Abstract
The Nramp1 (natural-resistance-associated macrophage protein 1) locus (Bcg, Ity, Lsh) controls the innate resistance or susceptibility of mice to infection with a group of unrelated intracellular parasites which includes Salmonella, Leishmania, and Mycobacterium. Nramp1 is expressed exclusively in professional phagocytes and encodes an integral membrane protein that shares structural characteristics with ion channels and transporters. Its function and mechanism of action remain unknown. The intracellular localization of the Nramp1 protein was analyzed in control 129/sv and mutant Nramp1− /− macrophages by immunofluorescence and confocal microscopy and by biochemical fractionation. In colocalization studies with a specific anti-Nramp1 antiserum and a panel of control antibodies directed against known cellular structures, Nramp1 was found not to be expressed at the plasma membrane but rather localized to the late endocytic compartments (late endosome/lysosome) of resting macrophages in a Lamp1 (lysosomal-associated membrane protein 1)-positive compartment. Double immunofluorescence studies and direct purification of latex bead–containing phagosomes demonstrated that upon phagocytosis, Nramp1 is recruited to the membrane of the phagosome and remains associated with this structure during its maturation to phagolysosome. After phagocytosis, Nramp1 is acquired by the phagosomal membrane with time kinetics similar to Lamp1, but clearly distinct from those of the early endosomal marker Rab5. The targeting of Nramp1 from endocytic vesicles to the phagosomal membrane supports the hypothesis that Nramp1 controls the replication of intracellular parasites by altering the intravacuolar environment of the microbe-containing phagosome.
In mice, resistance or susceptibility to infection with a number of antigenically and taxonomically unrelated intracellular parasites is determined by alleles of the chromosome 1 locus Bcg, also known as Ity or Lsh. Infections under the control of Bcg include several mycobacterial species (Mycobacterium bovis, Mycobacterium avium, Mycobacterium lepraemurium), Salmonella typhimurium, and Leishmania donovani (1–5). The genetic control is expressed phenotypically as a rapid microbial replication during the early phase of infection in reticuloendothelial (RE)1 organs of susceptible (Bcgs) mice, as opposed to absence of such multiplication in resistant (Bcgr) animals (2). While in the case of nonvirulent infections (Mycobacterium, Leishmania), onset of specific immune response clears the infection in susceptible animals, the highly virulent pathogen Salmonella leads to a fulminant and rapidly lethal infection in susceptible animals. In vivo experiments have shown that the cell population(s) responsible for phenotypic expression of Bcg is bone marrow derived, radiation resistant, and sensitive to the phagocyte poison silica (6). Furthermore, explanted macrophages from Bcgr and Bcgs mice show different capacities to restrict the growth of Mycobacteria, Salmonella, and Leishmania in vitro (7–11). Together, these results indicate that the macrophage is the cell type expressing the genetic difference at Bcg, in agreement with the intracellular nature of infectious agents affected by Bcg. The physiological role of Bcg within phagocytes is unknown, but its ability to affect the replication of such a divergent group of parasites supports a pivotal role in antimicrobial defenses of these cells.
Using positional cloning, we have recently isolated a candidate for Bcg designated Nramp1 (natural resistance-associated macrophage protein 1) (12). Nramp1 mapped within the minimal genetic and physical intervals defined for Bcg, and the expression of its mRNA was restricted to RE organs and to phagocytic cells derived from them. The predicted amino acid sequence of Nramp1 identifies an integral membrane protein composed of 12 predicted transmembrane (TM) domains, a glycosylated extracytoplasmic loop, and several putative phosphorylation sites in predicted intracellular loops. Sequence analysis of Nramp1 shows that susceptibility to infection in Bcgs inbred strains is associated with a single nonconservative G169D substitution in the predicted TM4 of the protein (13). In addition, creation of a null allele at Nramp1 by homologous recombination abrogates the natural resistance of Bcgr mice to infection with Mycobacterium, Salmonella, and Leishmania (14). Finally, transfer of the G169 allele of Nramp1 in transgenic animals of susceptible background (Bcgs; Nramp1D169) restores resistance to infection with intracellular parasites (15). Together, these results demonstrate that Nramp1, Bcg, Ity, and Lsh are the same gene.
Recently, we have isolated a second mammalian Nramp gene, Nramp2, that encodes a highly similar protein (77% similarity) (16, 17). As opposed to its phagocyte-specific Nramp1 counterpart, Nramp2 is expressed fairly ubiquitously in most tissues tested. Database searches and additional cloning experiments have shown that Nramp comprises a very ancient family of proteins with highly conserved members in invertebrates (Caenorhabditis elegans, Drosophila melanogaster), plants (Oryza sativa, Arabidopsis thaliana), fungi (Saccharomyces cerevisiae), and even bacteria (Mycobacterium leprae) (18, 19). This family is defined by a highly conserved hydrophobic core composed of 10 TM domains, including several invariant charged residues in TM domains, and helical periodicity of sequence conservation which predicts a helical bundle within the membrane with a conserved charged interior and a semi-conserved hydrophobic exterior. This type of structural organization is typical of families of ion transporters and channels (18). In addition, an invariant sequence motif in the Nramp family shows striking similarity with the ion permeation path of mammalian voltage-gated K+ channels (TMT-4X-G-D/Q-4X-GF; reference 20). Together, these observations suggest that Nramp1 may be a macrophage-specific ion transporter, and that its substrate may play a key bacteriostatic or bactericidal role in these cells. Interestingly, the yeast Nramp homologue SMF1 was recently proposed to encode a manganese transporter (21).
Some of the key unresolved issues concerning Nramp1 and its role in host resistance to infection include its unknown biochemical function, putative substrate, and how its action affects the intracellular survival of microbes ingested by professional phagocytes. It would also be important to understand how Nramp1 affects the replication of antigenically unrelated microbes (Mycobacterium, Salmonella, Leishmania) that have devised different strategies to evade the microbicidal arsenal of phagocytes (see Discussion; for review see reference 22), and why mutations at Nramp1 are seemingly without effect on the replication of other intracellular infections such as Listeria and Legionella (Gros, P., unpublished data).
In this study, we have used specific anti-Nramp1 antibodies to analyze the subcellular localization of the Nramp1 protein in macrophages by immunofluorescence and confocal microscopy, using a series of markers corresponding to known membranous compartments within this cell. We have determined that Nramp1 is not present in the plasma membrane of these cells but is rather found in the late endosome fraction. Moreover, upon phagocytosis, Nramp1 gets recruited to the membrane of the phagosome during the course of its maturation from early plasma membrane– derived phagosome to phagolysosome, and therefore becomes intimately associated with the membranous compartment containing the ingested parasites.
Materials and Methods
Isolation and Culture of Macrophages.
129/sv mice were purchased from Taconic Farms (Germantown, NY), and 129/sv mice bearing a null mutation at the Nramp1 locus (129/sv Nramp1− /−) were created as described by Vidal et al. (14). Mice were maintained and handled according to regulations of the Canadian Council on Animal Care. Resident peritoneal macrophages were isolated from either 129/sv mice or from 129/sv Nramp1− /− mutants by peritoneal lavage as described previously (23). In brief, mice were killed and the peritoneal cavity washed with 10 ml of warm RPMI (GIBCO BRL, Gaithersburg, MD) using a 10-ml syringe fitted with a 18-gauge needle. Resident cells were pelleted (1,000 g, 5 min), resuspended in complete RPMI supplemented with 10% heat-inactivated (56°C, 30 min) fetal bovine serum (GIBCO BRL), and plated onto glass coverslips. After incubation overnight at 37°C in 5% CO2, nonadherent cells were eliminated by three washes with warm PBS (137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, pH 7.4), and adherent macrophages were placed in complete RPMI medium. Macrophages were used for immunofluorescence or phagocytosis assays no longer than 24–48 h after isolation.
Antibodies.
A rabbit anti–mouse Nramp1 polyclonal antiserum was raised against a fusion protein containing amino acids 514 to 548 of Nramp1 (COOH-terminal domain) fused in frame to a 27-kD segment of glutathione S transferase (GST), and expressed in Eschericha coli, as described elsewhere (24). The immunoglobulin fraction of the hyperimmune rabbit antiserum was concentrated by ammonium sulfate precipitation; further purification of the anti-Nramp1 antibody was achieved by absorption of the anti-GST fraction of the antiserum to Sepharose beads coupled to GST. Coupling of GST to cyanogen bromide activated Sepharose beads, absorption of the antiserum, and further concentration of the anti-Nramp1 antibody were essentially as described (23). The rat anti-Lamp1 (lysosomal-associated membrane protein 1) monoclonal antibody (late endosome, early lysosome) has been described previously (25); the rabbit anti-calnexin (endoplasmic reticulum; 26) and anti-MG160 (27, 28) polyclonal antibodies were gifts of Dr. J.J.M. Bergeron (McGill University, Montreal, Quebec, Canada); crude or affinity purified rabbit anti-cathepsin D (lysosome) and anti-cathepsin B (lysosome, 29) polyclonal antisera were gifts of Dr. J. Mort (Shriner's Hospital, Montreal, Quebec, Canada); crude or affinity-purified rabbit polyclonal antiRab5 (early endosome; 30) and anti-Rab7 (late endosome; 31) were gifts from Drs. P. Chavrier and S. Meresse, respectively (Centre d'Immunologie, Institut National de la Santè et de la Recherche Mèdicale–Centre National de Recherche Scientifique, Marseille, France); the mouse monoclonal antibody 9E10 (32) directed against a short antigenic epitope of the c-Myc protein was purchased from BABCO (Berkeley, CA). Secondary antibodies Texas red–conjugated goat anti–rabbit, FITC-conjugated goat anti– rat, and Rhodamine-conjugated goat anti–mouse were purchased from Jackson ImmunoResearch Laboratories, Inc. (Bio/Can Scientific, Mississagua, ON). All antibodies were used at dilutions indicated in the figure legends.
Immunofluorescence.
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in PBS for 30 min at 4°C. After three washes in PBS, cells were then permeablized by treatment with 0.05% NP-40 in PBS with 1% BSA (fractionV, Boehringer Mannheim, Indianapolis, IN) and 5% normal goat serum (GIBCO BRL). Cells were washed again with PBS, and blocked for 1 h at room temperature in PBS containing 1% BSA, 10% normal goat serum, and 10% normal mouse serum (omitted for experiments with the mouse monoclonal antibody 9E10). The normal goat and mouse sera were heat inactivated (50°C) 30 min before use. The cells were incubated with the primary antibody diluted in blocking solution (dilutions are indicated in figure legends) for 1 h at room temperature, and then were washed three times in PBS containing 1% BSA and 0.2% Tween 20. Incubations for the secondary antibodies were done in a similar fashion, and were followed by a final wash in PBS. The coverslips were then mounted onto glass slides in ImmuMount (Shandon Inc., Pittsburgh, PA). Immunofluorescence was analyzed with either a Zeiss Axiophot microscope using the 63X oil immersion objective or an Olympus fluorescence microscope using the 40 and 100× oil immersion objectives. Certain colocalization studies were performed using a Bio Rad scanning confocal fluorescence microscope and digitizing equipment (Bio Rad Labs., Hercules, CA).
Phagocytosis and Kinetic Studies of Phagosome Maturation.
For phagocytosis experiments, resident peritoneal macrophages were fed a meal of latex beads (LBs) (3-μm diam, diluted 1:25 in warm RPMI medium from stock suspension; Sigma Chemical Co., St. Louis, MO), and the internalized LBs were used to follow the steady state or kinetics of association of various protein markers with the maturing phagosome. For colocalization studies of Nramp1 and Lamp1 proteins at steady state, macrophages were incubated with medium containing beads for 1 h, washed in PBS, and incubated in bead-free medium for 1 h to allow maturation of the phagosome into phagolysosome. Cells were then processed for immunofluorescence as described above. For kinetic experiments, macrophages were incubated with LB-containing medium for 5 min at 37°C, followed by five washes with PBS (at 4°C) to remove uninternalized beads. The chase period was initiated by incubating the cells with RPMI medium prewarmed at 37°C, and at predetermined times during the incubation at 37°C, cells were fixed, stained with specific antibodies, and processed for immunofluorescence. To determine the percentage of phagosomes positive for the markers analyzed, macrophages were initially examined under phase contrast to locate cell-associated LB. These cell-associated beads were then examined under fluorescence for the presence or absence of immunospecific label around the bead-containing phagosome. A minimum of 100 beads were scored for the presence of pairs of endosomal or lysosomal markers for each time point. At least two independent experiments were carried out for each pair of markers and averages were calculated and shown.
Isolation of Phagosomes from RAW 264.7 Macrophages.
The mouse monocyte–macrophage cell line RAW 264.7 was obtained from the American Type Culture Collection (Rockville, MD) and grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated FCS (GIBCO BRL), 20 mM Hepes pH 7.6, and 2 mM L-glutamine. An expression plasmid was constructed using the mammalian expression vector pCB6 (33) with an insert containing the entire coding region of mouse Nramp1 cDNA modified by the addition of four consecutive antigenic peptide epitopes (EQKLISEEDL) derived from the human c-Myc protein, fused in frame at the COOH terminus of Nramp1 (pCB6– Nramp1). pCB6-Nramp1 was introduced in RAW264.7 macrophages by electroporation; 700 μl of cells at a density of 2 × 107 cells/ml in complete medium was mixed with 40 μg of the purified plasmid in an electroporation cuvette. Cells were electroporated at a setting of 960 μF and 300 mV on a GenePulser (Bio Rad). Electroporated cells were plated in complete growth medium and selection in geneticin (G418, 0.5 mg/ml final concentration; GIBCO BRL) was initiated 48 h later. Stable transfectants (G418R) were isolated after 14 d of selection. G418R colonies were individually picked, expanded in culture, and tested for expression of the c-Myc epitope tagged Nramp1 protein by immunofluorescence, using either the anti-Nramp1 antibody GST54N (24) or the anti–c-Myc epitope monoclonal antibody 9E10 (32). Expression of the c-Myc–tagged Nramp1 protein in these cells was further verified by immunoprecipitation, using the antiNramp1 antiserum GST-54N according to a protocol and experimental conditions previously described by our group (24). One RAW264.7 G418R clone positive for Nramp1 protein expression and showing levels of expression comparable to those observed for the wild-type protein in resident macrophages was selected for further immunofluorescence studies and biochemical characterization of purified phagosomes.
Phagosomes were isolated from the RAW cells and transfectants by a modification of a method described previously (34). 10 subconfluent 150-mm dishes of each cell line were fed with a 1:200 dilution of blue-dyed LBs (0.8 μm, Sigma Chemical Co.) in culture medium for 1 h at 37°C in 5% CO2. The cells were then washed in PBS and incubated for 1 h in complete culture medium at 37°C in 5% CO2 to allow phagosome maturation. Cells were then scraped into PBS + 0.5% BSA + protease inhibitors (1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 100 μg/ml PMSF, all from Boehringer Mannheim) and recovered by centrifugation (2,000 g, 5 min). The cell pellets were washed and resuspended in homogenization buffer (8.5% sucrose, 3 mM imidazole, pH 7.4) and homogenized by passage through a 22-guage needle until 90% of the cells were broken, with most of the nuclei remaining intact as monitored by light microscopy. Nuclei and unbroken cells were pelleted and the supernatent was loaded onto a sucrose step gradient as follows. The supernatant was brought up to 40% sucrose by addition of 62% sucrose, and loaded on top of a 1 ml 62% sucrose cushion. Layers of 2 ml of 35, 25, and finally 10% sucrose (all sucrose solutions wt/wt in 3 mM imidazole, pH 7.4, + protease inhibitors) were sequentially added to the top of the tube, and the gradients were centrifuged at 100,000 g for 1 h at 4°C (SW41; Beckman Instrs., Inc., Fullerton, CA). Phagosomes were recovered from the 10–25% sucrose interface, washed with PBS containing protease inhibitors, and recovered by a final centrifugation at 40,000 g in an SW41 rotor at 4°C. The final pellets were resuspended in 2× Laemmli sample buffer (35). Phagosomes prepared by this protocol have been previously shown to be free of endoplasmic reticulum (endoplasmin, BiP, and calnexin) and Golgi apparatus (galactosyl transferase) contaminants (34).
Immunoblotting Analysis of Phagosomes.
Equal amounts of phagosomal proteins from each cell line were separated by SDS-PAGE on 7.5% gels and transferred onto nitrocellulose filters as described previously (36). The filters were blocked in TBST (100 mM Tris/Cl, pH 8, 0.9% NaCl, 0.1% Tween 20) + 1% BSA and Nramp1 was revealed using the anti-Myc mouse monoclonal antibody 9E10. Controls in this experiment included the antiLamp1 rat monoclonal antibody, and a rabbit anti Rab7 antiserum followed by incubation with goat secondary antibodies conjugated to horseradish peroxidase (Amersham Intl., Buckinghamshire, UK). Chemiluminescence was used for detection of immune complexes on the filter (ECL; Amersham Intl.).
Results
Nramp1 Is Localized to an Intracellular Compartment in Macrophages.
As a first step towards elucidating the biochemical function of Nramp1 and how it may affect replication of intracellular parasites, we set out to establish the subcellular localization of the protein. Studies of mRNA distribution in normal tissues and cell types identified expression restricted to RE organs and to mature macrophages derived from them (12). Consequently, peritoneal macrophages from 129/sv mice, a strain that bears the wild-type allele at Nramp1 (Nramp1G 169, Bcgr) were chosen for these studies. Peritoneal macrophages express many markers of fully mature phagocytes (37), and are positive for Nramp1 mRNA (Govoni, G., and P. Gros, unpublished data). In addition, experiments in vitro show that mutations at Nramp1 affect the capacity of these cells to control the replication of intracellular parasites (7–9). Peritoneal macrophages are also easy to obtain in small numbers. Finally, we have created a 129/sv mouse strain that bears a null mutation at Nramp1 (Nramp1− /−), providing an ideal control for subcellular localization studies in 129/sv macrophages (14).
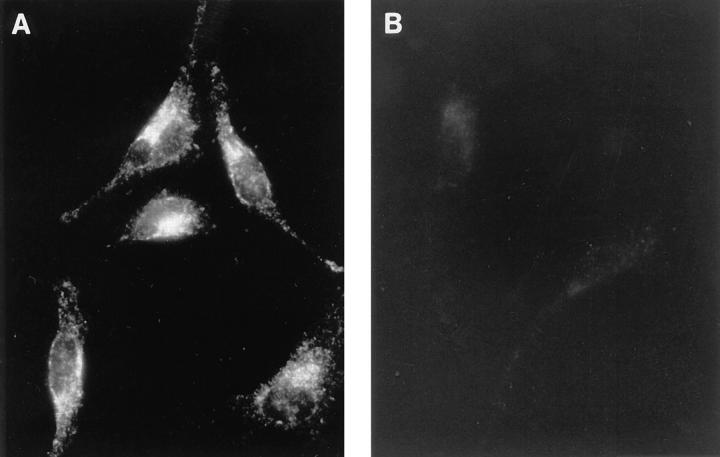
We have previously reported the production of a series of specific rabbit anti-Nramp1 polyclonal antisera (24), including the GST-35C serum raised against a protein comprising the COOH-terminal 35 residues of Nramp1 fused to GST. In immunoprecipitation and immunoblotting experiments using extracts from macrophages, this serum identified Nramp1 as an integral membrane phosphoglycoprotein of 90–95 kD (24), in agreement with structural and functional features predicted from the sequence of Nramp1 cDNA (12). GST-35C was used to localize the Nramp1 protein in macrophages by indirect immunofluorescence (Fig. 1). In 129/sv macrophages (Fig. 1 A), we observed a strong intracellular vesicular staining pattern that was intense in the perinuclear region but also extended throughout the length of the long cellular processes. This staining was specific and absent in macrophages from control Nramp1− /− mutants (Fig. 1 B). A similar staining pattern was observed using an unrelated anti-Nramp1 antiserum directed against the 53 NH2-terminal residues of the protein (data not shown). Finally, we did not detect any Nramp1 staining associated with either the plasma membrane or the nuclear membrane, indicating that Nramp 1 expression is restricted to a subcellular, probably membranous, compartment.
Figure 1.
Subcellular localization of the Nramp1 protein in macrophages. Peritoneal macrophages from normal 129/sv mice (A) and from 129/sv Nramp1− /− mutants (B) were harvested by peritoneal lavage, cultured for 48 h, and analyzed by indirect immunofluorescence with an anti-Nramp1 rabbit polyclonal antibody (GST-35C, at 1:50 dilution) raised against the 35 COOH-terminal residues of Nramp1. The secondary antibody was a goat anti–rabbit antiserum conjugated to Texas red (1:200 dilution). Cells in A and B were processed identically, and equal exposure times were used for photography.
Colocalization of Nramp1 with the Late Endosomal/Lysosomal Marker Lamp1.
We set out to identify the subcellular compartment of macrophages positive for Nramp1 protein expression. For this, we compared the pattern of Nramp1 staining obtained by immunofluorescence to that produced in the same cells by markers of discrete intracellular membrane compartments. The fluorescent signal produced by GST-35C (Nramp1) was found to be distinct from those produced by antibodies against the membrane protein MG160 (Golgi apparatus medial cisternae; perinuclear and asymmetric staining; Fig. 2 E), the integral membrane protein, and molecular chaperone calnexin (endoplasmic reticulum; cytoplasmic reticulated network and nuclear envelope; Fig. 2 H), or the soluble cysteine proteases cathepsin B and D (lysosomes; strong punctate staining uniformly distributed; Fig. 2, G and J, respectively). On the other hand, the Nramp1 staining shared some similarity with that produced by antibodies against the membrane-associated small GTP binding proteins Rab5 (early endosome; punctate reticular with small vesicles; Fig. 2 F), and in particular Rab7 (late endosome; punctate reticular; Fig. 2 I). However, we observed an even more striking similarity between the Nramp1 pattern and that of a marker for the late endosomal and early lysosomal compartments, Lamp1 (Fig. 2, C and D). The possible colocalization of Lamp1 and Nramp1 in this compartment was further investigated by double immunofluorescence on the same preparations of macrophages from either wild type 129/sv (Fig. 2, A and C), or Nramp1− /− mutant mice (Fig. 2, B and D), using rat antiLamp1 (Fig. 2, C and D) and rabbit anti-Nramp1 antibodies (Fig. 2, A and B). We observed complete colocalization of the Nramp1 and Lamp1 proteins to the same type of subcellular structures in the 129/sv macrophages (Fig. 2, A and C). Control macrophages from Nramp1− /− mice stained normally for Lamp1, but did not show any staining for Nramp1, establishing that the observed colocalization of Nramp1 and Lamp1 was not due to cross-reactivity of the secondary antibodies or other procedural artifacts (Fig. 2, B and D). Together, these results indicate that Nramp1 is expressed in the late endosomal/early lysosomal compartment of macrophages.
Figure 2.
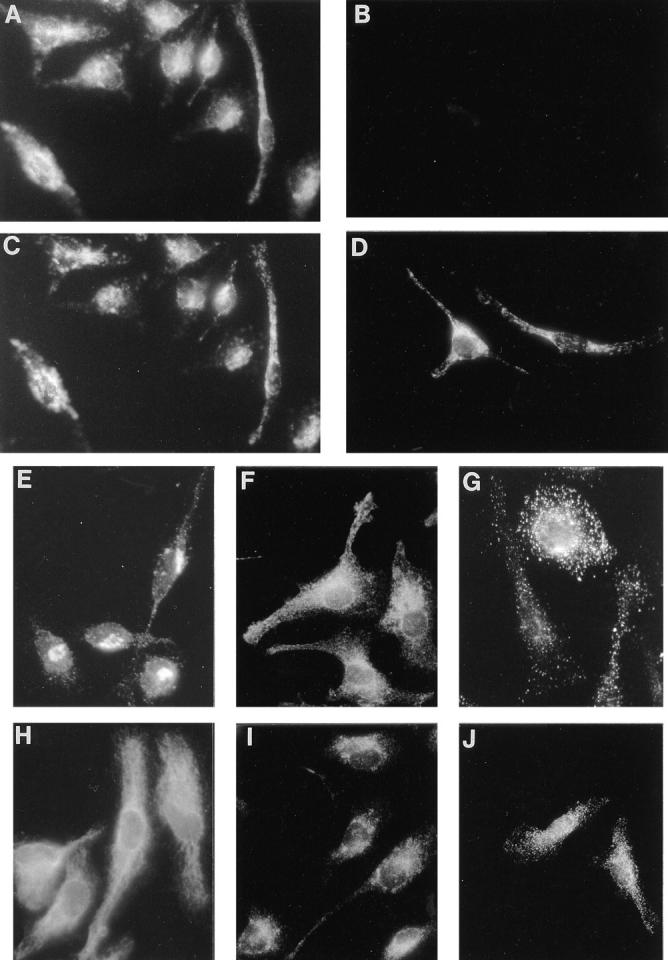
Colocalization of the Nramp1 and Lamp1 proteins in macrophages. Peritoneal macrophages were isolated from normal 129/sv (A, C, and E–J) and 129/sv Nramp1− /− mutants (B and D) and processed for double indirect immunofluorescence with the rabbit anti-Nramp1 antiserum GST-35C (A and B; 1:50 dilution) and a rat anti-Lamp1 monoclonal antibody (C and D; 1:10 dilution). Both cell populations were processed identically and equal exposure times were used for photography. Macrophages from 129/sv mice were also reacted with antibodies directed against MG160, a medial Golgi marker (E; 1:500 dilution) Rab5, an early endosomal marker (F; 1:200 dilution), cathepsins B and D, lysosomal proteases (G and J, respectively; 1:200 dilution), calnexin, a protein expressed in the endoplasmic reticulum and nuclear envelope (H; 1:100 dilution), and Rab7, a late endosomal marker (I; 1: 200 dilution).
Nramp1 Becomes Associated with the Phagosome After Phagocytosis.
Our localization of Nramp1 to the late endocytic compartment is exciting, since this compartment plays a major role in the ultimate destruction of internalized microbial targets by macrophages. Indeed, most intracellular parasites enter the macrophage by active phagocytosis; the resulting plasma membrane–derived phagosome then acquires various cytocidal and cytostatic properties (low pH, oxygen radicals, proteolytic enzymes) through a maturation process consisting of a series of complex fusion events involving endosomal and lysosomal partners. Consequently, our localization of Nramp1 to the late endocytic/lysosomal compartment would suggest that Nramp1 may become associated with the phagosomal membrane during maturation, and be intimately associated with invading parasites. To test this prediction, we monitored a possible association of Nramp1 with LB-containing phagosomes. LBs are inert spherical particles of defined size that are readily phagocytosed by macrophages; they serve as excellent phagosome markers for microscopy analysis and for biochemical purification of these organelles. LB phagosomes show normal fusogenic properties and have been extensively used to establish the kinetics of delivery of various endosomal and lysosomal markers to the maturing phagosome (34, 38, 39). Normal (129/sv) and Nramp1− /− mutant macrophages were harvested and fed a meal of LBs for 1 h. The cell monolayers were washed extensively to eliminate unphagocytosed beads, followed by a further hour of incubation to allow phagosome maturation. The cells were then fixed and analyzed for subcellular distribution of Nramp1 protein; individual fields were photographed under phase contrast (Fig. 3, A and C), and examined by immunofluorescence for Nramp1 staining (Fig. 3, B and D). In 129/sv macrophages showing internalized beads (Fig. 3, A and B), Nramp1 was concentrated primarily around the beads with a striking ring-like staining, strongly suggesting that Nramp1 was localized in the phagosome membrane. Beads located outside the cells (arrow, Fig. 3 A), and a small percentage of cell-associated beads remained negative for Nramp1. This suggests that the striking Nramp1 staining found associated with the majority of internalized beads was due to the presence of Nramp1 protein acquired during LB phagosome maturation, as opposed to an optical artifact of the beads on the previously noted intracellular Nramp1 staining background (Fig. 1 A). Cells without beads showed typical Nramp1 staining. Finally, Nramp1− /− macrophages showed no obvious defects in their ability to phagocytose LBs (Fig. 3 C), but the cells and LB phagosomes remained negative for Nramp1 (Fig. 3 D).
Figure 3.
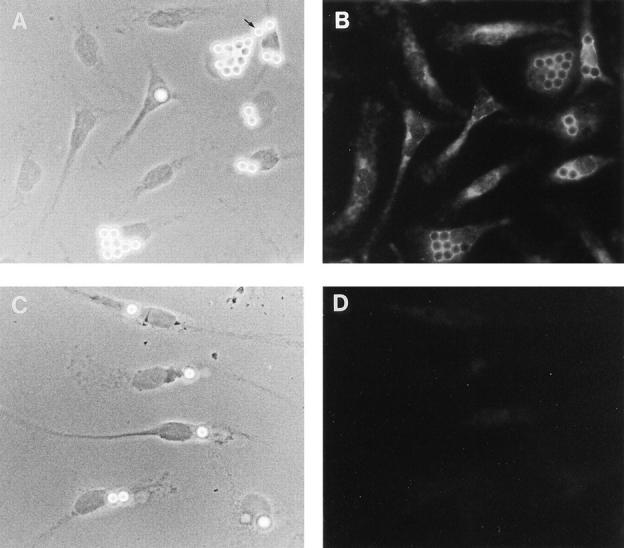
Nramp1 association with LB-containing phagosomes. Normal 129/sv (A and B) and mutant 129/sv Nramp1− /− (C and D) macrophages were harvested, cultured for 48 h, and fed a meal of LBs for 1 h at 37°C. The cells were washed free of unphagocytosed beads, and further incubated for 1 h to allow phagosome maturation. The cells were then fixed and subjected to indirect immunofluorescence with the anti-Nramp1 antiserum (see legend to Fig. 1). Phase contrast (A and C) and immunofluorescence micrographs (B and D) of the same fields of cells are shown. The position of an uninternalized LB is indicated by the arrow in A.
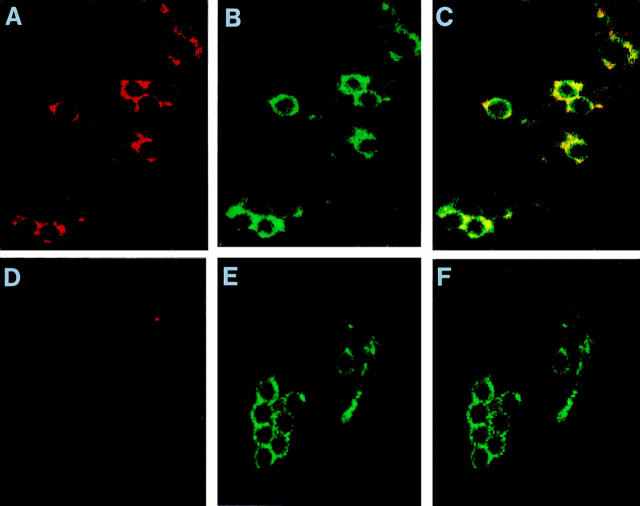
In the next set of experiments, double immunofluorescence and confocal microscopy were used to further validate the initial results of immunofluorescence, and to ascertain that Nramp1 is delivered to the phagosomal membrane during phagocytosis. Confocal microscopy permits microscopic analysis of individual cell sections, allowing accurate localization of proteins to subcellular membranous compartments (for review see reference 40). Normal 129/sv and Nramp1− /− mutant macrophages were fed a meal of LBs as above, and phagosome maturation was allowed to take place. Cells were then fixed and stained with both anti-Nramp1 (coupled to Texas red, red signal), and antiLamp1 antiserum (coupled to FITC; green signal). Cells were examined by confocal microscopy, and separate images from the same field were created for the Nramp1 staining (red; Fig. 4, A and D) and the Lamp1 staining (green; Fig. 4, B and E). In 129/sv macrophages, a striking ring-like staining concentrated at the periphery of individual LBs was observed for both markers (Fig. 4, A and B); similar images were obtained at different planal sections of the same cells (data not shown). These results indicate that both proteins become associated with the phagosomal membrane after phagocytosis and maturation. Superimposition of the Nramp1 and Lamp1 images in 129/sv macrophages indicate almost complete overlap of both markers in individual LB phagosomes (yellow; Fig. 4 C), suggesting similar kinetics of delivery of both proteins to the phagosomes. LB phagosomes from control Nramp1− /− macrophages were positive for Lamp1 but remained negative for Nramp1 (Fig. 4, D–F).
Figure 4.
Nramp1 and Lamp1 colocalization to the membrane of LB-containing phagosomes. Macrophages from normal 129/sv mice (A–C) and from 129/sv Nramp1− /− mutants (D–F) were fed a meal of LBs and further incubated to allow phagosome maturation (see legend to Fig. 3). The samples were then subjected to double indirect immunofluorescence with the rabbit anti-Nramp1 antibody (GST-35C) revealed by a Texas red coupled secondary antibody, and with the rat anti-Lamp1 antibody revealed by an FITC coupled secondary antibody. Slides were analyzed by confocal laser scanning microscopy on Bio Rad equipment, for optical sections of 0.2 μm. Red identifies Nramp1 positive structures (A and D), and green identifies Lamp1 positive structures (B and E). In C and F, images in A + B and D + E have been superimposed; the yellow color identifies colocalization of the Nramp1 and Lamp1 proteins to the same structures.
Detection of Nramp1 in Purified Phagosomes.
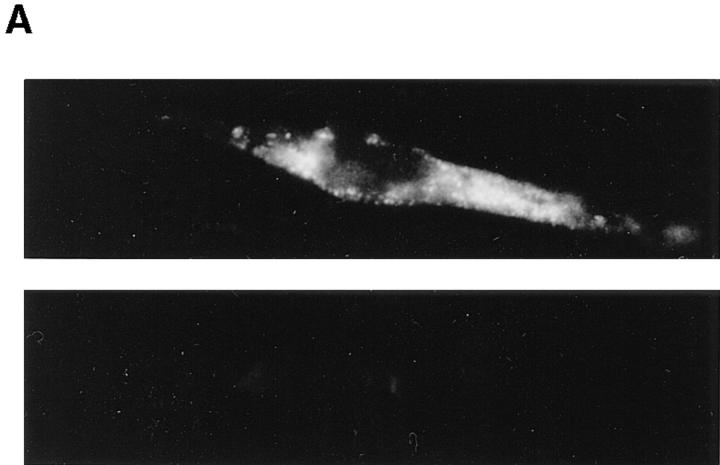
To provide direct biochemical evidence for the association of Nramp1 with the phagosomes, we proceeded to purify these organelles from macrophages, followed by immunoblotting for detection of Nramp1 protein. We have previously described a method for the isolation of LB phagosomes by subcellular fractionation and discontinuous density gradient centrifugation; such LB phagosomes are free of contaminants from the Golgi (galactosyl transferase) or the endoplasmic reticulum (endoplasmin, BiP, and calnexin; 34, 38). Since purification of phagosomes by this method requires a large number of macrophages (1–2 × 108), mouse peritoneal macrophages cannot be used as starting material (1–5 × 106/mouse). For these studies, we created a mouse macrophage cell line that expresses high levels of a transfected wild-type Nramp1G 169 allele. RAW264.7 is a mouse monocyte-macrophage cell line derived from BALB/c (Bcgs), and is thus homozygous for the Nramp1D 169 mutation which is phenotypically expressed as the absence of mature protein caused by improper maturation resulting in its rapid degradation (24). RAW264.7 macrophages were transfected with the mammalian expression pCB6 which contains a neo gene and an expression cassette that uses cytomegalovirus regulatory elements to direct high levels expression of cloned cDNAs (33). Wild-type Nramp1G 169 cDNA was modified at its COOH terminus before insertion in pCB6, by the in-frame addition of four copies of the antigenic c-Myc epitope EQKLISEEDL (32). This construct was transfected into RAW264.7 cells. Transfectants were selected in geneticin and initially screened for Nramp1 protein expression by immunoprecipitation using the mouse monoclonal 9E10 directed against the c-Myc epitope present in the tagged protein (data not shown). Several positive clones were identified and one of them showed levels of Nramp1 protein expression similar to that seen for the endogenous protein in wildtype 129/sv peritoneal macrophages. Immunofluorescence with the 9E10 antibody on transfected RAW264.7 cells expressing the tagged Nramp1 cDNA showed bright intracellular signals with a vesicular and pseudo-reticular staining (Fig. 5 A, top), similar to that seen for 129/sv macrophages analyzed with the Nramp1 antiserum 35C-GST (Fig. 1 A). This signal was specific and absent from untransfected RAW264.7 cells (Fig. 5 A, bottom). The RAW264.7 Nramp1 transfectants and RAW264.7 controls were then used as starting cells for the production and isolation of LB phagosomes (34). Phagosomes were then analyzed for expression of the Nramp1/c-Myc tagged protein by immunoblotting with the 9E10 antibody (Fig. 5 B, top). Phagosomes from Nramp1 transfectants expressed a single broad immunoreactive species of molecular mass 85–90 kD, that was absent from phagosomes of untransfected RAW cells. The molecular mass and electrophoretic mobility of this tagged protein are in agreement with that of Nramp1 expressed in wild-type peritoneal macrophages (24). Control immunoblotting experiments with an antiserum directed against the late endosomal marker Rab7 identified equal amounts of an immunoreactive 23-kD protein in phagosome preparations from control RAW cells and from RAW cells expressing the Nramp1/c-Myc protein (Fig. 5 B, bottom). Similar amounts of Lamp1 were also detected in both preparations by immunoblotting (data not shown). These control experiments indicate equal protein loading in each lane and equal transfer of endosomal and lysosomal markers to latex phagosomes prepared from both types of cells.
Figure 5.
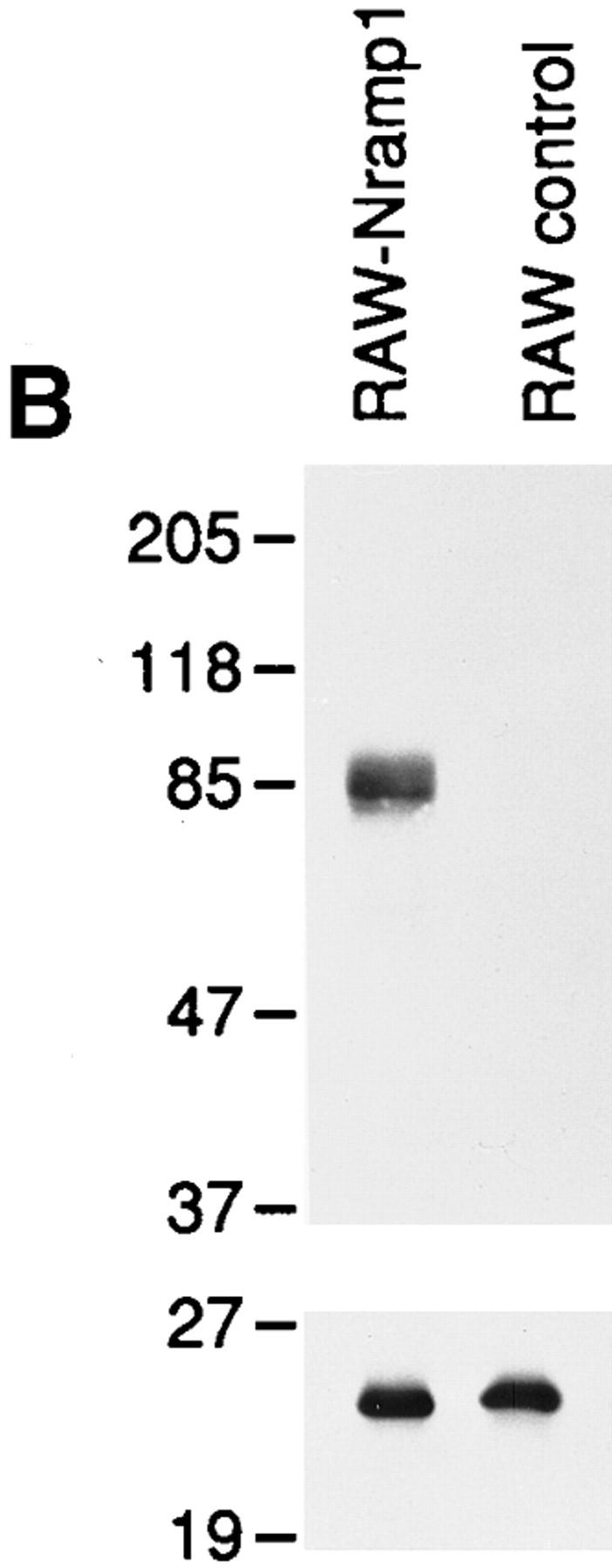
(A) Characterization of RAW264.7 macrophages expressing a transfected Nramp1G169 cDNA. RAW-Nramp1 is a clone of the macrophage cell line RAW 264.7 that has been transfected with a pCB6 expression vector containing a full length Nramp1G 169 modified by the in-frame addition of four antigenic c-Myc epitopes of sequence EQKLISEEDL. RAW-Nramp1 cells (top) and their untransfected RAW264.7 counterparts (bottom) were analyzed by indirect immunofluorescence using the mouse monoclonal 9E10 directed against the introduced c-Myc epitope (1:50 dilution). Both cell populations were treated identically and equal exposure times were used for photography. (B) Immunoblotting of LBcontaining phagosomes isolated from RAW264.7 cells and from RAWNramp1 transfectants. LB-containing phagosomes were purified from cell homogenates by subcellular fractionation on sucrose density gradients as described in Materials and Methods. Equal amounts of phagosomal proteins from each cell line were separated by SDS-PAGE on a 7.5% gel. Proteins were transferred to nitrocellulose and the Nramp1–c-Myc fusion protein was revealed using the anti-c-Myc epitope monoclonal antibody 9E10 (top). Equal loading of proteins on the gel, equal transfer to the membrane, and delivery of late endosomal markers to the latex phagosomes were verified by immunoblotting with polyclonal antisera against Rab7 (bottom) and Lamp1 (data not shown). The position of molecular mass markers (in kD) is indicated on the left side of the immunoblot.
Together, results presented in Figs. 3–5 establish that the Nramp1 protein is initially present in the late endosomal/ lysosomal compartment and is recruited to the phagosome upon maturation of this organelle which occurs after the initial phagocytic event.
Kinetics of Nramp1 Delivery to the Maturing Phagosome.
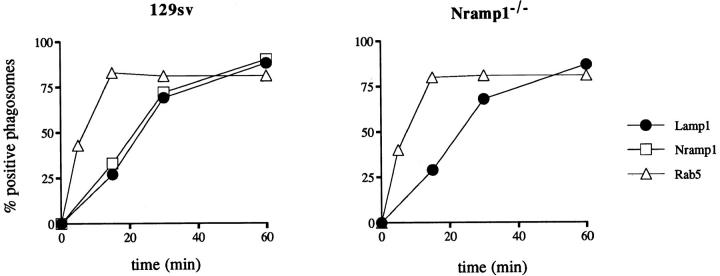
Next we wished to establish the kinetics of delivery of the Nramp1 protein to the phagosome and initiate studies to determine if the absence of Nramp1 protein in macrophages may affect the fusogenic properties of this organelle. Therefore, we determined the kinetics of Nramp1 delivery to the phagosome and compared it to that of an early endosomal marker Rab5 and that of a late endosomal/early lysosomal marker Lamp1. These kinetics were then compared for normal 129/sv macrophages and for Nramp1− /− mutants. Peritoneal macrophages were first incubated with LBcontaining medium for 5 min at 37°C to allow phagocytosis, followed by extensive washing of the monolayer at 4°C to eliminate nonphagocytosed beads and synchronize subsequent maturation of the LB phagosomes. Cells were then returned to 37°C to initiate maturation, and at predetermined times, cells were fixed and analyzed by immunofluorescence. To determine the percentage of phagosomes positive for the markers analyzed, macrophages were initially examined under phase contrast to locate cell-associated LBs (assumed to be phagosomes). These cell-associated beads were then examined under fluorescent light for the presence or absence of immunospecific signals at the periphery of the bead (LB phagosome). In 129/sv macrophages, acquisition of Nramp1 staining by phagosomes was linear over the first 30 min with 63% of phagosomes labeled, and ultimately 85% of phagosomes becoming positive after 60 min (Fig. 6 A). The kinetics of Nramp1 association (rate and final percentage) were found to be identical to that independently determined for Lamp1 (also plotted in Fig. 6 A). Additional experiments where phagosomes were immunostained for both Nramp1 and Lamp1 simultaneously showed that the vast majority of individual phagosomes were either positive for both Nramp1 and Lamp1 or negative for both markers, and this at all times examined (data not shown). By contrast, the kinetics of association of the early endosomal marker Rab5 with LB phagosomes were very different. Rab5 was delivered much more rapidly, with 40% of the LB phagosomes positive after 5 min of maturation, and with a maximum plateau reached at 15 min where 81% of the phagosomes were positive for this marker (Fig. 6 A). Together, these results indicate that Nramp1 and Lamp1 are delivered to the phagosome membrane concurrently.
Figure 6.
Kinetics of Nramp1 protein delivery to the maturing phagosome. Macrophages from normal 129/sv mice (left) and from 129/sv Nramp1− /− mutants (right) were fed a meal of LBs for 5 min, washed at 4°C, and further incubated to initiate phagosome maturation. At predetermined times, cells were fixed and analyzed by immunofluorescence for subcellular localization of Nramp1 (□), the late endosomal/early lysosomal marker Lamp1 (•), and the early endosomal marker Rab5 (▵). The percentage of phagosomes positive for each marker was determined after examination of the cells, first under phase contrast to locate cell-associated LBs, and then under fluorescence for the presence or absence of immunospecific signal at the periphery of the bead. A total of 100 beads were counted for each marker and at each time point. The average of values from two independent experiments are shown.
When these studies were repeated with macrophages from Nramp1− /− mutant mice, no significant differences were observed between the 129/sv and Nramp1− /− mice with respect to the acquisition of Rab5 and Lamp1 during phagosome maturation (Fig. 6 B). These results together with those shown in Fig. 5 B suggest that at this level of resolution, the absence of Nramp1 protein does not grossly affect the fusogenic properties of the phagosome to either Rab5, Rab7, or Lamp1 positive structures.
Discussion
At the cellular level, mutations at Nramp1 cause a loss of natural resistance to infections which is phenotypically expressed as uncontrolled intracellular replication of these microbes in macrophages of the RE system during the early phase of infection (2). We have previously established that Nramp1 mRNA expression is restricted to phagocytes (12); since Nramp1 mutations affect the intracellular growth of antigenically and taxonomically unrelated microbes, the Nramp1 protein must therefore play a key role in the microbicidal activity of these cells. This role, however, has so far remained elusive. Identifying the subcellular localization of the Nramp1 protein in macrophages may provide important clues on the physiological process associated with Nramp1 and underlying resistance/susceptibility but also on the puzzling lack of immediate relationship between microbial infections afflicting these cells and controlled by Nramp1.
Although the highly hydrophobic nature of the Nramp1 polypeptide predicted from cDNA sequencing was clearly suggestive of a membrane protein (12), the nature of the cellular membrane compartment expressing Nramp1 has remained controversial. While some have predicted that Nramp1 may localize to the nuclear envelope and regulate translocation of DNA binding proteins into the nucleus (41), others have proposed that it may exert its action at the plasma membrane playing a role in signal transduction leading to macrophage activation (42, 43). On the other hand, we had proposed that Nramp1 may be found at the phagosome membrane, possibly interacting with different intracellular parasites transiting through this compartment (12, 18). To address these issues, we have recently generated a series of specific anti-Nramp1 antibodies, and shown that Nramp1 behaves as an integral membrane protein, resistant to urea extraction (24). Nramp1 is phosphorylated in macrophages and is also heavily glycosylated with up to 40% of its mass accounted for by the posttranslational addition of two complex carbohydrate side chains of the tri- or tetraantennary type (24). In the present study, we have used these antibodies for double immunofluorescence and confocal microscopy to analyze the subcellular distribution of Nramp1 in populations of macrophage from normal mice (129/sv) and from animals bearing a null Nramp1 allele (14). These experiments have established that Nramp1 is expressed neither at the plasma membrane nor at the nuclear membrane but is rather found in an intracellular compartment. Colocalization studies using antibodies directed against known markers of specific subcellular membranous compartments have identified the Nramp1 positive compartment as late endosome/lysosome. Additional cell fractionation experiments and immunofluorescence analyses using phagosomes containing LBs further showed that Nramp1 is recruited to the phagosomal membrane during the phagosome maturation which follows the initial phagocytic event. The time kinetics of Nramp1 acquisition by maturing phagosomes is similar to kinetics of acquisition of Lamp1, another late endosomal/lysosomal marker, and are clearly distinct from that of Rab5, an early endosomal marker.
Having localized Nramp1 to the late endosomal/early lysosomal compartment of resting macrophages, and having established that Nramp1 is recruited to the phagosome, we can focus on the known physiological role of these subcellular compartments in pathogenesis of intracellular infections and evaluate the possible site and mechanism of action of Nramp1 during these events. A large body of data indicates that these compartments are essential effectors in the intracellular destruction of ingested parasites by macrophages. Most intracellular parasites enter host macrophages via a phagocytic event, resulting in initial encapsulation of the microbe in a plasma membrane–derived structure, the early phagosome (44). Phagosomes themselves have little microbicidal activity, and this activity is delivered to the phagosome through a maturation process that involves a series of complex fusion events, with ultimate fusion to terminal lysosomes to form the phagolysosome (34, 45, 46). Plasma membrane molecules are removed from the early phagosome via recycling, while new soluble or membrane-associated proteins are provided either directly by the biosynthetic pathway, or indirectly by fusion to endocytic organelles including early and late endosomes and the lysosome. The acquisition or loss of specific phagosomal membrane proteins such as Rabs and SNAREs is believed to be responsible for selection of the correct fusogenic partner for the subsequent steps in maturation (for review see reference 47). Phagosome maturation results in strong intravesicular acidification caused by recruitment of the membrane bound subunits of the vacuolar H+/ATPase, appearance of microbicidal function through delivery of the lysosome proteolytic arsenal, generation of reactive oxygen radicals via activation of the NADPH-dependent oxidase system, and release of lactoferrin and other bactericidal or bacteriostatic molecules (for review see reference 48).
On the other hand, intracellular parasites have developed competing mechanisms to circumvent or resist the cytocidal response of macrophages, and the dynamic balance between the two competing systems determines either successful destruction of the invading microbe or intracellular survival with successful replication and parasitism. Microbial tactics for intracellular survival include lysis of the phagosomal membrane and escape to the cytoplasm, inhibition or delay in phagosome maturation and/or acidification, and survival within the fused phagolysosome (for review see reference 22). In fact, the detailed characterization of the strategy used by specific microbes for intracellular survival in macrophages has proven important to elucidate normal cytocidal mechanisms of these cells. Likewise, mutations in host genes that affect intracellular survival of a selected group of parasites (such as Nramp1) can uncover a key macrophage effector mechanism particularly effective against this selected group. Mutations at Nramp1 have a dramatic effect on the growth rate of L. donovani, several species of Mycobacterium (M. bovis, M. intracellulare, M. avium, M. lepraemurium), S. typhimurium, and Brucella abortus in RE organs. However, they do not affect the growth of other intracellular parasites such as Listeria monocytogenes and Legionella pneumophila (in vivo infections and in vitro measurements in isolated macrophages; Gros, P., unpublished data). A rapid review of the intracellular survival strategies adopted by these microbes, contrasted with the subcellular localization and kinetics of association with the phagosome determined in this study for Nramp1, may provide clues on the temporal and site-specific mode of action of Nramp1.
L. donovani phagosomes mature to phagolysosome in a seemingly normal fashion, with the acquisition of lysosomal markers and strong acidification of the parasitophorous vacuole. L. donovani survives this harsh intracellular environment through synthesis of superoxide dismutase (SOD) that neutralizes reactive oxygen species, acid phosphatase, and proteases that inactivate or degrade lysosomal enzymes when activated by acidic pH (for review see reference 49). On the other hand, Mycobacterium tuberculosis blocks acidification of the phagosome by preventing fusion of the phagosome to vacuolar H+/ATPase positive vesicles (50). The exclusion mechanism is controversial but appears selective as M. tuberculosis–containing phagosomes do become positive for lysosomal glycoproteins (lgps) (51). Finally, S. typhimurium are taken up within specialized “spacious phagosomes” similar to macropinosomes (52). These phagosomes acquire lgps and lysosomal acid phosphatase, yet have no mannose-6-phosphate receptors (late endosome marker) and show decreased levels of the lysosomal protease Cathepsin D (53). It is believed that the S. typhimurium-containing phagosome acquires its lgps and lysosomal acid phosphatase through fusion of vesicles distinct from lysosomes, perhaps arising directly from the trans-Golgi network. Phagosomes containing S. typhimurium have also been shown to have greatly decreased fusion with late endocytic compartments and to exhibit delayed and attenuated acidification (for review see reference 22). Thus, although these three types of intracellular parasites have evolved different mechanisms to evade macrophage effector functions, they all transit through and remain associated with the phagosome, and this phagosome seems to fuse to some, but not all, endosomal or lysosomal vesicles. One common characteristic of phagosomes containing these parasites is that they all seem to acquire, at some stage, the late endosomal/early lysosomal marker Lamp1 (49, 50, 53–55). As we have shown that Nramp1 and Lamp1 colocalize within the cell (Figs. 2 and 3), and are delivered to LB-containing phagosomes with identical kinetics (Fig. 6), we predict that the phagosomal membrane enveloping these parasites also acquires Nramp1. This would place Nramp1 in close proximity to those intracellular parasites under its genetic control.
Conversely, L. pneumophila enters the macrophage through the formation of a unique “coiled phagosome” and replicates within a characteristic ribosome-dotted vacuole that does not fuse with endosomes or lysosomes. It does not acidify, nor does it become positive for endosomal or lysosomal markers, including Lamp1 (55). Hence, it appears that Nramp1 may not be delivered to the specialized vacuole containing intracellular L. pneumophila. In the case of L. monocytogenes, once in the phagosome, L. monocytogenes secretes an enzyme (listeriolysin) that lyses the phagosomal membrane within 20 min of phagocytosis, thus allowing escape to and replication in the cytosol (56). Interestingly, we have observed that 20 min after phagocytosis, only ∼30% of the phagosomes examined by immunofluorescence are positive for Nramp1 (Fig. 6). Thus, it seems that L. monocytogenes may escape from the phagosome before a significant amount of Nramp1 is delivered to that site. Therefore, the lack of effect of Nramp1 mutations on infections with L. monocytogenes and L. pneumophila is consistent with the unique intracellular behavior of these bacteria and of the phagosomes containing them.
We propose that Nramp1 is targeted to the membrane of the maturing phagosome and either directly or indirectly modifies the intraphagosomal environment to affect replication of intracellular parasites. Nramp1 could do this indirectly by affecting the fusogenic properties of the maturing phagosome; however, results shown in Figs. 5 and 6 indicate that LB-containing phagosomes from wild-type 129/sv or Nramp1− /− mutants show very similar kinetics of acquisition for early and late endosomal markers. Therefore, we favor a more direct transport mechanism that might involve the delivery of a cytocidal or cytostatic agent to the phagosome or the elimination of a factor that is essential for proliferation of the parasite at that site.
The nature of the putative transport mechanism of Nramp1 and its possible substrate(s) remain unknown. However, the discovery and characterization of Nramp homologues in phylogenetically distant organisms have pointed at possible candidate transport activities. The high degree of sequence similarity amongst Nramp family members (D. melanogaster, 70% similarity; C. elegans, 67%; O. sativa, 61%; S. cerevisiae, 41%; reference 18) suggests parallel functional conservation. Detailed analyses of the primary and secondary sequence features of the Nramp family has identified as the common structural unit of this family, a core hydrophobic domain that shares characteristics previously noted in families of ion transporters and channels. These include (a) high degree of sequence conservation of the 10 TM domains forming this core, (b) helical periodicity of sequence conservation in TM segments, predicting a helical bundle inserted in the membrane with a conserved polar interior and a semi-conserved hydrophobic exterior, (c) direct primary amino acid sequence similarity between the most highly conserved segments of the Nramp family (TM8-TM9 intracellular loop) and the highly conserved region of voltage-gated K+ channels of the shaker type (TMT-4X-G-D/Q-4X-GF; reference 20). Therefore, structural considerations suggest that the Nramp family may form a new group of ion transporters or channels.
Null mutations have been obtained and characterized for the Nramp homologues of D. melanogaster and the yeast S. cerevisiae. Drosophila with a nonfunctional Nramp homologue malvolio (mvl) gene have defects in the pathway involved in integration and processing of gustatory information. The mvl gene is expressed in the central nervous system, peripheral neurons, and macrophages (57). No information is currently available on the molecular mechanism of action of mvl. The yeast Nramp homologues SMF1 and SMF2 were originally isolated in a screen for suppressors of the mif mutation (mitochondrial import factor), a lethal mutation that causes a defect in protein import and translocation across the mitochondrial membrane. SMF1/SMF2 complement only the Ts allele but not a null allele of mif, suggesting indirect complementation as opposed to functional redundancy between mif and SMF1/SMF2 (58). Independently, SMF1 was recently identified in a screen for survival to otherwise lethal concentrations of the metal chelating agent EGTA (21). It was shown that deletion of SMF1 results in decreased cellular uptake of Mn2+ whereas overexpression of the gene results in increased Mn2+ uptake by cells. The protein encoded by SMF1, Smf1p, is located on the yeast plasma membrane, and was proposed to function as a Mn2+ transporter. (Complementation of mif mutant is explained by the fact that Mif encodes a Mn2+-dependent signal peptidase). It is tempting to suggest that other Nramp family members may also be involved in the transport of divalent cations such as Mn2+. Mn2+, Mg2+, Cu2+, Zn2+ or other divalent cations are essential cofactors for many metabolic enzymes, and alterations in their availability could have pleiotropic effects. Considering our localization of Nramp1 to the phagosomal membrane, it is tempting to speculate that Nramp1 protein could eliminate Mn2+ or other divalent cations from the phagosomal interior, as suggested by Supek et al. (21). Mn2+ is an essential cofactor for certain isoforms of SOD, an enzyme that neutralizes reactive oxygen species. Indeed, S. typhimurium, M. tuberculosis, M. bovis and L. donovani all encode their own SOD, suggesting that this enzyme plays an important role in the intracellular survival strategies of these microbes. Eliminating this response through removal of an essential cofactor would result in a net enhancement of the bactericidal activity of the macrophage. Finally, the recent discovery of Nramp homologues in several species of Mycobacterium (Gros, P., unpublished data) including M. leprae (19) suggest that bacteria may have evolved a parallel transport system, possibly competing for the same substrate as the phagosomal Nramp1. The proposal that mammalian Nramp1 and bacterial homologues function as transporters of similar types of substrates (such as Mn2+) is currently being tested.
Acknowledgments
We thank Susan Gauthier (McGill University) and Diane Gingras (Universite de Montreal) for expert technical assistance, Paul Walton (McGill University) for the use of his microscope, David Housman (Massachusetts Institute of Technology), and John Bergeron (McGill University) for helpful discussions. We also thank John Bergeron, John Mort (Shriner's Hospital, Montreal), Philippe Chavrier, and Stephane Meresse (Centre d'Immunologie, INSERM-CNRS, Marseille) for their gifts of antibodies.
This work was supported by National Institutes of Health grant No. 1 R01 A1 35237-01 to P. Gros, and by the Medical Research Council of Canada (MRC) to M. Desjardins. P. Gros is supported by a career scientist award from the MRC, M. Desjardins by a scholarship from Le Fonds de la Recherche en Sante du Quebec, and S. Gruenheid by a studentship from the MRC.
Footnotes
1 Abbreviations used in this paper: Bcgr, Bcg resistant; Bcgs, Bcg susceptible; GST, glutathione S transferase; Lamp1, lysosomal-associated membrane protein 1; LB, latex beads; lpgs, lysosomal glycoproteins; mif, mitochondrial import factor; mvl, malvolio; Nramp1, natural resistance-associated macrophage protein 1; RE, reticuloendothelial; SOD, superoxide dismutase; TM, transmembrane.
References
- 1.Appelberg R, Sarmento AM. The role of macrophage activation and of Bcg-encoded macrophage function(s) in the control of Mycobacterium aviuminfection in mice. Clin Exp Immunol. 1990;80:324–331. doi: 10.1111/j.1365-2249.1990.tb03288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gros P, Skamene E, Forget A. Genetic control of natural resistance to Mycobacterium bovis(BCG) in mice. J Immunol. 1981;127:2417–2421. [PubMed] [Google Scholar]
- 3.Plant J, Glynn AA. Genetics of resistance to infection with Salmonella typhimuriumin mice. J Infect Dis. 1976;133:72–78. doi: 10.1093/infdis/133.1.72. [DOI] [PubMed] [Google Scholar]
- 4.Skamene E, Gros P, Forget A, Patel PJ, Nesbitt MN. Regulation of resistance to leprosy by chromosome 1 locus in the mouse. Immunogenetics. 1984;19:117–124. doi: 10.1007/BF00387854. [DOI] [PubMed] [Google Scholar]
- 5.Bradley DJ. Regulation of Leishmania populations within the host. II. genetic control of acute susceptibility of mice to Leishmania donovaniinfection. Clin Exp Immunol. 1977;30:130–140. [PMC free article] [PubMed] [Google Scholar]
- 6.Gros P, Skamene E, Forget A. Cellular mechanisms of genetically controlled host resistance to Mycobacterium bovis(BCG) J Immunol. 1983;131:1966–1972. [PubMed] [Google Scholar]
- 7.Goto Y, Buschman E, Skamene E. Regulation of host resistance to Mycobacterium intracellulare in vivo and in vitro by the Bcggene. Immunogenetics. 1989;30:218–221. doi: 10.1007/BF02421210. [DOI] [PubMed] [Google Scholar]
- 8.Stach JL, Gros P, Forget A, Skamene E. Phenotypic expression of genetically-controlled natural resistance to Mycobacterium bovis(BCG) J Immunol. 1984;132:888–892. [PubMed] [Google Scholar]
- 9.Lissner CR, Swanson RN, O'Brien AD. Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the Ity gene in peritoneal and splenic macrophages isolated in vitro. . J Immunol. 1983;131:3006–3013. [PubMed] [Google Scholar]
- 10.Denis M, Forget A, Pelletier M, Gervais F, Skamene E. Killing of Mycobacterium smegmatisby macrophages from genetically susceptible and resistant mice. J Leukocyte Biol. 1990;47:25–30. doi: 10.1002/jlb.47.1.25. [DOI] [PubMed] [Google Scholar]
- 11.Crocker PR, Blackwell JM, Bradley DJ. Expression of the natural resistance gene Lshin resident liver macrophages. Infect Immun. 1984;43:1033–1040. doi: 10.1128/iai.43.3.1033-1040.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. . Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 13.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P. Haplotype mapping and sequence analysis of the mouse Nrampgene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 14.Vidal S, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus—genetic transfer of resistance to infections in C57bl/6J mice transgenic for the Nramp1(Gly169) allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal S, Belouchi AM, Cellier M, Beatty B, Gros P. Cloning and characterization of a second human NRAMPgene on chromosome 12q13. Mamm Genome. 1995;6:224–230. doi: 10.1007/BF00352405. [DOI] [PubMed] [Google Scholar]
- 17.Gruenheid S, Cellier M, Vidal S, Gros P. Identification and characterization of a second mouse Nrampgene. Genomics. 1995;25:514–525. doi: 10.1016/0888-7543(95)80053-o. [DOI] [PubMed] [Google Scholar]
- 18.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nrampdefines a family of membrane proteins. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cellier M, Belouchi A, Gros P. Resistance to intracellular infections—comparative genomic analysis of Nramp. . Trends Genet. 1996;12:201–204. doi: 10.1016/0168-9525(96)30042-5. [DOI] [PubMed] [Google Scholar]
- 20.Wood MW, VanDongen HM, VanDongen AM. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc Natl Acad Sci USA. 1995;92:4882–4886. doi: 10.1073/pnas.92.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. . Proc Natl Acad Sci USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-del Portillo F, Finlay BB. The varied lifestyles of intracellular pathogens within eukaryotic vacuolar compartments. Trends Microbiol. 1995;3:373–380. doi: 10.1016/s0966-842x(00)88982-9. [DOI] [PubMed] [Google Scholar]
- 23.Coligan, J.E., A.M. Kruisbeek, D.H. Margulies, E.M. Shevach, and W. Strober. 1995. Current Protocols in Immunology. R. Coico, editor. John Wiley and Sons, Inc., New York. 2.4.1.–2.7.11.
- 24.Vidal S, Pinner E, Lepage P, Gauthier S, Gros P. Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- 25.Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJD, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJM. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- 27.Croul S, Mezitis SG, Stieber A, Chen YJ, Gonatas JO, Goud B, Gonatas NK. Immunocytochemical visualization of the Golgi apparatus in several species, including human, and tissues with an antiserum against MG-160, a sialoglycoprotein of rat Golgi apparatus. J Histochem Cytochem. 1990;38:957–963. doi: 10.1177/38.7.2355176. [DOI] [PubMed] [Google Scholar]
- 28.Gonatas JO, Mezitis SG, Stieber A, Fleischer B, Gonatas NK. MG-160. A novel sialoglycoprotein of the medial cisternae of the Golgi apparatus. J Biol Chem. 1989;264:646–653. [PubMed] [Google Scholar]
- 29.Mort JS, Poole AR, Decker RS. Immunofluorescent localization of cathepsins B and D in human fibroblasts. J Histochem Cytochem. 1981;29:649–657. doi: 10.1177/29.5.6788835. [DOI] [PubMed] [Google Scholar]
- 30.Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 31.Meresse S, Gorvel JP, Chavrier P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J Cell Sci. 1995;108:3349–3358. doi: 10.1242/jcs.108.11.3349. [DOI] [PubMed] [Google Scholar]
- 32.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canfield VA, Levenson R. Transmembrane organization of the Na,K-ATPase determined by epitope addition. Biochemistry. 1993;32:13782–13786. doi: 10.1021/bi00213a005. [DOI] [PubMed] [Google Scholar]
- 34.Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutherford MS, Witsell A, Schook LB. Mechanisms generating functionally heterogeneous macrophages: chaos revisited. J Leukocyte Biol. 1993;53:602–618. doi: 10.1002/jlb.53.5.602. [DOI] [PubMed] [Google Scholar]
- 38.Desjardins M, Celis JE, van Meer G, Dieplinger H, Jahraus A, Griffiths G, Huber LA. Molecular characterization of phagosomes. J Biol Chem. 1994;269:32194–32200. [PubMed] [Google Scholar]
- 39.Rabinowitz S, Horstmann H, Gordon S, Griffiths G. Immunocytochemical characterization of the endocytic and phagolysosomal compartments in peritoneal macrophages. J Cell Biol. 1992;116:95–112. doi: 10.1083/jcb.116.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurent M, Johannin G, Gilbert N, Lucas L, Cassio D, Petit PX, Fleury A. Power and limits of laser scanning confocal microscopy. Biol Cell . 1994;80:229–240. doi: 10.1111/j.1768-322x.1994.tb00934.x. [DOI] [PubMed] [Google Scholar]
- 41.Blackwell JM. Structure and function of the natural resistance associated macrophage protein (Nramp1), a candidate for infectious and auto-immune disease susceptibility. Mol Med Today. 1996;1:205–211. doi: 10.1016/1357-4310(96)88773-9. [DOI] [PubMed] [Google Scholar]
- 42.Kishi F, Nobumoto M. Identification of natural resistance-associated macrophage protein in peripheral blood lymphocytes. Immunol Lett. 1995;47:93–96. doi: 10.1016/0165-2478(95)00070-l. [DOI] [PubMed] [Google Scholar]
- 43.Barton CH, White JK, Roach TI, Blackwell JM. NH2-terminal sequence of macrophage-expressed natural resistance-associated macrophage protein (Nramp) encodes a proline/serine-rich putative Srchomology 3-binding domain. J Exp Med. 1994;179:1683–1687. doi: 10.1084/jem.179.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller WA, Steinman RM, Cohn ZA. Membrane proteins of the vacuolar system. III. Further studies on the composition and recycling of endocytic vacuole membrane in cultured macrophages. J Cell Biol. 1983;96:29–36. doi: 10.1083/jcb.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jahraus A, Storrie B, Griffiths G, Desjardins M. Evidence for retrograde traffic between terminal lysosomes and the prelysosomal/late endosome compartment. J Cell Sci. 1994;107:145–157. doi: 10.1242/jcs.107.1.145. [DOI] [PubMed] [Google Scholar]
- 46.Pitt A, Mayorga LS, Schwartz AL, Stahl PD. Transport of phagosomal components to an endosomal compartment. J Biol Chem. 1992;267:126–132. [PubMed] [Google Scholar]
- 47.Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 48.Buttler, E., M.A. Lichtman, B.S. Coller, and T.J. Kipps. 1995. Williams Hematology. McGraw Hill Inc., New York. 869–875.
- 49.Titus RG, Theodos CM, Shankar AM, Hall LR. Interactions between Leishmania majorand macrophages. Immunol Ser. 1994;60:437–459. [PubMed] [Google Scholar]
- 50.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacteriumphagosomes produced by exclusion of the vesicular proton-ATPase. Science (Wash DC) 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 51.Xu S, Cooper A, Sturgill-Koszycki S, van Heyningen T, Chatterjee D, Orme I, Allen P, Russell DG. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium–infected macrophages. J Immunol. 1994;153:2568–2578. [PubMed] [Google Scholar]
- 52.Alpuche-Aranda CM, Racoosin EL, Swanson JA, Miller SI. Salmonellastimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-del Portillo F, Finlay BB. Targeting of Salmonella typhimuriumto vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6–phosphate receptors. J Cell Biol. 1995;129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonellainduces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc Natl Acad Sci USA. 1993;90:10544–10548. doi: 10.1073/pnas.90.22.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clemens DL, Horwitz MA. Characterization of the Mycobacterium tuberculosisphagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. . J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodrigues V, Cheah PY, Ray K, Chia W. Malvolio, the Drosophila homologue of mouse NRAMP-1 (Bcg), is expressed in macrophages and in the nervous system and is required for normal taste behavior. EMBO (Eur Mol Biol Organ) J. 1995;14:3007–3020. doi: 10.1002/j.1460-2075.1995.tb07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West AH, Clark DJ, Martin J, Neupert W, Hartl FU, Horwich AL. Two related genes encoding extremely hydrophobic proteins suppress a lethal mutation in the yeast mitochondrial processing enhancing protein. J Biol Chem. 1992;267:24625–24633. [PubMed] [Google Scholar]