Abstract
The genetic origins of CD8+ T cell–recognized unique antigens to which mice respond when immunized with syngeneic tumor cells are unknown. The ultraviolet light-induced murine tumor 8101 expresses an H-2Kb-restricted immunodominant antigen, A, that induces cytolytic CD8+ T cells in vivo A+ 8101 cells are rejected by naive mice while A− 8101 tumor cells grow. To identify the antigen H-2Kb molecules were immunoprecipitated from A+ 8101 cells and peptides were eluted by acid. The sensitizing peptide was isolated by sequential reverse-phase HPLC and sequenced using microcapillary HPLC-triple quadruple mass spectrometry. The peptide, SNFVFAGI, matched the sequence of the DEAD box protein p68 RNA helicase except for a single amino acid substitution, caused by a single nucleotide change. This mutation was somatic since fibroblasts from the mouse of tumor origin expressed the wild-type sequence. The amino acid substitution created an anchor for binding of the mutant peptide to H-2Kb. Our results are consistent with mutant p68 being responsible for rejection of the tumor. Several functions of p68, which include nucleolar assembly and inhibition of DNA unwinding, may be mediated through its IQ domain, which was altered by the mutation. This is the first description of a somatic tumor–specific mutation in the coding region of a nucleic acid helicase.
One of the oldest, most important and yet unresolved questions in tumor immunology is the nature of unique, or individually distinct, tumor antigens to which mice respond when immunized with tumor cells. Classical experiments (1, 2) showed that mice immunized with tumor cell lines rejected subsequent tumor cell transplants effectively, when the same tumor cell line was used for immunization and for challenge. Even cell lines of the same histologic type and induced by the same carcinogen, did not induce crossprotection (3). Shared cytolytic T cell–recognized antigens have been identified on experimental as well as on human tumors (4–9) and can, after active immunization, provide transient protection in experimental models when the dose of tumor cells used for challenge is small and the interval between immunization and challenge is short (10, 11, for review see reference 12). However, unique antigens provide strong and long-lived immunological protection against transplantation of the same experimentally induced cancer after active immunization with cancer cells (3), whereas shared or crossreactive antigens do not. For example the shared tumor antigen P1A (4) is expressed by multiple tumor lines as determined by Northern blots and by sensitivity to lysis by P1A-specific CTL. Even though this antigen induces crossreactive T cells that are cytolytic for multiple tumor lineages (13), protection is not provided by this shared antigen but by unique antigens (13).
Oncogenes such as ras and suppressor genes such as p53 can encode tumor antigens (14–18) but these antigens appear to be different from those which cause tumor rejection after immunization with tumor cells (19). A unique T cell–recognized antigen was found to be caused by a mutation in the ribosomal gene L9 (20). While lymph node cells specific for this antigen allowed SCID mice to reject a tumor challenge, it remains unknown whether this antigen is the natural rejection antigen of the tumor, particularly because progressor variants retained the antigen (20). Nevertheless, the availability of autologous normal and malignant controls yielded the first unequivocal evidence that a unique T cell–recognized antigen is caused by a somatic mutation and thus is tumor-specific. Mutant L9 encodes a peptide recognized by CD4+ T cells. However, in experimental tumor systems, CD8+ T cells are required for tumor rejection (21, see reference 12 for review), so antigens recognized by these T cells are prime candidates for rejection antigens. The genetic origins of several CD8+ cytolytic T cell–recognized unique tumor-specific antigens have been identified on human tumors (22–25); however not all unique antigens recognized by CD8+ cytolytic T cells may be capable of eliciting tumor rejection, and the functional significance of these human antigens in tumor rejection is unclear. The relevance of unique antigens for tumor rejection can be more readily studied in experimental systems than in human systems. Earlier studies in experimental systems have shown that mutagen treatment of cancer cells in vitro can result in variant tumor cells that are rejected by normal hosts and also induces mutations that encode CD8+ T cell–recognized antigens (26, 27). However, the genetic origins of unique antigens not caused by such manipulations on experimental tumors remained unknown. An interesting genetic alteration consisting of three consecutive nucleotide substitutions was reported to lead to a CTLdefined antigen on the murine 3LL tumor (28); but, this study and others (29, 30) lacked autologous controls. Therefore germline mutations could not be excluded.
In this study, we have determined the genetic origin of a unique CD8+ T cell–recognized, immunodominant antigen on a UV-induced regressor tumor, for which autologous controls are available. Expression of this antigen is correlated with tumor rejection, since progressor variants of this tumor do not express the antigen. We show that the antigen results from a somatic mutation which generates a single amino acid change in the murine p68 RNA helicase protein. This is also the first identification of a tumor-specific mutation in the coding region of a member of the family of DEAD-box proteins of putative RNA helicases.
Materials and Methods
Mice.
Female C57BL/6 (H-2b) mice, 5–6 wk old, were purchased from the Frederick Cancer Research Facility (Bethesda, MD). C57BL/6 nu/nu (H-2b) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained at the University of Chicago in a pathogen-free barrier facility, and fed autoclaved food and acidified sterile water.
Cell Lines.
The 8101 tumor was induced in our laboratory by chronic ultraviolet light irradiation of C57BL/6 mice, three times a week, as described (31). Tumor fragments were placed in vitro to establish a cell line. The heart and lungs of the mouse were harvested and chopped into fragments which were frozen in liquid nitrogen, and also adapted to in vitro culture to generate a heartlung fibroblast (HLF)1 cell line. TAP-deficient RMA-S (H-2b) (32) cells were used as targets after exogenous loading of peptides. RMA-S and RMA (32) cells were a gift of Dr. J.A. Bluestone (University of Chicago, IL). The BPV series of H-2b UV-induced tumors is a gift of Mr. Vijay Sreedhar and Dr. Margaret Kripke (University of Texas, M.D. Anderson Cancer Center, Houston, TX). Cytolytic T cell lines and clones were generated as described (31).
Purification of Sensitizing Activity.
Tumor cells were expanded in Nunc 10-chamber cell factories (Nunc, Thousand Oaks, CA), detached with trypsin-EDTA, washed once with PBS, quick-frozen as cell pellets and stored at −80°C in polypropylene tubes. To reduce peptide loss glass pipettes and polypropylene tubes were used throughout the purification procedure. A batch of 1010 cells was thawed, resuspended in lysis buffer (33) and rotated for 4–6 h at 4°C. The lysate was centrifuged at 3,500 g for 30 min, and the supernatant was rotated with 15–20 mg of purified monoclonal anti-H2-Kb Y-3 antibody coupled to protein A-Sepharose (Pharmacia, Uppsala, Sweden) for 4–6 h at 4°C, washed three times with PBS and three times with ddH2O (200 g, for 5 min). The antigen was eluted by vortexing the pelleted Sepharose with 3–4 ml of 0.2% TFA/H2O (vol/vol) for 15 min at room temperature. The eluate was divided into four 5,000 mol wt cutoff filters (Millipore UFC4LCC25; Marlborough, MA) and centrifuged for five h at 3,500 g, 4°C . The filtrate was concentrated to near-dryness by vacuum centrifugation, pooled into a final volume of 150–200 μl in 0.2% TFA/H2O and stored at −80°C in 1.5 ml polypropylene microfuge tubes (Sarstedt™, Inc., Newton, NC).
51Cr-release Assays.
Five thousand 51Cr-labeled targets were incubated with various numbers of T cells in flexible 96-well V-bottom microtiter plates (Dynatech, Chantilly, VA) for 4.5 h as described (31). The percentage of specific lysis was calculated by the formula: % cytolysis = [(experimental release-spontaneous release)/(maximum release − spontaneous release)] × 100. Spontaneous release was <15% of total release. To test HPLC fractions for sensitizing activity, RMA-S cells which had been pre-incubated at room temperature for at least 12 h, were 51Cr labeled and then added to 50 μl FCS in each well of a 96-well plate. These cells were then incubated with aliquots of HPLC fractions for 1.5 h at 37°C. T cells were added to each well in 50 μl CDMEM and the mixture was incubated for an additional 4 h at 37°C.
Micropore HPLC Separation.
Peptide fractionation was conducted on an Aquapore 18 column (2.1 mm × 3 cm). The peptide extract was concentrated to 200 μl, injected onto a narrow bore C18 column, and eluted with a 55-min binary gradient increasing from 0–60% B at the rate of 3% B for the first 5 min, then 0.9% B for the next 50 min. (Solvent A = 0.1% heptafluorobutyric acid (HFBA) in NANOpure water; solvent B = 0.085% HFBA in 60% acetonitrile; flow rate 200 μl/min). Fractions were collected into polypropylene tubes (Sarstedt™ 2 ml, Cat no. 72.692) at 1-min intervals and 0.3% of each fraction was tested for activity. Active fractions 36 and 37 were individually rechromatographed on the same column using a shallower gradient and TFA as the ionic modifier. The second dimension gradient increased from 0–60% B at the rate of 5% B for the first 5 min, then 0.7% B for the next 50 min (Solvent A = 0.1% TFA in NANOpure water; solvent B = 0.085% TFA in 60% acetonitrile; flow rate 200 μl/min). Fractions were collected into polypropylene tubes at 1-min intervals and 1.5% was tested for activity.
Identification of Candidate Peptide.
Candidate peptides were identified by combining mass spectrometry with a sensitive 51Cr-release assay as described previously (33). 60% of the second dimension fraction was loaded onto a C18 microcapillary HPLC column (100 μm i.d. × 25 cm) end-connected with a zero dead volume union to two capillaries of internal diameter 25 μm and 40 μm. Peptides were eluted with a 34-min gradient of 0-60% B (solvent A = 0.1 M acetic acid; solvent B = acetonitrile) at a flow rate of 1 μl/min. One-fifth of the eluent was deposited into each well of a 96-well microtiter plate containing 50 μl CRPMI while the remaining four-fifths was directed into the mass spectrometer. The m/z ratio of each peptide deposited in a particular well was recorded on the mass spectrometer. Peptides in individual wells of the 96-well plate were then tested for sensitizing activity. The ion abundance of a particular peptide is manually correlated with the sensitizing activity to identify the mass of the candidate peptide.
Sequence Analysis of the Tumor Antigen Candidate.
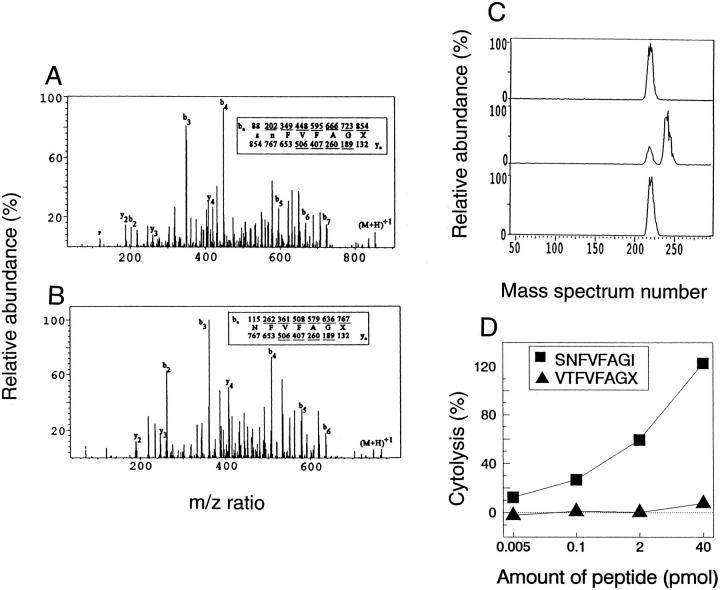
To determine the sequence of the tumor antigen an aliquot from the remaining 40% of subfraction 36-19 was loaded onto a C18 microcapillary column (75 μm i.d. × 12 cm) and eluted with a 12-min gradient (0–80% acetonitrile in 0.1 M acetic acid) directly into a triple quadruple mass spectrometer (Finnigan MAT, TSQ7000) essentially as described (34, 35). This instrument is equipped with an electrospray ionization source that was operated with a coaxial sheath (70% MeOH/H2O containing 0.12% acetic acid) flowing at 1.5 μl/min. A negative potential of 4.6 kV was applied to the heated capillary. Quadrupole one was set to pass a 2 mass unit window centered on 854, the m/z value corresponding to the (M + H)+1 ions of the tumor antigen. Ions of this mass were transmitted to quadrupole 2 where they suffered collision-activated dissociation (CAD). The resulting fragments were mass analyzed in quadrupole 3 to produce the CAD mass spectrum shown in Fig. 6.
Figure 6.
Structural characterization of the tumor epitope. (A) CAD mass spectrum recorded from the (M+H)+1 ions (m/z 854) of the tumor antigen. (B) CAD mass spectrum recorded on (M+H)+1 ions (m/z 767) from the tumor antigen after a single round of Edman degradation. The ions observed in each spectra are underlined. (C) Results of coelution experiments in which synthetic peptides SNFVFAGL or SNFVFAGI were added to the biologically active subfraction 37-19 containing the tumor antigen. (D) synthetic peptides SNFVFAGI and VTFVFAGX (X = L or I) were loaded onto RMA-S cells in the indicated concentrations, and tested for lysis by the anti-A CTL clone. The E/T ratio was 5:1. SNFVFAGI is specifically recognized by the anti-A CTL clone.
Manual Edman Degradation.
The mixture of peptides in an aliquot of HPLC fraction 36-19 was treated with 5 μl of 5% phenyl isothiocyanate (PITC) in pyridine. The resulting solution was overlaid with argon, vortexed, and incubated for 30 min at 45°C. This sample was lyophilized to dryness, resuspended in 15 μl sequencing grade TFA, overlaid with argon and incubated for 10 min at 37°C. TFA was removed under vacuum and the residue was dissolved in 5 μl H2O. The solution was vortexed, and extracted twice with 15 μl n-butyl acetate. For analysis by mass spectrometry, the aqueous layer was isolated, evaporated to dryness and resuspended in 1 μl acetic acid and 19 μl H2O. This modification removes the NH2-terminal amino acid from all peptides present in the mixture.
Identification of the COOH-terminal Amino Acid by Coelution with Synthetic Peptides.
An aliquot of the HPLC fraction containing the tumor antigen 854 was mixed with an equimolar amount of either synthetic peptide SNFVFAGL or SNFVFAGI and eluted from a microcapillary HPLC column directly into the mass spectrometer using a gradient of 0.1 M acetic acid and acetonitrile increasing at 2% per min. Mass spectra were acquired every 1.5 s over the mass range 300–1,400. The ion abundance at m/z 854 was plotted as a function of elution time.
Synthesis and Purification of Synthetic Peptides.
Peptides SNFVFAGI and VTFVFAGX were synthesized and purified either at the University of Virginia, or at the Oligopeptide Synthesis Facility at the University of Chicago by the solid phase method using standard fmoc chemistry and purified by HPLC. Peptide SNFVSAGI was synthesized and purified by Chiron Mimotopes (Raleigh, NC) and HPLC purified.
Amplification and Sequencing of cDNA and Genomic DNA.
PCR primers specific for the murine p68 RNA helicase cDNA were synthesized (IDT, Coralville, IA). 5′ primer Hel1, 5′-AATTAAGGTACCGGTCCTTGCCCTCGCAGCTCC-3 and 3′ primer Hel2A 5′-CGAGATCTCTGCACTGCAGTCATTTCTG-3′ amplify a 2.1-kb fragment that encompasses the coding region of the murine p68 RNA helicase cDNA. The cDNA was amplified using RT-PCR with the following conditions: 1.25 mM MgCl2, 25 pM of each primer, 70 U RNAsin (Promega, Madison, WI), 250 μM of each nucleotide, 1 μl RNA, and 25 U M-MLV reverse transcriptase (New England Biolabs, Beverly, MA), in a 100-μl reaction. The mixture was incubated at 38°C for 10 min to synthesize the cDNA, and 94°C for 5 min to inactivate the reverse transcriptase. 1 μl of Taq polymerase (Promega), was added to the reaction and the 2.1-kb cDNA was amplified by PCR for 40 cycles at 94°C for 1 min, 55°C for 2 min, 72°C for 3 min followed by a 5-min final extension at 72°C. The PCR product was cloned into the vector pcDNA3 (Invitrogen, San Diego, CA) using the KpnI site in the 5′ primer, and the BglII site in the 3′ primer. The subclones were sequenced using the Sequenase kit (Amersham, Arlington Heights, IL). A 465-bp PCR product including the region of the putative mutation was isolated from 8101 HLF genomic DNA and 8101-PRO genomic DNA using the same PCR conditions and the internal 5′ primer Hel1A 5′-CGGGGTACCACTCTGCAGGCAAAAGGGGTGGATT-3′. The PCR products were sequenced using the fmol sequencing system (Promega). Total RNA was isolated using either guanidinium isothiocyanate, or by using the TRI™ reagent (Molecular Research Center, Inc., Cincinnati, OH). Total RNA from 8101RE was passed over an oligo (dT) cellulose column to isolate poly (A)+ mRNA. Genomic DNA was isolated using the ONCOR non-organic DNA isolation kit (ONCOR, Gaithersburg, MD).
Results
Expression of the CD8+ T Cell–recognized A Antigen on the 8101 Tumor Correlates with Tumor Rejection by Naive Mice.
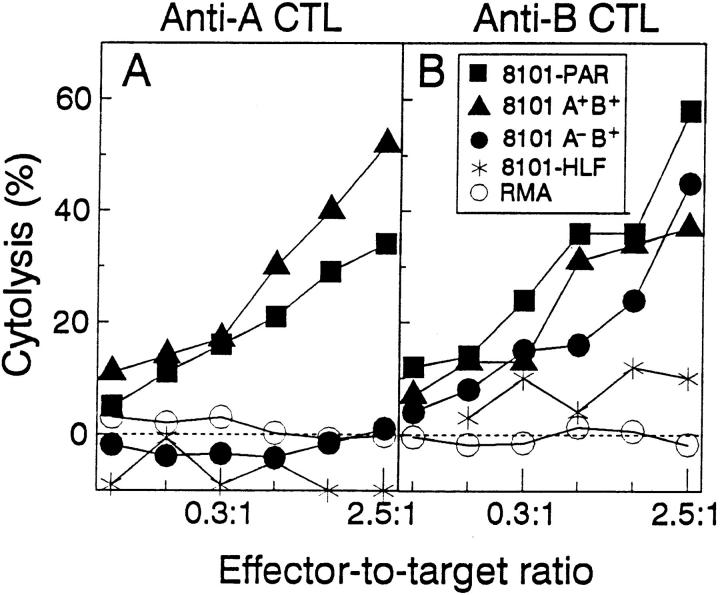
8101 was the first tumor isolated from a group of C57BL/6 mice that received chronic UV-irradiation. The tumor arose on the back of the mouse after 11 mo of irradiation. At this time the mouse was 14 mo old. As usually observed for UV-induced tumors, 8101 readily adapted to culture and the primary culture was cloned to study the antigenic diversity of the cells that adapted. CD8+ cytotoxic T lymphocyte (CTL) clones were generated from mixed lymphocyte tumor cell cultures (MLTCs) of spleen cells from mice immunized by i.p. injection of live uncloned tumor cells. The resulting T cell clones identified two types of tumor cell clones: one which expressed two antigens designated A and B, and a second that only expressed the B antigen. The parental uncloned cell line, designated 8101-PAR, expressed both antigens and was lysed by anti-A as well as anti-B CTL clones (Fig 1, A and B). The anti-A (Fig. 1 A) and anti-B (Fig. 1 B) CTL clones lysed only 8101 lineage tumor cells and did not lyse autochthonous normal fibroblasts, or RMA cells. We injected the original uncloned 8101-PAR tumor cell line and 8101 A+B+ and 8101 A−B+ clones into C57BL/6 nude mice to generate tumor fragments. The developing tumors were transplanted as fragments into naive normal C57BL/6 mice. Table 1 shows that tumors derived from the A+ clone were regularly rejected and thus were designated 8101-RE. Tumors derived from the A− clone grew progressively and were therefore designated as 8101-PRO. These results strongly suggested that the expression of the A antigen correlated with rejection of the A+ tumor in naive mice, and that absence of the A antigen led to progressive tumor growth. The B antigen was expressed on all 8101 lineage tumors but not other C57BL/6 UV tumors (Fig. 2 B), indicating that the 8101RE and 8101-PRO tumors were of the same clonal origin. The parental cell line 8101-PAR which, as suggested by clonal analysis, contained A+ as well as A− tumor cells, grew progressively when fragments were transplanted into naive normal mice. The reisolated tumor cells when readapted to culture were resistant to anti-A CTL, unlike the 8101-PAR tumor cells which had been used for the challenge (Fig. 1 A). These data indicated that normal mice usually selected against expression of the A antigen. Only one re-isolated tumor still showed sensitivity to lysis by anti-A CTL; this tumor may have been able to grow as A+ because of the simultaneous challenge with A−B+ progressor tumor cells which were also present in the 8101-PAR tumor. These A−B+ tumor cells may have prevented the establishment of an effective anti-A response. This suggestion is in agreement with our previous observation that progressor tumors can sometimes prevent an immune response to highly antigenic tumor cells (36).
Figure 1.
Antigenic differences between 8101 tumor cell clones. The B antigen is expressed on all three lines derived from the 8101 tumor while the A antigen is expressed on the uncloned 8101-PAR tumor cells, and on some of the 8101 tumor cell clones. Autologous non-malignant fibroblasts, 8101-HLF and a syngeneic lymphoma cell RMA are not lysed by either the anti-A or anti-B CTL clone. Target cells were tested in a 51Crrelease assay for lysis using the anti-A CTL ( A) and the anti-B CTL (B) as effector cells.
Table 1.
Growth In Vivo of Uncloned 8101 Tumor and 8101 Tumor Clones
| Tumor incidence* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype‡ | Tumor | Expt no. | Nude mice | Normal mice§ | Reisolate phenotype | |||||
| A+B+ | 8101-PAR | 1 | 1/1 | 15/15 | ND | |||||
| 2 | 1/1 | 6/6 | 2/3 A−, 1/3 A+ | |||||||
| 3 | 1/1 | 3/3 | 3/3 A− | |||||||
| A+B+ | 8101-RE | 1 | 1/1 | 0/4 | NA | |||||
| 2 | 1/1 | 0/4 | NA | |||||||
| A−B+ | 8101-PRO | 1 | 1/1 | 4/4 | ND | |||||
| 2 | 1/1 | 4/4 | ND | |||||||
Number of mice with progressively growing tumors per number challenged. Mice were followed for at least 4 wk, or until they became moribund at which point they were killed.
The phenotype, A+ or B+ is defined by the recognition of the tumor cells in vitro by an anti-A CTL clone or anti-B CTL clone. The phenotype A− is defined by the lack of recognition of the reisolated tumor cells in vitro by an anti-A CTL clone.
C57BL/6 mice were injected subcutaneously with C57BL/6 nude mouse tumor fragments using a trocar, except in experiment 1 (8101-PAR), where (B6C3) F1 mice were injected with the tumor fragments from a C3H/HeN nude mouse. Nude mice were always injected last as viability control. In experiment 1, mice were injected with 10 × 1 mm3 fragments. In experiment 2 and 3 mice were injected with 3 × 1 mm3 fragments. NA, not applicable. ND, not done.
Figure 2.
The anti-A and anti-B CTL clones are uniquely specific for the 8101 tumor lineage. The anti-A CTL clone ( A) lyses only the 8101-RE tumor cell but not five other C57BL/6-derived UV-induced tumors, or the H-2b haplotype lymphoma RMA. The anti-B CTL clone (B) lyses the 8101-RE and the 8101-PRO tumor cells, but not three other C57BL/6-derived UV-induced tumors, or RMA cells. Targets were tested for lysis in a 51Cr-release assay.
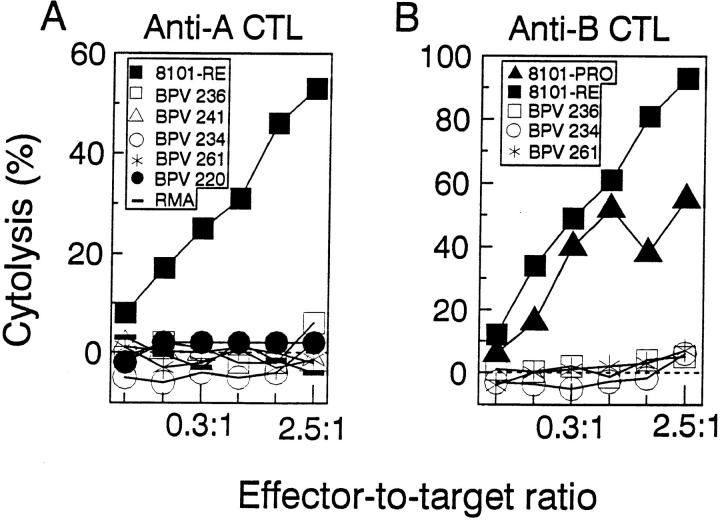
The A Antigen on the 8101-RE Tumor Is Unique, Immunodominant, Kb-restricted, and a Powerful Sensitizer of Anti-A Cytolytic T Cells.
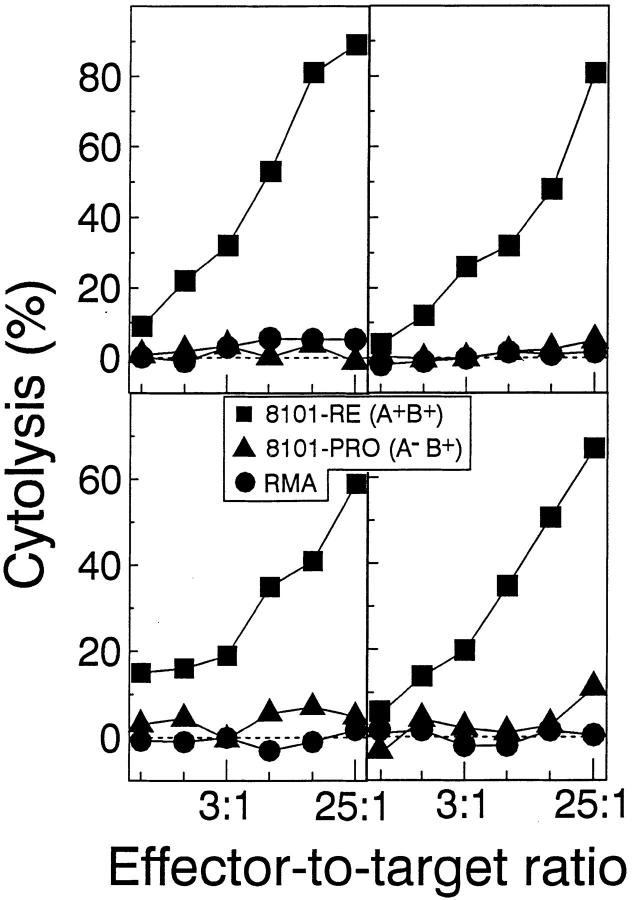
Since the expression of the A antigen correlated with rejection of the tumor challenge, we further characterized this antigen. Fig. 2 A shows that the anti-A CTL clone recognized only 8101-RE cells, but not any of 5 other UV-induced tumors of C57BL/6 origin that were tested. This result suggests that the 8101 A antigen is unique, i.e., individually distinct, for the 8101 tumor, as has been previously shown in our laboratory for other UVinduced tumors (31). To determine how commonly this antigen is recognized in vivo, four mice were injected repeatedly with 8101-RE cells, which express both the A and the B antigen. Peritoneal exudate cells (PEC) were isolated and directly tested for lytic activity (Fig. 3). PECs from all four mice lysed the A+B+ 8101-RE tumor cells but not the A−B+ 8101-PRO tumor cells, suggesting that the A antigen is regularly recognized and is immunodominant. Lysis by the anti-A CTL clone was inhibited by an anti-Kb but not an anti-Db antibody indicating that the A antigen was H-2Kb restricted (data not shown). To examine the possibility of biochemical identification of the A antigen, we determined whether peptides eluted from the H-2Kb molecules immunoprecipitated from 8101-RE cells would sensitize RMA-S cells to become a specific target for lysis by anti-A CTL. As little as 0.2% of an immunoprecipitate from 1 × 109 8101-RE cells contained sensitizing activity (data not shown).
Figure 3.
The A antigen is immunodominant in vivo. Each panel shows a different C57BL/6 mouse immunized i.p. on day 0, 3, 6, and 9 with 2–5 × 106 A+B+ 8101-RE tumor cells. The PEC were harvested on day 11 and directly tested in a 51Cr-release assay for lytic activity. Only the A+B+ 8101-RE cells, but not the A−B+ 8101-PRO cells, or RMA cells are lysed by the PEC.
Isolation and Sequencing of the A Antigen Peptide.
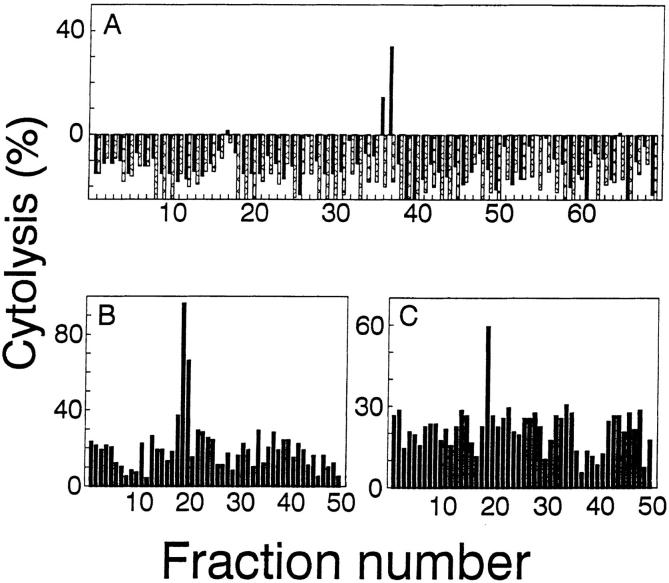
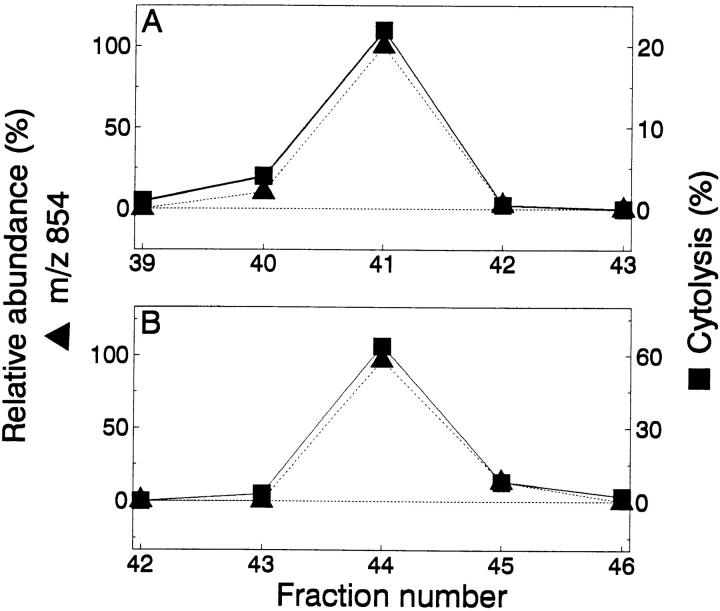
To isolate the naturally processed A antigen peptide, we immunoprecipitated the H-2Kb molecules from 5 × 1010 8101-RE tumor cells. Associated peptides were eluted from the MHC class I molecule with acid, and separated from high molecular mass proteins by filtration through a 5-kD molecular mass cutoff membrane. The unfractionated peptide extract sensitized RMA-S cells for lysis by the anti-A CTL clone (data not shown). The peptide extract was concentrated and then fractionated by reverse-phase high-performance liquid chromatography (RP-HPLC) using heptafluorobutyric acid (HFBA) as the ionic modifier. Aliquots from individual fractions were tested. Only two fractions, 36 and 37, sensitized RMA-S cells for lysis by an anti-A CTL clone (Fig. 4 A). Active fractions 36 and 37 were individually rechromatographed over the same HPLC column, using a shallower gradient and TFA as the ionic modifier. A peptide in subfraction 36-19, 36-20 (Fig. 4 B), and 37-19 (Fig. 4 C) sensitized RMA-S cells for lysis by the anti-A CTL clone. However, each subfraction contained more than 100 peptides, too many to sequence with the available material. Therefore, to identify the mass of the A antigen in these mixtures, 60% of each HPLC subfraction was analyzed separately (the remaining 40% was set aside for further analysis) with an on-line microcapillary column effluent splitter, as described previously (33). Four-fifths of the effluent is directed into the mass spectrometer for analysis, while the other one-fifth is simultaneously deposited into a 96-well plate for analysis of sensitizing activity. Since both pieces of data are acquired as a function of time, the ion abundance corresponding to a particular peptide can be correlated with the biological activity. The microcapillary split of subfraction 36-19 and 37-19 yielded activity (Fig. 5, A and B, respectively) but no sensitizing activity was observed from the peptides in subfraction 36-20 (data not shown). Therefore, the candidate peptide antigen should be present in wells 41 and 44, absent in neighboring wells of both microcapillary splits, and either absent or greatly reduced in abundance in subfraction 36-20 where no sensitizing activity was observed. Only a single peptide, that with m/z 854, fulfilled all the above criteria.
Figure 4.
Sequential HPLC separation of sensitizing peptides eluted from the H2-Kb molecule of 8101-RE cells. A shows the results of the firstdimension microbore HPLC separation of peptides eluted from 5 × 1010 8101-RE cells, using HFBA as the ionic modifier. A portion (0.3%) of each fraction was loaded onto RMA-S cells and tested for lysis by the anti-A CTL clone in a 51Cr-release assay. Sensitizing fractions 36 and 37 were subfractionated on the same microbore column using TFA as the ionic modifier (B and C). RMA-S cells were loaded with 1.5% of each subfraction and tested for lysis by the anti-A CTL clone. The E/T ratio was 5:1. Peptide loaded RMA-S cell with CTL (solid bars) or without CTL (hatched bars) as toxicity controls are shown in A. Toxicity controls were not done in B and C.
Figure 5.
Identification of the tumor antigen candidate peptide. The abundance of the peptide at m/z 854 correlates with the biological activity in the microcapillary HPLC split of subfractions 36-19 ( A) and 37-19 (B).
An aliquot of the remaining 40% of subfraction 36-19 was used to sequence the candidate peptide by mass spectrometry. Shown in Fig. 6 A is the CAD mass spectra on the (M+H)+1 ions at m/z 854. The observed fragmentation is sufficient to specify the sequence of residues 3-8 as FVFAGX, where X is leucine or isoleucine, and residues 1-2 as either SN or NS. Leucine and isoleucine, two amino acids of identical mass, cannot be differentiated on a triple quadrupole instrument. To determine the identity and order of the first two amino acids in the epitope, peptides in an aliquot of subfraction 36-19 were subjected to a single cycle of Edman degradation and then analyzed by microcapillary liquid chromatography-mass spectrometry (LC-MS). In the resulting mass spectra the (M+H)+1 ion for the antigen was observed at m/z 767, a shift of 87 daltons corresponding to the amino acid serine. The CAD mass spectra recorded on the (M+H)+1 ions at m/z 767 confirmed the seven-residue sequence, NFVFAGX. So, the complete amino acid sequence was SNFVFAGX.
To specify the COOH-terminal residue in the epitope as either Leu or Ile, we performed coelution experiments in which aliquots of the biologically active HPLC fraction 37-19 were analyzed by microcapillary LC-MS before and after being doped with synthetic peptides SNFVFAGI or SNFVFAGL. Results of this experiment are shown in Fig. 6 C. Analysis of the mixture doped with SNFVFAGL showed two discrete peptide components at m/z 854. In contrast, the mixture doped with SNFVFAGI showed only a single component at m/z 854. Therefore, coelution of SNFVFAGI with the tumor antigen confirms that the COOH-terminal residue in the epitope is isoleucine. The synthetic peptide SNFVFAGI sensitized RMA-S cells for lysis by the anti-A CTL clone, but control peptide VTFVFAGX did not (Fig. 6 D), nor did two additional H-2Kb-binding peptides tested in a separate experiment (data not shown). Half-maximal lysis of peptide loaded RMA-S cells occurred at 2 pmol peptide.
The Peptide SNFVFAGI Originates from a Somatic Tumorspecific Point Mutation in the p68 RNA Helicase Gene.
The peptide SNFVFAGI was analyzed for homology with known protein sequences using the BLAST program (37). The tumor-derived peptide matched the murine p68 RNA helicase sequence (38) except that the former had phenylalanine instead of serine at position five, suggesting that the tumor peptide might be encoded by a mutant p68 RNA helicase gene in the tumor cells. To confirm this hypothesis cDNA was synthesized and amplified from tumor cell (8101-RE) mRNA by RT-PCR and primers specific for the p68 RNA helicase. The amplified 2.1-kb product pooled from three independent RT-PCR reactions was cloned into the vector pcDNA3. Six cDNA clones were sequenced using primers for the 3′ end of the insert, that included the region of the putative mutation. Two of the six clones were identical to the wild-type sequence of murine p68 RNA (38) helicase while the other four had a T instead of a C at the nucleotide position 1812. This nucleotide substitution resulted in a change to phenylalanine from serine at amino acid 551 (Fig. 7). The C to T transition, which occurred at a dipyrimidine site, is a commonly observed UV-induced mutation (39).
Figure 7.
The mutant p68 RNA helicase peptide was generated by a single amino acid substitution that resulted from a single nucleotide change in 8101-RE. cDNA sequences of murine p68 RNA helicase are compared with p68 sequences of 8101-RE in the region of the mutation. Sequence identity is indicated by ---. RE: sequences from 4/6 cDNA clones from 8101-RE. HLF: sequence from 6 cDNA clones from 8101HLF. WT: published murine p68 RNA helicase cDNA sequence (reference 38). A single nucleotide substitution of C→ T at nucleotide 1812 was found in the 8101-RE tumor, but not in 8101-HLF.
The sequence data derived from the 8101-RE tumor cells suggests that the cloned tumor cell line is heterozygous for the mutation, and expresses both the wild-type and mutant forms of murine p68 RNA helicase. Sequencing of six cDNA clones from 8101-HLF (Fig. 7), and PCR sequencing of amplified genomic p68 DNA from autochthonous heart-lung fibroblasts and from 8101-PRO tumor cells (data not shown) revealed only wild-type sequences. These data indicated that the mutant peptide had been generated by a somatic mutation that was absent in the 8101PRO tumor cells and was consistent with our finding that the PRO tumor and HLF cells were resistant to lysis by the anti-A CTL clone (Fig. 1).
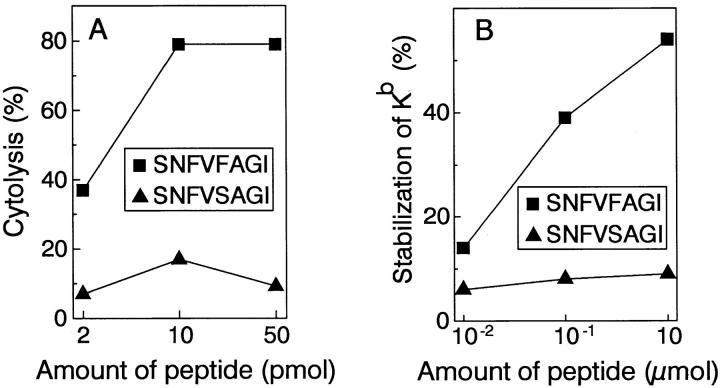
The H-2Kb-binding motif (40) predicts that, first, the anchor residue for binding to the molecule is at position five of the peptide, and is an aromatic residue, either phenylalanine or tyrosine, and second, that position eight of the peptide is either a leucine or isoleucine. This sequence motif predicts that the normal homologue of the A antigen peptide, which has serine at position five, would not bind to H-2Kb. Consistent with this prediction, we found that the wild-type peptide SNFVSAGI, in contrast to the mutant peptide SNFVFAGI, neither sensitized RMA-S cells for lysis by the anti-A CTL clone (Fig. 8 A) nor bound effectively to H2-Kb as measured by stabilization of H2-Kb on the surface of RMA-S cells (Fig. 8 B).
Figure 8.
The mutant p68 peptide sensitizes RMA-S cells for lysis by the anti-A CTL clone, and stabilizes MHC class I on the cell surface, but the corresponding wild-type peptide does not. The mutant peptide SNFVFAGI and the wild-type peptide SNFVSAGI were loaded onto RMA-S cells in the indicated amounts. (A) the loaded cells were tested for lysis by the anti-A CTL clone in a 51Cr-release assay at an E/T ratio of 2:1. (B) loaded cells were analyzed for cell surface expression of H-2Kb using fluorescence activated cell sorter analysis, by indirect immunofluorescence with the monoclonal anti-H2-Kb antibody Y-3 and a polyclonal goat anti– mouse IgG conjugated to fluorescein isothiocyanate as a second step. Percent stabilization of H-2Kb was calculated by subtracting the amount of H-2Kb present on RMA-S cells shifted to 37°C without added peptide, from the amount of H-2Kb present on RMA-S cells shifted to 37°C in the presence of peptide, and dividing by the amount of H-2Kb present on RMA-S cells which had been kept at room temperature throughout the experiment.
We found that three of four mice immunized repeatedly with 8101-RE cells intraperitoneally generated peritoneal exudate cells in vivo that recognized the mutant peptide loaded onto RMA-S cells (data not shown). In addition, the spleens from all 4 of these mice, after in vitro restimulation with the 8101-RE tumor, recognized the mutant p68 peptide (data not shown). These data confirm that SNFVFAGI indeed represents the immunodominant antigen of the regressor tumor 8101-RE.
Discussion
In this study, we have identified the genetic origin of the immunodominant A antigen of the ultraviolet light-induced regressor tumor 8101-RE. The antigenic peptide is SNFVFAGI. It is generated by a point mutation in the murine p68 RNA helicase gene, which changed a C to T, resulting in an amino acid substitution to phenylalanine from serine. The amino acid change also generated an anchor for binding of the peptide to the restricting molecule for the antigen, H-2Kb. Although other still unknown 8101-RE genes might also encode the same peptide, the fact that 8101PRO tumor cells which are not lysed by anti-A CTL also do not have the mutant p68, indicates that mutant p68 encodes the mutant peptide. In addition, the mutation is likely to have occurred in vivo since it was found in DNA of primary tumor cell cultures and thus is unlikely to be an artifact of in vitro culture. The mutation is of somatic tumorspecific origin, rather than representing a genetic polymorphism of germline origin, since autochthonous nonmalignant fibroblasts from the mouse which gave rise to the tumor did not harbor the mutation. Several unique CTL-recognized antigens have now been identified in human tumors and shown to be due to tumor-specific somatic mutations (22–25). However, this is the first identification of a unique tumor-specific CTL antigen in the murine system. We will now be able to evaluate the role of such an antigen in tumor rejection.
To our knowledge, our finding represents the first demonstration of a tumor-specific somatic mutation in the coding region of a member of the DEAD-box protein family of putative RNA helicases (41). A translocation into the 5′ non-coding region of a human putative RNA helicase has been reported earlier (42). In addition, two inherited syndromes in man, Bloom's syndrome (43) and Werner's syndrome (44), both of which show a predisposition to cancer development, have recently been discovered to be linked to DNA helicases. The p68 RNA helicase protein was first identified by Lane and Hoeffler in 1980 (45), because of its immunological cross-reactivity with an antibody that recognized the SV40 large T antigen. These investigators attempted to find a homologue of T antigen by searching for antibody-recognized determinants that cellular proteins might share with the T antigen (45). p68 is a nuclear protein (46), that was later discovered to be an RNA helicase (47, 48).
The primary amino acid sequence of the murine p68 protein is shown in Fig. 9. The first eight boxed motifs show the domains of homology of p68 with other DEAD box proteins, which play a central role in cell growth in a wide variety of organisms. p68 has been shown to undergo dramatic changes in nuclear localization during telophase, when it translocates from the nucleoplasma to the nucleoli (49). In addition, a stretch of amino acids, called the IQ domain (50) is located within the 139 carboxy-terminal amino acids that extend beyond the region of homology with other DEAD box proteins, and which distinguishes p68 from these proteins (41). This domain, which is also found in molecules such as neurogranin and neuromodulin, is subject in vitro to calmodulin (CaM) binding and phosphorylation by protein kinase C (PKC) (50). Experimental evidence suggests that CaM and/or PKC may regulate at least some of the activity of p68 during the cell cycle, through this domain (50). The mutation changes one of the two serines in the IQ domain to a phenylalanine (thick box in Fig. 9), but we do not yet know whether the mutation of S to F affects the physiologic function or localization of the protein. In addition to being an RNA helicase, p68 is also a powerful inhibitor of DNA helicases (51). This activity is quite similar to that of the p53 tumor suppressor gene which also prevents DNA helicase activity (51). It has been suggested that the general role of p53 is to safeguard the integrity of the genome by monitoring and stopping replication when DNA is damaged (52), and it is possible that p68 may serve a similar function as a tumor suppressor gene.
Figure 9.
Predicted primary amino acid sequence of wild-type murine p68 RNA helicase. The eight regions of homology to other DEAD-box proteins are outlined by the thin box. The IQ domain, found at the COOH-terminal end of the protein is outlined by the thick box. Within this IQ box, the eight–amino acid stretch from which the mutant peptide is derived is underlined. In the 8101-RE tumor, the second serine was changed to a phenylalanine in this eight–amino acid stretch.
Our study shows that the CD8+ T cell–recognized A antigen SNFVFAGI (a) is the immunodominant antigen of the 8101-RE tumor, which induces a powerful CD8+ T cell response in vivo when whole cells are used for vaccination, (b) sensitizes target cells at picomolar amounts for lysis by specific T cells and (c) is not expressed by the 8101PRO tumor. Conclusive evidence that this antigen leads to rejection of the 8101-RE tumor would come from demonstrating that expression of the A antigen after transfection of 8101-PRO converts the progressor to a regressor phenotype, i.e., that the progressor tumor is rejected by naive syngeneic mice after expression of the A antigen. We have not yet been able to detect expression of the mutant p68 protein after transfection despite using various eukaryotic expression vectors. It is possible that constitutive expression of this protein, which is tightly regulated during cell cycle, may be toxic to the cells. Nevertheless, the mutant p68 peptide is a strong candidate for a rejection antigen.
One critical question that bears investigation is whether the proteins from which unique tumor antigens are derived also play a role in the development of the malignant phenotype. The transformation of a cell from normal to malignant requires multiple genetic mutations, and it is hypothesized that each of these mutations confers a successive growth advantage upon the cell, which ultimately leads to malignancy (53). It is possible that the same mutations also generate unique tumor antigens. Alternatively, the mutations we observed may only generate the unique antigen but play no additional role in the tumorigenic process. Nevertheless, it is tempting to speculate on the role of p68 as a possible tumor suppressor gene which may be lost during tumor progression. Since two human syndromes are associated with both increased incidence of malignancy and defective helicase function (43, 44), it may be that p68 functions normally as a tumor suppressor, and loss of this protein function would then be associated with the malignant phenotype. Moreover, it is possible that the development of the A antigen is associated with defective function, and hence with the malignant phenotype. In contrast to the situation for tumor suppressor genes, other antigens may be mutant oncogenes which could be essential for maintaining the malignant phenotype, and thus would be expected to be retained by selection. These antigens may also serve as markers for the stages of tumor progression, and would be ideal targets for immunotherapy. Indeed, we have observed both retained and lost antigens on UV-induced tumors (20, 21). Studying the genetic origins of unique tumor antigens may identify genes that are functionally involved in malignancy, but which may not be identified by traditional approaches such as searching for chromosomal translocations or using subtractive libraries. Identifying the genetic origins of unique antigens encoding tumor-specific mutations could therefore contribute to a more complete understanding of the malignant process.
Acknowledgments
This work was supported by grants RO1CA37156, RO1CA22677 and RO1CA19266 and a gift from the Passis family to H. Schreiber, RO1AI33993 to D.F. Hunt and RO1AI20963 to V.H. Engelhard.
Footnotes
The authors deeply appreciate the generous gift of the BPV series of C57BL/6 tumors by Mr. Vijay Sreedhar and Dr. Margaret Kripke. We also thank Dr. Jeffrey Bluestone for his gift of the Y-3 hybridoma and RMA and RMA-S cells. We are grateful to Dr. Paola Rizzo for advice and assistance with the sequence analysis. We are also grateful to Mrs. Helene Auer and Ms. Julianne Liebsohn for excellent technical assistance. We would like to thank Drs. Donald Rowley, Maresa Wick, Paola Rizzo, Paul Monach, and Dominik Mumberg for critical reading of the manuscript.
P. Dubey and R.C. Hendrickson contributed equally to this paper.
This work was presented in abstract form at the FASEB meeting. June 1996. FASEB J. 10:A1437 (Abstr.).
1 Abbreviations used in this paper: CAD, collision-activated dissociation; CDMEM, complete DMEM; CRPMI, complete RPMI; HFBA, heptafluorobutyric acid; HLF, heart-lung fibroblast; LC-MS, liquid chromatography-mass spectrometry; MLTC, mixed-lymphocyte tumor cell culture; PEC, peritoneal exudate cell; PITC, phenylisothiocyanate; TAP, transporter associated with antigen processing.
References
- 1.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- 2.Klein G, Sjögren HO, Klein E, Hellström KE. Demonstration of resistance against methylcholanthreneinduced sarcomas of the primary autochthonous host. Cancer Res. 1960;20:1561–1572. [PubMed] [Google Scholar]
- 3.Basombrio MA. Search for common antigenicities among twenty-five sarcomas induced by methylcholanthrene. Cancer Res. 1970;30:2458–2462. [PubMed] [Google Scholar]
- 4.van den Eynde B, Lethe B, van Pel A, De Plaen E, Boon T. The gene coding for a major tumor rejection antigen of tumor P815 is identical to the normal gene of syngeneic DBA/2 mice. J Exp Med. 1991;178:489–495. doi: 10.1084/jem.173.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Bruggen P, Traversari C, Chomez P, Lurquin C, de Plaen E, van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science (Wash DC) 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 6.Gaugler B, van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, de Plaen E, Lethe B, Brasseru F, Boon T. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boël P, Wildmann C, Sensi M-L, Brasseur R, Renauld J-C, Coulie P, Boon T, van der Bruggen P. BAGE, a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 8.van den Eynde B, Peeters O, de Backer O, Gaugler G, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang AYC, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, Engelhard VH, Pasternack G, Cotter R, Hunt D, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci USA. 1996;93:9730–9735. doi: 10.1073/pnas.93.18.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coggin JH. Shared crossprotective OFAs on chemically induced rodent sarcomas. Immunol Today. 1989;10:76–77. doi: 10.1016/0167-5699(89)90229-6. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava PK, Old LJ. Reply to Coggins letter. Immunol Today. 1989;10:77. [Google Scholar]
- 12.Schreiber, H. 1993. Tumor immunology. In Fundamental Immunology. 3rd edition. W. Paul, editor. Raven Press, Ltd., New York. 1143–1178.
- 13.Ramarathinam L, Sarma S, Maric M, Zhao M, Yang G, Chen L, Liu Y. Multiple lineages of tumors express a common tumor antigen, P1A, but they are not crossprotected. J Immunol. 1995;155:5323–5329. [PubMed] [Google Scholar]
- 14.Jung S, Schluesener H-J. Human T lymphocytes recognize a peptide of single point-mutated, oncogenic ras proteins. J Exp Med. 1991;173:273–276. doi: 10.1084/jem.173.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peace DJ, Chen W, Nelson H, Cheever MA. T cell recognition of transforming proteins encoded by mutated ras proto-oncogenes. J Immunol. 1991;146:2059–2065. [PubMed] [Google Scholar]
- 16.Skipper J, Stauss H-J. Identification of two cytotoxic T lymphocyte-recognized epitopes in the ras protein. J Exp Med. 1993;177:1493–1498. doi: 10.1084/jem.177.5.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenton RG, Taub DD, Kwak LW, Smith MR, Longo DL. Cytotoxic T-cell response and in vivoprotection against tumor cells harboring activated ras proto-oncogenes. J Natl Cancer Inst. 1993;85:1294–1302. doi: 10.1093/jnci/85.16.1294. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi Y, Chen YT, Old L. A mouse mutant p53 product recognized by CD4+ and CD8+T cells. Proc Natl Acad Sci USA. 1994;91:3171–3175. doi: 10.1073/pnas.91.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carbone G, Borrello MG, Molla A, Rizzetti MG, Pierotti MA, Della G, Porta, Parmiani G. Activation of ras oncogenes and expression of tumor-specific transplantation antigens in methylcholanthrene-induced murine fibrosarcomas. Int J Cancer. 1991;47:619–625. doi: 10.1002/ijc.2910470423. [DOI] [PubMed] [Google Scholar]
- 20.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 21.Ward PL, Koeppen HK, Rowley DA, Schreiber H. Major histocompatibility complex class I and unique antigen expression by murine tumors that escaped from CD8+T cell-dependent surveillance. Cancer Res. 1990;50:3851–3858. [PubMed] [Google Scholar]
- 22.Coulie PG, Lehmann F, Lethé B, Herman J, Lurquin C, Andrawiss M, Boon T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, de Plaen E, Hankein T, meyer zum Bschenfelde K-H, Beach D. A p16INK4A-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science (Wash DC) 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 24.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, Rosenberg SA. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brändle D, Brasseur F, Weynants P, Boon T, van den Eynde B. A mutated HLA-A2 molecule recognized by autologous cytotoxic T lymphocytes on a human renal cell carcinoma. J Exp Med. 1996;183:2501–2508. doi: 10.1084/jem.183.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Plaen E, Lurquin C, Van Pel A, Mariamé B, Szikora J-P, Wölfel T, Sibille C, Chomez P, Boon T. Immunogenic (tum−) variants of mouse tumor P815: cloning of the gene of tum− antigen P91A and identification of the tum−mutation. Proc Natl Acad Sci USA. 1988;85:2274–2278. doi: 10.1073/pnas.85.7.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lurquin C, van Pel A, Mariamé B, de Plaen E, Szikora J-P, Janssens C, Reddehase MJ, Lejeune J, Boon T. Structure of the gene of tum− transplantation antigen P91A: the mutated exon encodes a peptide recognized with Ldby cytolytic T cells. Cell. 1989;58:293–303. doi: 10.1016/0092-8674(89)90844-1. [DOI] [PubMed] [Google Scholar]
- 28.Mandelboim O, Berke G, Fridkin M, Feldman M, Eisenstein M, Eisenbach L. CTL induction by a tumor-associated antigen octapeptide derived from a murine lung carcinoma. Nature (Lond) 1994;369:67–71. doi: 10.1038/369067a0. [DOI] [PubMed] [Google Scholar]
- 29.Uenaka A, Ono T, Akisawa T, Wada H, Yasuda T, Nakayama E. Identification of a unique antigen peptide pRL1 on BALB/c RL♂1 leukemia recognized by cytotoxic T lymphocytes and its relation to the Aktoncogene. J Exp Med. 1994;180:1599–1607. doi: 10.1084/jem.180.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Bergeyck V, de Plaen E, Chomez P, Boon T, van Pel A. An intracisternal A-particle sequence codes for an antigen recognized by syngeneic cytolytic T lymphocytes on a mouse spontaneous leukemia. Eur J Immunol. 1994;24:2203–2212. doi: 10.1002/eji.1830240941. [DOI] [PubMed] [Google Scholar]
- 31.Ward PL, Koeppen H, Hurteau T, Schreiber H. Tumor antigens defined by cloned immunological probes are highly polymorphic and are not detected on autologous normal cells. J Exp Med. 1989;170:217–232. doi: 10.1084/jem.170.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townsend A, Öhlen C, Bastin J, Ljunggren H-G, Foster L, Kärre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature (Lond) 1989;340:443–448. doi: 10.1038/340443a0. [DOI] [PubMed] [Google Scholar]
- 33.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science (Wash DC) 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 34.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science (Wash DC) 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 35.Hunt DF, Yates JR, III, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullen CA, Urban JL, van Waes C, Rowley DA, Schreiber H. Multiple cancers: tumor burden permits the outgrowth of other cancers. J Exp Med. 1985;162:1665–1682. doi: 10.1084/jem.162.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Lemaire L, Heinlein UAO. High-level expression in male germ cells of murine p68 RNA helicase mRNA. Life Sci. 1992;52:917–926. doi: 10.1016/0024-3205(93)90526-9. [DOI] [PubMed] [Google Scholar]
- 39.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden JP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA. 1991;90:4216–4220. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing of self peptides eluted from MHC molecules. Nature (Lond) 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–292. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 42.Akao Y, Marukawa O, Monkawa H, Nakao K, Kamei M, Hachiya T, Tsujimoto Y. The rck/p54 candidate proto-oncogene product is a 54-kilodalton D-E-A-D box protein differentially expressed in human and mouse tissues. Cancer Res. 1995;55:3444–3449. [PubMed] [Google Scholar]
- 43.Fellis NA, Groden J, Ye T, Straugen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to rec Q helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 44.Yu C, Oshima J, Fu Y, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J. Positional cloning of the Werner's syndrome gene. Science (Wash DC) 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 45.Lane DP, Hoeffler WK. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature (Lond) 1980;288:167–170. doi: 10.1038/288167a0. [DOI] [PubMed] [Google Scholar]
- 46.Ford MJ, Anton IA, Lane DP. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature (Lond) 1988;332:736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- 47.Hirling H, Scheffner M, Restle T, Stahl H. RNA helicase activity associated with the human p68 protein. Nature (Lond) 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 48.Iggo RD, Lane DP. Nuclear protein p68 is an RNA-dependent ATPase. EMBO (Eur Mol Biol Organ) J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iggo RD, Jamieson DJ, MacNeill SA, Southgate J, McPheat J, Lane DP. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeast. Mol Cell Biol. 1991;11:1326–1333. doi: 10.1128/mcb.11.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buelt MK, Glidden BJ, Storm DR. Regulation of p68 RNA helicase by calmodulin and protein kinase C. J Biol Chem. 1994;269:29367–29370. [PubMed] [Google Scholar]
- 51.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO (Eur Mol Biol Organ) J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lane DP. p53, guardian of the genome. Nature (Lond) 1992;350:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 53.Nowell PC. The clonal evolution of tumor cell populations. Science (Wash DC) 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]