Abstract
Recent studies have identified several coreceptors that are required for fusion and entry of Human Immunodeficiency Virus type 1 (HIV-1) into CD4+ cells. One of these receptors, CCR5, serves as a coreceptor for nonsyncytium inducing (NSI), macrophage-tropic strains of HIV-1, while another, fusin or CXCR-4, functions as a coreceptor for T cell line–adapted, syncytiuminducing (SI) strains. Using sequential primary isolates of HIV-1, we examined whether viruses using these coreceptors emerge in vivo and whether changes in coreceptor use are associated with disease progression. We found that isolates of HIV-1 from early in the course of infection predominantly used CCR5 for infection. However, in patients with disease progression, the virus expanded its coreceptor use to include CCR5, CCR3, CCR2b, and CXCR-4. Use of CXCR-4 as a coreceptor was only seen with primary viruses having an SI phenotype and was restricted by the env gene of the virus. The emergence of variants using this coreceptor was associated with a switch from NSI to SI phenotype, loss of sensitivity to chemokines, and decreasing CD4+ T cell counts. These results suggest that HIV-1 evolves during the course of infection to use an expanded range of coreceptors for infection, and that this adaptation is associated with progression to AIDS.
Primary isolates of HIV type 1 (HIV-1) show distinct differences in their biological properties in vitro, including differences in replication kinetics, tropism, and syncytium-inducing capacity (1–4). Macrophage-tropic, nonsyncytium-inducing (NSI)1 viruses are most commonly transmitted and predominate in the early stages of infection (4–7). Syncytium-inducing (SI) isolates generally appear later in infection (8, 9) and may acquire an expanded tropism in vitro, including the ability to replicate in CD4+ T cell lines.
HIV-1 tropism is largely controlled by the env gene of the virus (10–14) which encodes the surface (gp120) and transmembrane (gp41) envelope glycoproteins. Although not clearly defined, binding of gp120 to CD4 is believed to trigger a series of conformational changes, leading to exposure of the NH2-terminal domain of gp41 and subsequent fusion of the virus and host cell membranes (for review see reference 15). Despite high affinity binding to gp120, expression of human CD4 on the surface of nonpermissive cells is not sufficient to confer susceptibility to HIV-1 infection (16–18), indicating that additional factors are required for viral fusion and entry to take place.
Recent studies have identified several proteins that serve as cofactors for HIV-1 fusion with CD4+ T cells (19–24). These proteins are structurally related, G protein–coupled receptors characterized by seven transmembrane-spanning regions (25). One receptor, termed fusin (23) or CXCR-4 (26, 27), has been shown to mediate entry of T cell line– adapted (TCLA), SI strains of HIV-1 into CD4+ T cells, but does not permit entry of NSI isolates. A second molecule, CCR5, has recently been identified as a coreceptor for macrophage-tropic, NSI strains of HIV-1 (20, 21, 24). CCR5 is a member of a family of receptors that bind β-chemokines, including RANTES, MIP-1α, and -1β (28). Two additional β-chemokine receptors, CCR3 and CCR2b, have also been shown to function as coreceptors for some, but not all, primary strains of HIV-1 (19, 22). Taken together, these findings suggest that HIV-1 may use a broad range of coreceptors for infection of CD4+ cells.
While disease progression in many HIV-1–infected individuals is associated with a switch in viral phenotype from NSI to SI, the ability of HIV-1 isolates to induce syncytium formation is defined largely on the basis of in vitro assays (29) with no clear in vivo correlate. We hypothesize here that the molecular basis of the NSI to SI switch lies in a change in coreceptor usage from CCR5 to CXCR-4. Thus far, CXCR-4 has been shown to mediate entry of predominantly TCLA strains of HIV-1 (23). However, it is not yet clear whether primary strains of HIV-1 that use CXCR-4 emerge during the course of infection, and whether use of this coreceptor relates to the switch from NSI to SI phenotype.
Previously, we characterized in detail a series of sequential primary isolates of HIV-1 from three infected patients (5). Two of these patients experienced a switch in viral phenotype from NSI to SI followed by a marked drop in CD4+ T cell counts and the onset of AIDS. The third patient remained clinically stable with only NSI isolates for more than 10 yr after HIV-1 infection. Using sequential isolates from all three patients, we examined the coreceptor use and determined whether disease progression, marked by a switch from NSI to SI phenotype, was associated with the emergence of variants able to use CXCR-4 as a coreceptor, and whether this adaptation was associated with CD4+ T cell decline in vivo. We show here that HIV-1 uses primarily a single receptor, CCR5, in the early stages of infection. However, the virus expands its coreceptor use to include CCR5, CCR2b, CCR3, and CXCR-4 in patients with disease progression. The ability to use CXCR-4 as a coreceptor was only seen with viruses having an SI phenotype and was restricted by the env gene of the virus. The emergence of these viruses was associated with a drop in CD4+ T cell counts and clinical progression in the patients we studied.
Materials and Methods
Study Subjects.
Sequential isolates of HIV-1 were obtained from two individuals, B and C, both of whom progressed rapidly to AIDS after seroconversion. Each patient experienced a precipitous drop in CD4+ T cell counts that was preceded by an increase in viral load and a switch in viral phenotype from NSI to SI (5). Sequential isolates were also evaluated from patient D, who remained asymptomatic for more than a decade after HIV-1 infection (5). Isolates from this patient spanned a 5-yr period during which time he was clinically stable with a low viral load and normal CD4+ T cell counts. All of the isolates from patient D had an NSI phenotype (5).
Generation of Primary Isolates and Biological Clones.
Primary isolates of HIV-1 were obtained by a quantitative co-culture method as previously described (5, 30). In brief, cryopreserved patient PBMCs were thawed and the cells serially diluted fourfold in replicates of 4–10 (from 1 × 106 down to ∼103). The cells were added to wells containing 2 × 106 PHA-stimulated normal donor PBMCs and cultured in a final volume of 1.5 ml of RPMI medium supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), and interleukin-2 (10 U/ml). The culture supernatants were tested on days 7, 14, and 21 for the presence of HIV-1 p24 antigen by a commercially available ELISA assay (Abbott Labs., North Chicago, IL). A culture was considered positive if the p24 antigen level was >30 pg/ml. Virus present in the first well of each dilution series was pooled from the replicates and propagated by a single short-term (7–14 d) passage in PHA-activated normal donor PBMCs, while the biological clones from patient B were generated by similar propagation of virus present in the last positive well of each series. The resulting stocks were titered on normal donor PBMCs and the 50% tissue culture infectious dose (TCID50) was determined by the method of Reed and Muench (31). The syncytium-inducing properties of each isolate were assessed as previously described (5, 30) using the MT-2 cell assay of Koot et al. (29).
Cell Lines.
Cells lines stably expressing CCR1, CCR2b, CCR3, CCR4, CCR5, or CXCR-4 were generated as previously described (32). In brief, retroviral stocks were prepared using the expression vector pBABE-puro and the appropriate coreceptor. Human osteosarcoma cells expressing human CD4 (HOS.CD4) were infected with the pBABE-puro viruses, and 2 d later the cells were selected in 1.0 μg/ml puromycin. When the cells became confluent (after 5–7 days), they were maintained in DMEM containing puromycin, 10% FCS, penicillin (100 U/ml) and streptomycin (100 μg/ml).
Replication of Sequential HIV-1 Isolates.
To assess the coreceptor requirements of primary HIV-1 isolates and biologically cloned isolates, HOS.CD4 (104/well) expressing either CCR1, CCR2b, CCR3, CCR4, CCR5, or CXCR-4 were inoculated with 103 TCID50 of each isolate in a final volume of 1.0 ml of DMEM with added FCS, antibiotics, and 1.0 μg/ml puromycin. The cells were incubated with virus for 24 h at 37°C, washed three times, and samples of the culture supernatants were taken on days 0, 4, 7, 10, and 14 for HIV-1 p24 antigen measurements.
Chemokine Blocking.
Biologically cloned isolates of HIV-1 were used to infect PHA-stimulated, normal PBMCs in the presence of increasing concentrations of recombinant human RANTES, MIP-1α and -1β (R&D Sys., Inc., Minneapolis, MN). PBMCs (2 × 106) were infected with 200 TCID50 of each virus in 1.0 ml of medium. Chemokines were serially diluted twofold and added at the time of infection, such that the final concentration of each chemokine was 500 ng/ml in the highest well. Control cultures were set up in quadruplicate and infected in the absence of added chemokines. After overnight incubation at 37°C, the cells were washed and resuspended in medium containing the appropriate concentration of chemokines. HIV-1 p24 antigen was measured in the culture supernatants on day 7 after infection and the amount of chemokine blocking was calculated as the percent inhibition of HIV-1 p24 antigen production compared to controls.
Single Cycle Infectivity Assays.
HIV-1 env expression vectors were generated by direct DNA PCR amplification and cloning using sequential PBMC samples from patient B as previously described (33). Env clones expressing full-length gp160/120 were co-transfected with an env (−) luciferase reporter vector, pNL43LucE−, to generate env-pseudotyped virions (34). Reporter viruses pseudotyped with either macrophage-tropic, NSI (HIV-1JRFL) or TCLA SI (HIV-1HXB2) envelope glycoproteins were used as controls. HOS.CD4 cells stably transfected with either CCR1, CCR2b, CCR3, CCR4, CCR5, CXCR-4, or the retroviral expression vector (pBABE) were infected with each of the pseudotyped viruses (100 ng p24). Cells were lysed 4 d after infection using 100 μl of luciferase lysis buffer (Promega Corp., Madison, WI). The amount of luciferase activity in 20 μl of lysate was measured using commercially available reagents (Promega Corp.) in a luminometer (Top Count; Packard Instrs., Meriden, CT).
Results
Use of CCR5, CCR3, CCR2b, and CXCR-4 by Sequential HIV-1 Isolates.
In initial experiments, sequential HIV-1 isolates from patients B, C, and D were used to infect CD4+ cells expressing either CCR5, CCR3, CCR2b, or CXCR-4. HIV-1 isolates obtained early in the course of infection from all three patients replicated to low levels in CD4+ cells expressing CCR5, but failed to replicate to detectable levels in cells expressing either CCR2b, CCR3, or CXCR-4 (Table 1). A similar pattern of infection was seen with all subsequent isolates from patient D. However, sequential isolates from patients B and C showed a shift in receptor use which coincided with a switch from NSI to SI phenotype. While viruses isolated at early time points from these patients initially used only CCR5 for infection, viruses isolated later in the course of disease infected cells expressing either CCR5 or CXCR-4. In addition, several later isolates from patient C replicated in cells expressing CCR2b and CCR3 (Table 1). These results suggest that HIV-1 variants arise during the course of infection that use an expanded range of coreceptors for infection of CD4+ cells. In particular, adaptation to use CXCR-4 as a coreceptor was associated with a switch from NSI to SI phenotype and a drop in CD4+ T cell counts.
Table 1.
Replication of Sequential Primary Isolates
| Patient | Date | CD4 cells/mm3 | Phenotype‡ | p24 antigen* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCR2b | CCR3 | CCR5 | CXCR-4 | |||||||||||
| pg/ml | ||||||||||||||
| B | 5/85 | 528 | NSI | <30 | <30 | 200 | <30 | |||||||
| 11/86 | 677 | NSI | <30 | <30 | 26,200 | <30 | ||||||||
| 6/88 | 603 | SI | <30 | <30 | 29,500 | 400 | ||||||||
| 11/88 | 117 | SI | <30 | <30 | 33,320 | 7,820 | ||||||||
| C | 5/84 | 1,233 | NSI | <30 | <30 | 1,024 | <30 | |||||||
| 1/85 | 1,038 | NSI | <30 | <30 | 72,400 | <30 | ||||||||
| 2/86 | 1,049 | SI | <30 | <30 | 84,800 | 1,666 | ||||||||
| 7/86 | 485 | SI | <30 | <30 | 60,200 | 65,600 | ||||||||
| 12/86 | 346 | SI | 16,240 | 12,700 | 31,000 | 43,100 | ||||||||
| 5/87 | 141 | SI | 1,200 | 1,586 | 28,800 | 54,800 | ||||||||
| 6/88 | 186 | SI | <30 | <30 | 30,200 | 19,460 | ||||||||
| 11/89 | 38 | SI | 6,600 | 11,240 | 31,200 | 24,000 | ||||||||
| D | 3/86 | 725 | NSI | <30 | <30 | 140 | <30 | |||||||
| 6/87 | 690 | NSI | <30 | <30 | 181 | <30 | ||||||||
| 9/90 | 700 | NSI | <30 | <30 | 157 | <30 | ||||||||
| 3/91 | 750 | NSI | <30 | <30 | 155 | <30 | ||||||||
HIV-1 p24 antigen was measured in culture supernatants on days 0, 4, 7, 10, and 14 after infection. Values represent the peak p24 levels measured on either day 10 or 14. A value of <30 pg/ml was considered negative.
Because primary HIV-1 isolates may consist of a mixture of distinct variants with different phenotypes (3, 4), we tested a panel of biologically cloned primary HIV-1 isolates for their ability to infect CD4+ cells expressing CXCR-4 or the β-chemokine receptors, CCR1, CCR2b, CCR3, CCR4, and CCR5. These isolates were generated by limiting dilution and co-culture of PBMCs from patient B; five had an NSI phenotype (2–5, 3-1, 3–4, 4-1, 5-2), and two were SI (5–7, 5–8). As shown in Table 2, all five of the NSI viruses replicated in cells expressing CCR5. They did not replicate to detectable levels in cells expressing the other coreceptors, but replicated efficiently in control cultures of PHA-stimulated normal donor PBMCs. Similarly, the two SI viruses replicated well in control PBMC cultures. However, their coreceptor use was restricted to cells expressing CXCR-4; no replication was detected in cells expressing CCR1, CCR2b, CCR3, CCR4, or CCR5. It is not clear why some of the NSI biological clones replicated to lower levels in cells expressing CCR5 compared with others (Table 2; clones 2-5 and 5-2); however, these isolates may be characteristic of viruses with a “slow/low” phenotype (3), or alternatively, may be restricted at a postentry step in HOS.CD4 cells expressing CCR5.
Table 2.
HIV-1 Coreceptor Use by Sequential Biologically Cloned Isolates
| Sample date | Isolate | Phenotype | PBMC | p24 antigen* | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCR1 | CCR2b | CCR3 | CCR4 | CCR5 | CXCR-4 | |||||||||||||
| pg/ml | ||||||||||||||||||
| 5/85 | 2-5 | NSI | 69,200 | <30 | <30 | <30 | <30 | 146 | <30 | |||||||||
| 11/86 | 3-1 | NSI | 51,400 | <30 | <30 | <30 | <30 | 3,540 | <30 | |||||||||
| 3-4 | NSI | 70,800 | <30 | <30 | <30 | <30 | 19,600 | <30 | ||||||||||
| 6/8 | 4-1 | NSI | 81,400 | <30 | <30 | <30 | <30 | 39,240 | <30 | |||||||||
| 11/88 | 5-2 | NSI | 26,600 | <30 | <30 | <30 | <30 | 308 | <30 | |||||||||
| 5-7 | SI | 53,800 | <30 | <30 | <30 | <30 | <30 | 1,202 | ||||||||||
| 5-8 | SI | 89,200 | <30 | <30 | <30 | <30 | <30 | 2,950 | ||||||||||
HIV-1 p24 antigen was measured supernatants on days 0, 4, 7, 10, and 14 after infection. Values represent the peak p24 levels measured on day 10 or 14. A value of <30 pg/ml was considered negative.
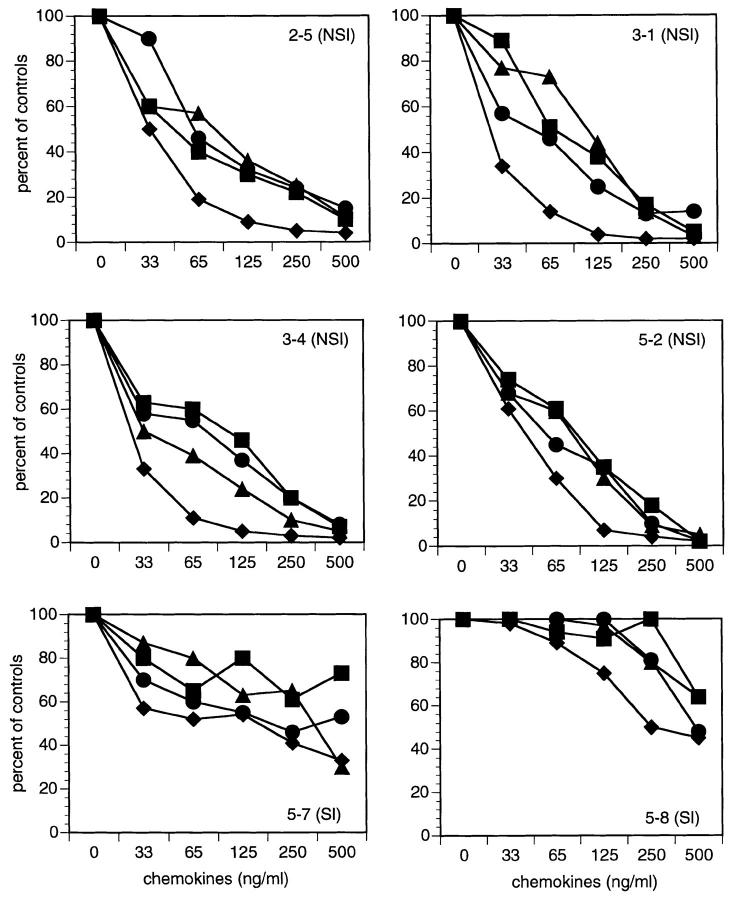
To determine whether coreceptor use was associated with sensitivity to chemokine blocking, we tested the biologically-cloned viruses for blocking by either RANTES, MIP-1α, -1β, or a combination of all three chemokines. As shown in Fig. 1, the NSI viruses (2–5, 3-1, 3–4, 5-2) were inhibited in a dose-dependent manner by addition of RANTES, MIP-1α, and -1β, either alone or in combination with an IC90 of 75–119 ng/ml. In contrast, the two SI isolates (5–7, 5–8) were only partially blocked, even at the highest chemokine concentration (500 ng/ml). These results suggest that there is a loss of sensitivity to the blocking effects of β-chemokines that is associated with acquisition of the ability to use CXCR-4 as a coreceptor for infection. An NSI isolate from late stage infection (5-2) remained sensitive to chemokine blocking consistent with its use of the CCR5 coreceptor (Table 2).
Figure 1.
Inhibition of sequential HIV-1 isolates by β-chemokines. Biologically cloned isolates of HIV-1 were used to infect PHA-activated normal donor PBMCs in the presence of increasing concentrations of RANTES (▪), MIP-1α (•), MIP-1β (▴), or all three combined (♦). HIV-1 p24 antigen was measured in culture supernatants on day 7 after virus inoculation. The percent inhibition was calculated based on control cultures infected without added chemokines.
Taken together, our results suggest that HIV-1 predominantly uses a single coreceptor, CCR5, in the early stages of infection. Variants using CXCR-4 appeared later and were associated with a loss of sensitivity to chemokine blocking in vitro. The presence later in infection of an isolate using CCR5 as a coreceptor (5-2), is consistent with the persistence of macrophage-tropic, NSI variants throughout the course of infection (4).
Env-mediated Interaction.
Fusion of HIV-1 with CD4+ T cells is mediated by interaction of the viral envelope glycoproteins with coreceptors expressed on the cell surface (15). Considerable variation in the env gene of HIV-1 occurs throughout the course of infection and is associated with changes in the tropism and SI properties of the virus. It follows that these changes may reflect the efficiency with which different viral envelope glycoproteins interact with specific coreceptors.
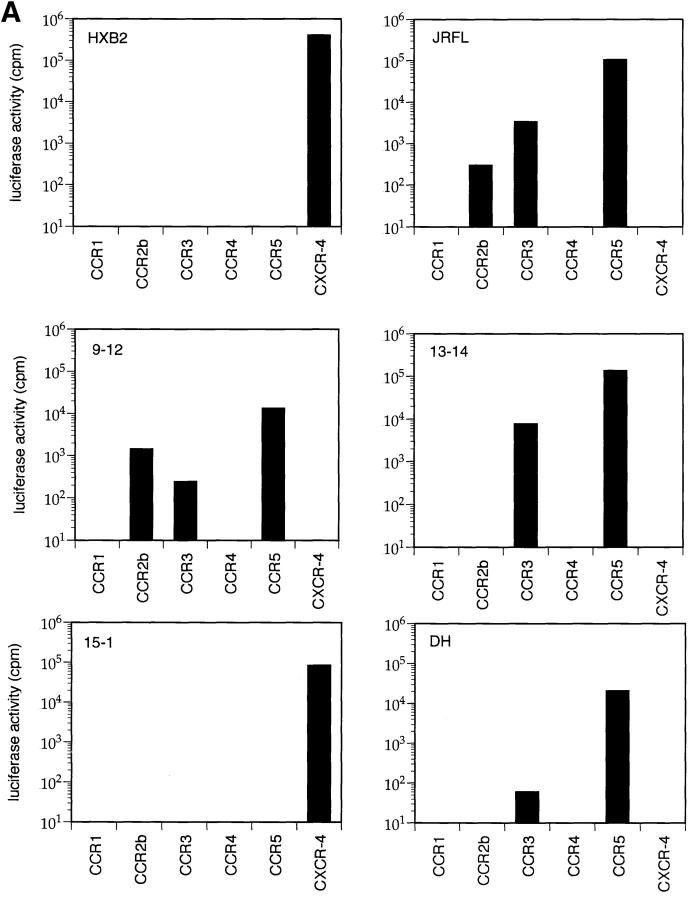
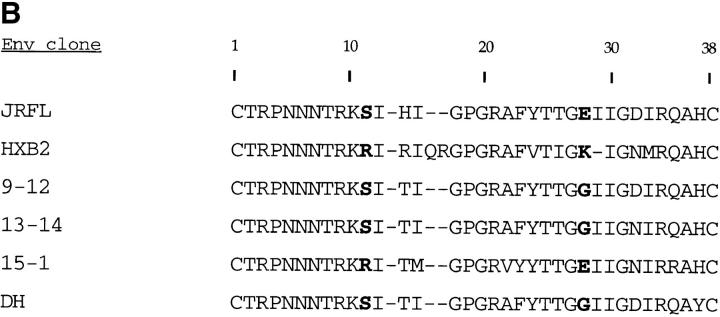
To test this hypothesis, we generated full-length env clones by direct DNA PCR amplification from sequential PBMC samples from patient B (33). Env clones 9-12 and 13-14 were derived from early NSI viruses (Table 1; 5/85 and 11/86, respectively), whereas env clone 15-1 was derived from an SI isolate (Table 1; 6/88). HIV-1 virions containing a luciferase reporter gene were pseudotyped with the cloned envelope glycoproteins and used in single cycle infectivity assays to evaluate viral entry into CD4+ cells expressing each of the β-chemokine receptors or CXCR-4. As shown in Fig. 2, luciferase activity was detected in cells expressing CCR5, CCR3, and CCR2b after infection with control viruses pseudotyped with envelope glycoproteins from a macrophage-tropic, NSI strain of HIV-1 (HIV-1JRFL) (Fig. 2 A). In contrast, a TCLA, SI virus (HIV-1HXB2) was only able to infect cells expressing CXCR-4. Virions pseudotyped with envelope glycoproteins from two of the primary env clones from patient B (9–12, 13–14) infected cells expressing CCR5, and to a lesser extent, CCR2b and CCR3, while a third clone (15-1) only infected cells expressing CXCR-4. Another primary HIV-1 env clone derived from a long-term survivor (DH) used CCR5 and CCR3 for infection. Sequencing of the V3 loop of env clones 9-12, 13-14, and DH revealed the presence of uncharged amino acids at positions 11 and 28 (Fig. 2 B), characteristic of NSI viruses, while clone 15-1 had a positively charged residue at position 11, characteristic of SI viruses (35, 36).
Figure 2.
Coreceptor use by primary HIV-1 env-pseudotyped virions. (A) HOS.CD4 expressing either CCR1, CCR2, CCR3, CCR4, CCR5, or CXCR-4 were infected with HIV-1 virions carrying a luciferase reporter gene and pseudotyped with envelope glycoproteins from either control viruses (HIV-1JRFL, HIV-1HXB2) or from primary virus env clones. Three of the primary env clones came from a patient with rapid disease progression (9–12, 13–14, 15-1), while the fourth clone was from a patient with long-term asymptomatic infection (DH). Luciferase activity was calculated by subtracting the background measurements made using env(−) virions and HOS.CD4 cells expressing only the retroviral vector, pBABE. (B) Deduced amino acid sequences of the V3 domain of gp120 from cloned primary HIV-1 env genes. Bold type indicates amino acids at positions 11 and 28 which are associated with the SI phenotype of the virus (35, 36).
Discussion
In this study, we examined coreceptor use by sequential primary isolates of HIV-1 obtained throughout the course of infection. Our results indicate that disease progression is associated with the appearance of viral variants able to use an expanded range of coreceptors for infection of CD4+ cells. HIV-1 isolates initially used a single coreceptor, CCR5, for infection of CD4+ cells in vitro. Later in the course of disease, the virus expanded its coreceptor use to include CCR5, CCR3, CCR2b, and CXCR-4. In particular, the appearance of viruses using CXCR-4 as a coreceptor was associated with a decline in CD4+ T cell counts and clinical progression to AIDS. In contrast, only viruses capable of using CCR5 could be isolated from a third patient, who remained asymptomatic for over 10 yr after HIV-1 infection with a low viral burden and normal CD4+ T cell counts (5). These results indicate that the expanded coreceptor use is not simply the result of increasing env diversity over time, but instead suggest that there may be a selective advantage for viruses that can use coreceptors in addition to CCR5.
Recent studies have identified CCR5 as an essential determinant of sexual transmission (32). CD4+ T-cells from two multiply exposed/uninfected individuals were shown to resist infection with macrophage-tropic, NSI strains of HIV-1 (32, 37, 38) due to a 32–base pair deletion in the gene encoding CCR5 (32). Our results indicate that viruses using CCR5 persist during asymptomatic infection, and in one patient, were present as long as 7 yr after seroconversion. These viruses are typical of many primary NSI isolates in that they exhibit a dual tropism for both CD4+ T lymphocytes and macrophages. Both cells types are known to express CCR5 and may be targets for these viruses in vivo. The persistence of viruses using CCR5 suggests they may escape immune surveillance mechanisms, possibly by sequestration in long-lived cell populations such as macrophages or dendritic cells.
Selective pressure exerted by either specific or nonspecific immune mechanisms may lead to the evolution of HIV-1 variants with distinct coreceptor requirements. Because CCR5 binds several β-chemokines, including RANTES, MIP-1α, and -1β, selection may favor variants that can replicate in the presence of high concentrations of these factors. We identified several variants of HIV-1 that were resistant to the blocking effects of exogenously added β-chemokines. This resistance was associated with loss of the ability to use CCR5 for infection. This suggests that HIV-1 may evolve in vivo to use alternative coreceptors, such as CXCR-4, in response to selection pressure exerted by the production of β-chemokines. Given the high rate of HIV-1 turnover in vivo (39, 40) and the small number of amino acid changes necessary to alter the virus phenotype (35, 36), it is not yet clear why these viruses do not emerge earlier. However, they may be more sensitive to neutralization by HIV-1–specific antibodies due to changes in the envelope glycoproteins, or alternatively, they may be blocked by the presence of stromal cell–derived factor 1, which was recently identified as the ligand for CXCR-4 (26, 27).
Relatively subtle changes in the viral genome can influence both phenotype and coreceptor use. As few as two amino acid changes in the V3 loop of env have been shown to affect the ability of primary isolates to induce syncytium formation in vitro (35, 36). Moreover, substitutions at two amino acid positions in V3 were sufficient to overcome the resistant phenotype of CD4+ T cells from two exposed/ uninfected individuals to infection with SI strains of HIV-1 (38). This is presumably due to a change in coreceptor use from CCR5, which is defective in these individuals (32), to CXCR-4 or another functional coreceptor. Indeed, recent reports have shown that use of both CCR3 and CCR5 is dependent on the sequence of the V3 loop (19).
By cloning sequential env genes from an HIV-1 infected individual, we have shown that coreceptor use is controlled by the env gene of the virus, and have identified primary HIV-1 variants with distinct coreceptor requirements. Previously, we found amino acid substitutions in the V3 loop of these variants that were characteristic of an NSI to SI switch in phenotype (37), and have now shown that the corresponding env clones display a switch in coreceptor use from CCR5 to CXCR-4. Although the exact mechanism for this switch is unclear, it is possible that mutations in V3 may directly affect interaction of the virus with specific sites on the coreceptor molecule or may indirectly influence receptor binding by altering the global conformation of gp120.
The appearance of HIV-1 variants able to use CXCR-4 as a coreceptor was associated with an increase in viral load and a marked drop in CD4+ T cell counts in the patients we studied. It is not clear if these viruses are directly cytopathic for CD4+ T cells in vivo. The most straightforward explanation is that the SI phenotype reflects adaptation of HIV-1 to use CXCR-4 expressed on CD4+ target cells in vivo. Because the tissue distribution of CXCR-4 is much broader than CCR5, this may allow the virus access to a wider range of potential target cells, or alternatively, may permit fusion with more permissive target cells, thereby facilitating HIV-1 replication and spread. Further studies will be necessary to discriminate between these possibilities and delineate the pathogenic mechanisms associated with CD4+ T cell depletion in vivo and the role of HIV-1 coreceptor use in this process.
Acknowledgments
We would like to thank Cladd Stevens and Pablo Rubinstein for providing PBMC samples and clinical information from patients B, C, and D; E. Fenamore for technical assistance, and Simon Monard for FACS® analysis.
This work was supported by grants and contracts (CA72149, AI36057, and AI45218) from the National Institutes of Health, from the American Foundation for AIDS Research in memory of Florence Brecher, and from the Aaron Diamond Foundation.
Footnotes
1 Abbreviations used in this paper: HOS.CD4, human osteosarcoma cells expressing human CD4; NSI, nonsyncytium-inducing; SI, syncytium-inducing; TCID50, 50% tissue culture infectious dose; TCLA, T cell line–adapted.
References
- 1.Cheng-Mayer C, Seto D, Tateno M, Levy JA. Biologic features of HIV-1 that correlate with virulence in the host. Science (Wash DC) 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 2.Connor RI, Ho DD. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fenyö E, Morfeldt-Måson L, Chiodi F, Lind B, Von Gegerfelt A, Albert J, Åsjö B. Distinctive replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuitemaker H, Koot M, Kootsra NA, Wouter-Dercksen M, de Goede REY, Van Steenwijk RP, Lange JMA, Eeftink-Schattenkerk JKM, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor RI, Mohri H, Cao Y, Ho DD. Increased viral burden and cytopathicity correlate temporally with CD4+T-lymphocyte decline and clinical progression in HIV-1 infected individuals. J Virol. 1993;67:1772–1778. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roos MTL, Lange JMA, de Goede REY, Coutinho RA, Schellekens PTA, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 7.Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science (Wash DC) 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 8.Tersmette M, de Goede REY, Al BJM, Winkel IN, Gruters RA, Cuypers HT, Huisman HG, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tersmette M, Lange JMA, de Goede REY, deWolf F, Eeftink-Schattenkerk JKM, Schellenkens PTA, Coutinho RA, Huisman JG, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS mortality. Lancet. 1989;i:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Quiroga M, Tung JW, Dina D, Levy JA. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jong J-J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature (Lond) 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 13.Shioda T, Levy JA, Cheng-Mayer C. Macrophage and T cell–line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature (Lond) 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 14.Westervelt P, Trowbridge DB, Epstein LG, Blumberg BM, Li Y, Hahn BH, Shaw GM, Price BW, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:25772582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, J.P., B.A. Jameson, R.A. Weiss, and Q.J. Sattentau. 1993. The HIV-cell fusion reaction. In Viral Fusion Mechanisms. J. Betz, editor. CRC Press, Inc., Boca Raton, FL. 233–289.
- 16.Ashorn P, Berger E, Moss J. Human immunodeficiency virus envelope glycoprotein/CD4–mediated fusion of nonprimate cells with human cells. J Virol. 1990;64:2149–2156. doi: 10.1128/jvi.64.5.2149-2156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesebro B, Buller R, Protis J, Wehrly K. Failure of human immunodeficiency virus entry and infection in CD4-positive human brain and skin cells. J Virol. 1990;64:215–221. doi: 10.1128/jvi.64.1.215-221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 19.Choe H, Farazan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 20.Deng HK, Choe S, Ellmeier W, Liu R, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of C–C chemokine receptor 5 as the major coreceptor for entry of macrophage-tropic human immunodeficiency virus type-1. Nature (Lond) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Moore JP, Koup RA, Paxton WA. β-chemokines inhibit HIV-1 entry into CD4+cells via the C-C CKR5 co-receptor. Nature (Lond) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 22.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper S, Parmentier M, Collman RG, Doms RW. A dual-tropic, primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR5, CKR3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science (Wash DC) 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: A RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science (Wash DC) 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 25.Horuk R. Molecular properties of the chemokine receptor family. Trends Pharmacol Sci. 1994;15:159–165. doi: 10.1016/0165-6147(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 26.Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer TA. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature (Lond) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 27.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, Schwartz O, Heard JM, ClarkLewis I, Legler DF, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell line adapted HIV-1. Nature (Lond) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 28.Samson M, Labbe O, Mollereau C, Vassert G, Parmentier M. Molecular cloning and functional expression of a new human C-C-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 29.Koot M, Vos AHV, Keet RPM, de Goede REY, Dercksen MW, Terpstra FG, Coutinho RA, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS (Phila) 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Connor RI, Notermans DW, Mohri H, Cao Y, Ho DD. Biological cloning of functionally diverse quasispecies of HIV-1. AIDS Res Hum Retroviruses. 1993;9:541–546. doi: 10.1089/aid.1993.9.541. [DOI] [PubMed] [Google Scholar]
- 31.Dulbecco, R. 1988. End-point method measurement of the infectious titer of a viral sample. In Virology. 2nd ed. J.P. Lippincott, Philadelphia, PA. 22–25.
- 32.Liu R, Paxton WA, Choe S, Ceradini D, Martin SA, Horuk R, Macdonald ME, Stuhlmann H, Koup RA, Landau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some mutiply-exposed individuals to HIV-1 infection. Cell. 1996;87:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 33.Connor RI, Sheridan KE, Lai C, Zhang L, Ho DD. Characterization of the functional properties of envgenes from long-term survivors of human immunodeficiency virus type-1 infection. J Virol. 1996;70:5306–5311. doi: 10.1128/jvi.70.8.5306-5311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 35.De Jong J-J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouchier RAM, Groenink M, Kootstra NA, Tersmette M, Huisman HG, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connor RI, Paxton WA, Sheridan KE, Koup RA. Macrophages and CD4+T-lymphocytes from two multiply exposed, uninfected individuals resist infection with primary NSI isolates of human immunodeficiency virus type-1. J Virol. 1996;70:8758–8764. doi: 10.1128/jvi.70.12.8758-8764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paxton WA, Martin SR, Tse D, O'Brien TR, Skurnick J, VanDevanter NL, Padian N, Braun JF, Kotler DP, Wolinsky SM, Koup RA. Relative resistance of HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposures. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 39.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature (Lond) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 40.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Duestch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, Shaw GM. Viral dynamics in human immunodeficiency virus type 1 infection. Nature (Lond) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]