineage specificity of Tα1,2 implies that in at least a fraction of thymocytes, Tα1,2 is activated before the endogenous Eα. This would result in the activation of at least some minilocus VD to J rearrangement in as yet uncommitted thymic precursors, some of which would give rise to γδ cells. The uncommitted thymocyte population in which Tα1,2-dependent minilocus VD to J rearrangement occurs is a matter of speculation, but should be positive for RAG-1 and RAG-2. One candidate would be the CD44lowCD25+ subset of DN cells, which expresses high levels of RAG-1 and RAG-2, and is actively rearranging endogenous TCR-β, -γ, and -δ genes (7, 14). However, CD44lowCD25− DN and ISP thymocytes may also be candidates if their reduced levels of RAG-1 and RAG-2 maintain permissiveness for at least low level VDJ recombination (7, 13).
The potential for premature activation of Eα directed by factors that interact with Tα1,2 is apparently normally held in check by additional cis-elements of Eα. Although the identities of the cis-elements that mediate this effect are uncertain at present, we speculate that they might map to the previously defined protein binding sites Tα3 and Tα4 (26, 27). Although little is known about protein binding to these sites, it is interesting that they contain two E box motifs. Restriction of the activity of the immunoglobulin heavy-chain enhancer to B cells has been shown to be due, at least in part, to negative regulation involving two E boxes, μE5 and μE4, present within the enhancer (50–52). In addition, an E box has been implicated in the negative regulation of CD4 gene expression during T cell development by the CD4 transcriptional silencer (53). It is therefore tempting to speculate that the Tα3 and Tα4 E boxes may play similar roles in the developmental activation of Eα, and, as a consequence, the developmental activation of Vα to Jα rearrangement. We are currently investigating this possibility.
Acknowledgments
We thank Drs. C. Hernandez-Munain and Y. Zhuang for their critical comments on the manuscript, and Cheryl Bock and Wendy Callahan of the Duke University Comprehensive Cancer Center Transgenic Mouse Shared Resource for the production of transgenic mice.
Footnotes
This work was supported by National Institutes of Health grant GM41052. M.S. Krangel is the recipient of American Cancer Society Faculty Research Award FRA-414. J.L. Roberts was supported in part by Public Health Service Training grant CA09058.
1 Abbreviations used in this paper: DN, double negative; DP, double positive; Eα, α enhancer; Eδ, δ enhancer; HMG, high mobility group; ISP, immature single positive.
References
- 1.Fowlkes BJ, Pardoll DM. Molecular and cellular events of T cell development. Adv Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg EV. The development of functionally responsive T cells. Adv Immunol. 1992;51:85–214. doi: 10.1016/s0065-2776(08)60487-3. [DOI] [PubMed] [Google Scholar]
- 3.Kisielow P, von Boehmer H. Development and selection of T cells: facts and puzzles. Adv Immunol. 1995;58:87–209. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Scollay R, Egerton M, Pearse M, Spangrude GJ, Shortman K. CD4 expressed on earliest T-lineage precursor cells in the adult murine thymus. Nature (Lond) 1991;349:71–74. doi: 10.1038/349071a0. [DOI] [PubMed] [Google Scholar]
- 5.Pearse M, Wu L, Egerton M, Wilson A, Shortman K, Scollay R. A murine early thymocyte developmental sequence is marked by transient expression of the interleukin 2 receptor. Proc Natl Acad Sci USA. 1989;86:1614–1618. doi: 10.1073/pnas.86.5.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8−triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 7.Wilson A, Held W, MacDonald HR. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179:1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godfrey DI, Kennedy J, Mombaerts P, Tonegawa S, Zlotknik A. Onset of TCR-β gene rearrangement and role of TCR-β expression during CD3−CD4−CD8−thymocyte differentiation. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 9.Groettrup M, Ungeweiss K, Azogui O, Palacios R, Owen MJ, Hayday AC, von Boehmer H. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor β chain and a 33 kd glycoprotein. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- 10.Saint-Ruf C, Ungeweiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science (Wash DC) 1994;266:1208–1212. doi: 10.1126/science.7973703. [DOI] [PubMed] [Google Scholar]
- 11.Dudley EC, Petrie HT, Shah LM, Owen MJ, Hayday AC. T cell receptor β chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1994;1:83–93. doi: 10.1016/1074-7613(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 12.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature (Lond) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman ES, Passoni L, Crompton T, Leu TMJ, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 14.Wilson A, de Villartay J-P, MacDonald HR. T cell δ receptor gene rearrangement and T early α (TEA) expression in immature αβ lineage thymocytes: implications for αβ/γδ lineage commitment. Immunity. 1996;4:37–45. doi: 10.1016/s1074-7613(00)80296-4. [DOI] [PubMed] [Google Scholar]
- 15.Chien Y-H, Iwashima M, Kaplan KB, Elliot JF, Davis MM. A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. Nature (Lond) 1987;327:677–682. doi: 10.1038/327677a0. [DOI] [PubMed] [Google Scholar]
- 16.Hockett RD, de Villartay J-P, Pollock K, Poplack DG, Cohen DI, Korsmeyer SJ. Human T-cell antigen receptor (TCR) δ-chain locus and elements responsible for its deletion are within the TCR α-chain locus. Proc Natl Acad Sci USA. 1988;85:9694–9698. doi: 10.1073/pnas.85.24.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satyanarayana K, Hata S, Devlin P, Roncarolo MG, de Vries JE, Spits H, Strominger JL, Krangel MS. Genomic organization of the human T-cell antigen-receptor α/δ locus. Proc Natl Acad Sci USA. 1988;85:8166–8170. doi: 10.1073/pnas.85.21.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley EC, Girardi M, Owen MJ, Hayday AC. αβ and γδ T cells can share a late common precursor. Curr Biol. 1995;5:659–669. doi: 10.1016/s0960-9822(95)00131-x. [DOI] [PubMed] [Google Scholar]
- 19.Livak F, Petrie HT, Crispe IN, Schatz DG. In frame TCR δ gene rearrangements play a critical role in the αβ/γδ T cell lineage decision. Immunity. 1995;2:617–627. doi: 10.1016/1074-7613(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima PB, Menetski JP, Roth DB, Gellert M, Bosma MJ. V-D-J rearrangements at the T cell receptor δ locus in mouse thymocytes of the αβ lineage. Immunity. 1995;3:609–621. doi: 10.1016/1074-7613(95)90132-9. [DOI] [PubMed] [Google Scholar]
- 21.Petrie HT, Scollay R, Shortman K. Commitment to the T cell receptor-αβ or -γδ lineages can occur just prior to the onset of CD4 and CD8 expression among immature thymocytes. Eur J Immunol. 1992;22:2185–2188. doi: 10.1002/eji.1830220836. [DOI] [PubMed] [Google Scholar]
- 22.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 23.Sleckman BP, Alt FW, Gorman JR. Accessibility control of antigen receptor variable region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 24.Lauzurica P, Krangel MS. Enhancer-dependent and -independent steps in the rearrangement of a human T cell receptor δ transgene. J Exp Med. 1994;179:43–55. doi: 10.1084/jem.179.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauzurica P, Krangel MS. Temporal and lineage-specific control of T cell receptor α/δ gene rearrangement by T cell receptor α and δ enhancers. J Exp Med. 1994;179:1913–1921. doi: 10.1084/jem.179.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho I-C, Yang L-H, Morle G, Leiden JM. A T-cell–specific transcriptional enhancer element 3′ of Cαin the human T-cell receptor α locus. Proc Natl Acad Sci USA. 1989;86:6714–6718. doi: 10.1073/pnas.86.17.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho I-C, Leiden JM. Regulation of the human T-cell receptor α gene enhancer: multiple ubiquitous and T-cell–specific nuclear proteins interact with four hypomethylated enhancer elements. Mol Cell Biol. 1990;10:4720–4727. doi: 10.1128/mcb.10.9.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1–induced DNA bending and multiple protein–protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 29.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 30.Van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO (Eur Mol Biol Organ) J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the T cell receptor Cαenhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 32.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 33.Ho I-C, Bhat NK, Gottschalk LR, Lindsten T, Thompson CB, Papas TS, Leiden JM. Sequencespecific binding of human Ets-1 to the T cell receptor α gene enhancer. Science (Wash DC) 1990;250:814–818. doi: 10.1126/science.2237431. [DOI] [PubMed] [Google Scholar]
- 34.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene (Amst) 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 35.Wigler M, Sweet R, Sim GK, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 36.Schlissel MS, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 37.Dialynas DP, Wilde DB, Marrack P, Pierres A, Wall KA, Havran W, Otten G, Loken MR, Pierres M, Kappler J, Fitch FW. Characterization of the murine antigenic determinant, designated L3T4a, recognized by monoclonal antibody GK1.5: Expression of L3T4a by functional T cell clones appears to correlate primarily with class II MHC antigen-reactivity. Immunol Rev. 1983;74:29–56. doi: 10.1111/j.1600-065x.1983.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 38.Shen, F.-W. 1981. Monoclonal antibodies to mouse lymphocyte differentiation alloantigens. In Monoclonal Antibodies and T-cell Hybridomas. Perspectives and Technical Advances. G.J. Hammerling, U. Hammerling, and J.F. Kearney, editors. Elsevier/North Holland Biomedical Press, Amsterdam. 25–31.
- 39.Hernandez-Munain C, Lauzurica P, Krangel MS. Regulation of T cell receptor δ gene rearrangement by c-Myb. J Exp Med. 1996;183:289–293. doi: 10.1084/jem.183.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Genderen C, Okamura RM, Farinas I, Quo R-G, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1–deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 41.Verbeek, S., D. Izon, F. Hofhuis, E. Robanus-Maandag, H. te Riele, M. Van de Wetering, M. Oosterwegel, A. Wilson, H.R. MacDonald, and H. Clevers. 1995. An HMG-box– containing T-cell factor required for thymocyte differentiation. Nature (Lond.). 374:70–74. [DOI] [PubMed]
- 42.Clevers H, Oosterwegel M, Georgopoulos K. Transcription factors in early T-cell development. Immunol Today. 1993;14:591–596. doi: 10.1016/0167-5699(93)90198-T. [DOI] [PubMed] [Google Scholar]
- 43.Su GH, Ip HS, Cobb BS, Lu M-M, Chen H-M, Simon MC. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J Exp Med. 1996;184:203–214. doi: 10.1084/jem.184.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhat NK, Komschlies KL, Fujiwara S, Fisher RJ, Mathieson BJ, Gregorio TA, Young HA, Kasik JW, Ozato K, Papas TS. Expression of ets genes in mouse thymocyte subsets and T cells. J Immunol. 1989;142:672–678. [PubMed] [Google Scholar]
- 45.Kola I, Brookes S, Green AR, Garber R, Tymms M, Papas TS, Seth A. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc Natl Acad Sci USA. 1993;90:7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bories J-C, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature (Lond) 1995;377:635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 47.Musuthamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature (Lond) 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 48.Melet F, Motro B, Rossi DJ, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene-targeting leads to a defect in thymus development and a delay in Friend Virus–induced erythroleukemia. Mol Cell Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C-Y, Petryniak B, Ho I-C, Thompson CB, Leiden JM. Evolutionarily conserved Ets family members display distinct binding specificities. J Exp Med. 1992;175:1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen L, Lieberman S, Eckhardt LA. The octamer/μE4 region of the immunoglobulin heavy-chain enhancer mediates gene repression in myeloma × T-lymphoma hybrids. Mol Cell Biol. 1993;13:3530–3540. doi: 10.1128/mcb.13.6.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruezinsky D, Beckmann H, Kadesch T. Modulation of the IgH enhancer's cell type specificity through a genetic switch. Genes Dev. 1991;5:29–37. doi: 10.1101/gad.5.1.29. [DOI] [PubMed] [Google Scholar]
- 52.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box–binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duncan DD, Adlam M, Siu G. Asymmetric redundancy in CD4 silencer function. Immunity. 1996;4:301–311. doi: 10.1016/s1074-7613(00)80438-0. [DOI] [PubMed] [Google Scholar]