Abstract
Recombinant HLA-A2, HLA-B8, or HLA-B53 heavy chain produced in Escherichia coli was combined with recombinant β2-microglobulin (β2m) and a pool of randomly synthesised nonamer peptides. This mixture was allowed to refold to form stable major histocompatability complex (MHC) class I complexes, which were then purified by gel filtration chromatography. The peptides bound to the MHC class I molecules were subsequently eluted and sequenced as a pool. Peptide binding motifs for these three MHC class I molecules were derived and compared with previously described motifs derived from analysis of naturally processed peptides eluted from the surface of cells. This comparison indicated that the peptides bound by the recombinant MHC class I molecules showed a similar motif to naturally processed and presented peptides, with the exception of the peptide COOH terminus. Whereas the motifs derived from naturally processed peptides eluted from HLA-A2 and HLA-B8 indicated a strong preference for hydrophobic amino acids at the COOH terminus, this preference was not observed in our studies. We propose that this difference reflects the effects of processing or transport on the peptide repertoire available for binding to MHC class I molecules in vivo.
The MHC class I molecule consists of a variable heavy chain noncovalently associated with an invariant β2microglobulin (β2m) molecule and a short 8–10 amino acid peptide. Studies of the crystal structure of MHC class I molecules have shown that the peptide lies in a peptidebinding groove of the MHC molecule and interacts with it via a number of peptide binding pockets (1). These pockets accommodate specific residues of the peptide and may allow only one or a few closely related amino acids to bind at these positions. Studies of the peptide binding specificity of different MHC molecules have utilized analysis of peptides naturally bound to MHC class I molecules on the cell surface. Sequencing of these peptides has revealed requirements for specific amino acids at particular positions of the peptide (2). Motifs for HLA-A2, HLA-B8, and HLA-B53 have been derived by this method (3).
The peptides presented by MHC class I molecules are derived from intracellular sources. Endogenous proteins or proteins derived from viruses or intracellular pathogens are degraded within the cytoplasm to form short peptides. These peptides are then transported into the endoplasmic reticulum by the transporter associated with antigen processing (TAP) molecule, where they encounter MHC class I heavy chain and β2m and promote assembly of these into a trimolecular complex. Therefore, peptide binding motifs of MHC class I molecules that are derived from analysis of peptides eluted from the cell surface include information on not only what has been selected by the MHC class I molecule, but also on what peptides have been made available to the class I molecule by the processing machinery of the cell and the peptide transporter. Therefore, it is important to determine the relative contributions of these factors to the observed motifs. We have used a method that involves assembly of the MHC class I molecule in the absence of peptide processing and transport, and therefore measures only the specificity of the class I molecule itself. By comparison of peptide binding motifs derived from the two methods, we provide information on the possible contribution of selective transport or processing to the peptide binding motifs observed within eluted peptides.
Materials and Methods
Random Peptide Library.
Peptides were synthesized manually using standard fmoc chemistry. Equimolar amounts of each of the naturally occurring amino acids was used to a total of 10-fold molar excess. Cysteine was not included in this mix and arginine was used at 1.5× molar concentration to compensate for previously observed low incorporation of this amino acid (4). The randomness of the peptide mixture was then analyzed by HPLC and laser-desorption time of flight mass spectrometry.
Assembly and Purification of MHC Class I Complexes.
HLA-B53 and HLA-B8 were produced using vectors pGMT7B53HIS (5) and pGMT7B8, respectively. HLA-A2 and β2m were produced using the vectors pHN1A2 and pHN1β2m, respectively (a gift from D. Garboczi, Harvard University, Cambridge, MA). HLAB53 and HLA-A2 were refolded with a random mix of peptides using a dilutional method as previously described (6). In brief, ∼30 mg of random peptide pool was dissolved in a small volume of 8 M urea and added to a solution of 1 μM heavy chain, 2 μM β2m in refolding buffer (0.4 M l-arginine, 0.1 M Tris [pH = 7.5], 2 μM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 0.5 mM PMSF). After incubation for 36–48 h at 4°C, the solution was concentrated and the refolded complex purified by gel filtration.
HLA-B8 was refolded using a dialysis method of refolding. In brief, peptide, β2m, and heavy chain were mixed with 20 mM Tris (pH = 7.5) and 150 mM NaCl and then dialysed against refolding buffer. Additional heavy chain was added during incubation at 4°C for 48 h, then the solution was concentrated and the refolded complex purified by gel filtration. In both methods, random peptides were used at a final concentration of ∼200-fold molar excess over heavy chain in the refolding reaction. Self-peptides from HLA-B53 were purified using a W6-32 affinity column as previously described (7).
Peptide Elution and Sequencing.
Bound peptides were eluted from the MHC class I molecules as previously described (7). In brief, MHC class I–peptide complexes were first concentrated on a Centricon-3 microconcentrator (Amicon, Beverly, MA) before the addition of 0.1% trifluoroacetic acid. The eluate was collected and separated by HPLC before sequencing with an Applied Biosystems 473A protein sequencer (Applied Biosystems, Foster City, CA). Samples of the random peptide library were also sequenced. Individual self-peptides eluted from affinity-purified HLA-B53 were sequenced as previously described (8).
Results and Discussion
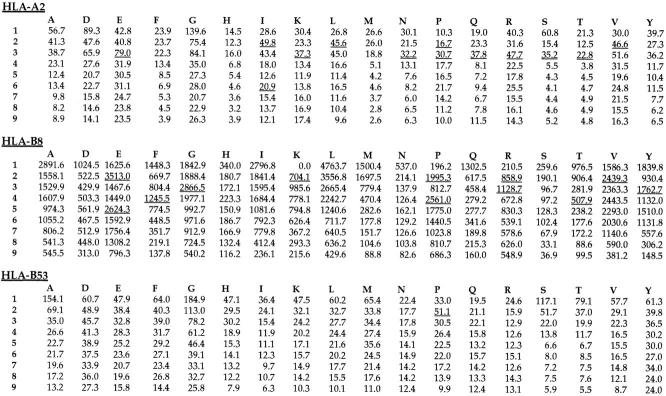
Analysis of the random peptide library by HPLC and laser desorption-mass spectrometry showed that the library was a homogeneous mix of peptides without any obvious peaks of abundant peptides and conformed to the mass range expected for nonamer peptides. Refolding of MHC class I molecules with the random peptide mixture resulted in the formation of stable complexes of heavy chain, peptide, and β2m, which migrated at the correct mass using gel filtration. A final concentration and wash step before peptide elution ensured that only peptides that formed complexes stable enough to remain associated during these procedures were analyzed. Peptides were eluted from ∼500 μg of MHC class I. The results of pool sequencing of these peptides and of the random peptide library showed that while there were no significant enrichments for any amino acids in the random library, such enrichments were found in the MHC class I–associated pool. Positions where the yield of a particular amino acid was increased to 150% or more of the previous cycle were considered significant (Fig. 1) (2). The motif identified using this method could be compared with that identified by previous studies (Fig. 2). It can be seen that although the motifs were not identical, the same anchor residues were observed by both methods with the exception of the COOH-terminal hydrophobic anchor previously observed in studies of HLA-A2 and HLA-B8. In addition, in some cases in which enrichment for anchor residues did not reach the 150% level, the same patterns of enrichment were seen for both the naturally bound peptides and the peptides bound by the recombinant class I. It is also interesting that the strength of motifs for different MHC class I alleles is conserved using both methods, so that, for example, whereas HLA-A2 shows strong enrichment for several possible minor anchors in both pool sequences, HLA-B53 shows a relatively weak proline anchor at position 2 using both methods (Fig. 2). In addition, whereas analysis of four individual self-peptides eluted from HLA-B53 and three T cell epitopes suggests a preference for hydrophobic COOH termini (Fig. 3), this was not observed in the results from pool sequencing. Thus the amino acid preferences observed from pool sequencing of HLA-B53 seem somehow less strong than those of, for example, HLA-A2.
Figure 1.
Results of pool sequencing of peptides bound to recombinant HLA class I molecules. The yield of each amino acid (indicated by single letter code) for each position of the peptide is shown for HLA-A2, HLA-B8, and HLA-B53. Positions where the yield of amino acid is 150% greater compared with the previous cycle are underlined.
Figure 2.
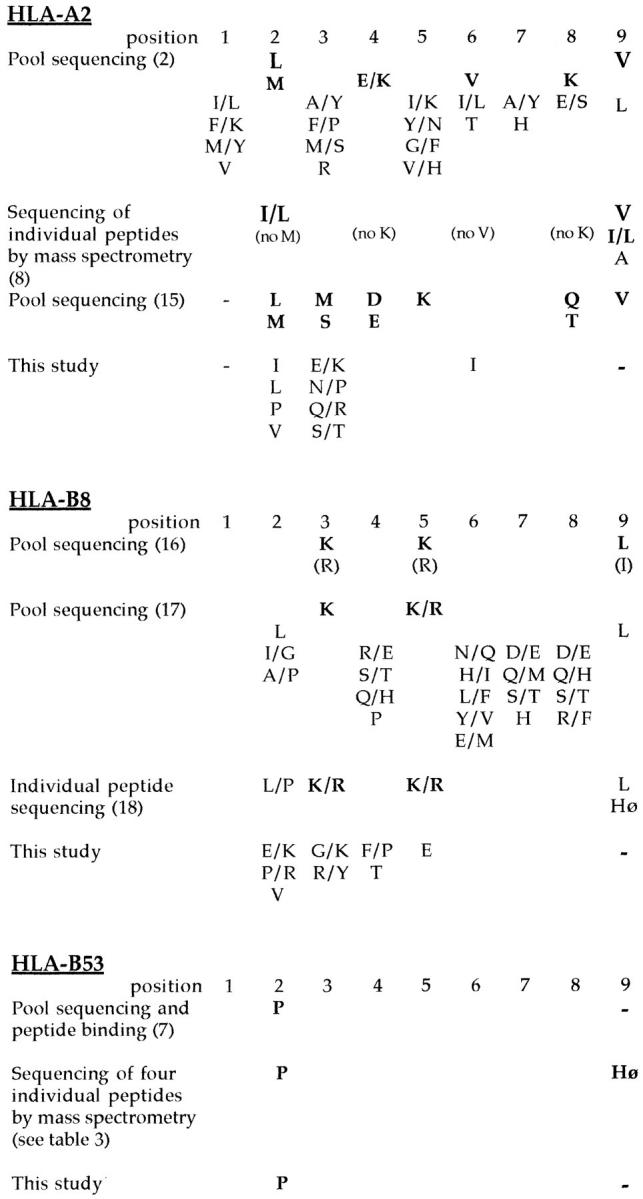
Comparison of the peptide binding motifs for HLA-A2, HLA-B8, and HLA-B53 derived by different methods. Motifs are indicated according to the convention established by Falk et al. (2), namely, dominant or strong anchor residues are indicated in bold type, and listed vertically in order of descending importance. It should be noted that different criteria were used to assign anchors in some studies, particularly when individual peptide as opposed to pool sequences were analyzed. Hø, hydrophobic residue.
Figure 3.
Sequences of individual self-peptides eluted from HLA-B53 and sequenced using tandem mass spectrometry. The isobaric residues isoleucine and leucine could not be distinguished in the unknown peptide. The sequences of three known HLA-B53 T cell epitopes are shown.
Previous studies of the peptide binding motifs of MHC class I molecules have indicated that most HLA class I alleles show a preference for a hydrophobic COOH-terminal amino acid, with a small number favoring a positive charge at this position (reviewed in reference 3). The majority of these motifs are derived from analysis of the peptides bound to MHC class I on the surface of B cells and thus reflect both the specificity of the MHC class I molecule itself and the effects of processing and transport. Direct binding of synthetic peptides to MHC class I has also been used to study the peptide binding specificity of these molecules. Although a number of methods have been used (reviewed in reference 3), it is difficult to reflect the dynamic interactions of MHC and peptide using single peptides. The number of MHC peptide complexes on the cell surface will be determined both by the ability of peptides to compete for binding and also by the stability of the subsequent complex. Parker and colleagues (9) have described experiments in which they measured the stability of complexes formed between recombinant HLA-A2 and synthetic peptides. Interestingly, when the requirements of the peptide COOH terminus were analyzed, these did not conform with the expected restrictions.
The motifs observed in this study differ from those previously reported because they show no preference for a COOH-terminal hydrophobic or positive charge. A trivial explanation for this would be that the solutions and the temperature at which binding was carried out may interfere somehow with peptide–MHC interactions. However, since preferences for both hydrophobic and other peptide anchors are conserved elsewhere in the motifs, this seems unlikely. One simple mechanism to explain the observed differences in peptide COOH termini is that naturally processed peptides are subject to selection for transport into the endoplasmic reticulum by the TAP transporter, whereas our random mixture is not. Studies of the substrate specificity of the TAP protein in human, mouse, and rat have suggested that whereas the rat TAP2u and murine TAP were highly selective for hydrophobic or aromatic COOH termini, the human TAP and the rat TAP2a transporters were only weakly selective for hydrophobic or positive charge and against negative charge (10). Therefore, this bias would tend to favor hydrophobic COOH termini in most MHC class I alleles. Those MHC class I molecules that appear to exhibit a preference for positively charged COOH termini instead may simply be those which are less able to tolerate hydrophobic COOH termini and thus bind the next most abundant species, those with positively charged COOH termini. However, analysis of HLA-A2-associated peptides in a mutant cell line lacking the TAP transporter suggests that the preference for a hydrophobic COOH terminus is still observed (11). Therefore, it is likely that other mechanisms of peptide processing may act independently of or synergistically with the TAP transporter to affect the COOH terminus of the peptides. Experiments to determine the alterations in substrate specificity conferred by the MHC-encoded LMP2 and LMP7 subunits of the proteasome have suggested that the presence of these subunits enhances proteolysis after hydrophobic and positively charged amino acids and decreases proteolysis after negatively charged amino acids (12, 13). In addition, we cannot exclude the possibility that other processes involved in the generation and assembly of MHC–peptide complexes, such as the involvement of molecular chaperones, may also act to bias the repertoire of peptides available for binding to MHC class I.
The preference of both the putative proteolytic enzymes and transporter molecules involved in class I processing for hydrophobic or positively charged COOH termini of peptides appears to accord with the observation that most MHC class I motifs have hydrophobic COOH termini, and the few that do not have positively charged COOHtermini. This has been interpreted as suggesting that the antigen processing machinery has evolved to supply the MHC class I molecule with peptides optimal for binding. However, because most MHC class I motifs are derived from the analysis of peptides eluted from MHC class I molecules on the cell surface, these motifs include the effects of both antigen processing and MHC binding. Therefore, we favor an alternate explanation for the observed preference for hydrophobic or positively charged COOH termini in MHC class I motifs, namely that this is largely an effect of processing and that in the absence of such processing this preference is not found.
This observation does not affect the use of currently available MHC class I motifs in the prediction of T cell epitopes, since a hydrophobic COOH terminus will still be required for natural processing and presentation of antigens. On the other hand, these observations necessitate a reevaluation of the role of processing in the selection of peptides for binding to MHC class I. A preference for COOH-terminal hydrophobic residues for both the processing machinery and class I molecules would imply that the system had evolved to optimize peptide delivery to class I. If class I does not select for hydrophobic COOH termini in the absence of processing, then it would seem that the selection for COOH-terminal hydrophobic anchors, in fact, acts to restrict the repertoire of peptides available to bind MHC class I. Although this may seem counterproductive, a precedent for such a limitation of peptides is already known. The TAP transporter of human, mouse, and rat appears to be a poor transporter of peptides with proline at positions 2 and 3 (14). However, a number of HLA-B alleles (including HLA-B53) appear to bind a proline anchor at position 2 of the peptide. Thus, it would seem that either the selectivity of the transporter is acting to reduce the number of peptides available to bind these, or that there is some advantage in longer peptides being transported and then cleaved to generate the MHC binding peptide within the endoplasmic reticulum. If the former is the case one can only speculate as to the possible benefits to the host of reducing the repertoire of endogenous peptides presented on MHC class I. However, it is possible that this acts to decrease the effective hole in the T cell repertoire caused by negative selection of T cells in the thymus.
The results of this work demonstrate that recombinant MHC class I molecules produced in E. coli retain their peptide specificity during refolding with peptide and β2m molecules in vitro. In addition, the observed differences between the peptide binding motifs derived from naturally processed peptides and peptides bound to MHC class I in vitro suggest that peptide processing and transport play a dominant role in determining which peptides will ultimately be presented in association with MHC class I.
Footnotes
Thanks to H. Reyburn for helpful comments.
M.P. Davenport is supported by the Lionel Murphy Foundation and the Wellcome Trust. A.V.S. Hill is a Wellcome Trust Principal Research Fellow. D. Barouch is supported by a Marshall Scholarship.
References
- 1.Bjorkman PJ, Saper MA, Samraoui B, Bennet WS, Strominger JL, Wiley DC. Structure of human class I histocompatibility antigen, HLA-A2. Nature (Lond) 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 2.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature (Lond) 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 3.Davenport, M.P., and A.V.S. Hill. 1996. Peptides associated with MHC class I and class II molecules. In HLA and MHC: Genes, Molecules and Function, M. Browning and A. McMichael, Editors. Oxford. βios Scientific. 277–308.
- 4.Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide binding specificity of the molecular chaperone BiP. Nature (Lond) 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 5.Smith, K.J., S.W. Reid, K. Harlos, A. McMichael, D.I. Stuart, J. Bell, and E.Y. Jones. Bound water structure and polymorphic amino acids act together to allow binding of different peptides to MHC class I HLA-B53. Immunity. 4:215–228. [DOI] [PubMed]
- 6.Garboczi D, Hung D, Wiley D. HLA-A2 peptide complexes: Refolding and crystalisation of molecules expressed in Escherichia coliand complexed with single antigenic peptides. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill AVS, Elvin J, Willis AC, Aidoo M, Allsopp CEM, Gotch FM, Gao XM, Takiguchi M, Greenwood BM, Townsend ARM, et al. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature (Lond) 1992;360:434–439. doi: 10.1038/360434a0. [DOI] [PubMed] [Google Scholar]
- 8.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH. Characterization of peptides bound to class I MHC molecule HLA-A2.1 by mass spectrometry. Science (Wash DC) 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 9.Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, Biddison WE, Coligan JE. Sequence motifs important for peptide binding to the human class I molecule, HLA-A2. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 10.Momburg F, Roelse J, Howard JC, Butcher GW, Hammerling GJ, Neefjes JJ. Selectivity of MHCencoded peptide transporters from human, mouse and rat. Nature (Lond) 1994;367:648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- 11.Henderson RA, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt DF, Engelhard VH. HLAA2.1 associated peptides from a mutant cell line: a second pathway of antigen presentation. Science (Wash DC) 1992;255:1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 12.Driscoll J, Brown MG, Finley D, Monaco JJ. MHC-linked LMPgene products specifically alter peptidase activities of the proteasome. Nature (Lond) 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 13.Gaczynska M, Rock KL, Goldberg AL. γ-interferon and expression of MHC genes regulates peptide hydrolysis by proteasomes. Nature (Lond) 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 14.Neefjes J, Gootfried E, Roelse J, Gromme M, Obst R, Hammerling GJ, Momburg F. Analysis of the fine specificity of rat, mouse, and human TAP peptide transporters. Eur J Immunol. 1995;25:1133–1136. doi: 10.1002/eji.1830250444. [DOI] [PubMed] [Google Scholar]
- 15.Kubo RT, Sette A, Grey HM, Appella E, Sakaguchi K, Zhu N-Z, Arnott D, Sherman N, Shabanowitz J, Michel H, et al. Definition of specific peptide motifs for four major HLA-A alleles. J Immunol. 1994;152:3913–3924. [PubMed] [Google Scholar]
- 16.Sutton J, Rowland-Jones S, Rosenberg W, Nixon D, Gotch F, Gao X-M, Murray N, Spoonas A, Driscoll P, Smith M, et al. A sequence pattern for peptides presented to cytotoxic T lymphocytes by HLA-B8 revealed by analysis of epitopes and eluted peptides. Eur J Immunol. 1993;23:447–453. doi: 10.1002/eji.1830230222. [DOI] [PubMed] [Google Scholar]
- 17.Malcherek G, Falk K, Rötzschke O, Rammensee H-G, Stevanovic S, Gnau V, Jung G, Melms A. Natural peptide ligand motifs of two HLA molecules associated with myasthenia gravis. Int Immunol. 1993;5:1229–1237. doi: 10.1093/intimm/5.10.1229. [DOI] [PubMed] [Google Scholar]
- 18.DiBrino M, Parker KC, Shiloach J, Turner RV, Tsuchida T, Garfield M, Biddison WE, Coligan JE. Endogenous peptides with distinct amino acid anchor residue motifs bind to HLA-A1 and HLA-B8. J Immunol. 1994;152:620–631. [PubMed] [Google Scholar]
- 19.Gotch F, McAdam SN, Allsopp CE, Gallimore A, Elvin J, Kieny MP, Hill AV, McMichael AJ, Whittle HC. Cytotoxic T cells in HIV2 seropositive Gambians. Identification of a virus-specific MHC restricted peptide epitope. J Immunol. 1993;151:3361–3369. [PubMed] [Google Scholar]
- 20.Koziel MJ, Dudicy D, Afdhal N, Grakoui A, Rioc CM, Houghton M, Walker BD. HLA class I–restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterisation of patterns of cytokine release. J Clin Invest. 1995;96:2311–2321. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]