Abstract
There is little known about the regulation of gene expression during TCR-mediated differentiation of immature CD4+8+ (double positive) thymocytes into mature T cells. Using the DPK CD4+8+thymocyte precursor cell line, we demonstrate that the early growth response-1 gene (Erg-1), encoding a zinc finger transcription factor, is rapidly upregulated after TCR stimulation. We also report that Egr-1 is expressed by a subset of normal double positive thymocytes in the thymic cortex, as well by a majority of medullary single positive thymocytes. Expression of Egr-1 is dramatically reduced in the thymus of major histocompatibility complex knockout mice, but can be induced by anti-CD3 antibody stimulation of isolated thymocytes from these animals. These and other data suggest that high level expression of Egr-1 in the thymus is a consequence of selection. A similar pattern of expression is found for family members Egr-2 and Egr-3. Using the DPK cell line, we also demonstrate that expression of Egr-1, 2, and 3 is dependent upon ras activation, as is the initiation of differentiation to a single positive cell. In contrast, the calcineurin inhibitor cyclosporin A, which inhibits DPK cell differentiation as well as positive selection, inhibits expression of Egr-2 and Egr-3, but not Egr-1. The identification of the Egr family in this context represents the first report of a link between the two known signaling pathways involved in positive selection and downstream transcriptional regulators.
The development of immature CD4+8+(double positive) thymocytes into mature CD4+ or CD8+ (single positive) T cells is initiated by the TCR-mediated recognition of MHC molecules expressed by thymic stroma. Although this process, termed positive selection, is accompanied by a number of easily observed changes in expression of cell surface proteins, most notably the loss of either CD4 or CD8 coreceptor, there is little known about control of this receptor mediated differentiation event at the level of gene regulation. Certainly, TCR-generated intracellular signals must rapidly change the activity and/or expression of transcriptional regulators which then elicit a cascade of cell type specific changes in gene expression necessary for establishment of the mature T cell phenotype. Immediate early genes, defined as those genes whose expression is not dependent upon de novo protein synthesis, are likely candidates as rapid response mediators between cell surface receptor generated signals and downstream changes in gene expression.
Early growth response-1 (Egr-1 1; also known as zif268, Krox-24, tis-8, NGFI-A, and pAT 225) is one such immediate early gene that was independently isolated from fibroblasts activated by serum or phorbol ester (1–5), from PC12 cells that were induced to undergo neuronal differentiation by nerve growth factor treatment (6), and from mitogen and phorbol ester activated peripheral blood T lymphocytes (7). The Egr-1 protein is a transcriptional regulator containing three zinc finger structural domains of the Cys2His2 type (8), with each zinc finger interacting with two guanines in a three base pair subsite of a minimum nonamer binding sequence (9–11). In addition to the DNA binding domain, both activation and inhibitory regions have been identified in Egr-1 (12–14), the latter of which binds a recently identified repressor protein (15). Based upon conservation of DNA binding domains, Egr-1 belongs to a family of transcription factors which also include Krox-20/ Egr-2/pAT 591 (16–19), Egr-3 (20), and Egr-4 /NGFI-C/ pAT133 (20–22). Members of the Egr-1 family share ∼90% amino acid identity in the zinc finger domain.
Egr-1 is expressed by diverse cell types in response to mitogens, differentiation signals, tissue or radiation injury, or neuronal excitation (23). The widespread expression of Egr-1 suggests that this transcriptional regulator may play a role in coupling common biochemical signaling pathways to rapid changes in gene expression. Although the function of Egr-1 remains to be elucidated in most systems, expression of Egr-1 in cultured hematopoietic precursor cell lines has been shown to block granulocytic differentiation although allowing differentiation to the macrophage lineage (24, 25), demonstrating the potential for this transcription factor to impact cell lineage commitment. Recently, it has also been reported that Egr-1 regulates a panel of genes involved in the growth factor and clotting response to endothelial cell injury (26, 27).
In lymphocytes, Egr-1 and other family members are induced in both B and T cells upon antigen receptor crosslinking or in response to mitogens or phorbol esters (1, 28– 32). Interestingly, in B lymphocytes, inducible expression of Egr-1 has been correlated with the developmental stage of the cell. Thus, mature but not immature B cells can be induced to express Egr-1 upon stimulation with phorbol esters or antiIgM antibodies (33). In addition, Fc receptor cross-linking, which can negatively regulate anti-immunoglobulin mediated B cell activation, inhibits Egr-1 induction (34, 35). Together, these results suggest a developmentally regulated positive role for Egr-1 in B cell activation. However, IL-4 can overcome Fc receptor mediated inhibition of B cell proliferation, without increasing Egr-1 mRNA (34). Thus, the role of Egr-1 in mature lymphocyte function may depend upon the specific means of cell activation. In the adult mouse, high levels of Egr-1 mRNA are found in the brain, thymus, heart muscle, and lung as well as lower levels in other tissues (2–4, 36, 37). The source of thymic Egr-1 expression has not been previously investigated.
We have described an in vitro culture system, using the DPK cell line, that allows us to analyze the TCR mediated transition of a double positive T cell to a CD4 single positive T cell (38). Using a PCR based subtractive hybridization method, we have identified Egr-1 as a gene that is rapidly induced during the differentiation of DPK cells. Based upon this finding, we have analyzed the expression of Egr-1 in normal thymus. We report here that high level Egr-1 mRNA, protein and DNA binding activity is associated with thymic selection, and may represent one of the earliest markers of TCR engagement in the thymus. We also present evidence that expression of the Egr family in this cell type is dependent upon ras and calcineurin signaling pathways, as is immature T cell differentiation. Our identification of the Egr family in this context represents an important link between signaling pathways involved in positive selection and downstream regulators of gene expression.
Materials and Methods
Mice.
Mice were obtained from the Scripps Research Institute rodent breeding colony. MHC-deficient mice were produced by breeding class II MHC knockout mice (39) with β2 microglobulin knockout mice (40) and were obtained from Dr. J. Sprent (The Scripps Research Institute).
Cells, Assays, and Antigens.
The derivation, maintenance and differentiation of the DPK cell line has been described previously (38). DPK cell lines expressing H-rasN17, a dominant negative mutant of p21ras, were generated by retroviral mediated gene transfer as previously described (41) using the pZip-RasH(17N) construct generously provided by Dr. C. Der (University of North Carolina at Chapel Hill) (42). In brief, DPK cells were infected by co-culture with a PA317 retroviral packaging cell line that had been previously transfected with pZip-RasH(17N). After 2 d, DPK cells were transferred off of the packaging line into complete medium containing G418. The resulting G418 resistant DPK cell lines were analyzed phenotypically and functionally as described in the text. DPK cells that expressed RasH(17N) were grown under identical conditions to wild-type DPK, and no alterations in growth rate or morphology were noted. DCEK-ICAM is a fibroblast cell line transfected with class II MHC Ek and ICAM-1 genes (43).
DPK cells were activated by pigeon cytochrome c peptide 88104 (synthesized at The Scripps Research Institute) and DCEKICAM cells as described previously (38). In some experiments, DPK cells or thymocytes were treated with 2C11 anti-CD3ε mAb (PharMingen, San Diego, CA) pre-adsorbed to plastic tissue culture dishes (10 μg/ml) or 5-μm polystyrene latex beads (10 μg 2C11 per 107 beads) (Interfacial Dynamics Corporation, Portland, OR). For isolation of RNA, 1 × 107 cells were cultured in a 2C11-coated 100-mm culture dish, or with 2C11-coated beads at a cell/bead ratio of 1:1.
Western Blot.
DPK cells were lysed in 1% NP-40 in PBS containing 2 μg/ml aprotinin (Sigma Chem. Co., St. Louis, MO), 2 μg/ml leupeptin (Sigma) and 1 mM PMSF (Sigma). Cell lysates were incubated on ice for 15 min and the post-nuclear supernatant recovered by microcentrifugation. 20 μg of total protein was subjected to SDS-PAGE (12%) and transferred to PVDF membrane (Millipore Corp., Bedford, MA). The membrane was blocked with 5% nonfat milk in wash buffer (0.15 M NaCl, 10 mM Tris, pH 7.4, 2.5 mM MgCl2/0.5% Tween 20) and probed with a monoclonal anti-Ras antibody (Transduction Laboratories, Lexington, KY) diluted in 2.5% nonfat milk in wash buffer. The blot was extensively washed and developed with the addition of peroxidase-conjugated goat anti–mouse IgG antibody (Bio-Rad Laboratories, Hercules, CA) and visualized using the Enhanced Chemiluminescence Detection System (Amersham, Arlington Heights, IL).
Antibodies and Staining.
Phycoerythrin-conjugated anti-CD4 (GIBCO BRL, Gaithersburg, MD), Red613-conjugated (GIBCO BRL) or cychrome-conjugated (PharMingen) anti-CD8α, biotinylated anti-CD69 or anti-CD3 (PharMingen) mAbs were used in FACS® analysis. Cell surface staining was performed as described previously (38), and stained cells were analyzed on a FACScan® or FACSort® using CellQuest software (Becton Dickinson, Mountain View, CA). Shown is the log fluorescence of 5,000–20,000 viable cells, gated according to their sideways and forward light scatter.
For four-color FACS® analysis, thymocytes (3 × 106) from a young adult B10.BR mouse were surface stained by standard procedures with anti-CD4-PE and anti-CD8-cychrome in conjunction with biotinylated anti-CD69 or anti-CD3 followed by APCstreptavidin (Biomedia, Foster City, CA). To stain for Egr-1, cells were washed in staining buffer (PBS, 3%, FCS, 0.1% azide) and fixed in 200 μl of 3% formaldehyde in PBS for 30 min at 4°C. Cells were washed in staining buffer, resuspended in 100 μl of 0.5% Triton X-100 in PBS and incubated for 10 min at room temperature. After a wash in staining buffer, cells were resuspended in 100 μl blocking buffer (5% goat serum in 0.1 M Tris, pH 7.2, 0.01% Triton X-100) and incubated for an additional 10 min. Anti-Egr-1 peptide antiserum (C-19) (Santa Cruz Biotechnology, Santa Cruz, CA) was added to cells at a final dilution of 1:1,200 and incubation continued for an additional 30 min at room temperature. Cells were then washed and incubated with FITC-conjugated F(ab′)2 anti-rabbit IgG (Jackson ImmunoResearch Labs., West Grove, PA) for 30 min at room temperature. After a final wash, stained cells were analyzed on a FACSort® upgraded to a FACSCaliber® with the addition of a second diode laser and FL4 detector.
For immunohistology, cryostat thin sections (8–10 μ) of thymus were fixed in 3% formaldehyde in PBS, incubated with 0.5% Triton X-100 for 10 min, and blocked for 10 min in 5% goat serum in 0.1 M Tris, pH7.2, containing 0.01% Triton X-100. Sections were stained with a specific affinity-purified rabbit anti-Egr-1 peptide antiserum (Santa Cruz Biotechnology) for 1 h followed by peroxidase-coupled F(ab′)2 donkey anti–rabbit IgG antiserum (Jackson ImmunoResearch Labs.) for 30 min. For control staining, the anti-Egr-1 peptide antiserum was preincubated for 2 h at room temperature with a 10-fold excess by weight of the immunizing peptide (Santa Cruz Biotechnology). Staining was developed with 3,3′-diamino benzidine tetrahydrochloride (Pierce, Rockford IL) and sections were lightly counterstained in Gill's Hematoxylin Solution Number 2 (Sigma) followed by acid alcohol and sodium bicarbonate treatments. Sections were dehydrated in increasing concentrations of alcohol, cleared with Hemo-De (Fisher, Pittsburgh, PA) and coverslips mounted in DPX mounting media (BDH Laboratory Supplies, Poole, UK).
Representational Difference Analysis.
TRIzol Reagent (GIBCO BRL) was used to prepare total RNA from DPK cells 6 h after incubation with FcR+ CHO cells (a gift from Dr. I. Mellman, Yale University) in the presence or absence of soluble 2C11 antiCD3ε mAb. Poly(A)+ RNA was isolated from total RNA using oligo-dT magnetic beads (Dynal, Lake Success, NY). Double stranded cDNA was prepared using SuperScript reverse transcriptase (GIBCO BRL) according to the manufacturer's instructions and digested with DpnII (New England Biolabs). Representational difference analysis was performed as described by Hubank and Schatz (44). cDNA prepared from DPK cells incubated with FcR+ CHO cells was used as driver, and cDNA prepared from identical cultures but with the addition of anti-CD3ε mAb was used as tester. Final products were purified from agarose gels, cloned into the TA cloning vector (Invitrogen, San Diego, CA) and sequenced using Taq cycle sequencing (Perkin-Elmer, Norwalk, CT). Sequence similarity searches identified four fragments of 705, 671, 434, and 280 bp as DpnII fragments of the murine Egr-1 gene.
RT-PCR.
Total RNA was prepared from DPK cells or thymocytes using TRIzol Reagent (GIBCO BRL) or RNeasy RNA Kit (Qiagen, Chatsworth, CA). Oligo-dT primed first strand cDNA was prepared from 1-4 μg of total RNA using the SuperScript Preamplification System (GIBCO BRL) according to the manufacturer's instructions. PCR was performed in a total volume of 25 μl containing 10 pmol of each primer, 1× PCR buffer (GIBCO BRL), 0.2 mM dNTPs, 2 mM MgCl2, 1 U Taq polymerase (GIBCO BRL), and 1–5 μl appropriately diluted cDNA. Cycle conditions were: 94°C for 4 min followed by 28–30 cycles of 94°C for 30 s, 58 or 61°C (for Egr-3) for 40 s and 72°C for 1 min. After a final incubation at 72°C for 2 min, loading dye was added and the reactions were run on a 2.0% agarose gel. PCR products were visualized by ethidium bromide staining.
Competitive RT-PCR was performed as described above but with the addition of various concentrations of the E4 competitor construct. E4 contains both Egr-1 and CD4 primer sites flanking an irrelevant DNA sequence. Amplification of E4 with appropriate primers results in PCR products of 395 bp (CD4) and 447 bp (Egr-1), while CD4 and Egr-1 cDNA yields PCR products of 486 bp and 356 bp, respectively. To measure the concentration of specific cDNA in a sample, a constant amount of cDNA was added to reaction tubes containing known concentrations of E4 competitor. Separate PCR reactions were run with CD4 and Egr-1 primers. Competitor and cDNA-derived PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. Fluorescence images were captured to disk using an ImageStore 7500 system (UVP, Inc., Upland, CA) and band intensities were quantitated using NIH Image software. The ratio of competitor to cDNA-derived PCR product was calculated and plotted versus E4 concentration. The concentration of specific cDNA in the sample was taken as the concentration of E4 to yield a band ratio of 1. A minimum of two independent competitive PCR assays were run to determine the specific cDNA concentration of an individual sample.
Upstream and downstream primer sequences used in RT-PCR were derived from different exons. Primer pairs were as follows: CD4, 5′-CTGATGTGGAAGGCAGAGAAGGATTC/5′-CAG CACGCAAGCCAGGAACACTGTCT; Egr-1, 5′-AATCCTCAAGGGGAGCCGAGCGAACA / 5 ′ - GAGTAGATGGGACTGCTGCTGTCGTTGGA; N-Ras, 5′-GGTGGTGGTTGGAGCAGGTGGTGTTG/5′-CCATGGGGACATCATCAGAAT C TTTC; Egr-2, 5′-CCCCTTTGACCAGATGAACGGAGTGG/ 5′-TGGATGGCGGCGATAAGAATGCTGAA; Egr-3, 5′-CGACTCGGTAGCCCATTACAATCAGA/5′-GAGATCGCCGCAGTTGGAATAAGGAG; CD69, 5′-CTACCTGCAAGAATGAGTGGATTTCA/5′-TTTTGTGGTTCACGGACACGCACCTC.
Electrophoretic Mobility Shift Assay.
Oligonucleotides containing an Egr-1 consensus site (underlined), 5′-CCCGGCGCGGGG GCGATTTCGAGTCA and 5′-TGACTCGAAATCGCCC or overlapping Egr-1/SP1 sites (SP1 site in bold) 5′-GGAGGAGCGGCGGGGGCG GGCGCCGG and 5′-CCGGCGCCCGCCCCGC, were annealed and labeled in a fill-in reaction using [α32P]dCTP (ICN, Costa Mesa, CA) and Klenow fragment of DNA polymerase (GIBCO BRL) as previously described (26). Labeled oligonucleotide probes were separated from free nucleotides using spin columns (Qiagen) and ∼3 × 105 cpm probe (2–10 × 105 cpm/ ng) was used in each binding reaction. Nuclear extracts from 5 × 106 cells were obtained by resuspending cells in 100 μl of Buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 300 mM Sucrose, 1.5 mM MgCl2, 0.5 mM DTT, 0.5% NP-40, 10 μg/ml pepstatin, antipain, chymostatin, aprotinin, and 0.1 μg/ml leupeptin, 0.5 mM PMSF). After a 5-min incubation at 4°C, nuclei were pelleted at 14,000 rpm for 10 s, washed once with 100 μl of Buffer A, then resuspended in 50 μl of Buffer B (20 mM Hepes, pH 7.9, 20% glycerol, 100 mM KCl, 100 mM NaCl, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT, protease inhibitors as in buffer A). The nuclear extracts were frozen in liquid nitrogen, thawed on ice, sonicated, and stored in aliquots at −70°C. For the gel shift assay, 4–5 μl of nuclear extract was incubated with a radiolabeled DNA fragment for 20 min at room temperature in a 20 μl reaction containing 1× binding buffer (20 mM Hepes, pH 7.9, 50 mM KCl, 5.0% glycerol, 1 mM DTT, 1 mg/ml BSA, 0.1% Nonidet P-40, and 50 μg/ml poly dI-dC). Protein-DNA complexes were separated from unhybridized probe on 6% non-denaturing acrylamide gels (Novex, San Diego, CA) in 0.5× Tris borate-EDTA buffer.
Results
Induction of the Egr-1 Transcription Factor in the DPK Thymocyte Cell Line.
The DPK cell line was derived from a CD4+8+ thymic lymphoma of an AND TCR transgenic mouse (38, 45). This cell line possess the phenotype of immature thymocytes, including expression of RAG-1,2 and TdT (41), and can be triggered to differentiate into CD4+8− cells upon activation by a pigeon cytochrome c peptide and Ek bearing antigen-presenting cells, or alternatively, by Ek bearing thymic epithelial cells in vivo or in vitro in the absence of antigen (38, 46). Although anti-CD3 mAb alone is a relatively poor inducer of DPK cell differentiation, it is active at high concentration or in conjunction with accessory molecule stimulation (41, 47). To identify genes involved in the early phase of TCR mediated double positive thymocyte differentiation, we used representational difference analysis to compare mRNA derived from DPK cells that were cultured in the presence or absence of anti-CD3ε mAb (see Materials and Methods). This approach allowed us to identify the Egr-1 immediate early gene, encoding a zinc finger transcription factor, as one such candidate gene. Subsequent RT-PCR analysis confirmed that DPK cells express little Egr-1 mRNA before activation, but express high levels as early as 1 h after anti-CD3ε mAb stimulation (Fig. 1 A). Levels of Egr-1 mRNA remain high for at least 5 h but decline by approximately fivefold 1 d after initial stimulation (Fig. 1 A). To ascertain whether the induction of Egr-1 mRNA resulted in DNA binding activity, we performed an electrophoretic mobility shift assay using nuclear extracts derived from DPK cells activated by antiCD3ε mAb. Nuclear extracts from DPK cells in the absence of activation had no detectable complexes binding to an oligonucleotide probe containing an Egr-1 consensus binding sequence (Fig. 1 B), consistent with the low level of Egr-1 mRNA. In contrast, a single binding complex was observed in activated DPK cells (Fig. 1 B). This complex has an identical mobility to that obtained using recombinant Egr-1, and supershifts with an anti-Egr-1 antibody (not shown). The inducibility of Egr-1 DNA binding activity is in contrast to Sp1 DNA binding activity which is constitutively expressed in DPK cells and is not affected by stimulation (Fig. 1 B).
Figure 1.
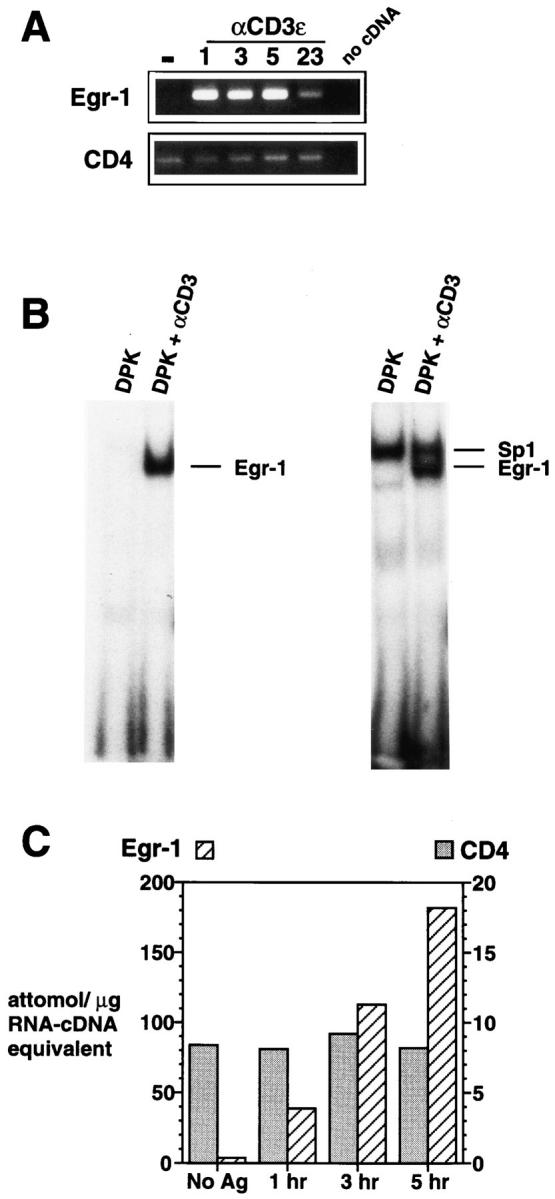
The Egr-1 gene is rapidly induced after TCR-mediated activation of the DPK double positive cell line. (A) Total RNA isolated from DPK cells cultured with immobilized anti-CD3ε mAb for the indicated times (shown in hours), was subjected to RT-PCR analysis using Egr-1 or CD4 primers. (B) Electrophoretic mobility shift assay using nuclear lysates prepared from DPK cells 8 h after activation by immobilized antiCD3ε mAb. Probes contained a single Egr-1 binding site (left) or overlapping Egr-1 and Sp1 sites (right). (C) DPK cells were cultured with DCEK-ICAM fibroblast antigen presenting cells and 1 μm pigeon cytochrome c peptide for the indicated times. Total RNA was isolated and subjected to a competitive RT-PCR assay (see Materials and Methods). Note the different scales for Egr-1 and CD4 mRNA expression.
Rapid and high level expression of Egr-1 mRNA is also observed upon activation of DPK cells by pigeon cytochrome c peptide and transfected fibroblast antigen-presenting cells (Fig. 1 C), a potent inducer of DPK cell differentiation (38). In this instance, a competitive PCR assay was used to quantify and compare levels of Egr-1 and CD4 mRNA. CD4 gene expression was used for comparison because expression of this gene is maintained throughout DPK cell differentiation (38). DPK cells activated by antigen and antigen-presenting cells or anti-CD3ε mAb express 20–30-fold higher levels of Egr-1 than CD4 mRNA (Fig. 1 C).
Egr-1 is one of a family of transcription factors that share a highly conserved DNA binding domain consisting of three zinc finger motifs. To determine whether gene expression of other family members was also upregulated upon activation of immature T cells, we examined gene expression of Krox-20, the murine homologue of human Egr-2 (16, 18), and Egr-3 (20) in DPK cells. Neither Krox20 (Egr-2) nor Egr-3 mRNA was detected by RT-PCR in unactivated DPK cells, while both genes were induced after anti-CD3ε mAb stimulation (Fig. 2 C). Both Krox-20 (Egr-2) and Egr-3 RT-PCR products were sequenced to confirm identity of the expressed genes.
Figure 2.
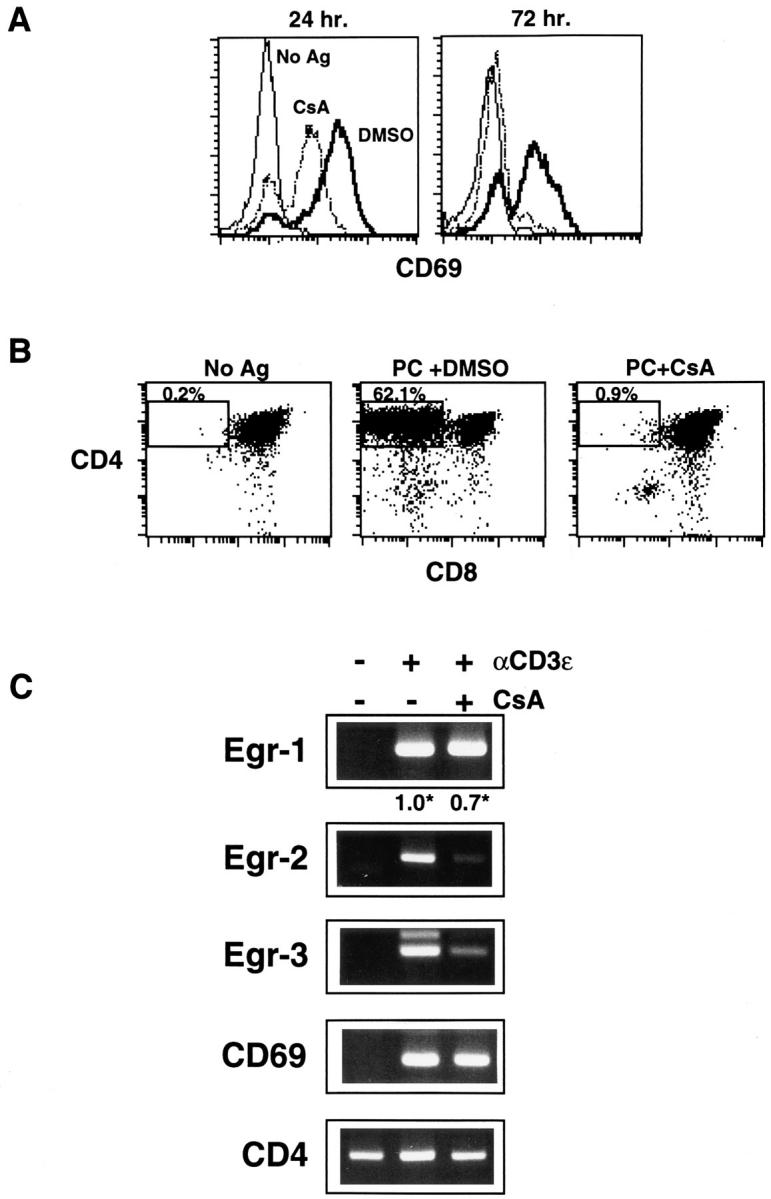
DPK cell differentiation and Egr-2,3 mRNA induction is cyclosporin A sensitive, while Egr-1 mRNA induction is cyclosporin A resistant. (A, B) DPK cells were cultured with DCEK-ICAM fibroblast antigen presenting cells and 2 μM pigeon cytochrome c peptide in the presence or absence of 100 ng/ml cyclosporin A, or appropriate dilution of solvent (DMSO) as indicated. Cells were harvested and stained for CD69 after 1 or 3 d in culture (A) or stained for CD4 and CD8 after 3 d in culture (B). (C) RT-PCR analysis of total RNA derived from DPK cells activated for 6 h with immobilized anti-CD3ε mAb in the presence or absence of 300 ng/ml cyclosporin A, using Egr-1, Egr-2 (Krox-20), CD4, CD69 or Egr-3 primers. (*) Also shown for the indicated samples is the relative level of Egr-1 cDNA normalized to expression of CD4 cDNA as determined by competitive RT-PCR assay. The identity of the lower major band in Egr-3 RT-PCR was verified by sequencing.
Cyclosporin A Inhibits DPK Differentiation and Egr-2,3, but not Egr-1 Gene Expression.
Our results have demonstrated that TCR engagement in the immature double positive DPK cell line leads to upregulation of members of the Egr gene family. However, it remained to be determined whether induction of these genes was downstream of signaling pathways that were required for immature T cell differentiation. Cyclosporin A (CsA), a potent inhibitor of calcineurin, has been reported to block positive selection (36, 48, 49). To investigate whether Egr gene expression in double positive cells was also dependent upon calcineurin activation, we tested the ability of CsA to block Egr induction in DPK cells. As observed for the production of single positive thymocytes, CsA inhibits the production of CD4 single positive DPK cells upon activation by antigen and antigen presenting cells (Fig. 2 B). Downregulation of cell surface CD8 as early as 24 h after activation was also blocked by CsA (not shown), indicating that calcineurin was involved in the initiation of this process and not simply single positive cell survival.
One of the earliest cell surface markers of positive selection in the thymus (50–52) and in DPK cells (38) is CD69. No significant effect of CsA on the early induction of CD69 mRNA was observed in activated DPK cells (Fig. 2 C). At 24 h after activation, CsA partially reduced but did not eliminate CD69 cell surface expression on DPK cells (Fig. 2 A). By 3 d, however, CD69 surface expression was absent from DPK cells, suggesting that calcineurin may be involved in maintenance but not induction of CD69 expression. In summary, although early CD69 gene induction is relatively resistant to the effects of CsA, the initiation of differentiation to a single positive cell is clearly CsA sensitive. CsA, however, had little effect on the induction of Egr-1 mRNA. Using expression of CD4 for normalization, DPK cells that were activated in the presence of CsA expressed ∼70% of the levels of Egr-1 mRNA expressed by cells activated in the absence of CsA (Fig. 2 C). In contrast, CsA almost completely blocks induction of Egr-2 (Krox20) and Egr-3 mRNA (Fig. 2 C). Thus, activation of some, but not all members of the Egr gene family are downstream of a signaling pathway required for positive selection.
Both DPK Differentiation and Egr Gene Expression are Blocked by a Dominant Negative Mutant of p21Ras.
It has been demonstrated that there is an inhibition of the production of single positive thymocytes in transgenic mice that express dominant negative mutants of p21ras or MEK (53, 54). Thus, we sought to determine the effect of a block in ras signaling upon expression of the Egr family in double positive cells. In addition, because positive selection may be a multi-step process, it was not clear from transgenic experiments whether ras activation was required for the initiation of the double positive to single positive transition or, alternatively, for thymocyte survival or other later events.
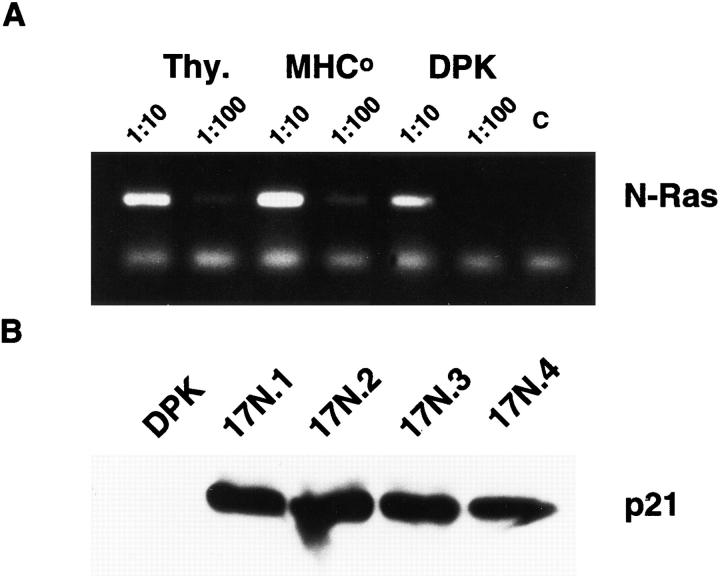
As shown in Fig. 3 A, N-ras mRNA can be detected in normal thymocytes, thymocytes derived from MHC-deficient mice (>90% double positive, Fig. 2 A), and DPK cells. This is consistent with previous reports of expression of both N-ras and K-ras in double positive thymocytes (53). To study the role of p21ras in Egr gene expression in immature T cells, retroviral mediated gene transfer was used to express a dominant negative mutant of p21ras, Haras N17 (with an asparagine substituted for serine at position 17) in DPK cells. This mutant blocks endogenous ras function by competing for guanine nucleotide exchange proteins, thereby preventing formation of ras-GTP complexes (42). Fig. 3 B shows expression of Ha-ras N17 in four independent DPK cell transformants that express high levels of this dominant negative mutant. The results obtained with line 17N4 are described here, although all four cell lines had similar responses.
Figure 3.
Expression of a dominant negative mutant of p21ras blocks DPK cell differentiation. (A) RT-PCR assay of N-ras expression in DPK cells or thymocytes derived from wild-type or MHC-deficient mice. (B) Cell lysates from DPK cells and four independent lines that express Ha-ras N17 were analyzed by Western blot and probed with anti-ras antibody. Endogenous p21ras is not visible in this exposure. (C) DPK or 17N4 cells were cultured with DCEK-ICAM fibroblast antigen presenting cells in the presence (bold lines) or absence (thin lines) of 2 μM pigeon cytochrome c peptide. After 3 d of culture, cells were collected, stained with mAb to CD69 and analyzed by flow cytometry. (D) DPK or 17N4 cells were cultured as in (C) except that cells were harvested on day 1 or 3 as indicated and stained with anti-CD4 and anti-CD8 mAbs. Shown are the percentages of CD4+8lo/- DPK cells in the designated regions.
One major difference between DPK cells and the majority double positive thymocyte population is that DPK cells are rapidly dividing. Given the potential role of ras as a protooncogene it was possible that a block in p21ras-mediated signaling would alter DPK cell growth and secondarily differentiation. However, the overexpression of rasN17 had no effect on the growth of DPK cells, nor were any changes in morphology observed (not shown). In addition, both DPK and 17N4 cells displayed similar abilities to mobilize calcium after TCR engagement (not shown). In Jurkat cells, activation of p21ras is both necessary and sufficient for induction of the early activation marker CD69 (55). Similarly, expression of rasN17 in DPK cells abrogates surface expression of CD69 upon activation (Fig. 3 C). In addition, RT-PCR analysis demonstrated that CD69 mRNA could be detected as early as 6 h after activation by antigen in wild-type but not 17N4 cells (not shown). This is in contrast to the effects of CsA, which had little effect on early CD69 induction. In response to antigen, DPK cells transit from a double positive to a CD4+8− single positive phenotype. Similar to normal thymocytes, the loss of cell surface CD8 in DPK cells is a gradual process, and transitional cells with low levels of CD8 are observed at early time points (Fig. 3 D). The production of CD4 single positive cells in response to antigen is almost completely inhibited in 17N4 cells, even at these early time points (Fig. 3 D). Together, these results indicate that p21ras signaling is required for initiation of DPK cell differentiation to a CD4 single positive cell.
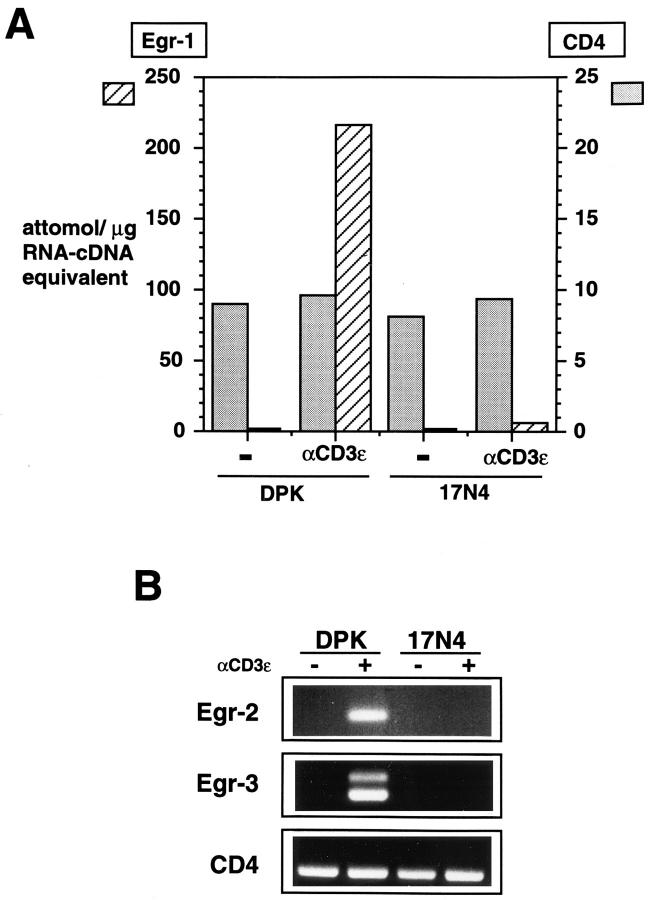
Having determined that DPK cell differentiation was dependent upon ras activation, we sought to determine whether the expression of the Egr gene family was also downstream of this signaling pathway. As shown in Fig. 4 A, induction of Egr-1 mRNA in DPK cells was blocked by expression of the dominant negative mutant of p21ras. Similarly, there was no detectable induction of Egr-2 (Krox-20) or Egr-3 mRNA in these cells after stimulation by anti-CD3ε mAb (Fig. 4 B). This is in contrast to CsA, which differentially affects the expression of the Egr family.
Figure 4.
Expression of Egr gene family is dependent upon ras signaling pathways. (A) Competitive RT-PCR was used to compare expression of Egr-1 and CD4 genes in DPK or 17N4 cells activated for 6 h with immobilized anti-CD3ε mAb. (B) RT-PCR analysis of total RNA derived from DPK or 17N4 cells activated for 6 h with immobilized anti-CD3ε mAb. Independent PCR reactions using Egr-2 (Krox-20), CD4, or Egr-3 primers were performed.
Egr-1 Is Expressed by Double Positive Thymocytes In Vivo as a Result of Recognition of MHC.
The neonatal and adult rat thymus has been reported to express Egr-1 mRNA (37). Our results in the DPK system suggested that Egr-1 expression in normal thymus might be the result of positive selection. To address this issue, we used competitive RT-PCR to analyze Egr-1 gene expression in normal thymocytes and in thymocytes derived from double gene knockout mice that do not express class I or class II MHC molecules (39, 40). These latter animals have normal numbers of double positive thymocytes but lack single positive thymocytes due to the inability of TCR bearing immature thymocytes to undergo positive selection (Fig. 5 A). Freshly isolated thymocytes from MHC-deficient animals expressed approximately 10–20-fold less Egr-1 mRNA than normal thymocytes (Fig. 5 B). In contrast, expression of the CD4 gene was similar in both groups of animals, consistent with the expression of this coreceptor by >90% of thymocytes in both normal and MHC-deficient mice (Fig. 5 A). To analyze functional Egr-1 protein expression, gel mobility shift assays were performed using nuclear extracts derived from freshly isolated thymocytes. Consistent with the pattern of Egr-1 mRNA expression, Egr-1 DNA binding activity was detected in nuclear lysates of thymocytes derived from wild-type but not MHC-deficient animals (Fig. 5 C). This complex has an identical mobility to that obtained with recombinant Egr-1 and supershifts with an anti–Egr-1 antibody (Fig. 5 C).
Figure 5.
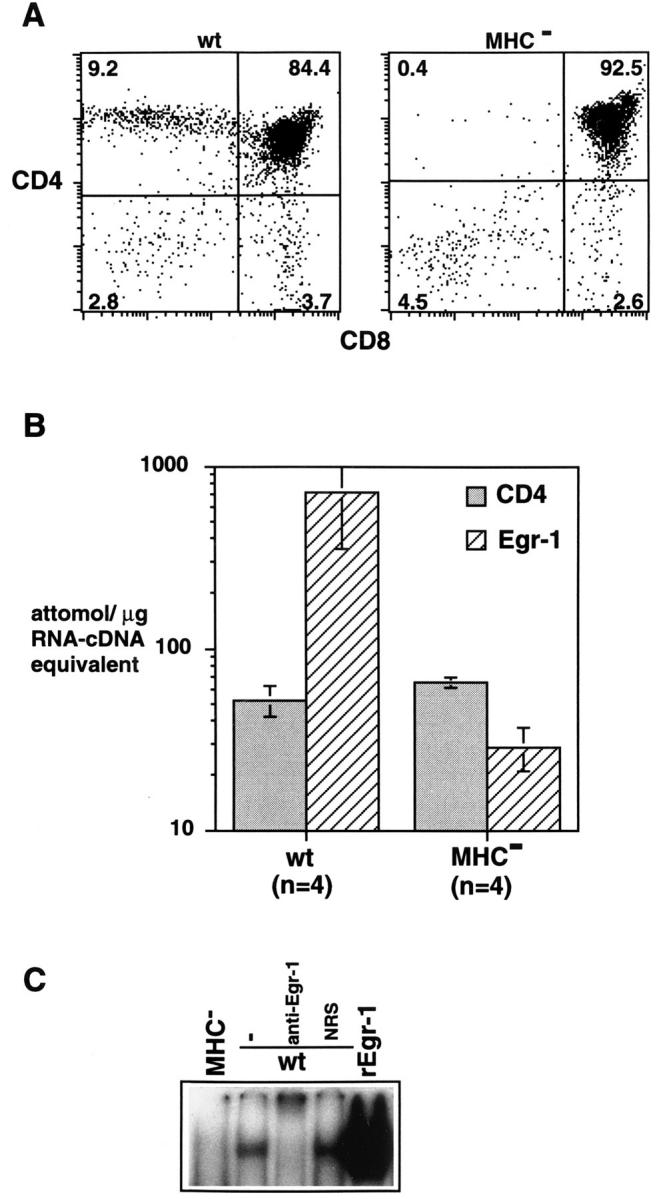
Egr-1 mRNA and DNA binding activity in the thymus is MHC dependent. (A) Thymocytes derived from wild-type or MHCdeficient mice were two color-stained for CD4 and CD8. (B) Competitive RT-PCR was used to determine the level of expression of CD4 and Egr-1 genes in thymocytes derived from wild-type or MHC-deficient mice. (C) Electrophoretic mobility shift assay using nuclear lysates prepared from freshly isolated thymocytes derived from wild-type or MHCdeficient mice using a probe containing an Egr-1 binding site. Nuclear extracts derived from 5 × 105 cells containing equivalent amounts of protein were used in binding reactions. In some instances as indicated, binding reactions contained anti-Egr-1 antibody or normal rabbit serum (NRS). For comparison, a binding reaction containing recombinant Egr-1 is shown.
These results suggested that activation of the Egr-1 gene occurred during MHC-dependent selection of double positive thymocytes. However, the data were also consistent with the possibility that Egr-1 was upregulated only in mature single positive thymocytes, subsequent to positive selection. To distinguish these possibilities we analyzed the expression of Egr-1 in purified double positive thymocytes derived from normal mice. Given the rapid induction of Egr-1 mRNA in DPK cells (Fig. 1) one would predict that if Egr-1 was induced during thymic selection, it would be expressed before loss of the double positive phenotype. This was found to be the case, and a significant amount of Egr-1 mRNA was detected in double positive thymocytes isolated by cell sorting (Fig. 6 A). Isolated double positive thymocytes also expressed Egr-1 DNA binding activity, as did CD4 single positive thymocytes (not shown). To demonstrate that the Egr-1 gene could be induced by TCR activation in double positive thymocytes, thymocytes derived from MHC-deficient mice were stimulated with immobilized anti-CD3ε mAb. As shown in Fig. 6 B, TCR activation results in rapid upregulation of Egr-1 mRNA in these cells. In contrast, Egr-1 is not upregulated in thymocytes derived from MHC-deficient mice upon treatment in culture with doses of dexamethasone or ionizing radiation that induce cell death (not shown).
Figure 6.
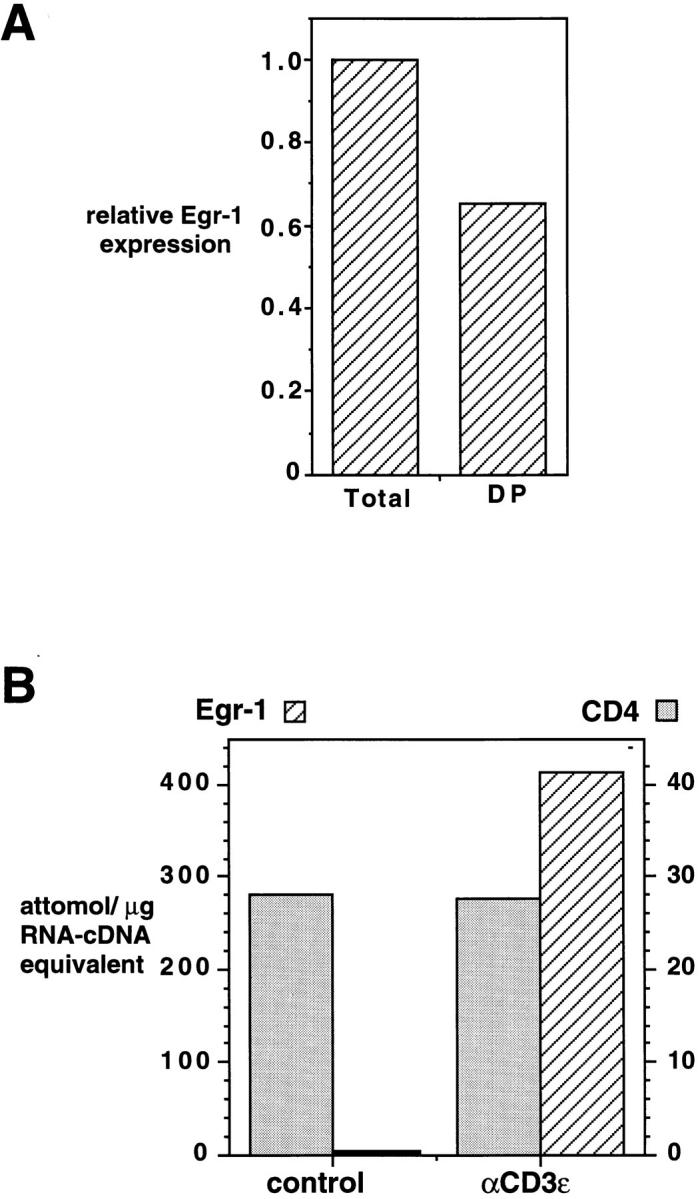
Expression of Egr-1 mRNA in double positive thymocytes. (A) Total thymocytes and CD4+8+ thymocytes (isolated by cell sorting, 95% DP) derived from the same animal, were assayed for expression of Egr and CD4 mRNA by competitive RT-PCR. Shown is the relative level of Egr-1 cDNA in the sample, normalized to the level of CD4 cDNA. (B) Total thymocytes derived from an MHC-deficient mouse were cultured with hamster immunoglobulin-coated or anti-CD3ε mAbcoated beads for 90 min before determination of Egr-1 and CD4 gene expression as in Fig. 1 C.
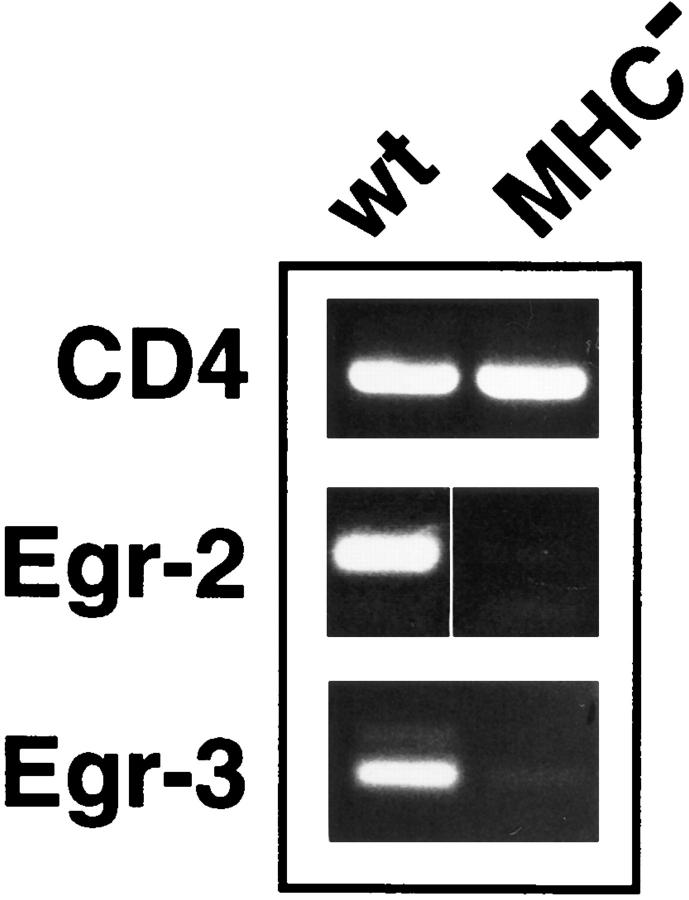
Similar to results obtained with DPK cells, both Krox-20 (Egr-2) and Egr-3 mRNA are also expressed in normal thymocytes, while there is no detectable expression of these genes in thymocytes derived from MHC-deficient animals (Fig. 7). These results are consistent with the coordinate upregulation of the Egr family of transcription factors during thymic selection.
Figure 7.
Expression of Egr-2 and Egr-3 mRNA in thymocytes is MHC dependent. Total RNA prepared from freshly isolated wild-type or MHC knockout thymocytes was subjected to RT-PCR analysis using CD4, Egr-2 (Krox-20), or Egr-3 primers as indicated.
To determine the location of cells within the thymus that express Egr-1 protein, thin sections of normal thymus were stained with a specific anti–Egr-1 peptide antiserum (Fig. 8). Isolated cells scattered throughout the cortex were found to express Egr-1. No enrichment of Egr-1 expressing cells in the subcapsular region or cortico-medullary junction was observed. These results are consistent with induction of Egr-1 in cortical double positive thymocytes as a result of TCR engagement. Most, but not all, medullary cells also stained, indicating that Egr-1 protein is expressed in a substantial number of single positive thymocytes as well (see below). In contrast, few cells stained in the thymic cortex of MHC-deficient mice, and in general the staining was weak, even in medullary regions (Fig. 8).
Figure 8.
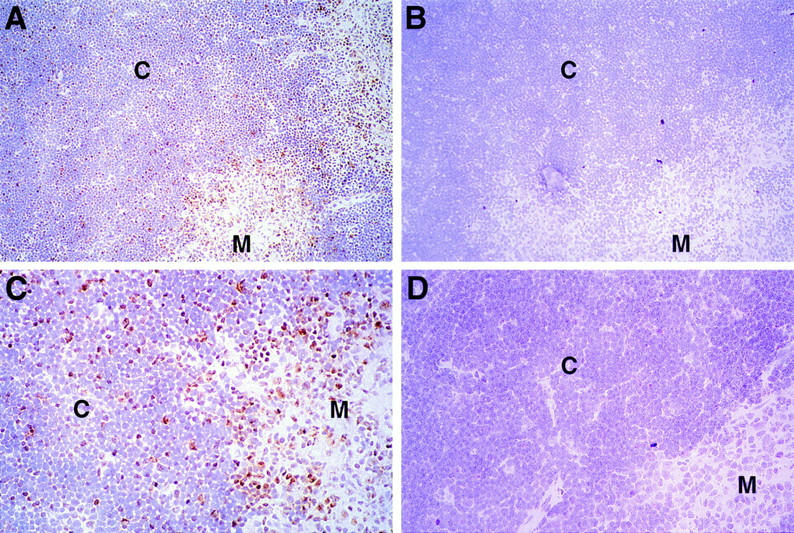
Expression of Egr-1 protein in the thymus. Thin sections of normal thymus (A–C) or MHC knockout thymus (D) were fixed in formaldehyde and stained with a specific rabbit anti-Egr-1 peptide antiserum (A, C, D) or the same antibody preincubated with specific peptide (B). Regions of cortex (C) and medulla (M) are indicated. Sections were counterstained with hematoxylin and photographed at ×20 (A, B) or ×40 (C, D). C shows a magnification of the same section photographed in A.
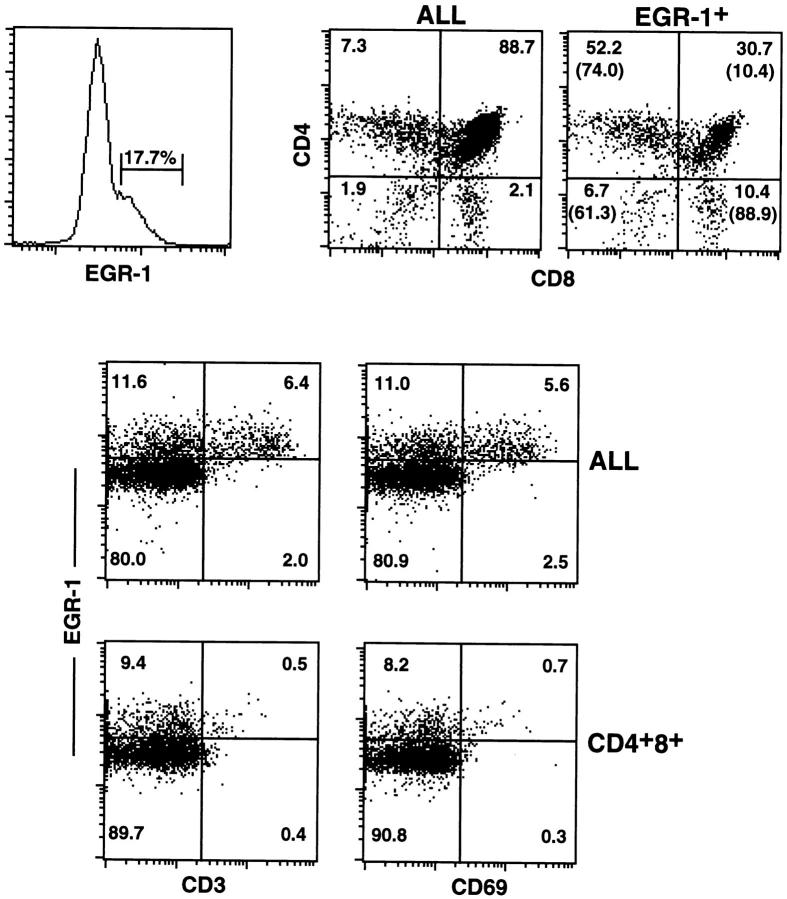
The expression of Egr-1 in thymocyte subsets was further analyzed by FACS® using three-color surface staining in conjunction with a fourth color for internal staining (Fig. 9). Using this method, a subset of thymocytes could be detected that stained with the anti–Egr-1 antiserum. This subpopulation included the majority of single positive thymocytes and ∼10% of double positive thymocytes. These results are consistent with the finding that isolated cortical thymocytes and most medullary thymocytes stained with this anti–Egr-1 antiserum. Interestingly, almost all the CD69+ or CD3hi thymocytes also stained for Egr-1, while ∼10% of thymocytes stained for Egr-1 but were CD3lo or CD69− (Fig. 9). Among double positive thymocytes, >90% of the cells that stained for Egr-1 were also CD3lo or CD69−. A majority of double negative thymocytes also stained for Egr-1, although the specificity of this staining remains to be confirmed by other means.
Figure 9.
Expression of Egr-1 protein in thymocyte subsets. Thymocytes from a young adult mouse were 4-color stained for expression of CD4, CD8, Egr-1, and CD69 or CD3, and analyzed by FACS® as described in Materials and Methods. Indicated in the dot plots are the percentage of thymocytes within each quadrant, or in parenthesis (upper right dot plot), the percentage of Egr-1+ thymocytes within each thymocyte subset. Where indicated, staining is shown for gated populations of thymocytes (either Egr-1+thymocytes as shown in histogram, or CD4+8+ thymocytes). Similar results were obtained from three other individual animals.
Discussion
Using representational difference analysis and the DPK culture system we sought to identify genes that play a role in the earliest stages of double positive thymocyte differentiation. Egr-1, an immediate early gene encoding a zinc finger transcription factor, was identified as one such candidate. We have demonstrated that the Egr-1 gene is rapidly induced after TCR mediated stimulation of the double positive DPK cell line or double positive thymocytes. In normal mice, Egr-1 mRNA, protein, and DNA binding activity is expressed by freshly isolated thymocytes, including the double positive subset. In contrast, thymocytes derived from MHC-deficient mice, which express a normal pattern of development through the double positive stage but which do not undergo positive or negative selection, express ∼10–20-fold lower levels of Egr-1 mRNA than wild-type mice. This correlates with the loss of detectable Egr-1 DNA binding activity and low level anti–Egr-1 antibody staining in thymocytes derived from MHC-deficient mice. Furthermore, Egr-1 expression is also low in thymocytes derived from TCR α chain knockout mice (not shown), demonstrating that a TCR–MHC interaction and not, for example, a coreceptor-MHC interaction, is required for high level Egr-1 expression in the thymus. These results are in contrast to expression of Sp1 DNA binding activity which is constitutively expressed in double positive thymocytes (56) and DPK cells before and after activation (Fig. 1).
The question then arises as to whether thymocytes that express Egr-1 are undergoing positive selection. Positive selection of double positive thymocytes is initiated by an interaction with MHC bearing epithelial cells that reside in the thymic cortex (57). If induction of Egr-1 in double positive cells is a consequence of positive selection, it would also be expected to take place in the cortex. This was found to be the case, and staining of thymic sections for Egr-1 revealed scattered thymocytes throughout the cortex that expressed the protein. Egr-1 is also expressed by many medullary (Fig. 8) and single positive thymocytes (Fig. 9). Whether Egr-1 expression is maintained or reinduced on these more mature thymocytes is uncertain, but Egr-1 upregulation has been associated with both early activation and late differentiation in other systems (28).
Whereas we can not directly determine the fate of Egr-1 expressing cells in the cortex, thymocytes that have upregulated CD3 or CD69, including CD4+8lo transitional thymocytes, also have upregulated Egr-1 (Fig. 9). This pattern of expression, coupled with our results in the DPK system, strongly suggests that Egr-1 is induced during positive selection. Approximately 10% of total thymocytes, the majority of which are also double positive thymocytes, also stained for Egr-1 by FACS® but were CD3lo or CD69− (Fig. 9). This may reflect the rapid induction of Egr-1 during selection, before upregulation of TCR or CD69. The observed rapid upregulation of Egr-1 in DPK cells and thymocytes as a result of TCR engagement is certainly consistent with this possibility. In the CD4 lineage, in fact, TCR upregulation occurs subsequent to the double positive stage and is coincident with downregulation of the CD8 coreceptor (58). Egr-1 expression may thus represent one of the earliest markers of positive selection in the thymus. Alternatively, some thymocytes that express Egr-1 at the double positive stage may fail to progress in their differentiation program or be in the process of negative selection (see below).
Although Egr-1 expression is associated with thymocyte differentiation, could upregulation of this transcription factor also occur during negative selection? In support of this, we have shown that double positive thymocytes stimulated with anti-CD3 antibody upregulate Egr-1 mRNA. The signals provided by CD3 cross-linking do not induce positive selection of isolated normal thymocytes, but can elicit cell death in conjunction with accessory signals (59). Expression of Egr-1, therefore, may not be limited to positive selection but may also be induced during negative selection. If indeed high level Egr-1 expression results from both positive and negative selection, the expression pattern of this transcription factor in the cortex may reveal the extent of specific TCR engagement in the normal thymus.
Although Egr-1 could be induced during negative selection, evidence suggests that induction of this transcription factor is not associated with thymocyte cell death per se. Although apoptotic thymocytes can be detected in the cortex of normal mice, there is no reduction in the frequency of such cells in MHC double knockout mice (60). Presumably, cell death in the latter instance is the result of death by neglect and not negative selection. In contrast, there is a dramatic reduction in Egr mRNA, protein and DNA binding activity in MHC knockout mice, suggesting that thymocyte death by neglect is not associated with Egr-1 upregulation. We also failed to detect induction of Egr-1 mRNA in MHC knockout thymocytes subjected to ionizing radiation or corticosteroids at doses that induce cell death. This further demonstrates that high level Egr-1 mRNA expression in the thymus is specifically associated with TCR activation.
It is unlikely that TCR-mediated selection events are the only source of Egr-1 gene expression in the thymus, because some Egr-1 mRNA can be detected in MHCdeficient animals. Double positive thymocytes may constitutively express the gene at very low levels, because Egr-1 mRNA can also be detected in DPK cells by RT-PCR if sufficient amounts of cDNA are used. Because the DNA binding activity of Egr-1 can be influenced by phosphorylation (61), both transcriptional and posttranslational mechanisms may contribute to the upregulation of functional Egr-1 in immature thymocytes. In addition, we have observed staining of double negative thymocytes using antiEgr-1 antiserum and FACS® analysis (Fig. 9). Because double negative cells tend to have higher background staining for many antibodies, this result needs to be confirmed. However, one intriguing possibility is that Egr-1 is also induced by pre-TCR activation (62) and plays a role in the differentiative and/or proliferative events that are involved in the double negative to double positive thymocyte transition.
Egr-1 belongs to a family of transcriptional regulators that contain closely related DNA binding domains that recognize similar sequence motifs. However, despite highly similar DNA binding domains, differences have been found in the ability of Egr family members to interact with a given promoter region (63). Thus, the degree of functional overlap between family members remains an open question. Egr-1, 2, 3, and 4 have all been reported to be upregulated upon activation of human T lymphocytes (19, 64). We have found that Egr-1, 2, and 3 are also coordinately upregulated in the thymus, and expression of mRNA for all three family members is dependent upon expression of MHC. Somewhat surprisingly given these results, only a single binding complex is detected in gel shift assays using thymocyte or DPK nuclear lysates. This complex runs identically to that obtained with recombinant Egr-1, and the complex supershifts with an anti–Egr-1 antibody. It is possible that the failure to detect Egr-2 and Egr-3 containing complexes is due to lower levels of expression of these transcription factors (19) or to the particular binding conditions of the assay.
Positive and negative selection are complex processes involving cell activation, cell differentiation and cell survival. How Egr expression specifically impacts these events is not known. Egr-1 knockout mice have no gross lymphocyte abnormality, including the production of mature T cells (65), while Egr-2 knockouts die as neonates due to hindbrain abnormalities (66). Whether there are more subtle changes in T cell development or T cell function in Egr-1 knockout mice, or compensatory effects of other family members remains to be determined. The critical gene targets for regulation by Egr family members in the context of T cell activation or thymocyte development are also unknown, although Egr-1 has been implicated in regulation of the IL-2 (63), tumor necrosis factor (67), ICAM-1 (68), and CD44 (69) genes. The CD69 gene promoter may also have an Egr-1 binding site (70). Egr-1 has also been reported to regulate delayed early expression of another immediate early gene, nur77 (71), which itself has been implicated in thymocyte cell death (72).
Despite the paucity of target genes known to be regulated by Egr-1, there is a general theme that has emerged as to its mode of action. Sp1, which shares homology with Egr-1 in the zinc finger domain, binds a GC-rich but distinct sequence from that of Egr-1. Egr-1 and Sp1 sites often overlap in gene promoters, allowing for competition. It has recently been shown that induction of Egr-1 as a result of endothelial cell injury can result in displacement of constitutively expressed Sp1, leading to activation of genes involved in wound repair or cell growth (27). A similar displacement of Sp1 by Egr-1 may also play a role in IL-2 gene activation (63) as well as regulation of the Egr-1 gene itself (73). As shown here, because Sp1 is constitutively expressed in double positive thymocytes, whereas Egr-1 is inducible, a similar process may occur in the thymus. Whether the result of displacement of Sp1 by Egr-1 in the context of T cell development would result in gene activation or repression, and how this would impact T cell development, must await identification of specific gene targets for Egr-1 in the thymus.
If indeed there is an induction of Egr family members during negative as well as positive selection, this does not imply that these transcription factors play identical roles in these distinct selection events. To address this issue, we have asked how signaling pathways impact both double positive cell differentiation and Egr family expression. Expression of dominant negative ras or MEK in transgenic animals has previously been shown to inhibit the appearance of single positive thymocytes (53, 54). Using the DPK system we have shown here that dominant negative ras also inhibits the initiation of the double positive to single positive transition of these cells. This correlates with an inhibition of the induction of Egr-1,2, and 3. Induction of Egr-1 in B cells has also been shown to be ras dependent (74). The fact that negative selection (53), and cell death of DPK cells (Shao, H. and J. Kaye, unpublished observation), is not impaired under conditions that disrupt ras signaling and Egr expression suggests that the Egr family is unlikely to play an obligatory role in negative selection.
We have also examined the effect of the calcineurin inhibitor CsA in this system. CsA has previously been shown to inhibit the production of single positive thymocytes in vivo (36, 48) and in vitro (49). CsA also blocks the double positive to single positive transition of DPK cells. In contrast to the effects of dominant negative ras, however, there was minimal effect of CsA on early induction of CD69. Induction of Egr-1 was found to be CsA resistant while expression of Egr-2 and Egr-3 were sensitive to CsA. Induction of Egr-1 in mature human T lymphocytes has also been reported to be CsA resistant (75), while induction of Egr-3 was found to be CsA sensitive (64). Similarly, induction of Egr-2 but not Egr-1 in a murine B cell line is inhibited by CsA (35). That expression of Egr-2 and 3 requires both ras and calcineurin signaling pathways is reminiscent of a similar dual requirement for IL-2 gene activation (76, 77), and the coordinate activity of these signaling pathways may be a common feature of many aspects of both mature T cell activation and immature thymocyte differentiation. There have been conflicting reports of whether CsA also blocks negative selection (36, 48, 49, 78), although the fact that CsA blocks positive selection is certainly a complicating factor. Nevertheless, it is clear that the Egr transcription factor family is specifically induced by TCR activation in immature thymocytes and lies downstream of the two known pathways that are required for positive selection.
Acknowledgments
The authors thank N. Mackman and G. Parry for assistance with gel shift assays, E. Adamson and G. Parry for recombinant Egr-1, C. Der for the ras17N construct, and J. Sprent for MHC-deficient mice. We also thank Drs. J. Sprent and A. Feeney for critically reviewing this manuscript. This is manuscript 10220-IMM from the Scripps Research Institute.
These studies were supported by National Institutes of Health grants AI33219 and AI31231 to J. Kaye.
Footnotes
1 Abbreviations used in this paper: CsA, cyclosporin A; Erg-1, early growth response.
H. Shao and D. Kono contributed equally to this work.
References
- 1.Sukhatme VP, Kartha S, Toback FG, Taub R, Hoover RG, Tsai-Morris C-H. A novel early growth response gene rapidly induced by fibroblast, epithelial cell and lymphocyte mitogens. Oncogene Res. 1987;1:343–355. [PubMed] [Google Scholar]
- 2.Sukhatme VP, Cao X, Chang LC, Tsai-Morris C-H, Stamenkovich D, Ferreira PCP, Cohen DR, Edwards SA, Shows TB, Curran T, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 3.Christy B, Nathans LF, Nathans D. A gene activated mouse 3T3 cells by serum growth factors encodes a protein with “zinc fingers” sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim RW, Varnum BC, Herschman HR. Cloning of tetradecanoyl phorbol ester-induced ‘primary response' sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1:263–270. [PubMed] [Google Scholar]
- 6.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcription factor. Science (Wash DC) 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 7.Irving SG, June CH, Zipfel PF, Siebenlist U, Kelly K. Mitogen-induced genes are subject to multiple pathways of regulation in the initial stages of T-cell activation. Mol Cell Biol. 1989;9:1034–1040. doi: 10.1128/mcb.9.3.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Koski RA, Gashler A, McKiernan M, Morris CF, Gaffney R, Hays RV, Sukhatme VP. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol Cell Biol. 1990;10:1931–1939. doi: 10.1128/mcb.10.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci USA. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science (Wash DC) 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 11.Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc-finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, and extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor EGR-1: Prototype of a zinc-finger family of transcription factors. Cell Biol. 1993;13:4556–4571. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo MW, Matheny C, Milbrandt J. Transcriptional activity of the zinc finger NGFI-A is influenced by its interaction with a cellular factor. Molec Cell Biol. 1993;13:6858–6865. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carman JA, Monroe JG. The EGR1 protein contains a discrete transcriptional regulatory domain whose deletion results in a truncated protein that blocks EGR1induced transcription. DNA Cell Biol. 1995;14:581–589. doi: 10.1089/dna.1995.14.581. [DOI] [PubMed] [Google Scholar]
- 15.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavrier P, Zerial M, Lemaire P, Almendral J, Bravo R, Charnay P. A gene encoding a protein with zinc fingers is activated during G0/G1transition in cultured cells. EMBO (Eur Mol Biol Organ) 1988;7:29–35. doi: 10.1002/j.1460-2075.1988.tb02780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavrier P, Janssen-Timmen U, Mattei M-G, Zerial M, Bravo R, Charnay P. Structure, chromosome location, and expression of the mouse zinc finger gene Krox20: multiple gene products and coregulation with the protooncogene c-fos . Mol Cell Biol. 1989;9:787–797. doi: 10.1128/mcb.9.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph LJ, Le Beau MM, Jamieson GA, Jr, Acharya S, Shows TB, Rowley JD, Sukhatme VP. Molecular cloning, sequencing, and mapping of EGR2, a human early growth response gene encoding a protein with “zincbinding finger” structure. Proc Natl Acad Sci USA. 1988;85:7164–7168. doi: 10.1073/pnas.85.19.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zipfel PF, Irving SG, Kelly K, Siebenlist U. Complexity of the primary genetic response to mitogenic activation of human T cells. Mol Cell Biol. 1989;9:1041–1048. doi: 10.1128/mcb.9.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patwardhan S, Gashler A, Siegel MG, Chang LC, Joseph LJ, Shows TB, Le Beau MM, Sukhatme VP. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene. 1991;6:917–928. [PubMed] [Google Scholar]
- 21.Crosby SD, Puetz JJ, Simburger KS, Fahrner TJ, Milbrandt J. The early response gene NGFI-C encodes a zinc finger transcriptional activator and is a member of the GCGGGGGCG (GSG) element-binding protein family. Mol Cell Biol. 1991;11:3835–3841. doi: 10.1128/mcb.11.8.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller H-J, Skerka C, Bialonski A, Zipfel PF. Clone pAT 133 identifies a gene that encodes another human member of a class of growth factor-induced genes with almost identical zinc-finger domains. Proc Natl Acad Sci USA. 1991;88:10079–10083. doi: 10.1073/pnas.88.22.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): Prototype of a zinc-finger family of transcription factors. Prog Nuc Acid Res. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen HQ, Hoffman-Liebermann B, Liebermann DA. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 25.Krishnaraju K, Nguyen HQ, Liebermann DA, Hoffman B. The zinc finger transcription factor Egr-1 potentiates macrophage differentiation of hematopoetic cells. Molec Cell Biol. 1995;15:5499–5507. doi: 10.1128/mcb.15.10.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui M-Z, Parry GCN, Oeth P, Larson H, Smith M, Huang R-P, Adamson ED, Mackman N. Transcriptional regulation of the tissue factor gene in human epithelial cells is mediated by Sp1 and EGR-1. J Biol Chem. 1996;271:2731–2739. doi: 10.1074/jbc.271.5.2731. [DOI] [PubMed] [Google Scholar]
- 27.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: A common theme in vascular injury. Science (Wash DC) 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 28.Sukhatme VP. Early transcriptional events in cell growth: the Egr family. J Am Soc Nephrol. 1990;1:859–866. doi: 10.1681/ASN.V16859. [DOI] [PubMed] [Google Scholar]
- 29.Seyfert VL, Sukhatme VP, Monroe JG. Differential expression of a zinc finger-encoding gene in response to positive versus negative signaling through receptor imunoglobulin in murine B lymphocytes. Molec Cell Biol. 1989;9:2083–2088. doi: 10.1128/mcb.9.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seyfert VL, McMahon S, Glenn W, Cao X, Sukhatme VP, Monroe J. Egr-1 expression in surface Ig-mediated B cell activation - Kinetics and association with protein kinase C activation. J Immunol. 1990;145:3647–3653. [PubMed] [Google Scholar]
- 31.Makover D, Cuddy M, Yum S, Bradley K, Alpers J, Sukhatme V, Reed JC. Phorbol ester-mediated inhibition of growth and regulation of proto-oncogene expression in the human T cell leukemia line JURKAT. Oncogene. 1991;6:455–460. [PubMed] [Google Scholar]
- 32.Perez-Castillo AP, Pipaon C, Garcia I, Alemany S. NGFI-A gene expression is necessary for T lymphocyte proliferation. J Biol Chem. 1993;268:19445–19450. [PubMed] [Google Scholar]
- 33.Seyfert VL, McMahon SB, Glenn WD, Yellen AJ, Sukhatme VP, Cao XM, Monroe JG. Methylation of an immediate-early inducible gene as a mechanism for B cell tolerance induction. Science (Wash DC) 1990;250:797–800. doi: 10.1126/science.2237429. [DOI] [PubMed] [Google Scholar]
- 34.Klaus SJ, Phillips NE, Parker DC. Effects of Il-4 and Fc gamma receptor II engagement on Egr-1 expression during stimulation of B lymphocytes by membrane immunoglobulin crosslinking. Molec Immunol. 1993;30:1553–1558. doi: 10.1016/0161-5890(93)90463-l. [DOI] [PubMed] [Google Scholar]
- 35.Gottschalk AR, Joseph LJ, Quintans J. FcgRII cross-linking inhibits anti-Ig induced egr-1 and egr-2 expression in BCL1 . J Immunol. 1994;152:2115–2122. [PubMed] [Google Scholar]
- 36.Jenkins MK, Schwartz RH, Pardoll DM. Effects of cyclosporin A on T-cell development and clonal deletion. Science (Wash DC) 1988;241:1655–1658. doi: 10.1126/science.241.4873.1655. [DOI] [PubMed] [Google Scholar]
- 37.Watson MA, Milbrandt J. Expression of the nerve growth factor-regulated NGFI-A and NGFI-B genes in the developing rat. Development. 1990;110:173–183. doi: 10.1242/dev.110.1.173. [DOI] [PubMed] [Google Scholar]
- 38.Kaye J, Ellenberger DE. Differentiation of an immature T cell line: a model of thymic positive selection. Cell. 1992;71:423–435. doi: 10.1016/0092-8674(92)90512-b. [DOI] [PubMed] [Google Scholar]
- 39.Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH. Depletion of CD4+T cells in major histocompatibility complex class II-deficient mice. Science (Wash DC) 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- 40.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta-microglobulin deficient mice lack CD4-8+cytolytic T cells. Nature (Lond) 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 41.DeKoning J, Carbone F, Kaye J. Contrast between class I-and class II-MHC mediated differentiation of a CD4+CD8+T cell line: implications for lineage commitment. Int Immunol. 1995;7:541–549. doi: 10.1093/intimm/7.4.541. [DOI] [PubMed] [Google Scholar]
- 42.Quilliam LA, Kato K, Rabun KM, Hisaka MM, Huff SY, Campbell-Burk S, Der CJ. Identification of residues critical for ras(17N) growth-inhibitory phenotype and for ras interaction with guanine nucleotide exchange factors. Molec Cell Biol. 1994;14:1113–1121. doi: 10.1128/mcb.14.2.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhlman P, Moy VT, Lollo BA, Brian AA. The accessory function of murine intercellular adhesion molecule-1 in T lymphocyte activation. J Immunol. 1991;146:1773–1782. [PubMed] [Google Scholar]
- 44.Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nuc Acids Res. 1994;22:5640–5649. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaye J, Hsu M-L, Sauron M-E, Jameson SC, Gascoigne NRJ, Hedrick SM. Selective development of CD4+T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature (Lond) 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 46.Poirier G, Lo D, Reilly CR, Kaye J. Discrimination between thymic epithelial cells and peripheral antigen presenting cells in the induction of immature T cell differentiation. Immunity. 1994;1:385–391. doi: 10.1016/1074-7613(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 47.DeKoning, J., and J. Kaye. Requirements for differentiation of an immature CD4+8+ T cell line. Dev. Immunol. In press. [DOI] [PMC free article] [PubMed]
- 48.Gao EK, Lo D, Cheney R, Kanagawa O, Sprent J. Abnormal differentiation of thymocytes in mice treated with cyclosporin A. Nature (Lond) 1988;336:176–179. doi: 10.1038/336176a0. [DOI] [PubMed] [Google Scholar]
- 49.Anderson G, Anderson KL, Conroy LA, Hallam TJ, Moore NC, Owen JT, Jenkinson EJ. Intracellular signaling events during positive and negative selection of CD4+CD8+thymocytes in vitro. J Immunol. 1995;154:3636–3643. [PubMed] [Google Scholar]
- 50.Bendelac A, Matzinger P, Seder RA, Paul WE, Schwartz RH. Activation events during thymic selection. J Exp Med. 1992;175:731–742. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swat W, Dessing M, von Boehmer H, Kisielow P. CD69 expression during selection and maturation of CD4+8+thymocytes. Eur J Immunol. 1993;23:739–746. doi: 10.1002/eji.1830230326. [DOI] [PubMed] [Google Scholar]
- 52.Brändle D, Muller S, Hengartner H, Pircher H. Regulation of RAG-1 and CD69 expression in the thymus during positive and negative selection. Eur J Immunol. 1994;24:145–151. doi: 10.1002/eji.1830240122. [DOI] [PubMed] [Google Scholar]
- 53.Swan KA, Alberola J -lla, J.A. Gross, M.W. Appleby, K.A. Forbush, J.F. Thomas, and R.M. Perlmutter. Involvement of p21ras distinguishes positve and negative selection in thymocytes. EMBO (Eur Mol Biol Organ) J. 1995;14:276–285. doi: 10.1002/j.1460-2075.1995.tb07001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alberola-Ila J, Forbush KA, Seger R, Krebs EG, Perlmutter RM. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature (Lond) 1995;373:620–623. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- 55.D'Ambrosio D, Cantrell DA, Frati L, Santoni A, Testi R. Involvement of p21rasactivation in T cell CD69 expression. Eur J Immunol. 1994;24:616–620. doi: 10.1002/eji.1830240319. [DOI] [PubMed] [Google Scholar]
- 56.Chen D, Rothenberg EV. Molecular basis for developmental changes in interleukin-2 gene inducibility. Molec Cell Biol. 1993;13:228–237. doi: 10.1128/mcb.13.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cosgrove D, Chan SH, Waltzinger C, Benoist C, Mathis D. The thymic compartment responsible for positive selection of CD4+T cells. Int Immunol. 1993;4:707–710. doi: 10.1093/intimm/4.6.707. [DOI] [PubMed] [Google Scholar]
- 58.Marodon G, Rocha B. Generation of mature T cell populations in the thymus: CD4 or CD8 down-regulation occurs at different stages of thymocyte differentiation. Eur J Immunol. 1994;24:196–204. doi: 10.1002/eji.1830240131. [DOI] [PubMed] [Google Scholar]
- 59.Punt JA, Osborne BA, Takahama Y, Sharrow SO, Singer A. Negative selection of CD4+CD8+thymocytes by T cell receptor-induced apoptosis requires a costimulatory signal that can be provided by CD28. J Exp Med. 1994;179:709–713. doi: 10.1084/jem.179.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Surh CD, Sprent J. T-cell apoptosis detected in situduring positive and negative selection in the thymus. Nature (Lond) 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 61.Huang R-P, Adamson ED. The phosphorylated forms of the transcription factor, Egr-1 bind to DNA more efficiently than non-phosphorylated. Biochem Biophys Res Comm. 1994;200:1271–1276. doi: 10.1006/bbrc.1994.1588. [DOI] [PubMed] [Google Scholar]
- 62.Fehling H J, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature (Lond) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 63.Skerka C, Decker EL, Zipfel PF. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J Biol Chem. 1995;270:22500–22506. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- 64.Mages HW, Stamminger T, Rilke O, Bravo R, Kroczek RA. Expression of PILOT, a putative transcription factor, requires two signals and is cyclosporin A sensitive in T cells. Int Immunol. 1993;5:63–70. doi: 10.1093/intimm/5.1.63. [DOI] [PubMed] [Google Scholar]
- 65.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 66.Schneider-Maunoury S, Topilko P, Seitanidou T, Levi G, Cohen-Tannoudji T, Pournin S, Babinet C, Charnay P. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- 67.Kramer B, Meichle A, Hensel G, Charnay P, Kronke M. Characterization of an Krox-24/Egr-1-responsive element in the human tumor necrosis factor promoter. Biochim Biophys Acta. 1994;1219:413–421. doi: 10.1016/0167-4781(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 68.Maltzman JS, Carmen CA, Monroe JG. Transcriptional regulation of the ICAM-1 gene in antigen receptor- and phorbol ester-stimulated B lymphocytes: role for transcription factor EGR1. J Exp Med. 1996;183:1747–1759. doi: 10.1084/jem.183.4.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maltzman JS, Carmen CA, Monroe JG. Role of EGR1 in regulation of stimulus-dependent CD44 transcription in B lymphocytes. Molec Cell Biol. 1996;16:2283–2294. doi: 10.1128/mcb.16.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez-Cabrera M, Munoz E, Blazquez MV, Ursa MA, Santis AG, Sanchez-Madrid F. Transcriptional regulation of the gene encoding the human C-type lectin leukocyte receptor AIM/CD69 and functional characterization of its tumor necrosis factor-α-responsive elements. J Biol Chem. 1995;270:21545–21551. doi: 10.1074/jbc.270.37.21545. [DOI] [PubMed] [Google Scholar]
- 71.Williams GT, Lau LF. Activation of the inducible orphan receptor gene nur77 by serum growth factors: Dissociation of immediate-early and delayed-early responses. Mol Cell Biol. 1993;13:6124–6136. doi: 10.1128/mcb.13.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calnan BJ, Szychowski S, Chan FK-M, Cado K, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 73.Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 74.McMahon SB, Monroe JG. Activation of the p21ras pathway couples antigen receptor stimulation to induction of the primary response gene egr-1 in B lymphocytes. J Exp Med. 1995;181:417–422. doi: 10.1084/jem.181.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunter KC, Irving SG, Zipfel PF, Siebenlist U, Kelly K. Cyclosporin A-mediated inhibition of mitogeninduced gene transcription is specific for the mitogenic stimulus and cell type. J Immunol. 1989;9:3286–3291. [PubMed] [Google Scholar]
- 76.Emmel EA, Verweij CL, Durand DB, Higgins KM, Lacy E, Crabtree GR. Cyclosporin A specifically inhibits function of nucelar proteins involved in T cell activation. Science (Wash DC) 1989;246:1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- 77.Woodrow MA, Rayter S, Downward J, Cantrell DA. p21ras function is important for T cell antigen receptor and protein kinase C regulation of nuclear factor of activated cells. J Immunol. 1993;150:3853–3861. [PubMed] [Google Scholar]
- 78.Vasquez NJ, Kaye J, Hedrick SM. In vivo and in vitro clonal deletion of double positive thymocytes. J Exp Med. 1992;175:1307–1316. doi: 10.1084/jem.175.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]