Abstract
A somatic process introduces mutations into antibody variable (V) region genes at a high rate in many vertebrates, and is a major source of antibody diversity. The mechanism of this hypermutation process remains enigmatic, although retrospective studies and transgenic experiments have recently suggested a role for transcriptional regulatory elements. Here, we demonstrate that mouse heavy (H) chain loci in which the natural VH promoter has been replaced by a heterologous promoter undergo hypermutation. However, while the distribution of mutation in such loci appears normal, the frequency of mutation does not. Conversely, moving the VH promoter 750 bp upstream of its normal location results in a commensurate change in the site specificity of hypermutation in H chain loci, and the foreign DNA inserted into the VH leader intron to produce this promoter displacement is hypermutated in a manner indistinguishable from natural Ig DNA. These data establish a direct mechanistic link between the IgH transcription and hypermutation processes.
Antibody variable (V)1 gene hypermutation results in diversification of the antibody repertoire in a variety of vertebrates (1). In mice and humans, this process is induced during T cell–dependent immune responses, is intimately associated with differentiation of memory B cells, and in combination with antigen affinity based selection, results in affinity maturation of serum antibodies (2). Past retrospective and descriptive studies in mice have shown that hypermutation introduces mainly single, untemplated nucleotide replacements at a rate estimated to be 10−3/bp/cell division (3–6), acts efficiently only in and immediately around fully rearranged L chain V and VH genes (i.e., in a “zone” from ∼300-bp 5′ to 1-kb 3′ of the V gene) (7–12), and is not mechanistically linked to isotype switch or V(D)J recombination (13–16).
Current data lend the most support to models for the mechanism of hypermutation that invoke a role for the transcription apparatus. Mutations are rare in regions 5′ of V gene promoters, suggesting a “boundary” (8, 11, 12, 17). The distribution of mutations in and around a V gene appears to be influenced by the location of regions flanking the 5′ side of the V coding sequence and not the V coding sequence itself (10, 17). Since 5′ regions distal to V coding sequence are clearly not required for hypermutation (18, 19), there may be an influence of the promoter region on distribution. More recent studies using transgenic technology have begun to shed light on the cis-acting elements necessary for hypermutation of Vκ L chain genes. A role for both the intronic and 3′ distal κ transcriptional enhancers in influencing the rate of hypermutation of the Igκ locus has been implicated (19), and the insertion of a Vκ promoter just 5′ of the Cκ exon in an Igκ transgene results in somatic mutation of this normally unmutated region (20). While it is not known whether transcription is required for hypermutation or whether the influence of promoters and enhancers on hypermutation is secondary to their role as transcriptional regulators, these data nevertheless suggest that common cis-acting elements are involved in these processes.
Materials and Methods
Transgenic Mice.
Transgenic mice were generated by standard methods (21) using C57BL/6 × C3H F1 mice and linearized plasmid constructs. Founder mice were backcrossed for two (“AA” mice), three (“3A” mice), or four (“4A” mice) generations to the A/J strain before use. All mice used in these experiments were heterozygous for multicopy transgenic arrays. Mice were housed under specific pathogen-free conditions and given autoclaved food and water. All experimental procedures on mice were conducted according to the National Institutes of Health guidelines.
Generation and Screening of Hybridomas.
All hybridomas were generated from the spleens of 8–10-wk-old transgenic mice immunized with the same preparation of p-azophenylarsonate (Ars)- KLH intraperitoneally using one of the following immunization protocols: 10 d after a single injection of 100 μg Ars-KLH in CFA (late primary), 3 d after secondary or tertiary boosting injections of 100 μg Ars-KLH in PBS at least 30 d after priming with 100 μg Ars-KLH in CFA, or 3 d after a regimen of priming with 100 μg Ars-KLH in CFA, a 7-d rest, and then four injections of 25–50 μg Ars-KLH in PBS spaced at 2-d intervals (fusion 16 d after initial priming, hyperimmunized primary). Supernatants from the hybridomas were screened by ELISA for those expressing the transgenic VH gene using the antiidiotypic antibodies E4 and 107 as previously described (15). E4 recognizes all canonical anti-Ars antibodies. 107 recognizes only canonical anti-Ars antibodies partially encoded by the 36-65 VH gene, due to the rare VH-D and D-JH junctional amino acids encoded by this gene (22, 23).
Reverse Transcriptase-PCR, Genomic PCR, and Nucleotide Sequencing.
Total RNA was prepared from hybridomas, reverse transcriptase-PCR performed, and PCR products directly sequenced as previously described (24). The hybrid loci were amplified from hybridoma genomic DNA by PCR, and bulk PCR products were directly sequenced also as described previously (18, 25). Since bulk PCR products were sequenced in all experiments, the contribution of Taq polymerase error to mutations in the final sequences was considered to be insignificant.
Results
If Ig promoters contain cis information necessary for the locus-specific action of hypermutation, replacement of the natural promoter in an Ig locus with a heterologous, but B cell–specific promoter should ablate hypermutation. Moreover, if the site specificity of mutation is regulated by the promoter, changing the location of this element relative to the VDJ coding sequence and downstream regulatory elements such as the intronic enhancer in an Ig locus should cause a corresponding change in the location of the characteristic “zone” of mutation. To test these predictions for the IgH locus and its promoter we created transgenic mice using two modified forms of a construct containing an antiArs VDJ gene called 36-65 from A/J mice, a minimal VH promoter (145 bp containing only the cap/initiation site, a TATA box, an octamer, a heptamer, and a purine rich region), and 1.5 kb of natural 3′ flanking sequence. In one construct, the natural VH promoter was replaced by the heterologous, but B cell–specific minimal B29 (Ig-β) gene promoter. In the second construct, the size of the intron between the VH leader exon and the main VH exon was enlarged 750 bp via insertion of a portion of a Drosophila melanogaster myosin heavy chain gene intron. Since all of the cis-acting elements necessary for hypermutation of VH transgenes present at ectopic chromosomal sites have yet to be defined (18, 26), we exploited a novel somatic recombination pathway (18, 25) to introduce our modified VH constructs into a natural context within the endogenous (i.e., nontransgenic) H chain locus (Fig. 1).
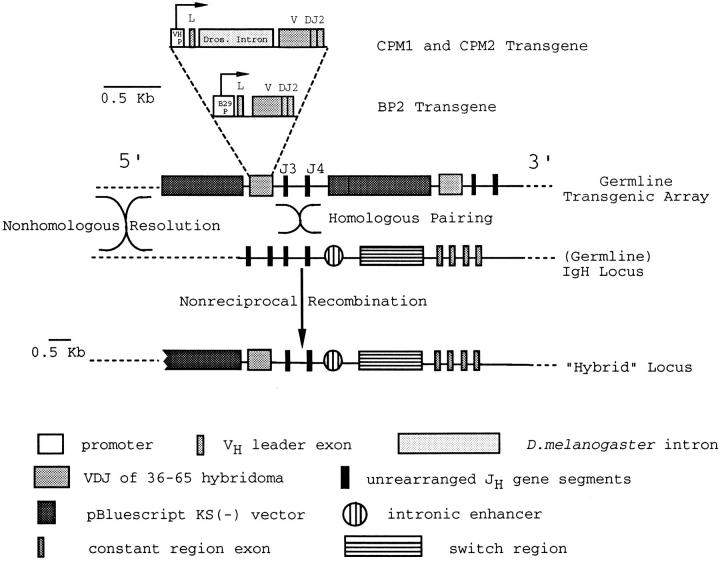
Figure 1.
Transgene structure and schematic representation of the nonreciprocal recombination pathway leading to the generation of hybrid H chain loci. CPM1 and CPM2 mice were produced using a plasmid containing the VH promoter, VH leader exon, the unmutated, rearranged VDJ from the antiarsonate hybridoma 36-65, and the unrearranged JH3-JH4 region of the H chain locus. Inserted 25 bp 3′ of the VH leader exon is a 750-bp piece of intron DNA from between exons 12 and 13 of the D. melanogaster myosin heavy chain gene. The BP2 mouse line was generated using a plasmid similar to the one described above except the 750-bp D. melanogaster intron DNA is not present and the minimal B29 promoter (38) was placed 43 bp 5′ of the ATG in the VH leader exon, replacing the natural VH promoter. All transgenic mice were generated as previously described (24). Nonreciprocal recombination (gene conversion) occurs as the result of homologous pairing between a region 3′ of the coding VDJ of one transgene copy in the transgenic array and the analogous region in the IgH locus. The result is one copy of the transgenic array being copied into the IgH locus at a natural position 5′of the intronic enhancer. This recombination takes place at a low frequency, so most B cells from the transgenic mice express a conventional H chain locus. DNA length scale bars are shown.
We have previously shown (18, 25) that transgenes derived from constructs containing the 36-65VH gene and various amounts of natural 5′ and 3′ flanking DNA (hereafter referred to as wild type) recombine with the endogenous IgH locus at a low frequency in B cells, giving rise to fully functional “hybrid” H chain genes that undergo normal hypermutation (Fig. 1). In such hybrid loci, the VDJ and its 5′ flanking sequences are transgene derived, and regions 3′ of the JH3–JH4 region are derived from the endogenous H chain locus. The kinetics and distribution of hypermutation are not altered in such hybrid loci, and the generation of these loci is not mutagenic or mechanistically linked to the hypermutation process (18, 24, 25).
Founder mice were crossed with A/J mice to introduce a “canonical” Vκ gene (Vκ10Jκ1), that normally is coexpressed with canonical VH genes, encoded by the same VH, D, and JH gene segments as the 36-65VH gene, in a major fraction of the B cells that respond to Ars immunization in the A/J mouse strain (13). One line of mice obtained from the B29 promoter construct, termed BP2, and two lines of mice created with the enlarged intron construct, termed CPM1 and CPM2, were immunized with Ars-KLH and hybridomas generated from late primary and secondary immune responses to sample the hypermutated B cell population. Hybridomas coexpressing H chains encoded by a hybrid locus and canonical endogenous κ L chains were identified using an antiidiotypic antibody specific for the 36-65 V region (18, 24). The presence of hybrid loci in hybridoma genomic DNA was confirmed by Southern blot analysis (data not shown).
Sequencing of the VH genes present in B29 promoterdriven hybrid loci expressed by late primary immune response hybridomas via reverse transcriptase–PCR revealed that these genes contained few or no somatic mutations. Analysis of VH genes in such hybrid loci expressed by secondary and tertiary hybridomas showed that they were mutated, but also at levels lower than expected (Table 1). However, the average frequency of mutation in expressed, canonical Vκ (Vκ10Jκ1) genes in these hybridomas was comparable to that observed in canonical Vκ genes expressed by control hybridomas (see legend to Table 1 and Materials and Methods for details). The low frequency of mutation observed in canonical Vκ genes was anticipated since previous studies on Ars-induced hybridomas derived from A/J mice have shown that the frequency of canonical Vκ mutation is two to threefold lower than of canonical VH mutation (10, 15). Chi-squared comparison of the frequency of mutations in the V genes expressed by hybridomas derived from BP2 transgenic mice revealed the frequency difference between endogenous canonical VH genes and B29 promoter-driven 36-65VH genes present in hybrid loci was highly significant (P <10−4). This difference in mutation frequency is particularly apparent among primary hybridomas. The significance of this difference is further supported by the fact that hybridomas expressing hybrid or endogenous H chain loci were often derived from the same mouse, or from littermates immunized with the same antigen preparation and from which fusions were performed at the same time (see legend to Table 1). In contrast, the mutation frequencies observed in the endogenous canonical Vκ genes expressed by these two groups of hybridomas were not significantly different (P ∼0.9). Thus, the hypermutation process had operated normally in trans in the precursors to the hybridomas expressing B29 promoter-driven hybrid loci.
Table 1.
Somatic Hypermutation in BP2, CPM1, and CPM2 Hybridomas
| Name of hybridomas* | Isotype | Mutations in VH | Mutations in Vκ | |||||
|---|---|---|---|---|---|---|---|---|
| BP2 hybridomas expressing hybrid H chain loci | ||||||||
| Primary | ||||||||
| 4ABP2-8 | G7 | IgG2b | 0 | 1 | ||||
| D10 | IgG2b | 0 | 0 | |||||
| E4 | IgG2b | 0 | 1 | |||||
| 4ABP2-16 | G6 | IgG1 | 1 | 3 | ||||
| A5 | IgG1 | 2 | 0 | |||||
| AABP2-2 | E3 | IgG2b | 2 | 3 | ||||
| 3APB2-7 | H3 | IgG2b | 0 | 2 | ||||
| Secondary and tertiary | ||||||||
| AABP2-28a | E5 | IgG1 | 1 | 1 | ||||
| 3ABP2-59 | A12 | IgG1 | 7 | 1 | ||||
| D7 | IgG2b | 2 | 1 | |||||
| 4ABP2-13 | H7 | IgG1 | 4 | 0 | ||||
| 3ABP2-44 | H8 | IgG1 | 1 | 2 | ||||
| BP2 hybridomas expressing endogenous H chain loci | ||||||||
| Primary | ||||||||
| 4ABP2-8 | E7 | IgM | 3 | 0 | ||||
| AABP2-10 | H7 | IgG1 | 5 | 1 | ||||
| 3ABP2-7 | B1 | IgG2b | 4 | 4 | ||||
| 4ABP2-23 | F10 | IgM | 1 | 3 | ||||
| D2 | IgG1 | 3 | 1 | |||||
| Secondary and tertiary | ||||||||
| 3ABP2-43 | D3 | IgG1 | 8 | 1 | ||||
| A12 | IgG1 | 11 | 2 | |||||
| 3ABP2-45 | G2 | IgG1 | 3 | 2 | ||||
| CPM hybridomas expressing hybrid H chain loci | ||||||||
| Primary | ||||||||
| 3ACPM2-23 | B2 | IgG1 | 3 | 1 | ||||
| G6 | IgG1 | 5 | 3 | |||||
| 4ACPM1-2 | D9 | IgG1 | 2 | 4 | ||||
| G3 | IgG1 | 0 | 4 | |||||
| C6 | IgG3 | 0 | 1 | |||||
| 4ACPM1-3 | D5 | IgG2b | 3 | 2 | ||||
| F1 | IgG3 | 12 | 2 | |||||
The first four–five characters before the dash in each hybridoma name indicate the name of the mouse from which that hybridoma was derived. Among the BP2 hybridomas, the groups (4ABP2-8 D10, E4, E7, G7); and (3ABP2-7 B1, H3) were derived from single mice. The groups (3APB2-44 H8, 3ABP2-45 G2); (AABP2-2 E3, AABP2-10 H7) and (4ABP2-16 G6, A5, 4ABP2-23 D3, A12) were derived from littermates immunized with the same dose and preparation of antigen and sacrificed for fusion on the same day.
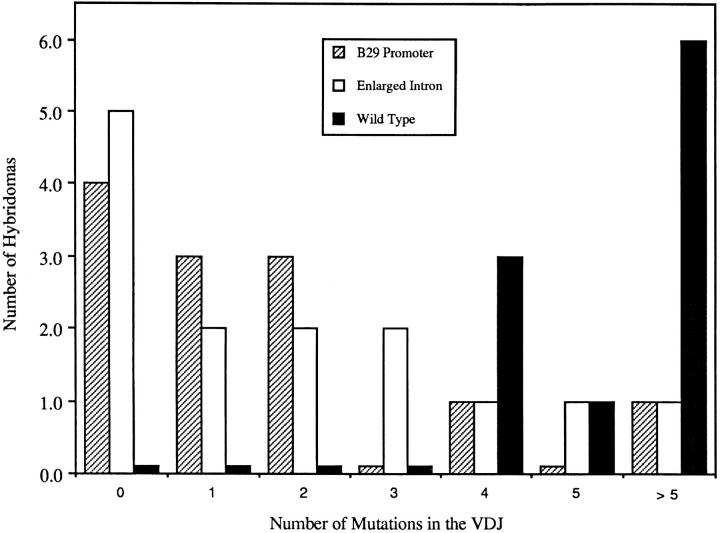
Low levels of VH mutation were also observed on average in enlarged leader intron hybrid loci expressed by late primary response hybridomas, and many such loci contained no VH mutations. As was the case for the hybridomas containing B29 promoter-driven hybrid loci, the canonical Vκ mutation frequency in hybridomas containing enlarged leader intron hybrid loci appeared normal (data from seven representative hybridomas are shown in Table 1). As summarized in Fig. 2, the mutation frequencies in the VH genes in both types of hybrid loci are well below that previously determined for wild-type 36-65VH hybrid loci at times late in the primary response.
Figure 2.
The total number of hybridomas expressing BP2 (B29 promoter-driven) and CPM (enlarged leader intron) hybrid loci with a given number of VH somatic mutations. Values for wild-type 36-65VH hybrid loci were obtained from previously published data (18). All hybridomas from CPM transgenic mice and wild-type 36-65VH transgenic mice were generated using the hyperimmunized primary protocol and 8–10-wk-old mice. For BP2 mice, primary, secondary, and tertiary immunization protocols were used (see Materials and Methods). Those hybrid locus expressing hybridomas from the B29 line that had the largest number of mutations (>2) were from secondary and tertiary immunizations.
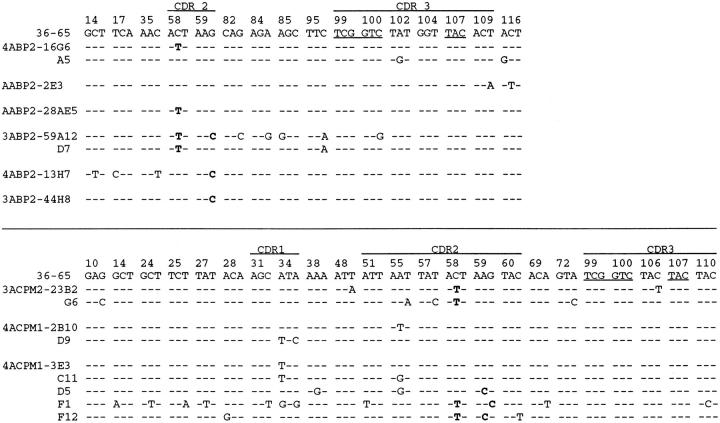
The frequency of VH gene mutation in hybrid loci is not directly reflective of their intrinsic rate of mutation, as many mutations in coding region can be selected in vivo. Fig. 3 shows the sequence of the expressed VH genes in the B29 promoter-driven and enlarged leader intron hybrid loci that contain at least one somatic mutation. Of the eight VH genes in B29 promoter-driven hybrid loci, six contain somatic mutations at positions 58 and 59 in CDR2 that are recurrently observed among canonical VH genes. Two such mutations have been shown to result in increased affinity for Ars (27). In three such VH genes, these recurrent mutations are the only mutations observed, and in one VH gene, a recurrent mutation is one of only two mutations. Of these four VH genes (those expressed by hybridomas 4ABP2-16G6, AABP2-28aE5, 3ABP2-59D7, and 3ABP2-44H8), three are expressed by hybridomas isolated from secondary or tertiary responses. In wild-type 36-65VH genes in hybrid loci and endogenous canonical VH genes, particularly those expressed by secondary and tertiary hybridomas, such mutations are usually accompanied by many other “selectively neutral” (e.g., silent) mutations (18, 28). Of the nine mutated enlarged leader intron hybrid loci, five contain recurrent mutations. These data indicate that the frequency of VH coding region mutation is an overestimate of the reduced intrinsic mutation rate in this region in both types of modified hybrid loci, particularly those driven by the B29 promoter.
Figure 3.
Nucleotide sequences of VH genes in hybrid loci expressed by hybridomas from BP2 (B29 promoter) transgenic mice (above solid line) and CPM (enlarged leader intron) transgenic mice (below solid line). Only those sequences containing at least one mutation are shown. The reference sequence is that of the unmutated 3665VH gene. Hybridoma names are listed to the left of each sequence. Dashed lines indicate sequence identity. Mutations are shown explicitly. Only those codons in which mutations were observed, as well as junctional codons, are shown. Recurrently observed nucleotide changes at codons 58 and 59 are indicated in bold. The rare junctional codons of the 36-65VH transgene are underlined. Codons are numbered sequentially from the mature amino terminus, and the location of complementarity determining regions (CDR) are shown. At codon 58, the recurrent mutation (ACT→ ATT) results in a change from threonine to isoleucine, demonstrated to increase affinity for Ars two- to threefold (27). A variety of amino acid substitutions have been recurrently observed at codon 59. A change from lysine to threonine (AAG→ ACG), observed in two enlarged leader intron hybrid loci, increases affinity for Ars two- to fourfold (27). The lysine to asparagine mutation (AGG→ AGC) observed at this position in 3 BP2 hybridomas and 1 CPM hybridoma is recurrently observed, but its effect on Ars affinity has not been tested.
Two explanations were considered for these observations: (a) the intrinsic rate of hypermutation throughout the modified hybrid loci was reduced; and (b) the site specificity of hypermutation was altered such that the VH gene no longer was present in the region of “peak” mutational activity. To distinguish between these possibilities, hybrid loci were cloned from hybridoma DNA and regions flanking the 5′ and 3′ sides of the VH gene were sequenced. Analysis of five B29 promoter-driven hybrid loci chosen to represent the entire range of VH mutation frequency (i.e., 0 –7 VH mutations, hybridomas AABP2-2E3, 3ABP2-59A12, and 4ABP2-13H7, -16A5, and -8D10) revealed that the distribution of mutation was similar to wild-type 36-65VH hybrid and endogenous canonical VH loci. However, mutation frequencies in both 5′ and 3′ flanking regions were low. In the 300 bp of DNA flanking the 5′ side of VDJ, only one mutation was observed in the 1.5 kb sequenced. In the 1.5 kb of DNA flanking the 3′ side of VDJ, only 23 mutations were observed in the 7 kb sequenced. However, 15 of these mutations were observed in the first 300 bp 3′ of the VDJ. In previous analyses of the distribution of mutation around endogenous canonical VH genes and 36-65VH genes present in wild-type hybrid loci, 12–14% of the total mutations were found 5′ of the VDJ and 86–88% were found 3′ of VDJ (17, 18). Moreover, the majority of 3′ mutations were observed within the first 300 bp 3′ of VDJ (see below). The chemical nature of mutations observed in B29 promoter-driven hybrid loci was also characteristic of bona fide hypermutation (29–32, and see below). Thus, the effect of replacing the natural VH promoter with the B29 promoter appears to only be a reduction in the rate of hypermutation.
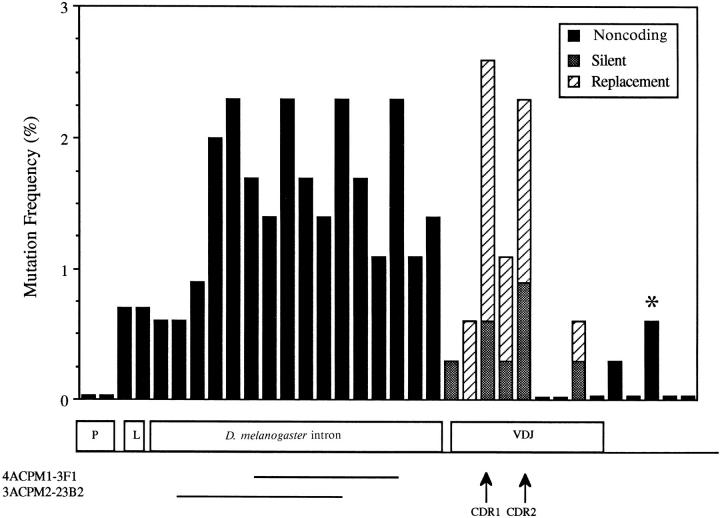
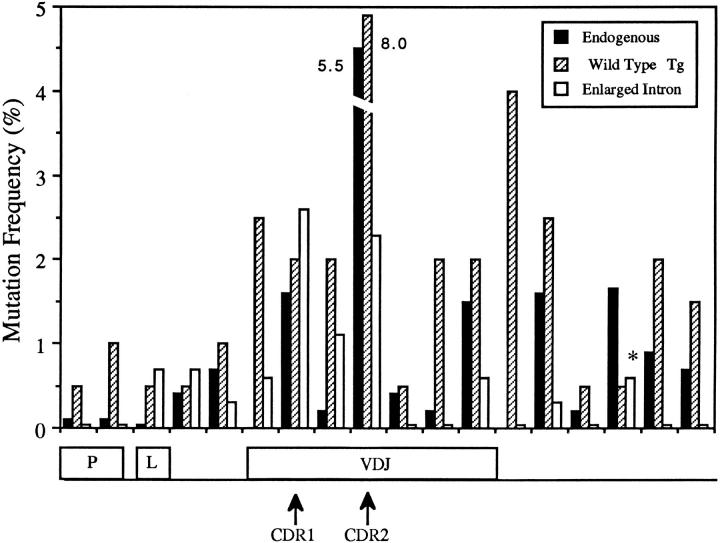
Sequencing of DNA flanking the 5′ side of the VH gene in seven enlarged leader intron hybrid loci revealed high frequencies of mutation in both the Drosophila intron and natural Ig intron regions (Fig. 4). However, few mutations were observed in the region flanking the 3′ side of VH in these hybrid loci, a region that, as discussed above, displays a high frequency of mutation in wild-type 36-65VH hybrid and endogenous canonical VH loci (Fig. 5). These data indicate that the distribution, but not the rate, of mutation was altered due to the lengthening of the leader intron.
Figure 4.
Mutation frequency distribution in the enlarged leader intron hybrid loci. Seven hybridomas from three different mice (two of the CPM1 line and one of the CPM2 line) representing the entire range of VH mutation observed (see Fig. 2 and Table 2) were analyzed. The mutation frequency was determined as follows: number of mutations in a 50-bp interval divided by 350 (the total number of bases in the interval sequenced from all the hybridomas) × 100. The first 50-bp interval began 8 bp 5′ of the purine rich motif in the VH promoter. The solid lines under the D. melanogaster intron indicate those intervals that were deleted from the hybrid loci in hybridomas 4ACPM1-3F1 and 3ACPM2-23B2. A total of 350 bp were deleted from the 4ACPM1-3F1 hybrid locus; in addition, this hybridoma also had the most mutations in its V(D)J coding sequence, 12, of any enlarged leader intron hybridoma analyzed. *, 3′ flanking interval in which two mutations were observed in the 4ACPM1-3F1 hybrid locus. A total of 470 bp were deleted from the 3ACPM2-23B2 hybrid locus. These large deletions were scored as one mutation per interval deleted. The mutation frequency in the VDJ was calculated using either those mutations that do not encode amino acid substitutions (silent mutations − stippled bars) or those mutations that encode amino acid changes (replacement mutations − diagonal striped bars).
Figure 5.
Mutation frequency distribution in enlarged leader intron hybrid loci, wild type 36-65VH hybrid loci, and canonical endogenous IgH loci from normal A/J mice. Hybridomas expressing all enlarged leader intron and wild-type 36-65VH hybrid loci, and most endogenous IgH loci were isolated using the hyperimmunized primary protocol described in Materials and Methods. Mutation frequency was determined as described in Fig. 4. Seven enlarged leader intron hybrid loci were analyzed. The data obtained from intervals in the D. melanogaster intron from these hybrid loci (Fig. 4) are not shown. Sequences of the four wild-type 36-65VH hybrid loci used to generate this graph have been previously described (18). The eighteen normal A/J VH locus sequences used were also previously described (17). For these endogenous A/J loci, data was not available for the first 5′ interval in the VDJ, and in the first interval flanking the 3′ side of VDJ. *, 3′ flanking interval containing the two mutations from the hybridoma 4ACPM1-3F1, which had a 350-bp intron deletion and the most VH mutations (12) of any hybrid locus expressing hybridoma analyzed.
In wild-type 36-65VH hybrid and endogenous canonical VH loci, the distribution of mutation encompasses ∼1 kb with mutation frequency increasing abruptly near the leader exon, peaking once in the main VH exon, and then gradually declining in 3′ flanking DNA (Fig. 5). In the enlarged leader intron hybrid loci, this distribution begins abruptly at a similar location, but is larger and bimodel with the 3′ peak encompassing the main VH exon (Fig. 4). We believe that the shape of this distribution is not reflective of a change in the intrinsic site specificity of hypermutation for the following reasons. First, a major fraction of mutations in the VH gene in these hybrid loci caused amino acid replacements in either CDR1 or CDR2, and could have been positively selected by antigen (Fig. 3, and replacement mutation frequency in the VDJ is indicated by striped bars in Fig. 4). Second, two such hybrid loci, those expressed by hybridomas 4ACPM1-3F1 and 3ACPM2-23B2, contain major deletions in the Drosophila intron DNA (Fig. 4). We speculate that these deletions took place early during the hypermutation process in vivo, subsequently allowing a higher frequency of mutation to take place in the VH gene, thus resulting in a higher probability of introduction of affinity enhancing mutations. Indeed, both of the enlarged leader intron hybrid loci with intron deletions also contain mutations in VH CDR2 known to be positively selected, and one contains a total of 12 VH mutations, seven more than any other enlarged leader intron hybrid locus examined. In total, these data argue that the transcriptional promoter region dictates the site specificity of hypermutation in the H chain locus.
Previous data obtained from the analysis of Igκ transgenes in which major portions of the Vκ exon were replaced by heterologous DNA showed that this DNA was hypermutated (33). In agreement with these findings, our results demonstrate that the Drosophila intron DNA is as mutable as Ig DNA. An analysis of the types of mutations in this DNA revealed that their chemical nature is similar to that previously shown to be characteristic of somatic mutation in Ig V region DNA and flanking sequence (29–32). Of the 67 mutations scored, 65 were nucleotide substitutions and 2 were single base insertions (Table 2). There were also the two large (350- and 470-bp) deletions mentioned above. 48% of the mutations were transitions and 58% of the changes were from germline A residues. 34% of the mutations occurred at either GC or TA dinucleotides and 22% occurred in the trinucleotides AGC or TAC (or their inverse repeats) previously identified as B cell somatic hypermutation “hotspots” (30, 32). 17% of the mutations occurred in the “RGYW” hotspot motif identified by Rogozen and Kolchanov (29). Finally, a pairwise comparison of the frequency of mutations that, had they occurred on different strands, would give rise to the same change observed on one strand (e.g., A to G versus T to C) indicated that the mutation took place with a strand bias of the same polarity as that previously indicated from the analysis of canonical endogenous VH genes (28).
Table 2.
Location and Type of Somatic Mutations in Enlarged Leader Intron Hybrid Loci
| Nucleotide | 3ACPM2-23 | 4ACPM1-2 | 4ACPM1-3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B2 | G6 | D9 | G3 | C6 | D5 | F1 | ||||||||
| Vector | ||||||||||||||
| 1 | ||||||||||||||
| 3′ of Promoter | ||||||||||||||
| 113 | GTC→ GGC | |||||||||||||
| 116 | CTC→ CAC | |||||||||||||
| Leader intron | ||||||||||||||
| 186 | TAA→ TCA | |||||||||||||
| 202 | GGT→ GCT | |||||||||||||
| 209 | ATT→ AAT | |||||||||||||
| Dros. intron | ||||||||||||||
| 247 | TAG→ TTG | |||||||||||||
| 249 | CA→ CCA | |||||||||||||
| 263 | TGT→ TCT | |||||||||||||
| 276 | start Δ | TGT→ TAT | ||||||||||||
| 306 | Δ | CAT→ CGT | ||||||||||||
| 324 | Δ | TAC→ TCC | ||||||||||||
| 367 | Δ | |||||||||||||
| 369 | Δ | CAC→ CTC | ||||||||||||
| 375 | Δ | AAC→ AGC | AAC→ AGC | |||||||||||
| 391 | Δ | GTA→ GAA | ||||||||||||
| 400 | Δ | ATG→ AGG | ||||||||||||
| 414 | Δ | GAA→ GCA | ||||||||||||
| 417 | Δ | TAT→ TGT | ||||||||||||
| 427 | Δ | AAG→ AGG | ||||||||||||
| 428 | Δ | AGT→ AAT | ||||||||||||
| 431 | Δ | TAT→ TTT | ||||||||||||
| 436 | Δ | CTT→ CAT | ||||||||||||
| 441 | Δ | AGT→ ACT | ||||||||||||
| 466 | Δ | CA→ CCA | ||||||||||||
| 472 | Δ | AGC→ AAC | ||||||||||||
| 479 | Δ | GAT→ GGT | ||||||||||||
| 489 | Δ | AAA→ ACA | ||||||||||||
| 509 | Δ | AAT→ ATT | ||||||||||||
| 519 | Δ | start Δ | ||||||||||||
| 520 | Δ | TAA→ TGA | Δ | |||||||||||
| 531 | Δ | TAA→ TTA | Δ | |||||||||||
| 565 | Δ | GAT→ GGT | Δ | |||||||||||
| 576 | Δ | AGT→ AAT | Δ | |||||||||||
| 579 | Δ | AAT→ ACT | Δ | |||||||||||
| 583 | Δ | TAC→ TGC | Δ | |||||||||||
| 592 | Δ | TAT→ TTT | Δ | |||||||||||
| 595 | Δ | GTA→ GCA | Δ | |||||||||||
| 613 | Δ | TAT→ TCT | Δ | |||||||||||
| 634 | Δ | TAC→ TTC | Δ | |||||||||||
| 638 | Δ | TAA→ TGA | Δ | |||||||||||
| 646 | Δ | TAC→ TCC | Δ | |||||||||||
| Nucleotide | 3ACPM2-23 | 4ACPM1-2 | 4ACPM1-3 | |||||||||||
| B2 | G6 | D9 | G3 | C6 | D5 | F1 | ||||||||
| 701 | Δ | TAT→ TTT | Δ | |||||||||||
| 703 | Δ | TAG→ TGG | Δ | |||||||||||
| 705 | Δ | GCA→ GTA | Δ | |||||||||||
| 706 | end Δ | Δ | ||||||||||||
| 712 | AGC→ AAC | Δ | ||||||||||||
| 715 | AAT→ AGT | Δ | ||||||||||||
| 721 | AGC→ ATC | Δ | ||||||||||||
| 726 | TTC→ TGC | Δ | ||||||||||||
| 734 | TTA→ TAA | Δ | ||||||||||||
| 735 | TAT→ TTT | Δ | ||||||||||||
| 751 | TAA→ TGA | Δ | ||||||||||||
| 756 | AAG→ AGG | Δ | ||||||||||||
| 763 | TTA→ TAA | Δ | ||||||||||||
| 773 | GCC→ GTC | Δ | ||||||||||||
| 776 | AAG→ AGG | Δ | ||||||||||||
| 794 | TTT→ TCT | Δ | ||||||||||||
| 826 | CGT→ CTT | Δ | ||||||||||||
| 845 | CGT→ CAT | Δ | ||||||||||||
| 855 | AAT→ ATT | Δ | ||||||||||||
| 857 | TTT→ TCT | Δ | ||||||||||||
| 860 | TAT→ TTT | TAT→ TGT | Δ | |||||||||||
| 868 | AAG→ AGG | Δ | ||||||||||||
| 869 | end Δ | |||||||||||||
| 905 | TAT→ TGT | TAT→ TGT | ||||||||||||
| 926 | GGT→ GAT | |||||||||||||
| 951 and 952 | ACC→ ATT | |||||||||||||
| 954 | CCT→ CAT | |||||||||||||
| 956 | TAA→ TCA | |||||||||||||
| 968 | AAT→ ATT | |||||||||||||
| Leader intron | ||||||||||||||
| 977 | AAG→ AGG | AAG→ AGG | ||||||||||||
| 983 | AAT→ ACT | |||||||||||||
| 1006 | TCC→ TAC | |||||||||||||
| FW 1 | ||||||||||||||
| 1068 | AGC→ ACC | |||||||||||||
| 1079 | GGC→ GAC | |||||||||||||
| 1110 | GCT→ GTT | |||||||||||||
| 1114 | CTG→ CAG | |||||||||||||
| 1119 | TAT→ TTT | |||||||||||||
| CDR 1 | ||||||||||||||
| 1132 | GCT→ GTT | |||||||||||||
| 1139 | TAT→ TTT | TAT→ TGT | ||||||||||||
| 1141 | TAA→ TCA | TAA→ TGA | ||||||||||||
| FW2 | ||||||||||||||
| 1152 | AAA→ AGA | |||||||||||||
| 1183 | TTG→ TAG | |||||||||||||
| Nucleotide | 3ACPM2-23 | 4ACPM1-2 | 4ACPM1-3 | |||||||||||
| B2 | G6 | D9 | G3 | C6 | D5 | F1 | ||||||||
| CDR2 | ||||||||||||||
| 1190 | TAT→ TTT | |||||||||||||
| 1203 | AAT→ AGT | |||||||||||||
| 1204 | ATG→ AAG | |||||||||||||
| 1210 | ATA→ ACA | |||||||||||||
| 1212 | ACT→ ATT | ACT→ ATT | ACT→ ATT | |||||||||||
| 1215 | AAG→ ACG | |||||||||||||
| 1216 | AGT→ ACT | |||||||||||||
| FW3 | ||||||||||||||
| 1246 | CAC→ CTC | |||||||||||||
| 1255 | TAG→ TCG | |||||||||||||
| CDR3 | ||||||||||||||
| 1357 | ACT→ ATT | |||||||||||||
| 1368 | TAC→ TCC | |||||||||||||
| JH2-JH3 | ||||||||||||||
| Intron | ||||||||||||||
| 1479 | TAT→ TTT | |||||||||||||
| 1595 | TTA→ TCA | |||||||||||||
| 1597 | ATA→ ACA | |||||||||||||
| 1694 | ||||||||||||||
The left hand column lists the nucleotide positions observed to be somatically mutated in the enlarged leader intron hybrid loci. Nucleotide 1 is 148 nucleotides 5′ of the initiation codon in the VH leader exon. The sequenced region ends 199 bases 3′ of the VDJ. Various subregions of the hybrid loci are labeled at the beginning of each subregion. The mutations observed in each of the seven hybrid loci are shown below the hybridoma names. In each case the germline nucleotide triplet (or doublet in the case of the insertion mutations) is shown, as is the mutant nucleotide triplet, with the mutant base underlined. The beginning and end of the large continuous deletions present in two of the hybrid loci are indicated by “start Δ” and “end Δ” with all intervening nucleotide positions listed also indicated by “Δ”.
Discussion
The experiments reported here provide a direct test of previously formulated hypotheses based on retrospective studies (8, 11, 17) proposing that the transcriptional promoter plays a mechanistic role in the hypermutation process. The data obtained demonstrate that the H chain promoter is the cis-acting element that establishes the 5′ boundary to hypermutation, and that hypermutation acts preferentially in a 1–1.5-kb region just 3′ of this element irrespective of the location of 3′ flanking regulatory elements (such as the intronic enhancer). Taken together with retrospective analyses of the distribution of mutations in Igκ and λ loci (9, 10, 12, 17), recent results suggesting an important role for the Vκ promoter in influencing location of somatic mutations in Igκ transgenes (20), and the compatibility of the β-globin promoter with hypermutation of Igκ transgenes (19), our data suggest that promoters perform a generic mutation directing function in all loci subjected to hypermutation. Since the heterologous B29 promoter is compatible with H chain locus hypermutation, albeit at what appears to be a reduced rate, the trans-acting factor(s) involved in regulating site specificity must interact with both VH and B29 promoters. One likely candidate for such a factor is RNA polymerase. Given this idea, the polarity of hypermutation relative to the promoter is explained by the directionality of transcription, and the rate of transcription is related to the rate of hypermutation.
The conclusion that the B29 promoter supports a reduced rate of hypermutation appears, at face value, discordant with the previous results of Betz et al. (19) who found that replacing the natural Vκ promoter with the human β-globin promoter in an Igκ transgene resulted in high levels of Vκ mutation. However, in their studies hypermutation was only evaluated in Peyer's patch germinal center B cells, a population known to undergo chronic antigen stimulation and prolonged hypermutation. In this approach, the frequency of transgene mutation cannot be compared to endogenous internal control V genes. Moreover, careful examination of the data presented by Betz et al. (19) suggests that the frequency of mutation observed in β-globin promoter driven κ transgenes is somewhat lower than in κ transgenes in which transcription is driven by the natural promoter.
If the rate of hypermutation is related to rate of transcription, promoters with different activities should result in different mutation rates. In this regard, the B29 promoter has an approximately fivefold lower activity in in vitro transfection experiments than the VH promoter/IgH intron enhancer combinations (Wall, R., personal communication). However, it appears that the activity of the B29 promoter in hybrid loci and of the VH promoter in canonical endogenous loci is not greatly different in hybridoma cells lines; Northern analysis of RNA from hybridomas derived from BP2 mice revealed similar steady state levels of H chain mRNA expressed from both types of loci (data not shown). Nevertheless, the activity of the B29 promoter in association with the H chain enhancers has not been assessed in hypermutating B cells, and could differ from the activity of VH promoters. Since quantitative analysis of gene transcription rates in antigen specific germinal center B cells is not yet technically feasible, further studies will be required to resolve these issues.
Interestingly, B cell clones expressing B29 promoterdriven hybrid loci appear to gain only limited access to the memory B cell compartment. Only 10% of the BP2 hybridomas expressing a canonical VH and Vκ isolated from secondary and tertiary fusions expressed a hybrid locus. In contrast, nearly 50% of such BP2 hybridomas from primary fusions expressed hybrid loci. Since affinity or specificity enhancing mutations appear to be prerequisites for clonal selection into memory (24, 34, 35), an attractive explanation for this observation is that a lower rate of mutation supported by the B29 promoter results in a lower probability of sustaining these necessary mutations. It remains to be determined whether the mutation rates supported by different VH and VL promoters differ substantially. If so, the structure of this portion of a V gene may be as important as the V(D)J coding region in determining whether a given clonotype contributes to the memory B cell compartment.
The influence of the promoter on hypermutation rate could result from indirect effects such as changes in DNA accessibility or topology mediated by transcription. However, more direct effects of promoter function on hypermutation rate (see below) could also be considered. How this element might regulate the location of the characteristic zone of hypermutation is less apparent. However, since in our experiments a minimal VH promoter was used in the enlarged leader intron transgene constructs, and replacement of this promoter with the minimal B29 promoter in the wild-type construct did not appear to alter the distribution of somatic mutation, this “mutation directing” activity of the promoter must overlap substantially, if not completely, with its activity in directing transcription.
Two models for the hypermutation process have been previously proposed that are particularly relevant in light of these conclusions. Peters and Storb (20) have suggested that a tissue-specific trans-acting mutator factor binds to RNA polymerase during transcription initiation in Ig loci, and that this factor-polymerase complex has a limited half-life during elongation, restricting the intact complex from transcribing DNA further than ∼1.5 kb from the promoter. In this model, an increased frequency of “pausing” of the polymerase induced by the mutator factor is thought to result in recruitment of enzymes involved in transcriptioncoupled DNA repair that subsequently act in an error prone fashion. Given the data presented here and by Betz et al. (19) demonstrating that heterologous promoters are compatible with hypermutation, an Ig locus–specific cisacting factor(s) other than the promoter would seem to be necessary for “loading” of the factor into the transcription initiation complex. Moreover, this model relies on the rather ad hoc assumption that the action of transcription coupled DNA repair, which normally repairs DNA damage efficiently in regions in which RNA polymerase has paused (36), is reversed, resulting in a substantially higher mutation rate in these regions.
Steele and Pollard (37) have suggested that an RNA intermediate is involved in hypermutation. In their model, error prone reverse transcription of this RNA intermediate, followed by recombination of the mutant cDNA into the template VDJ gene, results in mutational alteration of the VDJ and flanking sequence. This model is consistent with the nature of the promoter influencing the rate of hypermutation, since this rate should be directly related to the amount of RNA intermediate produced, which, in turn, should be related to the rate of transcription. However, this model attempts to account for the restricted distribution of mutation by invoking a reverse transcription primer site(s) between the V and C exons, thus resulting in mutant cDNAs being produced only from this primer site to the mRNA cap site. Our data are incompatible with this latter proposition, since moving the promoter 750 bp further upstream of the V-C intron dramatically reduces mutation frequency in this intron (Fig. 4).
Finally, neither of these models adequately accounts for the lower frequency of mutation observed in transcribed regions immediately 3′ of the promoter (Fig. 5), which our data suggest is a characteristic feature of the mutation mechanism that is not influenced by the location of the promoter or the type of DNA subjected to mutation (Fig. 4). Further evaluation of these models will require experimental evaluation of whether components of the Ig transcriptional apparatus differ in B cells that have and have not activated the hypermutation process, and whether hypermutating B cells contain mutant Ig cDNA intermediates. Such studies will undoubtedly provide new insight into the nature of the trans-acting factors involved in the hypermutation process.
Acknowledgments
We would like to thank Dr. Mary Beth Davis for the D. melanogaster intron clone, Dr. Randy Wall for the B29 promoter clone, Dr. Latham Claflin for suggesting the enlarged intron experiment, Judith Morgan for construction of transgenic founders, William Monsell for technical assistance, and all other members of the Manser lab for their indirect contributions.
Footnotes
This work was supported by grants from the National Institutes of Health (AI23739) and the Council for Tobacco Research (3763). K. Tumas-Brundage was supported by a National Institutes of Health training grant (CA09683).
1 Abbreviations used in this paper: Ars, arsonate; V, variable.
References
- 1.Weill J-C, Reynaud CA. Rearrangement/hypermutation/gene conversion: when, where and why? . Immunol Today. 1996;17:92–97. doi: 10.1016/0167-5699(96)80586-x. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky K, Forster I, Cumano A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science (Wash DC) 1987;238:1088–1094. doi: 10.1126/science.3317826. [DOI] [PubMed] [Google Scholar]
- 3.McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien NC, Pollack RR, Desaymard C, Scharff MD. Point mutations cause the somatic diversification of IgM and IgG2a. J Exp Med. 1988;167:954–973. doi: 10.1084/jem.167.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wysocki LJ, Gefter ML. Gene conversion and the generation of antibody diversity. Ann Rev Biochem. 1989;58:509–531. doi: 10.1146/annurev.bi.58.070189.002453. [DOI] [PubMed] [Google Scholar]
- 6.Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med. 1993;178:1293–1307. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roes J, Huppi K, Rajewsky K, Sablitsky F. V gene rearrangement is required to fully activate the hypermutation mechanism in B cells. J Immunol. 1989;142:1022–1026. [PubMed] [Google Scholar]
- 8.Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunglobulin genes: 5′ boundary is near the promoter and 3′ boundary is approximately 1 kb from V(D)J. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motoyama N, Okada HM, Azuma T. Somatic mutation in constant regions of mouse λ light chains. Proc Natl Acad Sci USA. 1991;88:7933–7937. doi: 10.1073/pnas.88.18.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber JS, Berry J, Manser T, Claflin JL. Position of the rearrranged Vκ and its 5′ flanking sequences determines the location of somatic mutations in the Jκ locus. J Immunol. 1991;146:3652–3655. [PubMed] [Google Scholar]
- 11.Rothenfluh HS, Taylor L, Bothwell ALM, Both GW, Steele EJ. Somatic hypermutation in 5′ flanking regions of heavy chain antibody variable regions. Eur J Immunol. 1993;23:2152–2159. doi: 10.1002/eji.1830230916. [DOI] [PubMed] [Google Scholar]
- 12.Rogerson BJ. Mapping the upstream boundary of somatic mutations in rearranged immunoglobulin transgenes and endogenous genes. Mol Immunol. 1994;31:83–98. doi: 10.1016/0161-5890(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 13.Manser T, Wysocki LJ, Margolies MN, Gefter ML. Evolution of antibody variable region structure during the immune response. Immunol Rev. 1987;96:141–162. doi: 10.1111/j.1600-065x.1987.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 14.Berek C, Milstein C. The dynamic nature of the antibody repertoire. Immunol Rev. 1988;105:5–26. doi: 10.1111/j.1600-065x.1988.tb00763.x. [DOI] [PubMed] [Google Scholar]
- 15.Manser T. Evolution of antibody structure during the immune response: the differentiative potential of a single B lymphocyte. J Exp Med. 1989;170:1211–1230. doi: 10.1084/jem.170.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan H, Shlomchik M, Weigert M. Heavychain class switch does not terminate somatic mutation. J Exp Med. 1990;172:531–536. doi: 10.1084/jem.172.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber JS, Berry J, Manser T, Claflin JL. Mutation in Ig V(D)J genes are distributed asymmetrically and independently of the position of V(D)J. J Immunol. 1994;153:3594–3602. [PubMed] [Google Scholar]
- 18.Giusti A, Manser T. Hypermutation is observed only in antibody H chain V region transgenes that have recombined with endogenous immunoglobulin H DNA: implications for the location of cis-acting elements required for somatic mutation. J Exp Med. 1993;177:797–809. doi: 10.1084/jem.177.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 20.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, B., F. Costantini, and E. Lacy. 1986. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 332 pp.
- 22.Marshak-Rothstein A, Siekevitz M, Margolies MN, Madgett-Hunter M, Gefter ML. Hybridoma proteins expressing the predominant idiotype of the anti-azophylarsonate response of A/J mice. Proc Natl Acad Sci USA. 1980;77:1120–1124. doi: 10.1073/pnas.77.2.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissman D, Rothstein TL, Marshak-Rothstein A. A rapid method for comparing monoclonal antibodies by limited proteolysis and electrophoresis. Hybridoma. 1985;4:329–339. doi: 10.1089/hyb.1985.4.329. [DOI] [PubMed] [Google Scholar]
- 24.Vora KA, Manser T. Altering the antibody repertoire via transgene homologous recombination: evidence for global and clone-autonomous regulation of antigen-driven B cell differentiation. J Exp Med. 1995;181:271–281. doi: 10.1084/jem.181.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giusti AM, Coffee R, Manser T. Somatic recombination of heavy chain variable region transgenes with the endogenous immunoglobulin heavy chain locus in mice. Proc Natl Acad Sci USA. 1992;89:10321–10325. doi: 10.1073/pnas.89.21.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sohn J, Gerstein RM, Hsieh CL, Lemer M, Selsing E. Somatic hypermutation of an immunoglobulin μ heavy chain transgene. J Exp Med. 1993;177:493–504. doi: 10.1084/jem.177.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharon J, Gefter ML, Wysocki LJ, Margolies MN. Recurrent somatic mutations in mouse antibodies to p-azophenylarsonate increase affinity for hapten. J Immunol. 1989;142:596–601. [PubMed] [Google Scholar]
- 28.Manser, T. 1991. Regulation, timing, and mechanism of antibody V gene somatic hypermutation: lessons from the arsonate system. In Somatic Hypermutation in V Regions. E.J. Steele, editor. CRC Press, Boca Raton, FL. 41–54.
- 29.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighboring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 30.Betz AG, Neuberger MS, Milstein C. Discriminating intrinsic and antigen-selected mutational hotspots in immunoglobulin V genes. Immunol Today. 1993;14:405–411. doi: 10.1016/0167-5699(93)90144-a. [DOI] [PubMed] [Google Scholar]
- 31.Gearhart, P. 1993. Somatic mutation and affinity maturation. In Fundamental Immunology. 3rd ed. W. Paul, editor. Raven Press, New York. 865–885.
- 32.Smith DS, Creadon G, Jena PK, Portanova JP, Kotzin BL, Wysocki LJ. Di- and trinucleotide target preferences of somatic mutagenesis in normal and autoreactive B cells. J Immunol. 1996;156:2642–2652. [PubMed] [Google Scholar]
- 33.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YU, Gonzalez-Fernandez A, Pannell R, Neuberger MS, Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature (Lond) 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 34.Weiss U, Rajewsky K. The repertoire of somatic antibody mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J Exp Med. 1990;172:1681–1689. doi: 10.1084/jem.172.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casson LP, Manser T. Random mutagenesis of two complementarity determining region amino acids yields an unexpectedly high frequency of antibodies with increased affinity for both cognate antigen and autoantigen. J Exp Med. 1995;182:743–750. doi: 10.1084/jem.182.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele EJ, Pollard JW. Hypothesis: somatic hypermutation by gene conversion via errror prone DNA→ RNA→ DNA information loop. Mol Immunol. 1987;24:667–673. doi: 10.1016/0161-5890(87)90049-6. [DOI] [PubMed] [Google Scholar]
- 38.Hermanson GG, Briskin M, Sigman D, Wall R. Immunoglobulin enhancer and promoter motifs 5′ of the B29 B cell specific gene. Proc Natl Acad Sci USA. 1989;86:7341–7345. doi: 10.1073/pnas.86.19.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]