Abstract
CD94 is a C-type lectin expressed by natural killer (NK) cells and a subset of T cells. Blocking studies using anti-CD94 mAbs have suggested that it is a receptor for human leukocyte antigen class I molecules. CD94 has recently been shown to be a 26-kD protein covalently associated with an unidentified 43-kD protein(s). This report shows that NKG2A, a 43-kD protein, is covalently associated with CD94 on the surface of NK cells. Cell surface expression of NKG2A is dependent on the association with CD94 as glycosylation patterns characteristic of mature proteins are found only in NKG2A that is associated with CD94. Analysis of NK cell clones showed that NKG2A was expressed in all NK cell clones whose CD16-dependent killing was inhibited by cross-linking CD94. The induction of an inhibitory signal is consistent with the presence of two immunoreceptor tyrosine-based inhibitory motifs (V/LXYXXL) on the cytoplasmic domain of NKG2A. Similar motifs are found on Ly49 and killer cell inhibitory receptors, which also transmit negative signals to NK cells.
NK cells exhibit cytolytic activity against tumors and virus-infected cells (1). The expression of MHC class I molecules on target cells confers resistance to NK-mediated lysis (2–4). A number of NK cell receptors have been identified that interact with MHC class I molecules and send inhibitory signals to the NK cell. In mice, these receptors are the Ly49 molecules, type II integral membrane proteins of the C-type lectin superfamily (5–7). In humans, the best characterized NK cell receptors for HLA class I molecules are members of the Ig superfamily and have been termed killer cell inhibitory receptors (KIR) (8–10). In addition, CD94, a C-type lectin (11) like Ly49, has also been suggested as a receptor for HLA class I molecules (12–14).
CD94 was originally described as a 70-kD disulfide bonded dimeric protein that was expressed primarily on NK cells and a subset of T cells (15). Cross-linking of CD94 in in vitro cytotoxicity assays leads to one of three effects on target cells depending on the NK cell clone (16, 17): increased target cell lysis, decreased target cell lysis or no substantial change in lysis. These different functional phenotypes, which divide NK cell clones into three distinct groups, were shown to correlate with different patterns of intracellular tyrosine phosphorylation (17). The protein sequence of CD94 cannot account for the diverse phenotypes observed after cross-linking CD94 as it has a cytoplasmic tail of only seven amino acids, lacking any well defined signaling motifs (11). Recently CD94 was shown to be a 25–30-kD protein covalently associated with an as yet unidentified protein(s) of 43 kD (14). An understanding of the basis for the phenotypic differences observed in NK clones after CD94 cross-linking probably requires the identification of the CD94 associated protein(s) and their signaling potential.
The NKG2 family of genes, designated NKG2A, C, D and E, was originally identified by screening a subtractive library enriched for NK- and T cell–specific transcripts (18). Chromosomal mapping and analysis of the cDNA sequences showed that like CD94, the NKG2 genes are located in the NK complex on chromosome 12 and the proteins encoded by these genes are members of the C-type lectin family (19). Moreover, Northern blot analysis showed that the NKG2 mRNAs have a similar tissue distribution to CD94 (19). These observations, coupled with the fact that NKG2A is ∼43 kD in size and contains two immunoreceptor tyrosine based inhibitory motifs (ITIM) in its cytoplasmic domain, prompted us to explore the possibility that NKG2A may be the unidentified, covalently-associated partner of CD94. In this report we show that NKG2A and CD94 form heterodimers which are expressed on the cell surface of NK cells. Furthermore, the transmission of an inhibitory signal after CD94 cross-linking correlates with the expression of NKG2A by NK cell clones. This suggests that the CD94/NKG2A heterodimer can deliver an inhibitory signal to NK cells, presumably mediated by the cytoplasmic domain of NKG2A.
Materials and Methods
Cells.
NK-92 cells (20) (a gift from Dr. H Klingemann, Terry Fox Laboratory, BC Cancer Agency, Vancouver, Canada) were maintained in Myelocult H5100 (Stemcell Technologies, Vancouver, Canada) supplemented with 20 U/ml of rIL-2 (a gift from Hoffmann-La Roche, Nutley, NJ). The NK cell line YT (a gift from Dr. J. Ortaldo, Laboratory of Experimental Immunology, NCI-FCRDC, Frederick, MD), the T cell line, Jurkat, and the murine mastocytoma line P815, were grown in RPMI 1640 (Biofluids, Rockville, MD), supplemented with 10% FCS (Hyclone, Logan, UT), 1 mM Hepes (Biofluids).
Polyclonal NK cells were obtained from PBMC from healthy donors using the MACS NK isolation kit (Miltenyi Biotec, Incorporated, CA). NK cell populations were always >92% CD56 positive and <6% CD3 positive.
NK cell clones were generated by seeding NK cells under limiting dilution into U-bottom 96-well plates in the presence of 2 × 105 irradiated allogeneic feeder cells. Additional feeder cells (105) from the same donor were added after 5 d in culture. NK clones were identified between days 15 and 21 and then expanded. All clones were CD3−, CD16+, CD56+, and CD94+.
Antibodies.
The anti-CD94-specific mAb (HP-3B1) was purchased from Immunotech (Westbrook, ME), the anti-Ly49C (5E6) and anti-CD16 (3G8) mAbs were purchased from PharMingen (San Diego, CA), and the anti-CD3 (UCH-T1), antiCD16 (B-E16) and anti-CD56 (B159) mAbs were purchased from Serotec (Washington, DC). FITC labeled goat anti–mouse Ig was purchased from Jackson ImmunoResearch Laboratories Inc. (Westgrove, PA). The NKG2A-specific mAb 1A12 was a gift from Dr. J.P. Houchins (University of Minnesota, Minneapolis, MN).
NKG2A-peptide-specific rabbit antiserum was obtained by immunization with a peptide containing the amino-terminal 25 amino acids of NKG2A coupled to KLH. Peptide-specific antibodies were purified from the rabbit antiserum using a recombinant form of the entire cytoplasmic domain of NKG2A made in Escherichia coli. (a gift from Dr. J.P. Houchins) coupled to Emphaze (3M) biosupport medium AB1 (Pierce, Rockford, IL).
Immunoprecipitations and Immunoblot Analysis.
Cells were harvested and washed three times in PBS and surface labeled by incubation with 0.5 mg/ml NHS-LC biotin (Pierce Chem. Co., Rockford, IL) for 30 min at room temperature. Cells were washed three more times in PBS and lysed in a buffer containing 2% Triton X-100, 50 mM Tris (pH 8.0), 0.15 M NaCl, 20 mM iodoacetamide, 5 mM EDTA at 2 × 107cells/ml. After preclearing with Sepharose CL4B (Pharmacia Biotech, Uppsala, Sweden), lysates were incubated for 2 h at 4°C with mAbs or polyclonal antisera which had previously been adsorbed to protein G agarose (GIBCO BRL, Gaithersburg, MD). Bound proteins were washed five times in a buffer containing 2% Triton X-100, 20 mM Tris, 450 mM NaCl and separated by SDS-PAGE. Proteins were transferred to Immobilon-P and biotinylated proteins were detected by incubation with streptavidin-peroxidase (Amersham Corp., Arlington Heights, IL) followed by ECL (Amersham). Immunoblot analysis was performed as previously described (21).
Glycosidase Digestions.
Whole cell lysates or immunoprecipitated proteins were digested with endoglycosidase Hf (New England Biolabs, Beverly, MA), or mock digested using conditions specified by the manufacturer. Digested proteins were separated by SDS-PAGE and analyzed by Western blot as described above.
Cytotoxicity Assays.
Standard chromium release assays were used to measure NK cytotoxicity (12). The anti-CD16 mAb 3G8 and the anti-CD94 mAb HP-3B1 were used at final concentrations of 10 μg/ml each.
Results and Discussion
CD94 was recently shown to be covalently associated with an unidentified 43-kD protein(s) (14). We hypothesized that the CD94-associated protein may be a member(s) of the NKG2 family of proteins as they have molecular masses consistent with that of the CD94-associated protein(s) and have a similar tissue distribution and structure to CD94 (19, 22).
We wanted to determine whether NKG2A was associated with CD94. Initially, three cell lines NK-92, YT and Jurkat were assessed for NKG2A expression by Western blot. Analysis with anti-NKG2A peptide Ab showed that no NKG2A reactive proteins could be detected in Jurkat cells (Fig. 1). Under reducing conditions, proteins with molecular masses of 40–45 kD and ∼35 kD could be detected in NK-92 and ∼40 kD and ∼35 kD in YT. Under nonreducing conditions, the major species detected in YT cells had a molecular mass of ∼40 kD similar to that observed under reducing conditions whereas in NK-92 cell lysates the major species had a molecular mass of ∼70 kD. A 35-kD protein with similar mobility to that observed under reduced conditions was again detected in both NK-92 and YT cell lysates. These data indicate that NKG2A is covalently associated with another protein which is present in NK-92 but absent in YT. Unlike NK-92, YT does not express CD94, as assessed by flow cytometry (data not shown), which suggested that CD94 could be the heterodimer partner of NKG2A.
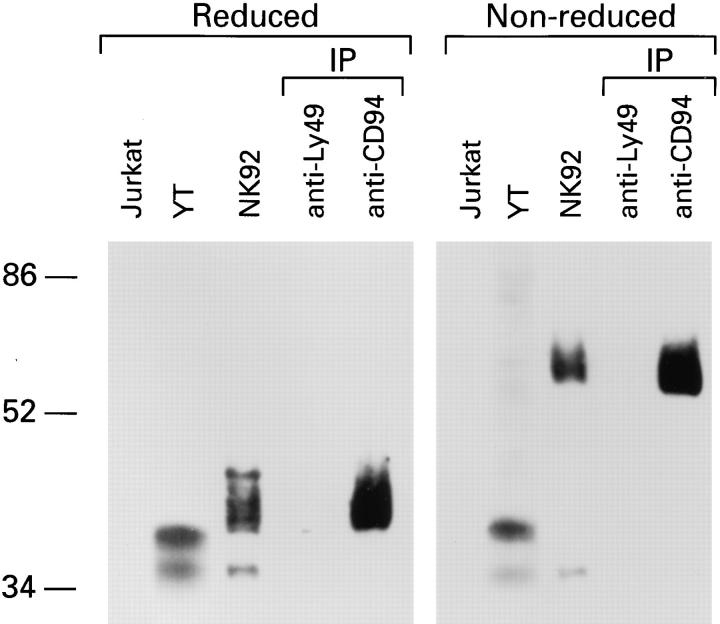
Figure 1.
NKG2A is covalently associated with CD94. Lysates from either Jurkat, YT or NK-92 cells, together with anti-CD94 or control (antiLy49) immunoprecipitates (IP) from NK-92 were separated by SDS-PAGE under reducing or non-reducing conditions and analyzed by Western blot with anti-NKG2A peptide-specific Ab.
To test directly whether CD94 is associated with NKG2A, proteins were immunoprecipitated with anti-CD94 mAb from NK-92 cell lysates and analyzed by Western blot with anti-NKG2A-peptide Ab (Fig. 1) or anti-NKG2 mAb (data not shown). Under reducing conditions, a 40–45-kD protein of similar mobility to the protein detected in the whole cell lysates of NK-92, was detected in anti-CD94 but not control (anti-Ly49) immunoprecipitates (Fig. 1). Under non-reducing conditions, a 70-kD protein, again of similar molecular mass to that observed in the NK-92 lysates, was detected by anti-NKG2A-peptide Ab in antiCD94 immunoprecipitates. These data indicate that NKG2A and CD94 form heterodimers.
The 35-kD NKG2A protein was always detected in whole cell lysates of NKG2A positive cells and is always less abundant than the 40–45-kD form (Fig. 1, also see Fig. 3). The fact that anti-peptide-Ab specific for the amino terminus and mAbs-specific for the lectin domains of NKG2A (data not shown) each react with the 35-kD protein, suggested that it may represent NKG2B. The existence of a putative NKG2B protein was postulated from the detection of transcripts (cDNAs) for an alternative splice variant of NKG2A lacking 54 bases (22). These bases encode 18 amino acids in the stalk or neck region of the protein which links the lectin domain to the transmembrane region and contains a potential N-linked glycosylation site. The absence of this lower molecular mass form in anti-CD94 immunoprecipitates suggests that it lacks the structural requirements for pairing with CD94.
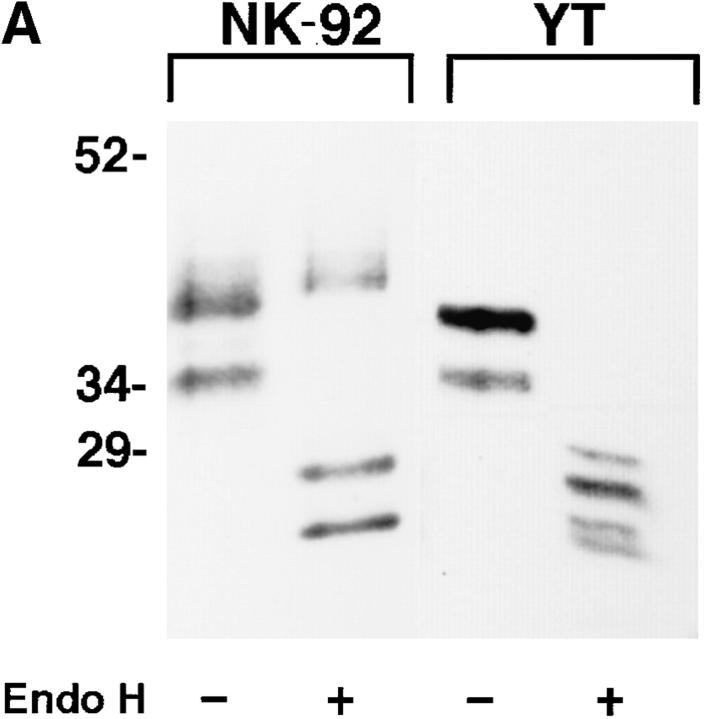
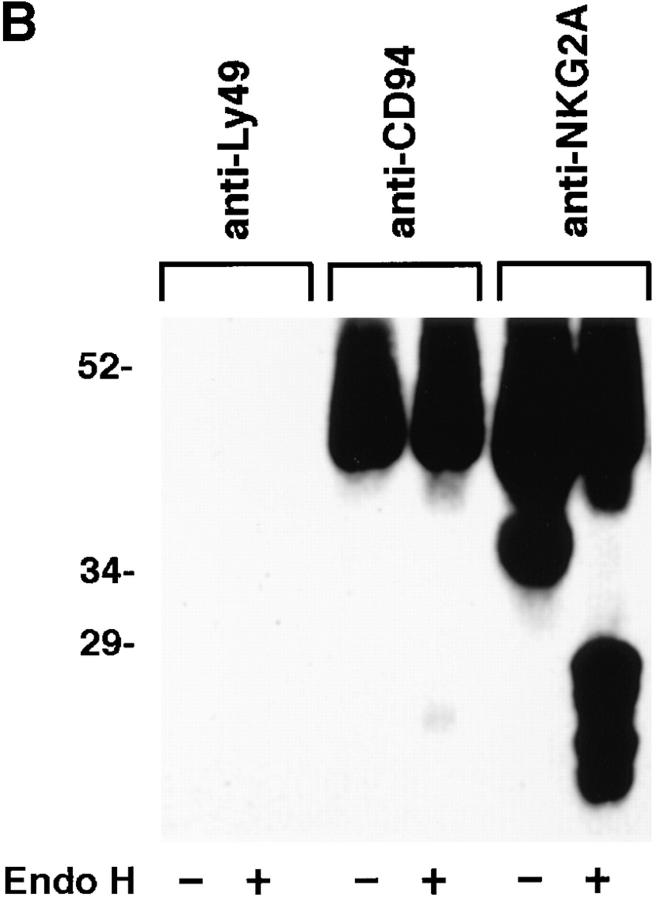
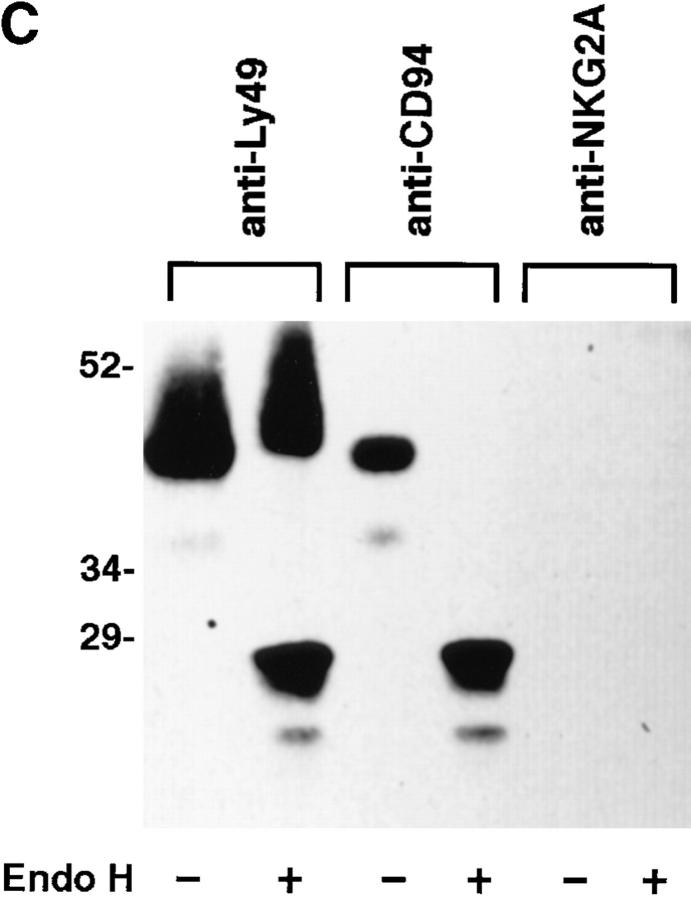
Figure 3.
NKG2A fails to mature in the absence of CD94. (A) Whole cell lysates of NK-92 or YT were either digested with Endoglycosidase Hf (+) or mock digested (−), separated by SDS-PAGE under reducing conditions and analyzed by Western blot with anti-NKG2A peptide-specific Ab. (B) Anti-CD94, anti-NKG2A or control (anti-Ly49), immunoprecipitates from NK-92 cells were either digested with Endoglycosidase Hf, (+) or mock digested (−), separated by SDS-PAGE under reducing conditions and analyzed by Western blot with anti-NKG2A peptide-specific Ab. (C) NK-92 cell lysates were exhaustively precleared with control (anti-Ly49), anti-CD94 or anti-NKG2A Ab. The remaining lysates were divided in two and either digested with Endoglycosidase Hf, (+) or mock digested (−), separated by SDS-PAGE under reducing conditions and analyzed by Western blot with antiNKG2A peptide Ab. The immunoblots in B and C were overexposed to detect trace amounts of immature CD94-associated NKG2A in B and to emphasize that no mature NKG2A could be detected in NK-92 lysates after removal of CD94 associated NKG2A in C.
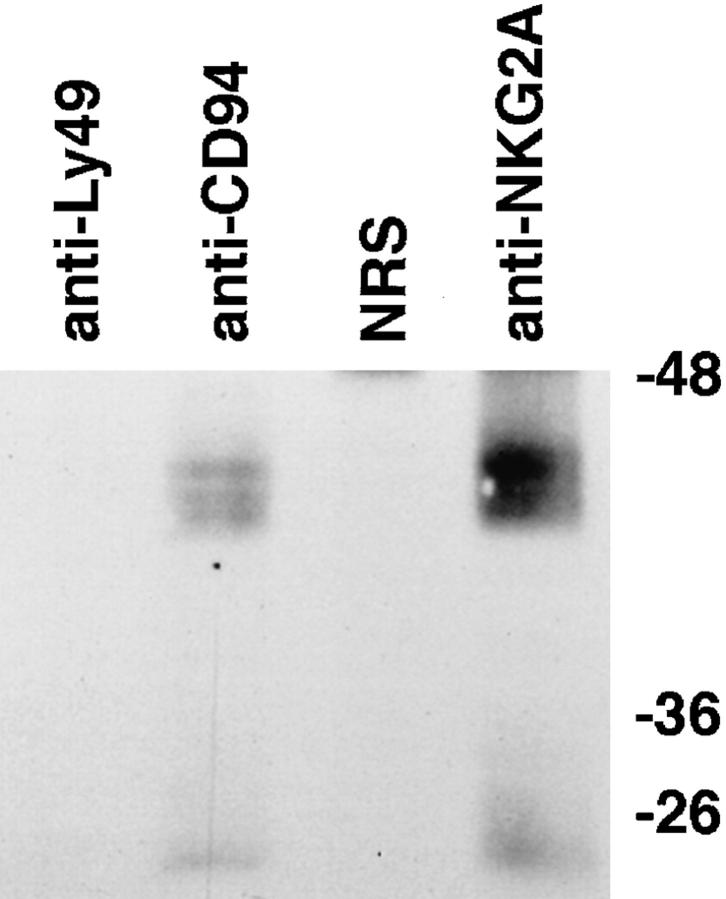
Given that NKG2A forms heterodimers with CD94, we next determined whether this heterodimer was expressed at the cell surface and thus was capable of interacting with target cell ligands. Cell surface proteins from polyclonal NK cells were labeled with biotin and proteins were immunoprecipitated with anti-CD94 mAb, anti-NKG2A-peptide Ab or control Ab. Detection with streptavidin-HRP revealed proteins with a molecular mass of ∼40–45 kD and a group of smaller proteins of molecular mass 25–28 kD in both the anti-CD94 and anti-NKG2A immunoprecipitates but not in control immunoprecipitates (Fig. 2). Western blot analysis of the immunoprecipitated proteins with antiNKG2A peptide Ab confirmed the identity of the 40–45-kD protein as NKG2A (data not shown). The 25–28-kD proteins observed in the anti-CD94 and anti-NKG2A immunoprecipitates correspond to the molecular mass of CD94 (14) and, as expected, these proteins did not react with anti-NKG2A Ab in Western blot analysis (data not shown). Thus, these data show for the first time that NKG2A is expressed on the cell surface of polyclonal NK cells and is complexed to CD94.
Figure 2.
NKG2A/CD94 heterodimers are expressed on the cell surface of polyclonal NK cells. Cell surface proteins of polyclonal NK cells were labeled with biotin and immunoprecipitated with anti-CD94, anti-NKG2A peptide-specific Ab, anti-Ly49 mAb or, normal rabbit serum (NRS). Immunoprecipitated proteins were reduced and separated on SDS-PAGE, transferred to PVDF, detected with streptavidin conjugated-HRP and visualized with enhanced chemiluminescence.
The fact that NKG2A derived from YT migrated as a monomer under non-reducing conditions (see Fig. 1), in conjunction with numerous immunoprecipitation experiments which had failed to show cell surface expression of NKG2A on YT cells (unpublished observations), suggested that CD94 is required for cell surface expression of NKG2A. These data also suggest that NKG2A may not fold correctly in the absence of CD94 and be retained intracellularly. To analyze the maturation state of NKG2A protein in YT and NK-92 cells, we made use of the enzyme endoglycosidase H (Endo H) which cleaves high mannose type polysaccharide side chains that are characteristic of immature glycoproteins, but not mature glycoproteins (23). YT and NK-92 cell lysates were either digested with Endo H or mockdigested and then analyzed by Western blot. This analysis showed the presence of Endo H–sensitive species of NKG2A migrating with apparent molecular masses of ∼28 kD and ∼25 kD (after digestion) in the lysates of both NK-92 and YT (Fig. 3 A). However, mature, Endo H–resistant species (M r ∼43–45 kD) were only observed in NK-92 cells. The finding that only Endo H–sensitive species of NKG2A were detected in YT cells indicates that NKG2A does not transit through the Golgi compartment and suggests that NKG2A requires the expression of an additional partner, such as CD94, in order to undergo normal maturation for cell surface expression.
To show that the maturation of NKG2A is dependent on the association with CD94, anti-CD94, anti-NKG2A and control immunoprecipitates from NK-92 cell lysates were either digested with Endo H or mock-digested and then subjected to Western blot analysis with anti-NKG2A peptide Ab. As expected, no NKG2A was detected in control antiLy49C immunoprecipitates (Fig. 3 B). The anti-NKG2A immunoprecipitates contained significant amounts of both Endo H–sensitive (∼28 kD and ∼25 kD) and –resistant species (∼43 kD), whereas NKG2A associated with CD94 (anti-CD94 immunoprecipitate) was greatly enriched for mature, Endo H–resistant NKG2A, such that only trace amounts of Endo H–sensitive NKG2A were detected (Fig. 3 B).
To confirm these results, NK-92 cell lysates were exhaustively precleared with either control (anti-Ly49C), anti-CD94 or anti-NKG2A peptide Ab. Precleared lysates were divided in half and digested with Endo H or mock digested and the remaining NKG2A analyzed by immunoblot. While both Endo H–resistant and sensitive forms of NKG2A were observed after preclearing with the control anti-Ly49C mAb, only Endo H–sensitive forms of NKG2A were observed after preclearing with anti-CD94 (Fig. 3 C). As expected, no NKG2A was observed after preclearing with anti-NKG2A. Collectively, these data show that in the absence of CD94, NKG2A is retained in the endoplasmic reticulum and unlike related proteins such as Ly49 and NKR-P1 is unable to homodimerize. Furthermore CD94 association is required for maturation and hence cell surface expression of NKG2A. Transfection studies have shown cell surface expression of CD94 in the absence of NKG2 proteins (11). It will be interesting to determine whether CD94 ever exists as a monomer or homodimer on the cell surface in vivo.
CD94 is expressed on virtually all NK cells, yet the transmission of an inhibitory signal after cross-linking CD94 has been shown to be restricted to particular NK cell clones (15–17). Western blot and mRNA analyses of NK cell clones (unpublished observations) had indicated that the expression of NKG2A was also restricted to a subset of NK cells. The cytoplasmic tail of NKG2A has two ITIMs similar to those found on KIR which can mediate an inhibitory signal through the association of hematopoietic cell phosphatase (24, 25). The presence of such motifs in NKG2A and the clonal distribution of NKG2A suggested that crosslinking of the NKG2A/CD94 heterodimer might transduce the inhibitory signal observed in some NK cell clones (17). Therefore, we assessed the effects of cross-linking CD94 on the activation signals delivered through CD16 in redirected lysis assays using a panel of 11 NK cell clones derived from two donors, 10 of which expressed NKG2A as detected by Western blot analyses (data not shown).
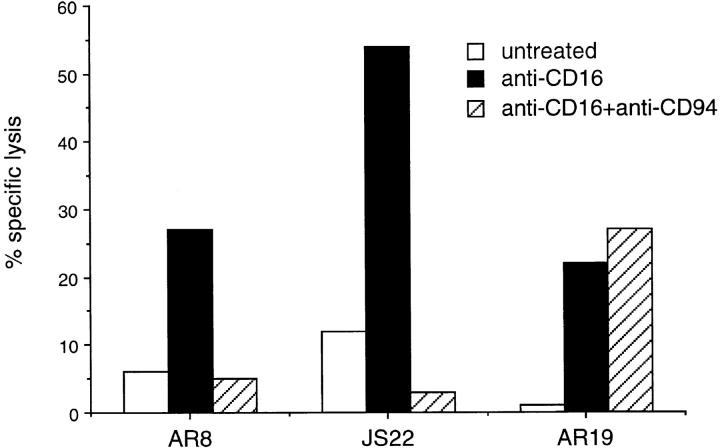
Data from three NK cell clones are shown in Fig. 4. Low levels of lysis of P815 target cells by both the NKG2A positive clones AR8 and JS22, and the NKG2A negative clone AR19 were observed in the absence of any mAb (Fig. 4). As expected, preincubation of P815 cells with anti-CD16 lead to a significant increase in target cell lysis by all clones. The presence of anti-CD94 had little effect on CD16dependent lysis of P815 cells by AR19. However crosslinking of CD94 on the NKG2A positive clones, AR8 and JS22, markedly inhibited anti-CD16-mediated lysis of P815 cells to levels comparable with the untreated controls. Our data suggest that NKG2A expression is required for the generation of an inhibitory signal after CD94 cross-linking. However, whether all NK cell clones expressing NKG2A are inhibited after CD94 cross-linking awaits a more extensive clonal analysis and is the subject of ongoing research.
Figure 4.
CD94 cross-linking inhibits CD16-mediated lysis of NKG2A positive NK cell clones. 51Cr-labeled P815 target cells were either preincubated with anti-CD16 (anti-CD16), or anti-CD16 and anti-CD94 (anti-CD16 + anti-CD94) or complete medium (untreated) prior to incubation with the NKG2A positive NK cell clones AR8 and JS22 or the NKG2A negative NK cell clone, AR19 for 4 h. 51Cr released was measured in a γ counter and the percentage of specific lysis calculated.
These data do not exclude the possibility that CD94 can covalently associate with other proteins. In particular, given the high degree of homology between NKG2A and NKG2C (22) and the phenotypic differences observed among NK cell clones after CD94 cross-linking (17), it would seem likely that CD94 can also associate with NKG2C. NKG2C has structural features in its transmembrane and cytoplasmic domains similar to those in the recently described p50 receptors which activate NK cells upon cross-linking (26, 27). Hence the expression of a putative CD94/NKG2C heterodimer may be responsible for the signal to engage the lytic machinery in NK clones which are activated after CD94 cross-linking.
We have shown that CD94 forms covalent heterodimers with NKG2A and that association with CD94 is required for cell surface expression of NKG2A. Furthermore the generation of an inhibitory signal after cross-linking with anti-CD94 antibodies correlates with the expression of the NKG2A protein. Whether the human NKG2A/CD94 protein represents a structurally related, functional counterpart of the murine Ly49 molecules remains to be determined.
Acknowledgments
We would like to thank Drs. J.P. Houchins for E. coli. expressing the cytoplasmic tail of NKG2A and the NKG2A-specific mAb 1A12, H-G. Klingemann for NK-92 cells and J. Ortaldo for YT cells. We thank the NIAID Peptide Synthesis Facility for peptide conjugates. We also thank Drs. E. Fernandez, E. Long, B. Passer, K. Parker, and J. Shuman for advice and comments on the manuscript.
Footnotes
Note added in proof: Since the original submission of this manuscript similar results including a description of the NKG2A/B-specific mAb 1A12 were reported by Lazetic et al. (Lazetic, S., C. Chang, J.P. Houchins, L.L. Lanier, and J.H. Phillips. 1996. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 157:4741–4745).
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimizu Y, DeMars R. Demonstration by class I gene transfer that reduced susceptibility of human cells to natural killer cell-mediated lysis is inversely correlated with HLA class I antigen expression. Eur J Immunol. 1989;19:447–451. doi: 10.1002/eji.1830190306. [DOI] [PubMed] [Google Scholar]
- 3.Storkus WJ, Alexander J, Payne JA, Dawson JR, Cresswell P. Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc Natl Acad Sci USA. 1989;86:2361–2364. doi: 10.1073/pnas.86.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science (Wash DC) 1991;253:199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 5.Smith HR, Karlhofer FM, Yokoyama WM. Ly-49 multigene family expressed by IL-2-activated NK cells. J Immunol. 1994;153:1068–1079. [PubMed] [Google Scholar]
- 6.Karlhofer FM, Ribaudo R, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+IL-2-activated natural killer cells. Nature (Lond) 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 7.Daniels BF, Karlhofer FM, Seaman WE, Yokoyama WM. A natural killer cell receptor specific for a major histocompatibility complex class I molecule. J Exp Med. 1994;180:687–692. doi: 10.1084/jem.180.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, Bottino C, Moretta L, Moretta A, Long EO. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 9.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science (Wash DC) 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 10.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1: a natural killer cell receptor for HLA-B allotypes. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 11.Chang C, Rodriguez A, Carretero M, Lopez-Botet M, Phillips J, Lanier LL. Molecular characterization of human CD94: a type II membrane glycoprotein related to the C-type lectin superfamily. EurJ Immunol. 1995;25:2433–2437. doi: 10.1002/eji.1830250904. [DOI] [PubMed] [Google Scholar]
- 12.Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, Barbaresi M, Pende D, Ciccone E, LopezBotet M, Moretta L. Human natural killer cell receptors for HLA-class I molecules. Evidence that the Kp43 (CD94) molecule functions as receptor for HLA-B alleles. J Exp Med. 1994;180:545–555. doi: 10.1084/jem.180.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbone E, Terrazzano G, Colonna M, Tuosto L, Piccolella E, Franksson L, Palazzolo G, Perez-Villar JJ, Fontana S, et al. Natural killer clones recognize specific soluble HLA class I molecules. Eur J Immunol. 1996;26:683–689. doi: 10.1002/eji.1830260326. [DOI] [PubMed] [Google Scholar]
- 14.Phillips JH, Chang C, Mattson J, Gumperz J E, Parham P, Lanier LL. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B and HLA-C allotypes. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 15.Aramburu J, Balboa MA, Ramirez A, Silva A, Acevedo A, Sanchez-Madrid F, DeLandazuri MO, Lopez-Botet M. A novel functional cell surface dimer (Kp43) expressed by natural killer cells and T cell receptor γ/δ+ T lymphocytes. I. Inhibition of the IL-2 dependent proliferation by anti-Kp43 monoclonal antibody. J Immunol. 1990;144:3238–3247. [PubMed] [Google Scholar]
- 16.Perez-Villar JJ, Melero I, Rodriguez A, Carretero M, Aramburu J, Sivori S, Orengo AM, Moretta A, Lopez-Botet M. Functional ambivalence of the Kp43 (CD94) NK cell-associated surface antigen. J Immunol. 1995;154:5779–5788. [PubMed] [Google Scholar]
- 17.Brumbaugh KM, Perez-Villar JJ, Dick CJ, Schoon RA, Lopez-Botet M, Leibson PJ. Clonotypic differences in signaling from CD94 (kp43) on NK cells lead to divergent cellular responses. J Immunol. 1996;157:2804–2812. [PubMed] [Google Scholar]
- 18.Houchins JP, Yabe T, McSherry C, Miyokawa N, Bach FH. Isolation and characterization of NK cell or NK/T cell-specific cDNA clones. J Mol Cell Immunol. 1990;4:295–306. [PubMed] [Google Scholar]
- 19.Yabe T, McSherry C, Bach FH, Fisch P, Schall RP, Sondell PM, Houchins JP. A multigene family on human chromosome 12 encodes natural killer-cell lectins. Immunogenetics. 1993;37:455–460. doi: 10.1007/BF00222470. [DOI] [PubMed] [Google Scholar]
- 20.Gong J-H, Maki G, Klingemann H-G. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8:652–658. [PubMed] [Google Scholar]
- 21.Brooks AG, Campbell PL, Reynolds P, Gautam A, McCluskey J. Antigen presentation and assembly by mouse I-Ak class II molecules in human APC containing deleted or mutated HLA DMgenes. J Immunol. 1994;153:5382–5392. [PubMed] [Google Scholar]
- 22.Houchins JP, Yabe T, McSherry C, Bach FH. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J Exp Med. 1991;173:1017–1020. doi: 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothman JE, Orci L. Movement of proteins through the Golgi stack: a molecular dissection of vesicular transport. FASEB J. 1990;4:1460–1468. doi: 10.1096/fasebj.4.5.2407590. [DOI] [PubMed] [Google Scholar]
- 24.Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet J-P, Long EO. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitory receptor. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry AM, Lanier LL, Weiss A. Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J Exp Med. 1996;184:295–300. doi: 10.1084/jem.184.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, Bottino C, Moretta L. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875–884. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, Conte R, Poggi A, Moretta A, Moretta L. The human leukocyte antigen (HLA)-C-specific “activatory” or “inhibitory” natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. 1996;183:645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]