Abstract
Immature thymocytes undergo a selection process within the thymus based on their T cell antigen receptor (TCR) specificity that results either in their maturation into functionally competent, self-MHC–restricted T cells (positive selection) or their deletion (negative selection). The outcome of thymocyte selection is thought to be controlled by signals transduced by the TCR that vary in relation to the avidity of the TCR–ligand interaction. The TCR is composed of four distinct signal transducing subunits (CD3-γ, -δ, -ε, and ζ) that contain either one (CD3-γ, -δ, -ε) or three (-ζ) signaling motifs (ITAMs) within their intracytoplasmic domains. A possible function for multiple TCR ITAMs could be to amplify signals generated by the TCR during selection. To determine the importance of the multiple TCR-ζ chain ITAMs in thymocyte selection, transgenes encoding α/βTCRs with known specificity were bred into mice in which ζ chains lacking one or more ITAMs had been genetically substituted for endogenous ζ. A direct relationship was observed between the number of ζ chain ITAMs within the TCR complex and the efficiency of both positive and negative selection. These results reveal a role for multiple TCR ITAMs in thymocyte selection and identify a function for TCR signal amplification in formation of the T cell repertoire.
During their development, thymocytes are subjected to a selection process that results in the survival of functionally competent, self-MHC–restricted cells (positive selection), and the deletion of potentially autoreactive cells (negative selection; reference 1). The fate of immature thymocytes is ultimately dictated by the specificity of their TCRs (2) and is presumably controlled by TCR-mediated signals that vary with the avidity of TCR-self ligand interactions within the thymus. The TCR complex contains multiple signal transducing subunits (CD3-γ, -δ, -ε, and ζ) that share a common functional sequence, the immunoreceptor tyrosine-based activation motif (ITAM)1 within their intracytoplasmic domains (3, 4). After TCR engagement, phosphorylation of ITAMs leads to the recruitment of SH2 domain–containing proteins (e.g., tyrosine kinases) to the TCR complex and initiation of the T cell activation cascade (4–7). The CD3 subunits each contain a single ITAM, whereas ζ contains three ITAMs within its longer cytoplasmic tail. ITAM sequences are conserved but nonidentical, and it remains unclear whether individual motifs perform unique or similar functions (8). It has been suggested that due to its singular configuration, ζ chain may function as the predominant TCR signaling structure and that its triplicated ITAMs may serve primarily to facilitate TCR signal amplification (9, 10).
ζ chain is required for normal T cell development as ζ-deficient (ζ−/−) mice have markedly reduced numbers of both CD4+CD8+ (double positive, DP) and CD4+CD8− and CD4− CD8+ (single positive, SP) thymocytes (11–14). However, the major developmental defects in ζ−/− mice were subsequently shown to be the result of impaired TCR surface expression rather than lack of ζ-mediated signals per se, as transgene-encoded ζ chain variants that lacked ITAM sequences but could still facilitate TCR surface expression (ζ-0 ITAM), restored T cell development in ζ−/− mice (15). These results demonstrated that ζ chain signals were not specifically required for T cell development as signal transduction by TCR complexes that contain the CD3 subunit ITAMs, but not ζ chain ITAMs, was sufficient for the generation of mature T cells.
A specific role for the multiple ITAM TCR structure could be in selection of the T cell repertoire during thymocyte development. Recent data support the idea that relatively low affinity TCR–ligand interactions can promote the selection of immature thymocytes, whereas higher affinity interactions are required for mature T cell activation (2, 16–20). Thus, the multiple ζ chain ITAMs could be required to amplify TCR signals originating from the relatively weak interactions that mediate thymocyte selection. Since this question could not be easily addressed in mice with a heterogeneous population of T cells with diverse TCR specificities, we fixed the specificity of the TCRs expressed on developing thymocytes by introducing transgenes that encode productively rearranged TCR-α and -β chains (α/β-TCR Tg) into ζ−/− mice (21, 22). We then bred into the α/β-TCR Tg/ζ−/− background transgenes encoding either full-length ζ chain (3 ITAM), ζ chains containing a single ITAM (1 ITAM), or a ζ chain lacking all three ITAMs (ζ- 0 ITAM). The results of these studies reveal that ζ chain signals influence both positive and negative thymocyte selection and identify a role for the multiple ITAM TCR structure in selection of the T cell repertoire.
Materials and Methods
Transgenic Mice.
Transgenes encoding full-length ζ chain (ζ-3 ITAM Tg) and ζ chain variants with intact extracellular and transmembrane domains, but containing only a single ITAM (either the membrane proximal ITAM or the membrane distal ITAM; [ζ-1 ITAM Tg]), or containing no ITAMs (ζ-0 ITAM Tg) were generated using the identical (human CD2) promoter/ enhancer cassette. The generation of ζ−/− mice, ζ transgenes, and genetic reconstitution of ζ−/− mice with ζ transgenes has been described previously (11, 15). The α/β-TCR transgenes used in these experiments encoded either an MHC class I–restricted TCR specific for male antigen (H-Y; 21) or an MHC class II–restricted TCR specific for pigeon cytochrome C (AND; 22). α/βTCR transgenes were introduced into the ζ−/− background by breeding and identified by Southern blotting or PCR analysis of tail DNA. All mice used for these studies were bred and maintained in a barrier (specific pathogen-free) facility.
Proliferation Assays.
Single cell suspensions were prepared from LNs from normal (ζ+/+) or transgene-reconstituted ζ−/− mice. T cells were purified by incubating cells on rabbit anti–mouse Igcoated plates. Accessory cells and APCs (either for FcR-mediated antibody cross-linking or allogeneic stimulator cells) were prepared from spleen cell suspensions of C57BL/6 or BALB/c mice. APCs were depleted of T cells with anti-Thy1.2 + C′ and irradiated with 3,000 rads. 105 responder T cells were combined with 5 × 105 accessory cells in flat-bottom 96-well plates in the presence or absence of the indicated stimulants. After the specified time, cells were pulsed for 8 h with 1 μCi [3H]thymidine and harvested. For mitogen stimulation, Con A at 2 μg/ml was added to the culture. For anti-CD3ε stimulation, 25% tissue culture supernatant from the 145-2C11 hybridoma was added to the culture. Cells were cultured in presence of syngeneic (C57BL/6) APCs and harvested after 48 h. For the mixed lymphocyte reaction, responder T cells were combined with either syngeneic (C57BL/6) or allogeneic (BALB/c) APCs. Cells were harvested after a total of 96 h.
Flow Cytometry.
For multicolor flow cytometry (FCM), thymocytes or LN cells were first incubated with antibody to the Fc receptor (mAb 2.4G2) to prevent FcR binding. For two- and threecolor FCM, cells were incubated with FITC-conjugated, PEconjugated, and biotinylated antibodies, followed by the addition of red 670 streptavidin (GIBCO BRL, Gaithersburg, MD). The FCM was performed on a Becton Dickinson Immunocytometry Systems FACScan® using standard Cell Quest software. Data were collected on 10–20 × 104 viable cells as determined by forward and side light scatter. The majority of monoclonal antibodies used for FCM analysis were purchased from PharMingen (San Diego, CA) and included biotinylated anti-CD4 (RM4.5) and PE–anti-CD8 (6.7), FITC and PE–anti-CD3 (145-2C11), FITC–anti-TCRVβ14 (14-2), and FITC–anti-Vα11 (RR8-1). Anti-TCRVβ8 (F23.1) and anti–H-Y clonotypic receptor (T3.70) were purified from cell culture supernatants and labeled with FITC.
Superantigen-induced Thymocyte Deletion.
4–8-wk-old mice were injected intraperitoneally with PBS alone, 10 μg staphylococcus enterotoxin B (SEB; Sigma Chemical Co., St. Louis, MO) in PBS, or 20 μg SEB in PBS, every other day for 1 wk (three doses). 2 d after the final injection, mice were killed and thymocytes were harvested. Thymocytes were stained with anti-Vβ8–FITC or anti-Vβ14–FITC, followed by anti-CD3ε–PE. Deletion was assessed by software gating on CD3high (SP cells) and determining the percentage of cells expressing individual TCRVβ chains. Percent deletion was determined by dividing the percentage of CD3high, TCRVβ8+ thymocytes remaining after SEB injection by the percentage of CD3high, TCRVβ8+ cells present in mice injected with PBS alone.
Results
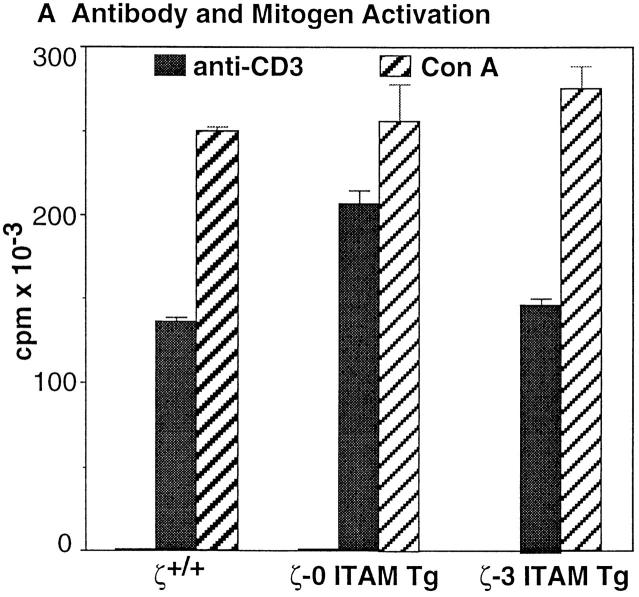
Reconstitution of ζ−/− mice with a transgene encoding a ζ variant chain lacking all three ζ chain ITAMs (ζ-0 ITAM) restores TCR surface expression and promotes the generation of large numbers of CD4+CD8− and CD4−CD8+ SP thymocytes and peripheral T cells (15). Significantly, the T cells generated in ζ-0 ITAM Tg mice are functionally competent as assessed by their ability to respond to TCR-dependent stimuli (Fig. 1; 23). These results demonstrate that TCR-ζ chain signals are not specifically required for either the generation or activation of mature T cells.
Figure 1.
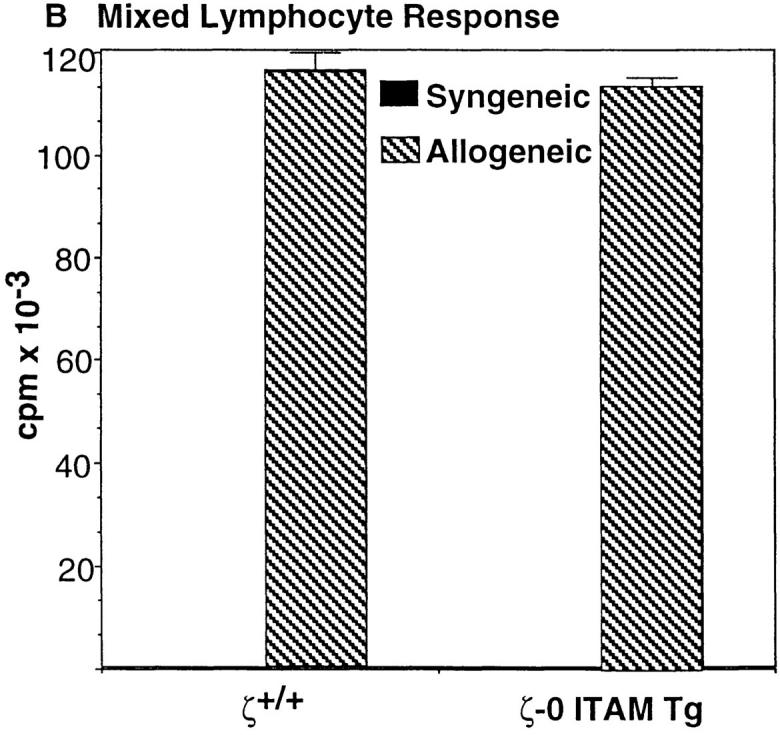
Role of TCR-ζ chain ITAMs in mature T cell function. (A) Proliferative response of LN T cells to anti-CD3ε and Con A stimulation. (B) Proliferative response of LN T cells to allogenic stimulator cells. LN T cells were obtained from 4–8-wk-old nontransgenic (ζ+/+/) mice, or ζ−/− mice that express transgenes encoding either full-length ζ chain (ζ-3 ITAM Tg) or ζ chain lacking ITAM sequences (ζ-0 ITAM Tg) and stimulated as described in Materials and Methods.
To examine the importance of the multiple TCR-ζ ITAMs in the selection of the T cell repertoire, we fixed the specificity of the TCRs expressed on developing thymocytes by introducing a transgene that encodes productively rearranged TCR-α and -β chains specific for the male H-Y antigen in the context of H-2Db (H-Y TCR; 21). This approach enabled us to examine both positive selection (H-2Db female mice) and negative selection (H-2Db male mice) of thymocytes bearing a single α/β-TCR.
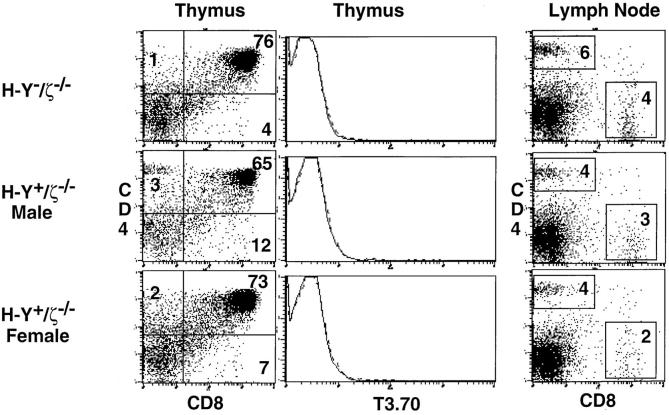
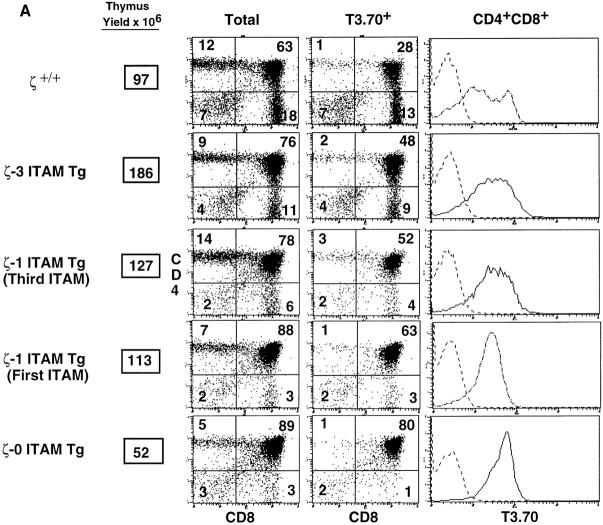
Examination of ζ−/− mice that express the H-Y transgene revealed that the developmental impairment observed previously in ζ−/− mice was also observed in both male and female H-Y+/ζ−/− mice (Fig. 2). These results were expected because of the extremely low level of TCR surface expression in the absence of ζ chain (11–14) and are consistent with a requirement for the TCR for normal thymocyte development and selection. To address the importance of the ζ chain ITAMs in thymocyte selection, we next examined H-Y+/ζ−/− mice reconstituted with transgenes encoding either full-length ζ chains (H-Y+/ζ-3 ITAM Tg mice), ζ chains containing a single ITAM (H-Y+/ζ-1 ITAM Tg mice), or ζ chains lacking all three ITAMs (H-Y+/ζ- 0 ITAM Tg mice) (15). Each of the ζ transgenes was capable of restoring TCR surface expression in mice lacking endogenous ζ chain (Fig. 3; 15). For these experiments transgenic founder lines were chosen that gave comparable levels of TCR surface expression that were equal to or greater than that found on thymocytes from ζ+/+ mice (Fig. 3; 15).
Figure 2.
Phenotype of H-Y−/ζ−/− and H-Y+/ζ−/− mice. Data show immunofluorescence and multicolor FCM analysis of thymocytes and LN T cells from adult (6–12-wk-old) H-2Db mice. Three-color FCM analysis was performed on cells stained with anti–H-Y clonotypic antibody (T3.70) conjugated to FITC, anti-CD8–PE and anti-CD4–biotin, followed by streptavidin–red 670 (16). CD4 versus CD8 two-color profiles are displayed on total thymocytes or total lymph node T cells. Single-color profiles (solid lines) depict T3.70 staining on total thymocytes. Dotted lines reflect staining with negative control antibody.
Figure 3.
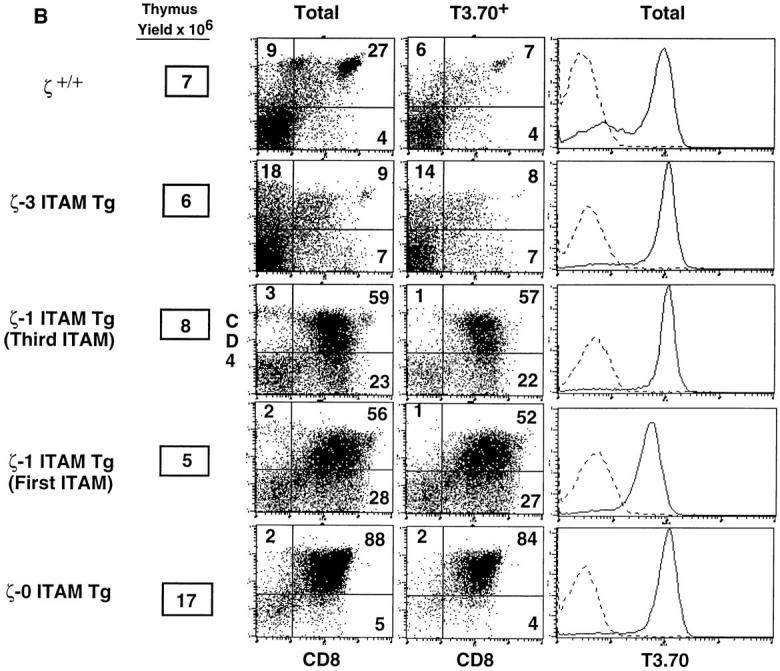
Role of TCRζ–mediated signals in thymocyte positive and negative selection. (A) Phenotype of thymocytes from H-Y+/ζ−/−; ζ Tg female mice. (B) Phenotype of thymocytes from H-Y+/ζ−/−; ζ Tg male mice. H-Y+/ζ−/− mice were reconstituted with transgenes encoding either full-length ζ chains (ζ-3 ITAM Tg), ζ chains that contain a single ITAM (either the first or third, ζ-1 ITAM Tg) or ζ chains that lack ITAMs (ζ-0 ITAM Tg). Data show immunofluorescence and multicolor FCM analysis of thymocytes from adult H-2Db mice. Three-color FCM was performed on cells stained with anti–H-Y clonotypic antibody (T3.70) conjugated to FITC, anti-CD8–PE, and anti-CD4– biotin, followed by streptavidin– red 670 (16). Two-color plots show total thymocytes or software-gated T3.70+ thymocytes. Numbers in the quadrants reflect the percentage of total thymocytes in that quadrant. Singlecolor profiles (solid lines) depict T3.70 staining on either CD4+CD8+ thymocytes (A) or total thymocytes (B). Dotted lines reflect staining with negative control antibody.
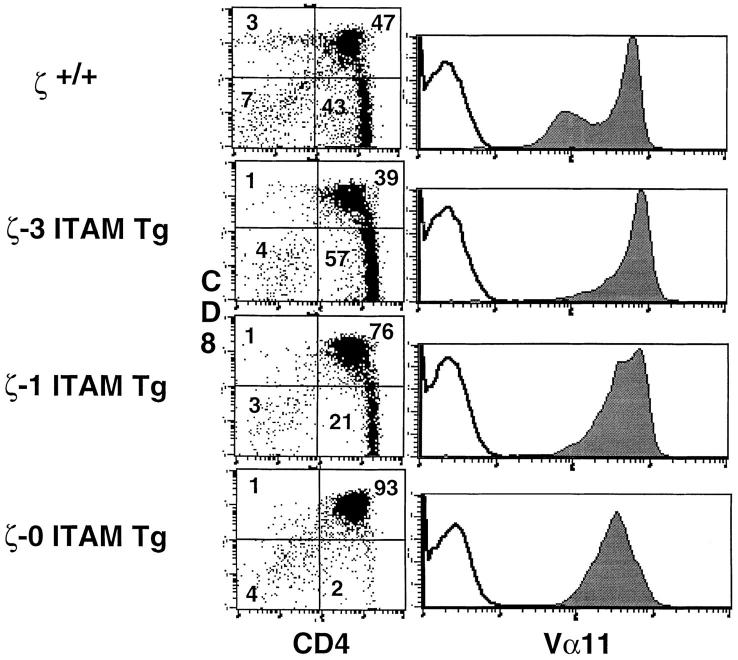
In female H-2Db, H-Y+/ζ+/+ mice, interaction of the H-Y TCR on immature thymocytes with an unidentified ligand results in positive selection and generation of CD8 SP thymocytes that express high surface levels of the H-Y TCR (detected by reactivity with the clonotype-specific antibody, T3.70; reference 24; Fig. 3 A, Table 1 A). Importantly, a phenotype similar to that of H-Y+/ζ+/+ mice was observed in female H-Y+/ζ−/− mice reconstituted with the full-length ζ transgene (H-Y+/ζ-3 ITAM Tg; Fig. 3 A, Table 1 A). These mice also had large thymi that contained a high percentage of T3.70+, CD8 SP cells indicating that positive selection was effectively restored in ζ−/− mice by expression of a transgene-encoded full-length (3 ITAM) ζ chain (Fig. 3). However, in female H-Y+/ζ−/− mice reconstituted with a transgene encoding the signaling-deficient ζ chain (H-Y+/ζ-0 ITAM Tg), positive selection of T3.70+ thymocytes was markedly impaired as evidenced by the extremely low percentage of T3.70+, CD8 SP thymocytes (Fig. 3 A, Table 1 A). Interestingly, H-Y+/ζ−/− females reconstituted with transgenic ζ chains that contain a single ITAM (either the first [membrane proximal] or third [membrane distal]) exhibited an intermediate phenotype (Fig. 3 A). Although there were subtle differences in the two H-Y+/ζ-1 ITAM Tg lines, the percentage of T3.70+, CD8 SP thymocytes in both lines consistently fell between those observed in mice reconstituted with either the ζ-3 ITAM or ζ-0 ITAM transgenes. The direct relationship between the number of ζ chain ITAMs and the generation of clonotypic (T3.70+) CD8 SP thymocytes was even more evident when absolute numbers of T3.70+, CD8+ thymocytes in the various transgenic lines were compared (Table 1 A). Examination of LNs from H-Y+/ζ-0 ITAM Tg females also revealed a lower percentage of T3.70+, CD8+ T cells relative to H-Y+/ζ-3 ITAM Tg females (Fig. 4). A similar relationship between the number of TCR-ζ ITAMs and the efficiency of positive selection was also observed with a class II–restricted TCR-αβ Tg (AND; reference 22; Fig. 5). Together, these results demonstrate that TCR ligand interactions that can generate signals that promote positive selection in the presence of full-length ζ chains are unable to generate these signals in the absence of ζ chain ITAMs.
Table 1.
Numbers of Thymocytes in H-Y+/ζ+ /+ Mice and H-Y+/ζ− /− Mice Reconstituted with Transgenes Encoding the Full-length ζ Chain (ζ-3 ITAM Tg), a ζ Chain Containing the Third ITAM (ζ-1ITAM Tg) or a ζ Chain lacking ITAMs (ζ-0 ITAM Tg)
| Females | ζ+/+ n = 3 | ζ-3 ITAM Tg n = 3 | ζ-1 ITAM Tg n = 6 | ζ-0 ITAM Tg n = 6 | ||||
|---|---|---|---|---|---|---|---|---|
| Total | ||||||||
| Thymocytes | 97 ± 42 | 166 ± 74 | 122 ± 45 | 56 ± 22 | ||||
| Total T3.70+ | 53 ± 25 | 117 ± 33 | 91 ± 51 | 47 ± 18 | ||||
| T3.70+ DN* | 8.2 ± 3 | 6.2 ± 3 | 3.3 ± 2 | 1.9 ± 2 | ||||
| T3.70+ DP* | 28 ± 10 | 89 ± 29 | 74 ± 26 | 42 ± 16 | ||||
| T3.70+ CD8 SP* | 15.5 ± 12 | 17 ± 6 | 9.8 ± 5 | 0.6 ± 0.2 | ||||
| Males | n = 4 | n = 5 | n = 6 | n = 7 | ||||
| Total | ||||||||
| Thymocytes | 6.0 ± 1.0 | 5.6 ± 2.7 | 7.6 ± 5.2 | 17 ± 10 | ||||
| Total T3.70+ | 5.1 ± 0.7 | 4.9 ± 2.4 | 6.6 ± 4.3 | 14.3 ± 9.3 | ||||
| T3.70+ DN* | 3.9 ± 0.4 | 2.6 ± 1.8 | 0.9 ± 0.6 | 0.8 ± 0.9 | ||||
| T3.70+ DP* | 0.5 ± 0.4 | 1.0 ± 0.7 | 4.0 ± 2.5 | 11.0 ± 7.6 |
Data are given in 106 cells as means ± SEM rounded to the nearest whole number. T3.70+ thymocyte subpopulations were calculated by multiplying the total T3.70+ thymocyte number by the percent of cells in a given quadrant as depicted in Fig. 2.
Double negative = CD4−CD8−; DP = CD4+CD8+; CD8 SP = CD4−CD8+.
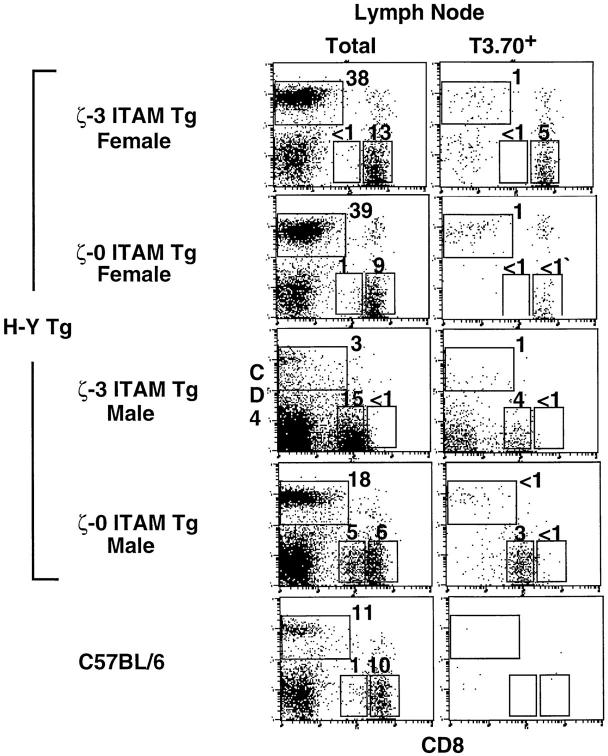
Figure 4.
Analysis of LN T cells from H-Y+/ζ Tg mice. Immunofluorescence and multicolor FCM analysis of LN cells from adult female and male H-Y+/ ζ-3 ITAM Tg or H-Y+/ζ-0 ITAM Tg mice. Also shown are LN cells from a nontransgenic C57BL/6 (H-Y−/ζ+/+) mouse. Threecolor FCM was performed on total LN cells after staining with FITC– anti–H-Y clonotypic antibody (T3.70), anti-CD8–PE and anti-CD4– biotin, followed by streptavidin–red 670. CD4 versus CD8 two-color profiles are displayed on total (left) or gated T3.70+ (right) cells. Numbers in the regions reflect the percentage of total cells found in that region.
Figure 5.
Positive selection in MHC class II–restricted α/βTCR transgenic mice. AND+/ζ−/− mice were generated by mating and then reconstituted with transgenes encoding either a full-length ζ chain (ζ-3 ITAM Tg), a ζ chain containing a single (third) ITAM (ζ-1 ITAM Tg), or a ζ chain that lacks ITAMs (ζ- 0 ITAM Tg). Data show immunofluorescence and multicolor FCM analysis of thymocytes from adult H-2Db mice. FCM was performed on cells stained with FITC-conjugated antibody specific for the transgenic Vα chain (Vα11), anti-CD8–PE and antiCD4–biotin, followed by streptavidin–red 670. Two-color plots show software-gated Vα11+ thymocytes. Numbers in the quadrants reflect the percentage of total thymocytes in that quadrant. Single-color profiles (solid lines) depict Vα11 staining on total thymocytes. Dotted lines reflect staining with negative control antibody.
Despite the near absence of T3.70+, CD8+ LN T cells, large numbers of T3.70−, CD8 SP thymocytes and T cells were detected in H-Y+/ζ-0 ITAM Tg females (Figs. 3 A and 4). Since these T3.70− CD8 SP cells expressed α/β-TCRs composed of transgenic TCR-β chains paired with non- transgenic, endogenously derived TCR-α chains, their TCR specificity was not restricted to the H-Y antigen. Therefore, the reduction in T3.70+, CD8 SP thymocytes and T cells in H-Y+/ζ-0 ITAM Tg mice specifically reflected the failure of the H-Y TCR to promote positive selection of T3.70+ DP thymocytes.
To assess the role of the ζ ITAMs in negative selection, we next examined H-Y+/ζ Tg male mice. Both H-Y+/ζ+/+ males and H-Y+/ζ-3 ITAM males had small thymi with few (<107) thymocytes which were predominantly CD4− CD8− (Fig. 3 B, Table 1 B). The marked reduction in DP thymocytes in these mice is consistent with previous data indicating that negative selection occurs before the DP stage in H-Y Tg males (25). In contrast to mice expressing full-length ζ chain, H-Y+/ζ-0 ITAM Tg males exhibited a very different phenotype. Thymi from these mice were three- to sixfold larger than H-Y+/ζ-3 ITAM Tg males (Table 1 B) and contained predominantly T3.70+, DP thymocytes (Fig. 3 B). Again, the phenotype exhibited by both lines of H-Y+/ζ-1 ITAM Tg mice was intermediate between H-Y+/ζ-3 ITAM Tg males and H-Y+/ζ-0 ITAM Tg males. For example, thymi from H-Y+/ζ-1 ITAM Tg males contained lower percentages of DP thymocytes than did H-Y+/ζ-0 ITAM Tg males, but contained higher percentages of CD4−CD8low cells which are thought to be the immediate precursors of DP thymocytes (26–28; Fig. 3 B, Table 1 B). Thus, the early negative selection of H-Y clonotypic (T3.70+) thymocytes was attenuated in H-Y+/ζ-0 ITAM Tg and H-Y+/ζ-1 ITAM Tg males.
We next examined LNs to determine the fate of the DP thymocytes observed in male H-Y+/ζ-0 ITAM Tg and H-Y+/ζ-1 ITAM Tg mice. Whereas LN from both H-Y+/ ζ+/+ males and H-Y+/ζ-3 ITAM Tg males contained almost no “conventional” CD4high or CD8high T cells, large numbers of both CD4high and CD8high T cells were present in H-Y+/ζ-0 ITAM Tg (Fig. 4) and H-Y+/ζ-1 ITAM Tg males (data not shown). These CD4high and CD8high T cells expressed high levels of TCR which were uniformly T3.70low/− indicating that they were composed primarily of nontransgenic TCR-α chains. Therefore, although negative selection of T3.70+ DP thymocytes was not abrogated in H-Y+/ζ-0 ITAM Tg and H-Y+/ζ-1 ITAM Tg males, some DP thymocytes were able to escape negative selection. Furthermore, these thymocytes were also capable of being positively selected on the basis of non–H-Y restricted TCRs generated from rearrangement and expression of endogenous TCR-α genes.
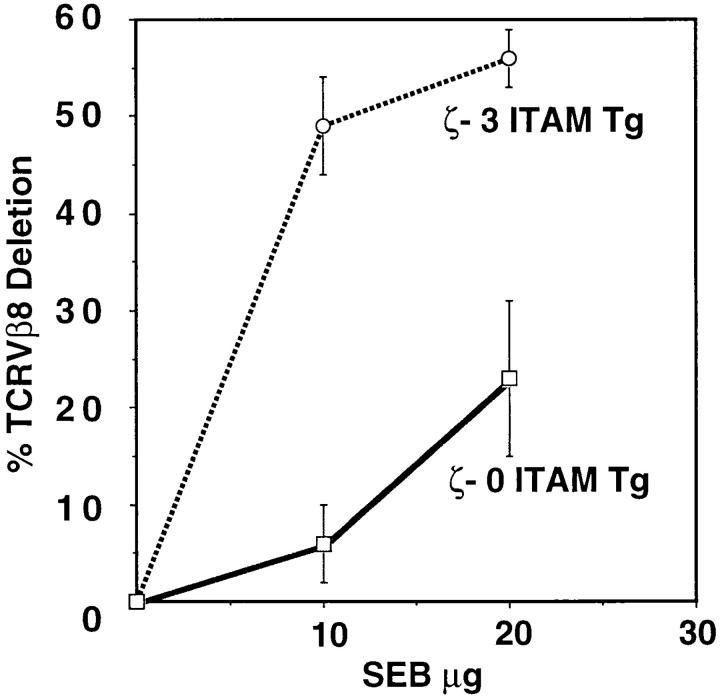
To study the role of ζ-mediated signals in deletion of more mature (SP) thymocytes, we examined superantigenmediated thymocyte deletion in the various transgenic lines. The superantigen SEB deletes SP thymocytes bearing TCRs that include Vβ8 (29). Adult mice were injected with SEB or PBS every other day for 1 wk and the percentage of TCRVβ8+ SP thymocytes remaining was determined by FCM analysis. At similar doses of antigen, the efficiency of SEB-induced thymocyte deletion in ζ-0 ITAM mice was significantly reduced relative to ζ-3 ITAM Tg mice (Fig. 6). Nevertheless, thymocyte deletion by SEB was detectable in ζ-0 ITAM Tg mice and increased with increasing SEB dosage (Fig. 6). The deletion of Vβ8+ thymocytes was specific as the percent TCRVβ14+ SP thymocytes (which are not deleted by SEB) was similar in ζ-0 ITAM Tg and ζ-3 ITAM Tg mice (data not shown). Therefore, in the absence of TCR-ζ signals, deletion of SP thymocytes by SEB was significantly impaired.
Figure 6.
Superantigen-mediated deletion of TCRVβ8+ thymocytes. Mice were injected with PBS alone, 10 μg SEB in PBS, or 20 μg SEB. Thymocytes were stained with anti-Vβ8–FITC or anti-Vβ14–FITC, followed by anti-CD3ε–PE. Deletion was assessed by software gating on CD3high (SP cells) and determining the percentage of cells expressing individual TCRVβ chains. Deletion was specific for cells expressing Vβ8 as cells expressing Vβ14 were not deleted (29). Results are given as mean ± SEM and are from at least six different SEB injected mice per group compiled from two to three separate experiments.
Discussion
In these studies, we investigated the role of the TCR-ζ chain ITAMs in thymocyte selection by fixing the specificity of the TCR and then varying the TCR signaling potential by altering the number of TCR-ζ chain ITAMs within the TCR complex. The results reveal a direct relationship between the number of ζ chain ITAMs in the TCR complex and the efficiency of both positive and negative thymocyte selection. In addition, they suggest that the multiple ζ chain ITAMs function primarily to amplify signals generated by the TCR during thymocyte selection. In this respect, our findings are consistent with in vitro results obtained from mature T cell clones which failed to identify a specific function for individual ζ ITAMs, but found that triplication of a single motif resulted in a quantitative enhancement of signaling (9, 10). However, our current results do not preclude a specific role for individual TCR-ζ ITAMs in more specialized T cell functions that have yet to be examined.
Our results with two α/β-TCR Tg systems (H-Y and AND) suggest that positive selection, which is thought to be mediated by low avidity TCR–ligand interactions (2, 16–19) is particularly dependent upon signal amplification by the TCR-ζ chain. In H-Y+/ζ-0 ITAM Tg female mice, most thymocytes that expressed only the H-Y–specific TCR failed to be positively selected, whereas a large number of thymocytes that expressed non–H-Y–specific TCRs were capable of being positively selected. We predict that the mature T cells generated in both non-α/β-TCR Tg/ζ-0 ITAM Tg (15) and H-Y+/ζ-0 ITAM Tg mice are selected on the basis of TCRs with relatively high affinity for self ligands, and that these TCRs might otherwise promote negative selection in ζ+/+ or ζ-3 ITAM Tg mice where the TCR contains the full complement of ITAMs. Signal amplification by the ζ chain ITAMs may therefore not be absolutely required for positive selection if the avidity of the positively selecting TCR–ligand interaction is sufficiently high. In these instances, signal transduction by the remaining ITAMs supplied by the CD3 chains (which are targeted to the cell surface in our model system) is apparently adequate for selection in the absence of ζ-mediated signals.
Interestingly, incremenatal reduction in the number of TCR-ζ ITAMs resulted in graded effects on positive selection. Graded or quantitative effects on positive selection have also been observed with increasing concentrations of positively selecting ligand (16–19). These and other (20) data suggest that a broad range of signaling responses below a certain threshold may be capable of mediating positive selection, albeit with different efficiency.
Our results demonstrate that TCR signal amplification also plays an important role in negative selection; however, in H-Y Tg mice, the impact of removing the ζ ITAMs from the TCR complex varied depending on the developmental stage. Negative selection of early “transtitional” (CD8lowCD4−) thymocytes was abrogated in the absence of ζ-mediated signals, presumably because low CD8 surface density lowers the avidity of the ligand interaction (21, 25). These cells could give rise to T3.70+ (CD4highCD8high) DP thymocytes, but DP thymocytes were more susceptible to negative selection as demonstrated by their low numbers in H-Y+/ζ-0 ITAM Tg males. Nevertheless, negative selection was also impaired at the DP stage in H-Y+/ζ-0 ITAM Tg mice to the extent that some DP thymocytes survived, rearranged, and expressed endogenous TCR-α, and were positively selected on the basis of non–H-Y clonotypic TCRs. Superantigen-induced deletion of SP thymocytes was also affected in ζ-0 ITAM Tg mice, demonstrating that negative selection which occurs at later stages of thymocyte developement is also impaired in the absence of ζ-mediated signals.
That reduction in the number of TCR ITAMs quantitatively affects both positive and negative selection is consistent with the hypothesis that the outcome (positive or negative) of thymocyte selection is dictated by quantitative rather than qualitative differences in the TCR signaling response (2). Although it might have been predicted, we did not observe positive selection of T3.70+ thymocytes in H-Y+ζ 0-ITAM Tg males. However, the signaling capacity of the H-Y TCR in H-Y+ζ 0-ITAM Tg males may not have been sufficiently reduced to observe such an outcome, as negative selection, though impaired, was not abrogated in these mice. Our findings suggest that signals generated from negative selecting interactions are more difficult to attenuate than those generated during positive selection, not only because higher avidity TCR–ligand interactions are thought to promote negative selection, but also because of the effect of TCR signal amplification. Therefore, these results provide an alternative interpretation to studies suggesting that positive and negative selection are biochemically distinguishable based primarily on the finding that approaches that disrupt positive selection fail to perturb negative selection (30, 31).
Finally, it has remained unclear how relatively small differences in the affinity or avidity of TCR self–ligand interactions can differentially lead either to positive or negative selection (32). Our data suggest that the ability of the TCR to elicit such developmentally distinct cell fate decisions stems from its unique structure. Multiple ITAMs may serve to greatly magnify TCR signals and thereby translate relatively small differences in avidity into large signaling differences. Thus, the unique configuration of the TCR, with its capacity for signal amplification, plays a pivitol role in determining the ultimate fate of developing thymocytes and in shaping the mature T cell repertoire.
Acknowledgments
We thank B.J Fowlkes, A.S. Rosenberg, A. Singer, and M. Vacchio for reading the manuscript and for helpful discussions.
Footnotes
1 Abbreviations used in this paper: DP, double positive; FCM, flow cytometry; ITAM, immunoreceptor tyrosine-based activation motif; SEB, Staphylococcus enterotoxin B; SP, single positive.
References
- 1.Robey E, Fowlkes BJ. Selective events in T cell development. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 2.Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 3.Reth M. Antigen receptor tail clue. Nature (Lond) 1989;338:383–384. [PubMed] [Google Scholar]
- 4.Samelson LE, Klausner RD. Tyrosine kinases and tyrosine-based activation motifs. Current research on activation via the T cell antigen receptor. J Biol Chem. 1992;267:24913–24916. [PubMed] [Google Scholar]
- 5.Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 6.Irving BA, Weiss A. The cytoplasmic domain of the T cell receptor ζ chain is sufficent to couple to receptorassociated signal transduction pathways. Cell. 1991;64:891–901. doi: 10.1016/0092-8674(91)90314-o. [DOI] [PubMed] [Google Scholar]
- 7.Romeo C, Amiot A, Seed B. Sequence requirements for induction of cytolysis by the T cell antigen/Fc receptor ζ chain. Cell. 1992;68:889–897. doi: 10.1016/0092-8674(92)90032-8. [DOI] [PubMed] [Google Scholar]
- 8.Letourneur F, Klausner RD. Activation of T cells by a tyrosine kinase activation domain in the cytoplasmic tail of CD3ε. Science (Wash DC) 1992;255:79–82. doi: 10.1126/science.1532456. [DOI] [PubMed] [Google Scholar]
- 9.Irving BA, Chan AC, Weiss A. Functional characterization of a signal transducing motif present in the T cell antigen receptor ζ chain. J Exp Med. 1993;177:1093–1103. doi: 10.1084/jem.177.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan AC, Desai DM, Weiss A. The role of protein tyrosine kinases and protein tyrosine phosphatases in T cell antigen receptor signal transduction. Annu Rev Immunol. 1994;12:555–592. doi: 10.1146/annurev.iy.12.040194.003011. [DOI] [PubMed] [Google Scholar]
- 11.Love PE, Shores EW, Johnson MD, Tremblay ML, Lee EJ, Grinberg A, Huang SP, Singer A, Westphal H. T cell development in mice that lack the ζ chain of the T cell antigen receptor complex. Science (Wash DC) 1993;261:918–921. doi: 10.1126/science.7688481. [DOI] [PubMed] [Google Scholar]
- 12.Ohno H, Aoe T, Taki S, Kitamura D, Ishida Y, Rajewsky K, Saito T. Developmental and functional impairment of T cells in mice lacking CD3ζ chains. EMBO (Eur Mol Biol Organ) J. 1993;12:4357–4366. doi: 10.1002/j.1460-2075.1993.tb06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malissen M, Gillet A, Rocha B, Trucy J, Viver E, Boyer C, Köntgen F, Brun N, Mazza G, Spanopoulou E, et al. T cell development in mice lacking the CD3-ζ/η gene. EMBO (Eur Mol Biol Organ) J. 1993;12:4347–4355. doi: 10.1002/j.1460-2075.1993.tb06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C-P, Ueda R, She J, Sancho J, Wang B, Weddell G, Loring J, Kurahara C, Dudley EC, Hayday A, et al. Abnormal T cell development in CD3-ζ−/−mutant mice and identification of a novel T cell population in the intestine. EMBO (Eur Mol Biol Organ) J. 1993;12:4863–4875. doi: 10.1002/j.1460-2075.1993.tb06176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shores EW, Huang K, Tran T, Lee E, Grinberg A, Love PE. Role of TCRζ chain in T cell development and selection. Science (Wash DC) 1994;266:1047–1050. doi: 10.1126/science.7526464. [DOI] [PubMed] [Google Scholar]
- 16.Ashton-Richardt PG, Bandeira A, Delaney JR, Van Kaer L, Pircher H-P, Zinkernagel RM, Tonegawa S. Evidence for a differential avidity model of T cell selection in the thymus. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 17.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 18.Sebzda E, Wallace VA, Mayer J, Yeung RSM, Mak TW, Ohashi PS. Positive and negative thymocyte selection induced by different concentration of a single peptide. Science (Wash DC) 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 19.Pircher H, Rohrer UH, Moskophidis D, Zinkernagel RM, Hengartner H. Lower receptor avidity required for clonal deletion than for effector T cell function. Nature (Lond) 1991;351:482–485. doi: 10.1038/351482a0. [DOI] [PubMed] [Google Scholar]
- 20.Ignatowicz, L., J. Kappler, and P. Marrack. 1996. The repertoire of T cells shaped by a single MHC/peptide ligand. 23: 521–529. [DOI] [PubMed]
- 21.von Boehmer H. Developmental biology of T cells in T cell receptor transgenic mice. Annu Rev Immunol. 1990;8:531–356. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 22.Kaye J, Hsu M-L, Sauron M-E, Jameson SC, Gascoigne NRJ, Hedrick SM. Selective development of CD4+T cells in transgenic mice expressing a class II MHC–restricted antigen receptor. Nature (Lond) 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 23.Combadiere B, Freedman M, Chen L, Shores EW, Love PE, Lenardo M. Qualitative and quantitative contributions of the T cell receptor ζ chain to mature T cell apoptosis. J Exp Med. 1996;183:2109–2117. doi: 10.1084/jem.183.5.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teh H-S, Kishi H, Scott B, von Boehmer H. Deletion of autospecific T cells in T cell receptor (TCR) transgenic mice spares cells with normal TCR levels and low levels of CD8 molecules. J Exp Med. 1989;169:795–806. doi: 10.1084/jem.169.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahama Y, Shores EW, Singer A. Negative selection of precursor thymocytes before their differentiation into CD4+ CD8+cells. Science (Wash DC) 1992;258:653–656. doi: 10.1126/science.1357752. [DOI] [PubMed] [Google Scholar]
- 26.Paterson DJ, Williams AF. An intermediate cell in thymocyte differentiation that expresses CD8 but not CD4 antigen. J Exp Med. 1987;166:1603–1608. doi: 10.1084/jem.166.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacDonald HR, Budd RC, Howe RC. A CD3− subset of CD4−8+ thymocytes: a rapidly cycling intermediate in the generation of CD4−CD8+cells. Eur J Immunol. 1988;18:519–523. doi: 10.1002/eji.1830180405. [DOI] [PubMed] [Google Scholar]
- 28.Shortman K, Wilson A, Egerton M, Pearse M, Scollay R. Immature CD4− CD8+murine thymocytes. Cell Immunol. 1988;113:462–479. doi: 10.1016/0008-8749(88)90042-1. [DOI] [PubMed] [Google Scholar]
- 29.Marrack P, Kappler J. The Staphylococcalenterotoxins and their relatives. Science (Wash DC) 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 30.Swan, K.A., J. Alberola-Ila, J.A. Gross, M.W. Appleby, K.A. Forbush, J.F. Thomas, and R.M. Perlmutter. Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO (Eur. Mol. Biol. Organ.) J. 14:276–285. [DOI] [PMC free article] [PubMed]
- 31.Alberola-Ila J, Hogquist K, Swan KA, Bevan MJ, Perlmutter RM. Positive and negative selection invoke distinct signaling pathways. J Exp Med. 1996;184:9–18. doi: 10.1084/jem.184.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NRJ. T cell receptor affinity and thymocyte positive selection. Nature (Lond) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]