Abstract
The nucleoporins Nup60p, Nup2p, and Nup1p form part of the nuclear basket structure of the Saccharomyces cerevisiae nuclear pore complex (NPC). Here, we show that these necleoporins can be isolated from yeast extracts by affinity chromatography on karyopherin Kap95p-coated beads. To characterize Nup60p further, Nup60p-coated beads were used to capture its interacting proteins from extracts. We find that Nup60p binds to Nup2p and serves as a docking site for Kap95p–Kap60p heterodimers and Kap123p. Nup60p also binds Gsp1p–GTP and its guanine nucleotide exchange factor Prp20p, and functions as a Gsp1p guanine nucleotide dissociation inhibitor by reducing the activity of Prp20p. Yeast lacking Nup60p exhibit minor defects in nuclear export of Kap60p, nuclear import of Kap95p–Kap60p-dependent cargoes, and diffusion of small proteins across the NPC. Yeast lacking Nup60p also fail to anchor Nup2p at the NPC, resulting in the mislocalization of Nup2p to the nucleoplasm and cytoplasm. Purified Nup60p and Nup2p bind each other directly, but the stability of the complex is compromised when Kap60p binds Nup2p. Gsp1p–GTP enhances by 10-fold the affinity between Nup60p and Nup2p, and restores binding of Nup2p–Kap60p complexes to Nup60p. The results suggest a dynamic interaction, controlled by the nucleoplasmic concentration of Gsp1p–GTP, between Nup60p and Nup2p at the NPC.
Keywords: nucleoporin; karyopherin; nuclear pore complex; nuclear import; nuclear export
Introduction
The translocation of macromolecules between the nucleus and cytosol of eukaryotic cells occurs through the nuclear pore complex (NPC)* and is facilitated by karyopherins (Kaps, importins, exportins, or transportins) (Gorlich and Kutay, 1999). Members of the karyopherin β family of proteins bind the nuclear import or export signals of molecules that need transport (herein referred to as cargo) and accomplish deposition of their cargo at the opposite side of the NPC via interactions with multiple proteins of the NPC (nucleoporins [Nups]). Nups that contain FG peptide repeats (FG Nups) are proposed to function as binding sites for karyopherins within the NPC, and translocation may proceed by the repeated association and dissociation of karyopherins with FG Nups strategically located throughout the NPC (Radu et al., 1995; Rexach and Blobel, 1995). Despite this and other models that attempt to explain the translocation process, the actual mechanisms of karyopherin movement within the NPC remain a mystery.
In Saccharomyces cerevisiae, 30 distinct Nups interact in multiple copies to form an NPC (Rout et al., 2000). Yeast NPCs are shaped as an octagonal ring measuring 95-nm in diameter and 35-nm in depth (Yang et al., 1998). A subset of Nups form filaments that extend into the cytoplasm and a different subset form a basket structure that extends into the nucleoplasm. All other Nups are distributed symmetrically about a central axis in the NPC (Rout et al., 2000), forming a ringed scaffold with spokes and a central transporter that serves as the conduit for macromolecular transport (Yang et al., 1998). The nuclear basket structure and the cytoplasmic filaments of the NPC contain Nups that are proposed to function in the initiation and termination of karyopherin-mediated transport reactions (Shah and Forbes, 1998; Kehlenbach et al., 1999; Ullman et al., 1999; Yaseen and Blobel, 1999).
Nup60p, Nup2p, and Nup1p form part of the nuclear basket structure of the yeast NPC. Nup60p and Nup1p are the only two Nups in S. cerevisiae that localize exclusively to the nucleoplasmic side of the NPC (Rout et al., 2000). A third Nup, Nup2p, is tethered to the nucleoplasmic side (Hood et al., 2000; Solsbacher et al., 2000), but is also present in the nucleoplasm (Loeb et al., 1993). Nup60p was identified recently (Rout et al., 2000) and there is little information about its function. Nup2p and Nup1p are well characterized and share redundant functions, as nup2Δ and some nup1Δ yeast strains are viable but nup2Δ nup1Δ double-knockout strains are nonviable (Loeb et al., 1993). Nup2p and Nup1p contain 16 and 23 FxFG peptide repeats, respectively (Davis and Fink, 1990; Loeb et al., 1993), which serve as docking sites for Kap95p–Kap60p heterodimers (karyopherin αβ or importin αβ) in vitro (Belanger et al., 1994; Rexach and Blobel, 1995; Solsbacher et al., 2000; Allen et al., 2001). Nup2p and Nup1p also interact with other karyopherin βs, such as Msn5p, Nmd5p, Kap121p, and Kap122p (Ryan and Wente, 2000). In addition to being docking sites for karyopherin βs, Nup2p and Nup1p also bind Kap60p (Srp1p) monomers directly (Floer et al., 1997; Booth et al., 1999; Solsbacher et al., 2000) and facilitate Cse1p-dependent export of Kap60p from the nucleus (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000). Nup2p may also have an ancillary role in nuclear export of tRNA as it interacts with Los1p, the exportin for tRNA (Hellmuth et al., 1998). Finally, Nup2p contains a COOH-terminal domain that binds Gsp1p, the yeast homologue of the Ran GTPase (Dingwall et al., 1995).
Gsp1p is a ras-like guanine nucleotide binding protein that governs the interaction of karyopherin βs (importins and exportins) with their cargoes and Nups (Nakielny and Dreyfuss, 1999; Allen et al., 2001). Upon binding importins, Gsp1p–GTP disrupts their interaction with cargo and Nups, thereby terminating import reactions. Conversely, Gsp1p–GTP binding to exportins enhances their binding to cargo and Nups, serving to initiate export reactions. Not surprisingly, maintenance of a Gsp1p–GTP gradient across the nuclear envelope is crucial for nuclear transport reactions and cell survival. The sequestration of Prp20p (a Gsp1p guanine nucleotide exchange factor [GEF]) and Rna1p (a Gsp1p GTPase activating protein [GAP]) to the yeast nucleoplasm and cytoplasm, respectively, likely generates a Gsp1p–GTP gradient where the concentration of Gsp1p–GTP is high in the nucleus and low in the cytoplasm.
Here we characterize the function of Nup60p: (a) as a Gsp1p–GTP-sensitive tether for Nup2p at the NPC; (b) as a regulator of Prp20p function; (c) as a novel Gsp1p binding protein; (d) as a docking site for Kap123p and Kap95p–Kap60p heterodimers; and (e) as a platform for the assembly and departure of Kap60p–Cse1p–Gsp1p–GTP export complexes from the nucleus. Most interestingly, we find that Gsp1p–GTP enhances the interaction between Nup60p and Nup2p 10-fold, possibly forming a bridge between the two Nups. Molecular mechanisms are described for the dynamic assembly and disassembly of Nup60p–Nup2p complexes and the in vivo role of Nup60p in nuclear import and export reactions.
Results
Identification of Kap95p binding proteins
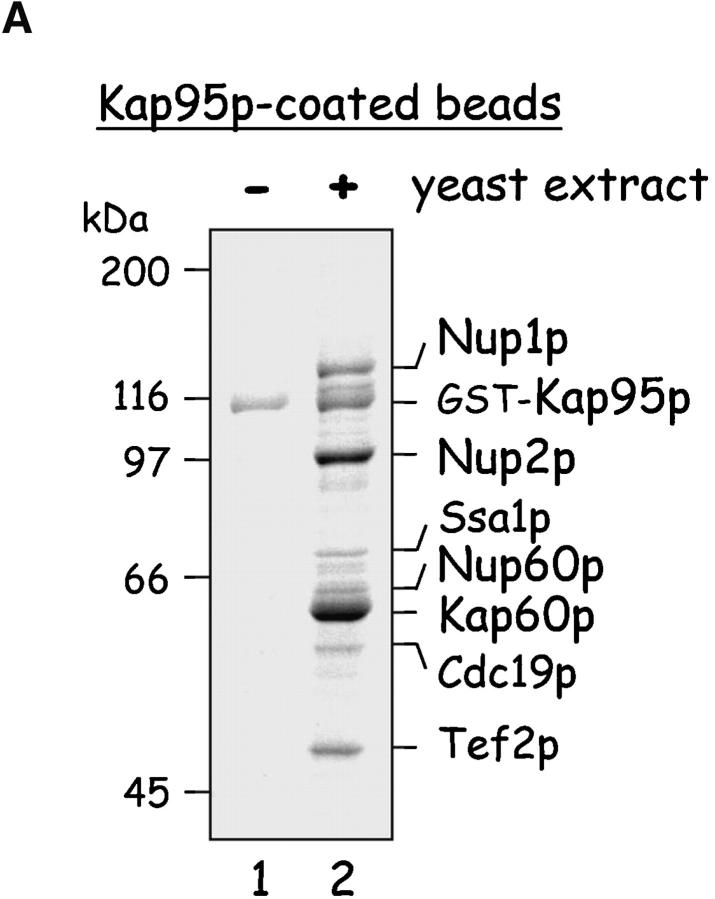
As part of our ongoing effort to characterize the mechanism of Kap95p–Kap60p-mediated transport across the NPC, we isolated and identified Kap95p (karyopherin/importin β) binding proteins from yeast extracts. Kap95p is the yeast homologue of vertebrate karyopherin β1/importin β (Enenkel et al., 1995). Glutathione S-transferase (GST)–Kap95p was immobilized on glutathione-coated Sepharose beads (beads) and added to yeast extracts to capture interacting proteins. After washing the beads, bound proteins were eluted with 250 mM MgCl2, resolved by SDS-PAGE, and identified using mass spectrometry (MALDI-TOF and LC-MS). The most abundant proteins bound to Kap95p were identified as Nup1p, Nup2p, and Kap60p (Fig. 1 A); this was not surprising as these proteins bind Kap95p directly (Rexach and Blobel, 1995). The less abundant proteins bound were Ssa1p, Cdc19p, and Tef2p. Ssa1p plays a stimulatory role in Kap60p-dependent import reactions (Shulga et al., 1996), so its association may be specific. Cdc19p (pyruvate kinase) and Tef2p (translation elongation factor) are possible contaminants due to their high abundance in the yeast cytoplasm. Notably, Nup60p is also among the less abundant proteins that bind Kap95p, but its association is specific as it binds Kap95p monomers directly (see Figs. 5 A and 6 B). All of the captured Nups (Nup60p, Nup1p, and Nup2p) are components of the nuclear basket structure of the yeast NPC (Hood et al., 2000; Rout et al., 2000). Although much is known about Nup1p and Nup2p and their functions (see Introduction), the recently identified Nup60p is less well characterized.
Figure 1.
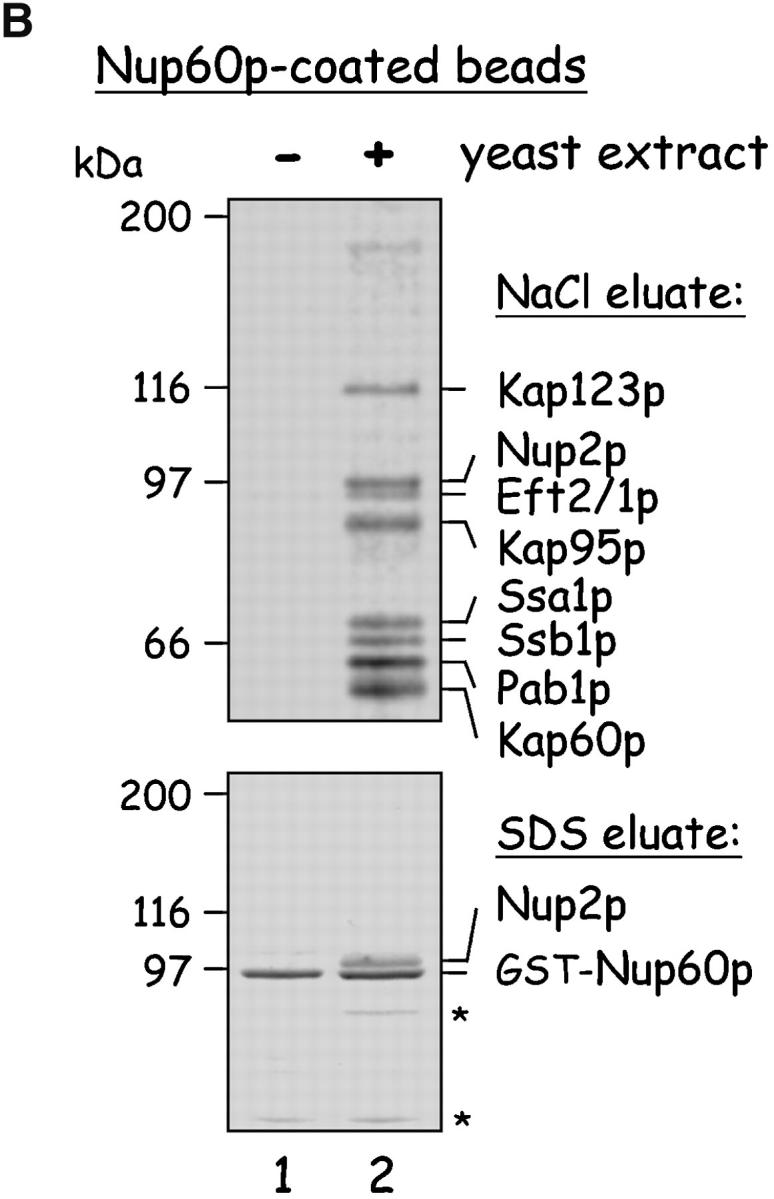
Yeast proteins that bind Nup60p and Kap95p. (A) Proteins in yeast extracts captured on Kap95p-coated Sepharose beads. GST-Kap95p (5 μg) was immobilized on glutathione-coated Sepharose beads (beads) and incubated with yeast extract (10 mg protein) or buffer as indicated. After washing beads, bound proteins were eluted with 250 mM MgCl2, collected by precipitation with trichloroacetic acid and deoxycholate, resolved by SDS-PAGE, and stained with Coomassie blue. Visible proteins were identified by mass spectrometry (see Materials and methods). Note that the three Nups captured by Kap95p are components of the nuclear basket structure of the yeast NPC. (B) Proteins in yeast extracts captured on Nup60p-coated beads. GST-Nup60p (1 μg) was immobilized on the beads and incubated with yeast extract (10 mg protein) or buffer as before. Bound proteins were eluted with 1 M NaCl (top) followed by SDS (bottom) and were identified as before. The asterisks mark a degradation product of GST–Nup60p. The identity of the ∼180-kD protein in the top panel could not be determined. Note that a portion of Nup2p remained tightly bound to Nup60p-coated beads even after incubation in 1 M NaCl.
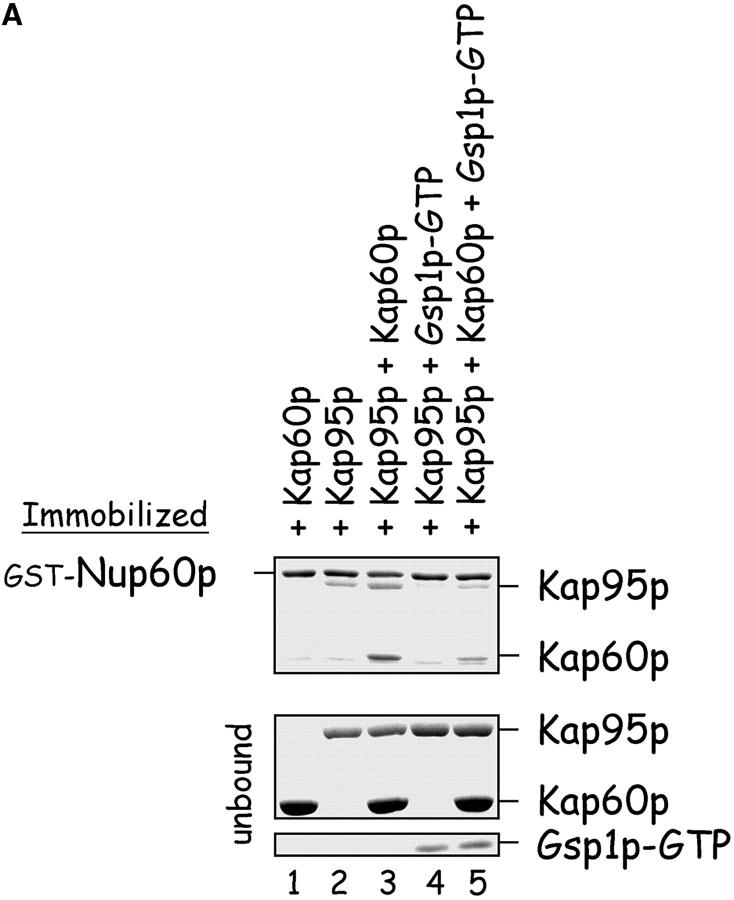
Figure 5.
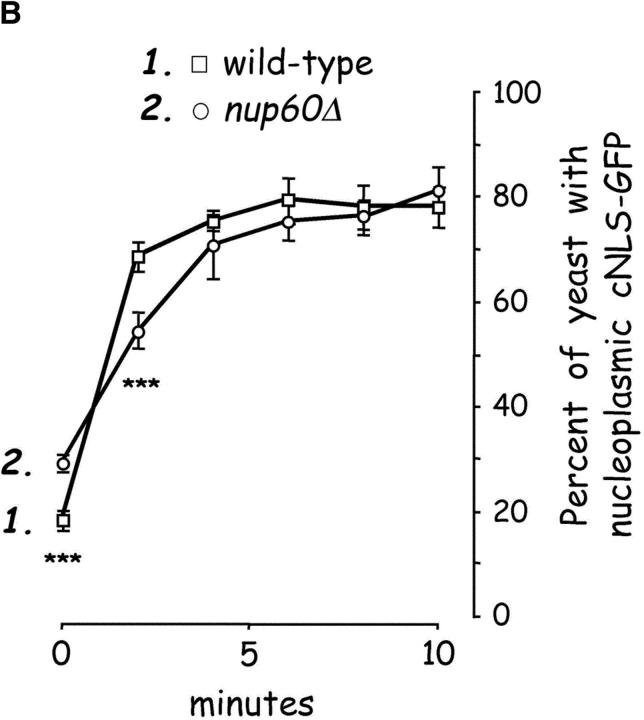
Nup60p is a docking site for Kap95p–Kap60p heterodimers. (A) Nup60p binds Kap95p, but only in the absence of Gsp1p–GTP. GST–Nup60p (1 μg) was immobilized on beads and incubated with Kap95p (2 μg), Kap60p (3 μg), and/or Gsp1p–GTP Q71L (0.5 μg). After 1 h at 4°C, unbound and bound proteins were collected, resolved by SDS-PAGE, and stained with Coomassie blue. Note that Kap95p monomers bind to Nup60p, that Kap60p enhances binding of Kap95p to Nup60p, and that Gsp1p–GTP blocks binding of Kap95p and Kap95p–Kap60p heterodimers to Nup60p. (B) Nup60p plays a minor role in Kap95p–Kap60p-dependent import of cNLS-bearing cargo into the nucleus. Wild-type and nup60Δ yeast expressing the SV-40 T-antigen NLS fused to GFP were metabolically poisoned to deplete intracellular ATP and assayed for recovery of nuclear import upon removal of the poison (see Materials and methods). Values plotted at the indicated time points represent the mean fraction of yeast with predominantly nucleoplasmic NLS-GFP from six separate experiments (error bars represent SEM). The asterisks (***) indicate P < 0.01 for comparison of the two values at the indicated time points (unpaired, two-tailed t test). Note that yeast lacking Nup60p exhibit a slower initial rate of cNLS–GFP nuclear import.
Identification of Nup60p-interacting proteins
To characterize Nup60p and its function, we first identified Nup60p-interacting proteins. GST–Nup60p was immobilized on beads and incubated with yeast extracts to capture interacting proteins. After washing the beads, bound proteins were eluted with 1 M NaCl, resolved by SDS-PAGE, and identified using mass spectrometry. Nup60p captured three karyopherins (Kap95p–Kap60p and Kap123p), one Nup (Nup2p), two heat shock proteins (Ssa1p and Ssb1p), and the mRNA binding protein Pab1p (Fig. 1 B, top). Interestingly, some Nup2p remained bound to the Nup60p beads even after salt extraction of all other proteins (Fig. 1 B, bottom); this suggests a strong and direct interaction between Nup60p and Nup2p.
Nup60p functions as a major anchoring site for Nup2p at the NPC
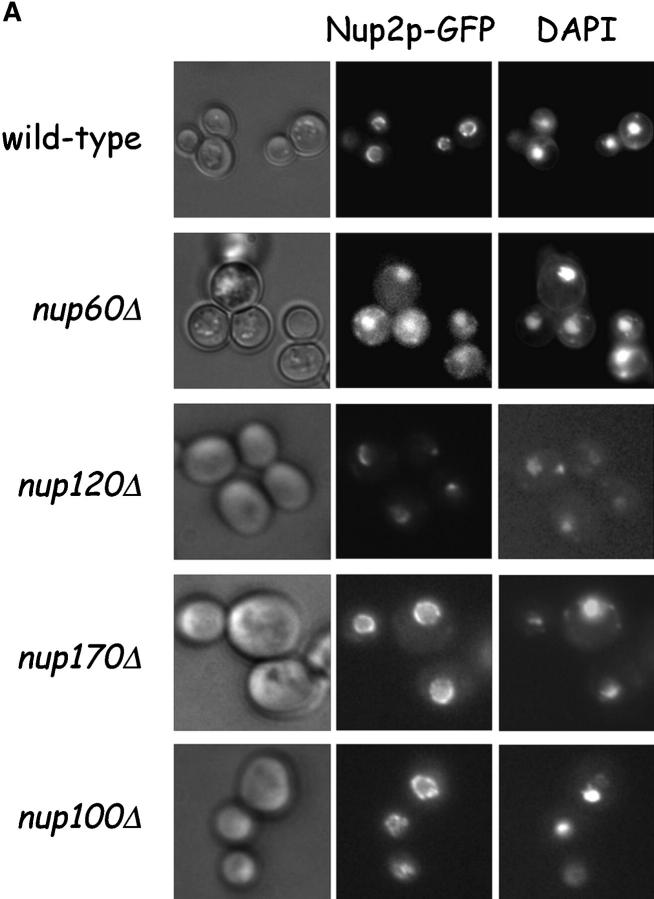
To test the proposed interaction between Nup60p and Nup2p in vivo, NUP2 was replaced with NUP2-GFP in wild-type yeast and yeast lacking Nup60p (nup60Δ). The Nup2p–GFP fusion was visualized by fluorescence microscopy. As expected for wild-type yeast, Nup2p–GFP localizes to the nuclear envelope in a punctate pattern typical of Nups (Fig. 2 A, top). In contrast, Nup2p–GFP clearly mislocalizes in nup60Δ yeast, and is observed primarily in the nucleoplasm and partially in the cytoplasm (Fig. 2 A, middle). A small fraction of Nup2p–GFP may have remained bound to the NPC in the nup60Δ yeast; however, as the majority of Nup2p–GFP fails to associate with the nuclear envelope in nup60Δ yeast, we conclude that Nup60p is the primary tethering site for Nup2p at the NPC (also see below).
Figure 2.
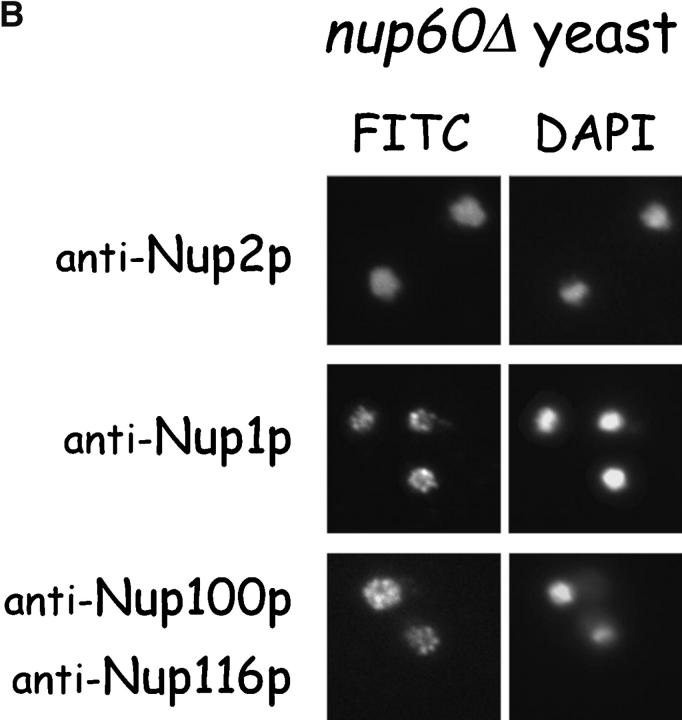
Nup2p is tethered to the NPC via Nup60p. (A) Direct visualization of Nup2p–GFP fusions in yeast. Various yeast strains that express NUP2-GFP from the NUP2 locus were grown in rich media at 30°C and were observed live under a fluorescence microscope. DAPI stain was used to visualize DNA in nuclei and pictures were taken using nuclei as the focal point. Note that in wild-type, nup170Δ, and nup100Δ yeast, Nup2p–GFP fusions accumulate in a punctate pattern at the nuclear periphery, but in nup60Δ yeast Nup2p–GFP is mislocalized to the nucleoplasm and cytoplasm. (B) Indirect immunofluorescence visualization of Nup2p, Nup1p, and Nup100p/Nup116p in nup60Δ yeast. nup60Δ yeast grown to early log phase in rich media at 30°C were fixed in 3.7% formaldehyde for 10 min and processed for immunofluorescence microscopy using affinity-purified anti-Nup antibodies and FITC-labeled secondary antibodies (left). DAPI was used to visualize nuclei (right). Note the mislocalization of Nup2p to the nucleoplasm in nup60Δ yeast in contrast to the normal punctate staining of Nup1p and Nup100p/Nup116p at the nuclear envelope.
It is possible that the absence of Nup60p from the NPC causes specific defects in the nuclear basket structure leading indirectly to the observed mislocalization of Nup2p. This scenario is unlikely as the NPC localization of Nup1p (another nuclear basket component) remains unaltered in nup60Δ yeast as visualized by indirect immunofluorescence (Fig. 2 B, middle). In additional control experiments, indirect immunofluorescence of nup60Δ yeast shows normal localization of Nup100p and Nup116p at the NPC (Fig. 2 B, bottom) and Nup2p mislocalization to the nucleoplasm (top) as before. The short fixation times used to prepare yeast for immunofluorescence microscopy precludes visualization of mislocalized Nup2p in the cytoplasm (Fig. 2 B, top), as is often observed in living nup60Δ yeast (Fig. 2 A)
It is also possible that the absence of Nup60p could cause general structural defects in the NPC, leading indirectly to the observed mislocalization of Nup2p. To test this possibility, Nup2p–GFP fusions were visualized in yeast lacking Nup100p (nup100Δ), Nup170p (nup170Δ), or Nup120p (nup120Δ). These Nups are located at various positions within the NPC and the corresponding deletion strains are viable, but display defects in NPC structure and distribution. In nup170Δ yeast, NPCs cannot maintain a normal stoichiometry of some FG Nups (Kenna et al., 1996) or a normal permeability barrier (Shulga et al., 2000), whereas in nup120Δ yeast, NPCs cluster at one side of the nucleus (Li et al., 1995; Pemberton et al., 1995). As Nup100p localizes near Nup2p in the nucleoplasmic side of the NPC (Hood et al., 2000; Rout et al., 2000; Solsbacher et al., 2000), the location of Nup2p–GFP was also examined in nup100Δ yeast. We find that the nuclear envelope localization of Nup2p–GFP is not affected by the absence of Nup100p or Nup170p (Fig. 2 A, bottom). In nup120Δ strains, Nup2p–GFP fluorescence appears clustered as expected (Fig. 2 A, middle). We conclude that the observed mislocalization of Nup2p in nup60Δ yeast is caused specifically by the absence of Nup60p.
Nup60p binds directly to Nup2p
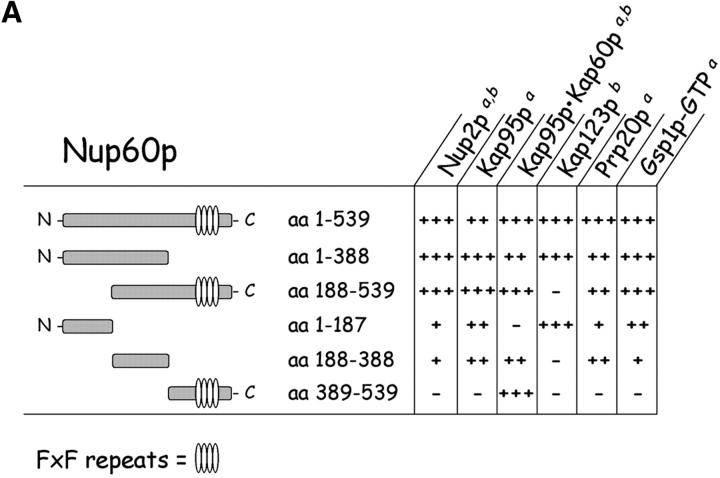
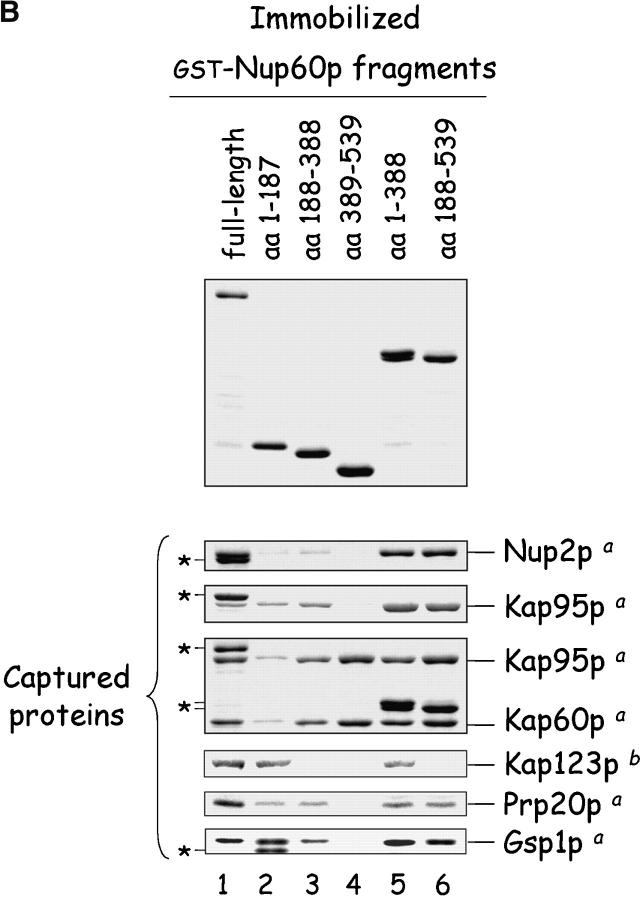
The cellular mislocalization of Nup2p in yeast lacking Nup60p (Fig. 2) and the fact that Nup2p remains bound to Nup60p even in 1 M NaCl (Fig. 1 B, bottom) imply a direct association of Nup2p and Nup60p at the NPC. To test for a direct interaction, immobilized GST–Nup60p was incubated with purified recombinant Nup2p in solution. Nup2p binds tightly to Nup60p (KD ∼396 nM) in the absence of other proteins (Fig. 3, A and D) . To map the region of Nup60p that binds Nup2p, several Nup60p fragments were expressed as GST fusions and were incubated with recombinant Nup2p. As illustrated in Fig. 6 , Nup2p binds weakly to a central region of Nup60p (amino acids [aa] 188–388) and did not bind the NH2 terminus or COOH terminus of Nup60p alone. However, Nup60p fragments containing the middle region and the NH2 or COOH terminus bound Nup2p to the same levels as full-length Nup60p (Fig. 6 B). The same Nup60p fragments also captured Nup2p from yeast extracts with similar results (data not shown). Other Nup60p-interacting proteins bind to different regions of Nup60p; for example, Kap123p in yeast extracts binds selectively to the NH2 terminus of Nup60p (aa 1–187) and all Nup60p fragments containing the NH2 terminus (Fig. 6).
Figure 3.
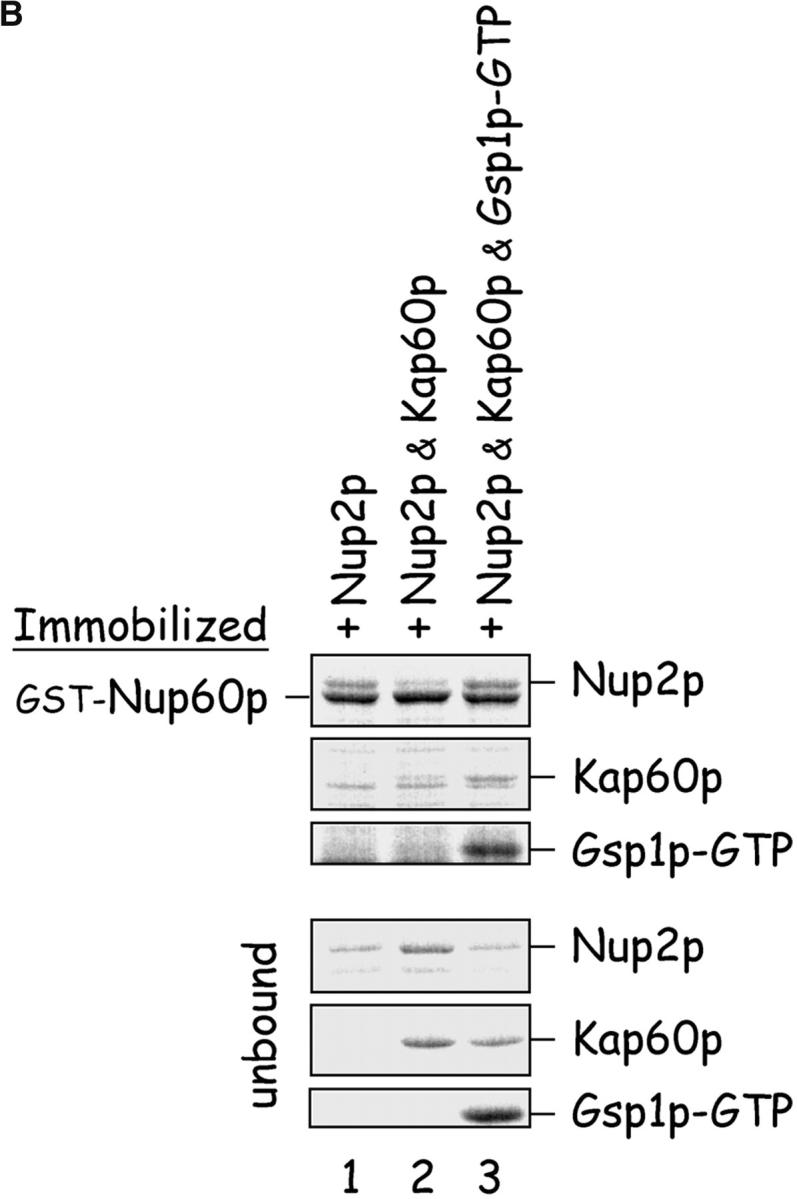
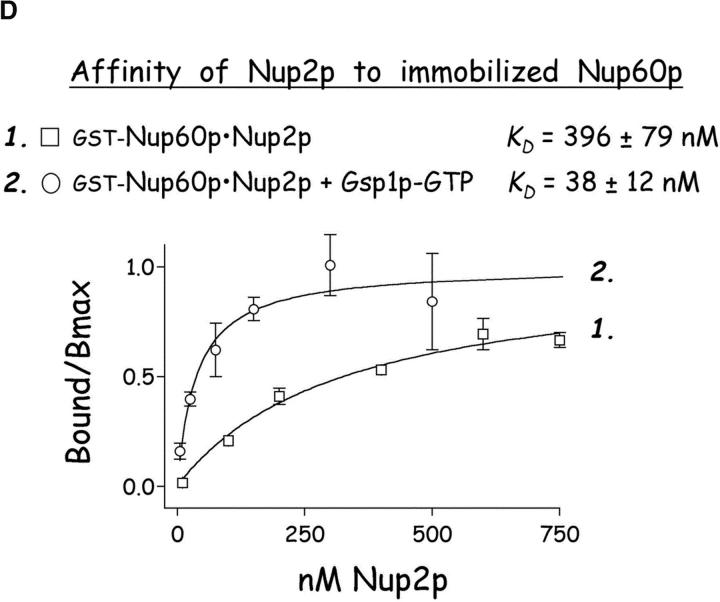
Gsp1p–GTP and Kap60p modulate the interaction between Nup60p and Nup2p. (A) The interaction between Nup2p and Nup60p, and the effect of Kap60p. GST–Nup60p (1 μg) was immobilized on beads and incubated with Nup2p (0.5 μg), Nup2pΔ (aa 1–50) (0.5 μg), or Kap60p (1 μg) as indicated. After 1 h at 4°C, unbound and bound proteins were collected, resolved by SDS-PAGE, and visualized with Coomassie blue. Note that Nup2p binds Nup60p, that Kap60p prevents the interaction, and that the NH2 terminus of Nup2p is not required for binding Nup60p. (B) Effect of Gsp1p–GTP on the interaction between Nup2p and Nup60p. GST–Nup60p (1 μg) was immobilized on beads and incubated with Nup2p (0.5 μg), Kap60p (1 μg), or Gsp1p–GTP (His-Gsp1p Q71L) (1 μg) as before. Note that Kap60p interferes with the interaction of Nup2p with Nup60p, but that the presence of Gsp1p–GTP restores binding and promotes formation of Nup60p–Gsp1p–Nup2p–Kap60p complexes. (C) Gsp1p–GTP enhances binding of Nup2p to Nup60p in yeast extracts. GST-Nup60p (1 μg) was immobilized on beads and was incubated with yeast extract (∼1 mg) supplemented with 1.25 μM recombinant Gsp1p–GTP (Q71L), 0.5 μM recombinant Kap60p, or no additional protein. The amount of Nup2p bound to Nup60p-coated beads was determined by quantitative Western blotting as described in Materials and methods. The amount of Nup2p was expressed as the ratio of Nup2p bound per unit of immobilized GST–Nup60p, using the incubation of extract without additions as baseline. Shown are the mean ratios for two samples with error bars representing the SEM; this experiment was performed three times with similar results. The asterisks (***) indicate a P < 0.05 for comparison of mean Nup2p captured from extracts supplemented or not with additional Gsp1p–GTP (unpaired, two-tailed t test). Note that addition of Gsp1p–GTP to yeast extract increases by ∼65% the amount of Nup2p bound to Nup60p-coated beads. (D) Gsp1p–GTP increases the affinity between Nup60p and Nup2p. Nup60p-coated beads were incubated with various concentrations of radiolabeled Nup2p for 2 h at 25°C in binding buffer with 10 mg/ml BSA and protease inhibitors. The concentration of GST–Nup60p within the beads was 25 nM and 150 nM for experiments with or without Gsp1p–GTP, respectively. The dissociation constant (KD) of the Nup60p–Nup2p complex in the presence and absence of 3 μM Gsp1p–GTP Q71L was calculated as described in Materials and methods. To facilitate comparison, the results were plotted as a fraction of maximal Nup2p bound versus Nup2p concentration. Each data point was performed in duplicate and error bars represent SEM. Note the 10-fold higher affinity between Nup60p and Nup2p in the presence of Gsp1p–GTP.
Figure 6.
Mapping of Nup, karyopherin, Prp20p, and Gsp1p–GTP binding sites on Nup60p. (A) The cartoon depicts the Nup60p fragments used in this study provides a summary of the binding interactions observed in B. The + and − designations provide a qualitative assessment of the binding avidity. The absence of detectable binding is denoted by −, whereas +, ++, and +++ represent relative degrees of binding. (B) Binding of karyopherins, Nup2p, Prp20p, and Gsp1p–GTP to various Nup60p fragments. Each GST–Nup60p fragment (1 μg each) was immobilized on beads and incubated with purified Nup2p, Kap95p, Kap95p–Kap60p heterodimers, Prp20p, or Gsp1p–GTP (1 μg each), as indicated. After 1 h at 4°C, the beads were washed and bound proteins were collected, resolved by SDS-PAGE, and visualized with Coomassie blue stain. Alternatively, each GST–Nup60p fragment (5 μg each) was immobilized on beads and incubated with 10 mg of yeast extract for 2 h at 4°C. After washing the beads, bound proteins were eluted with 1 M NaCl, collected by precipitation with trichloroacetic acid and deoxycholate, resolved by SDS-PAGE, and stained with Coomassie blue. Asterisks designate GST–Nup60p fragments used as bait. The superscripts “a” and “b” denote the source of proteins used in the experiments: “a” marks cases where purified recombinant proteins where used, and “b” marks cases where yeast extracts were used. Note the shift in binding site selection of Kap95p in the presence and absence of Kap60p.
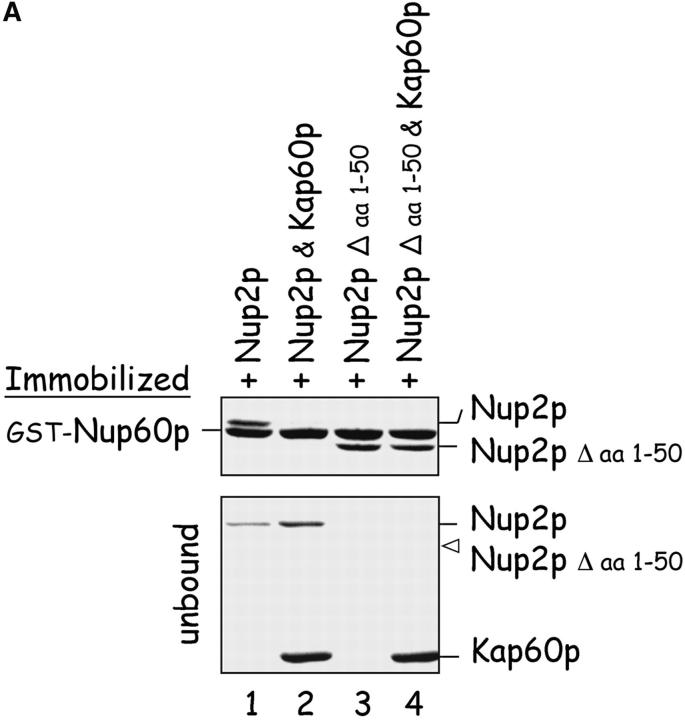
Kap60p modulates the interaction between Nup60p and Nup2p
The NH2 terminus of Nup2p binds Kap60p with high affinity (KD ∼0.2 nM; data not shown) and may exert an effect on the Nup60p–Nup2p interaction. To test that possibility, immobilized Nup60p was incubated with Nup2p in the presence or absence of Kap60p. We find that Kap60p prevents the association of Nup60p and Nup2p (Fig. 3 A, lane 2); this effect is due to Kap60p binding to Nup2p because Nup60p does not bind Kap60p monomers directly (Fig. 5 A, lane 1). Nup2p missing the NH2-terminal 50 amino acids (Nup2pΔaa 1–50) cannot bind Kap60p monomers (data not shown), but binds tightly to Nup60p (Fig. 3 A, lane 3), even in the presence of Kap60p (lane 4). This demonstrates that Nup2p has different binding sites for Nup60p and Kap60p. Notably, Nup60p binds Nup2pΔ(aa 1–50) better than full length Nup2p, as judged by the lack of unbound Nup2pΔ(aa 1–50) (Fig. 3 A, bottom, lanes 3 and 4) in comparison to Nup2p in the same assay (lanes 1 and 2). This suggests a role for the NH2 terminus of Nup2p in weakening the interaction between Nup60p and Nup2p. Kap95p monomers also bind the NH2 terminus of Nup60p (Fig. 6) and interfere with the association between Nup60p and Nup2p in the absence of additional proteins (data not shown).
Gsp1p–GTP strengthens the interaction between Nup60p and Nup2p
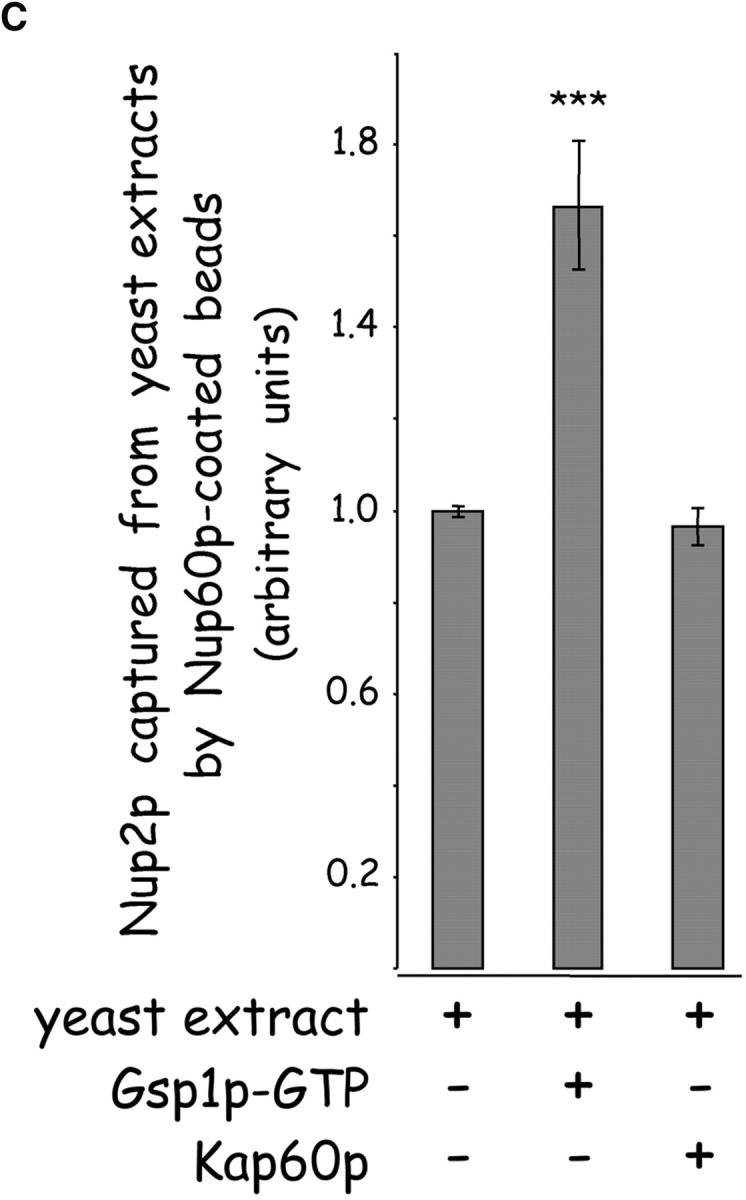
Nup2p contains a Gsp1p binding domain in its COOH terminus (Dingwall et al., 1995) that may exert an effect on the interaction between Nup2p and Nup60p. Therefore, immobilized Nup60p was incubated with purified Nup2p in the presence or absence of Gsp1p–GTP. We find that Gsp1p–GTP strengthens the affinity of interaction between Nup2p and Nup60p by 10-fold (from KD ∼396 nM to KD ∼38 nM) (Fig. 3 D), whereas Gsp1p–GDP has no effect (data not shown). Gsp1p–GTP also restores binding of Nup60p and Nup2p in the presence of Kap60p, forming Nup60p–Gsp1p–Nup2p–Kap60p complexes (Fig. 3 B, lanes 2 and 3). To test whether Gsp1p–GTP can modulate the stability of the Nup60p–Nup2p complex in the milieu of yeast extracts, Nup60p-coated beads were mixed with an extract supplemented (or not) with 0.5 μM Kap60p or 1.25 μM Gsp1p–GTP (Gsp1p–GTP Q71L). The amount of Nup2p bound was quantified via Western blotting using affinity-purified anti-Nup2p antibodies and 125I-protein A followed by quantitative phosphor imaging. We find that Gsp1p–GTP increases the yield of Nup2p bound to Nup60p by 65% when compared with incubations without additions (Fig. 3 C; P < 0.05). The addition of 0.5 μM Kap60p has no significant effect on the levels of Nup2p bound to Nup60p; the high abundance of Kap60p in yeast extracts may preclude observation of an effect by exogenously added Kap60p (Fig. 3 C). In contrast, the concentration of Gsp1p–GTP in the extracts appears to be limiting for the stabilization of Nup2p–Nup60p complexes. In vivo, depletion of Gsp1p–GTP may signal the release of Nup2p from its Nup60p anchor at the NPC.
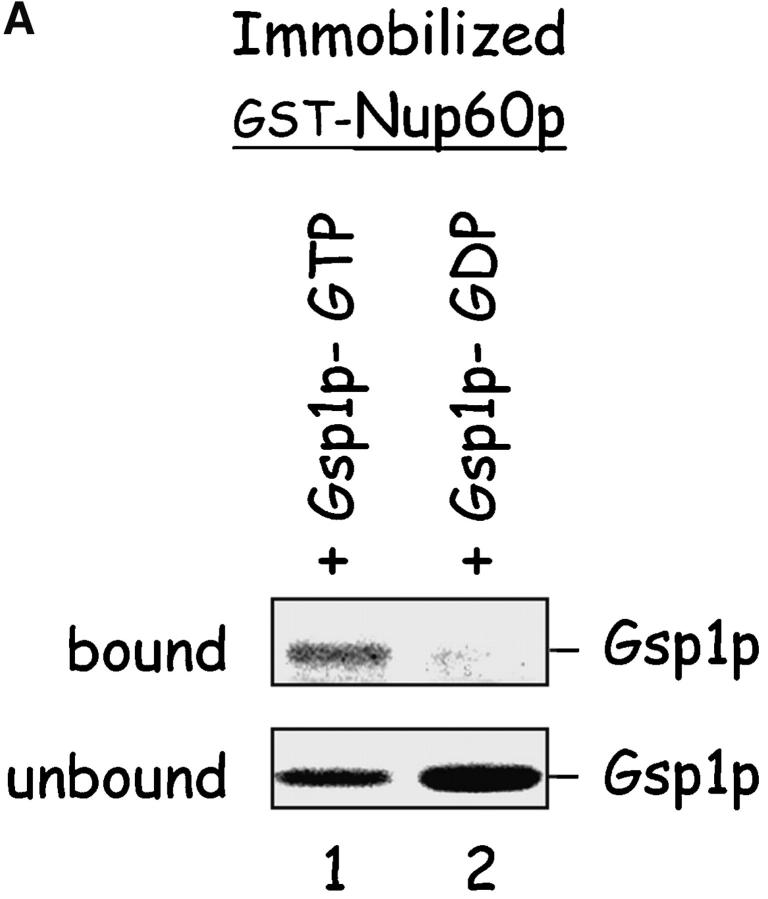
Nup60p is a Gsp1p–GTP binding protein
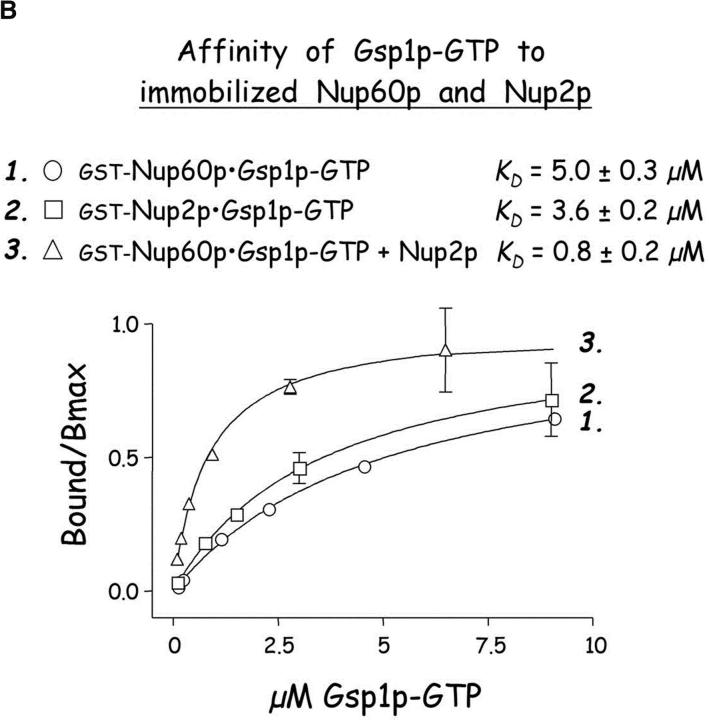
An unexpectedly large amount of Gsp1p–GTP bound to Nup60p-coated beads containing substoichiometric amounts of Nup2p (Fig. 3 B, lane 3), more than could be explained by Gsp1p–GTP binding to Nup2p alone. To test whether Nup60p binds Gsp1p directly, Nup60p-coated beads were incubated with purified Gsp1p–GTP or Gsp1p–GDP. Surprisingly, Gsp1p–GTP (but not Gsp1p–GDP) binds to Nup60p in the absence of additional proteins (Fig. 4 A). We calculated the affinity of Gsp1p–GTP towards Nup60p as KD ∼5 μM, a value that is similar to the calculated affinity of Gsp1p–GTP towards Nup2p (KD ∼3.6 μM) (Fig. 4 B). These low affinities are physiologically relevant, as we estimate the concentration of Gsp1p–GTP in yeast nuclei to be ∼1–10 μM based on the reported number of Gsp1p molecules per cell (26,300) (Gygi et al., 1999) and assuming that most Gsp1p is concentrated in the nucleoplasm in the GTP-bound form. Therefore, it is possible that Nup60p and Nup2p “sense” minor fluctuations in the concentration of Gsp1p–GTP at the nuclear basket structure of the NPC.
Figure 4.
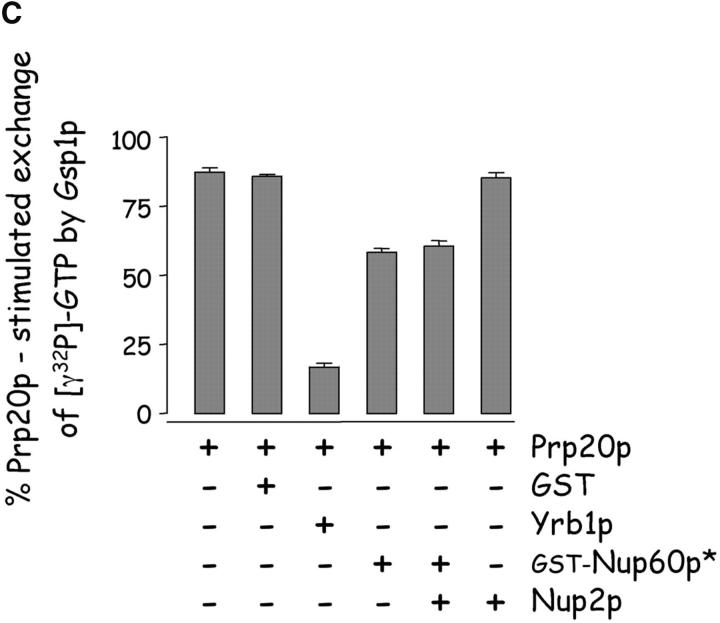
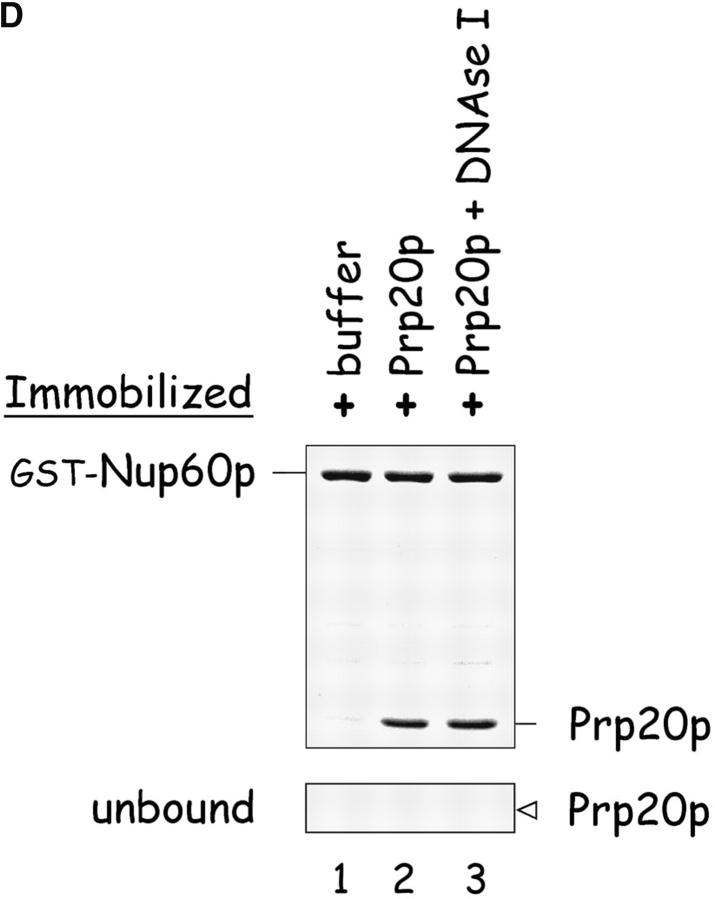
Nup60p binds Gsp1p–GTP and Prp20p, and functions as a Gsp1p GDI. (A) Nup60p binds Gsp1p–GTP. GST-Nup60p (1 μg) was immobilized on beads and incubated with His-Gsp1p (2 μg) preloaded with GTP or GDP. After 1 h at 4°C, bound and unbound proteins were resolved by SDS-PAGE and visualized with Coomassie blue. Note that Nup60p binds Gsp1p–GTP, but not Gsp1p–GDP. (B) Affinity of Gsp1p–GTP to Nup60p, Nup2p, and Nup60p–Nup2p complexes. GST–Nup-coated beads were incubated with various concentrations of His-Gsp1p–[γ-32P]GTP for 2 h at 4°C in binding buffer with 10 mg/ml BSA and protease inhibitors. The concentrations of GST–Nup60p and GST–Nup2p within beads were 800 nM and 1.5 μM, respectively. The dissociation constants (KD) of the Nup2p–Gsp1p–GTP complex and the Nup60p–Gsp1p–GTP complex in the presence and absence of 500 nM Nup2p were calculated as described in Materials and methods. Results were plotted as a fraction of maximal Gsp1p–GTP bound versus Gsp1p–GTP concentration. Each data point was performed in duplicate and the error bars represent SEM. Note that Nup60p and Nup2p cooperate to bind Gsp1p–GTP. (C) Nup60p inhibits the Prp20p-stimulated release of GTP from Gsp1p. His-Gsp1p–[γ-32P]GTP immobilized on nickel-coated agarose beads (15 nM Gsp1p–GTP within the beads) was incubated with 0.9 nM Prp20p and 1 mM GDP, plus 4 μM GST–Nup60p (aa 188–539), Yrb1p, Nup2p, Kap95p, or GST. GST–Nup60p (aa 188–539) (indicated by asterisk) was used instead of full-length Nup60p due to its superior solubility and protease resistance. After 10 min, Prp20p activity was stopped with ice-cold buffer, beads were washed, and the [γ-32P]GTP that remained bound to the beads was quantified by scintillation counting. Each data point was performed in duplicate and error bars represent SEM. Note that Nup60p reduces (but does not abolish) the activity of Prp20p. (D) Nup60p binds Prp20p. GST–Nup60p (1 μg) was immobilized on beads and incubated with purified Prp20p (1 μg) in the presence or absence of DNAse I and RNAse I (1 U and 1 μg, respectively). After 1 h at 4°C, unbound and bound proteins were resolved by SDS-PAGE and visualized with Coomassie blue staining. Note that purified Prp20p binds Nup60p.
The Gsp1p binding domain of Nup60p was mapped using purified Gsp1p–GTP and various fragments of Nup60p. Gsp1p–GTP binds to the NH2 terminus and middle regions of Nup60p, but not to the COOH terminus (Fig. 6). Sequence analysis of Nup60p shows no significant homology to typical Gsp1p binding domains similar to those in karyopherins, Yrb1p, or Nups (Dingwall et al., 1995; Gorlich et al., 1997). Nup60p exhibits no activity as a Gsp1p GAP or GEF (data not shown), does not protect Gsp1p–GTP from Rna1p-mediated hydrolysis in a manner similar to karyopherin βs (data not shown), nor does it stimulate Rna1p (Gsp1p GAP) activity. We conclude that Gsp1p–GTP binding to Nup60p serves the main purpose of stabilizing the interaction between Nup60p and Nup2p, perhaps by forming a bridge between the two proteins. Indeed, Gsp1p–GTP exhibits an affinity towards the Nup60p–Nup2p complex (KD ∼0.8 μM) that is fivefold stronger than its affinity towards Nup60p (∼5 μM) or Nup2p alone (∼ 3.6 μM) (Fig. 4 B). The data suggests a novel function for Gsp1p–GTP as a bridging factor between two Nups.
Nup60p binds Prp20p and exhibits Gsp1p GDI activity
Although Nup60p itself is not a Gsp1p GAP or GEF (data not shown), we observed that Nup60p inhibits the Prp20p-stimulated exchange of GTP bound to Gsp1p (Fig. 4 C), thus Nup60p functions as a guanine nucleotide dissociation inhibitor (GDI) for Gsp1p. The effect is mild in comparison to the effect of Yrb1p, which is a known inhibitor of Prp20p activity (Fig. 4 C). Interestingly, Nup60p reduces the activity of Prp20p but cannot abolish it, even at micromolar concentrations. Nup60p may inhibit Prp20p activity by sequestering Gsp1p–GTP away from Prp20p or by interacting directly with Prp20p. The first scenario is unlikely as Nup2p increases the affinity of Gsp1p–GTP to Nup60p by fivefold (Fig. 4 B), but does not contribute towards the inhibition of Prp20p activity by Nup60p (Fig. 4 C). In support of the second scenario, we find that Nup60p binds directly to Prp20p (Fig. 4 D). As Prp20p is a DNA binding protein, the specificity of the Nup60p–Prp20p interaction was confirmed in the presence of DNAseI to ensure that contaminating DNA did not facilitate the association (Fig. 4 D). We conclude that Nup60p functions as a mild Gsp1p GDI, most likely via its interaction with Prp20p. Other examples of Nup–Prp20p interactions include Nup1p–Prp20p (Floer et al., 1997) and vertebrate NUP98–RCC1 (Fontoura et al., 2000). We also find that Nup2p binds Prp20p (data not shown), but does not function as a Gsp1p GDI (Fig. 4 C). The possible GDI activity of Nup1p and NUP98 has not been tested. Interestingly, all the Nups that bind Prp20p/RCC1 are also components of the nuclear basket structure of the NPC.
Nup60p functions in the Kap95p–Kap60p-dependent nuclear import of cNLS-bearing cargo
Kap95p is among the proteins captured by Nup60p and, conversely, Nup60p is among the proteins captured by Kap95p (Fig. 1). Thus, we tested in a solution binding assay with all purified proteins whether Nup60p and Kap95p bind each other directly. We expected they would, as Nup60p contains four degenerate FxFG peptide repeats (FxF) similar to those in Nup1p and Nup2p that are known to bind Kap95p (Rexach and Blobel, 1995; Solsbacher et al., 2000). Indeed, immobilized Nup60p binds Kap95p monomers and Kap95p–Kap60p heterodimers (Figs. 5 A and 6), but not Kap60p monomers (Fig. 5 A, lane 1). Curiously, Kap95p alone does not bind the COOH terminus of Nup60p, which contains the FxF peptide repeats (aa 389–539) (Fig. 6). In contrast, the same Nup60p fragment binds strongly to Kap95p–Kap60p heterodimers (Fig. 6), indicating that Kap60p promotes Kap95p interaction with FxFG repeats, as shown previously for Nup1p (Rexach and Blobel, 1995). Since Nup60p is located in the nucleoplasmic face of the NPC, it is probably exposed to a high concentration of Gsp1p–GTP (∼1–10 μM). Gsp1p–GTP interferes with the interaction of Nup60p and Kap95p (Fig. 5 A, lanes 4 and 5), much like it does with many other Nup–karyopherin interactions (Allen et al., 2001).
Our reconstitution experiments suggest that Nup60p functions in vivo as a docking site for Kap95p–Kap60p heterodimers during import of cargo containing classical nuclear localization signals (cNLSs). However, the contribution of Nup60p in this process must be dispensable, as yeast lacking Nup60p (nup60Δ) are viable (Rout et al., 2000). Nevertheless, Nup60p may function to increase the efficiency of Kap95p–Kap60p import reactions. We examined whether nup60Δ yeast display defects in Kap95p-dependent nuclear import by visualizing a reporter protein (cNLS–green fluorescent protein [GFP]) in isogenic wild-type and nup60Δ strains. We find that the steady-state nucleoplasmic distribution of cNLS–GFP is not different in nup60Δ yeast when compared with wild-type (data not shown). However, differences in the rate of nuclear import were observed after the reversible poisoning of yeast with sodium azide and 2-deoxyglucose (Fig. 5 B). Metabolic poisoning of yeast depletes cellular ATP and inhibits nuclear import pathways (Shulga et al., 1996). This allows the cNLS–GFP reporter protein (which is <30 kD) to diffuse out of the nucleus and equilibrate between the cytoplasm and nucleoplasm (Shulga et al., 1996). Upon removal of the poison and introduction of fresh medium, yeast resume their metabolic activity and nucleocytoplasmic transport. The relative rate of nuclear import can be estimated by counting the fraction of yeast exhibiting predominantly nuclear fluorescence at various times (Shulga et al., 1996).
We find that wild-type and nup60Δ yeast effectively import cNLS–GFP after removal of the metabolic poison, with >80% of cells exhibiting nuclear cNLS–GFP fluorescence after 10 min (Fig. 5 B). However, a small but statistically significant difference between wild-type and nup60Δ yeast was observed at the two earliest time-points. At time 0 (after the 2 h metabolic poisoning), 28% of nup60Δ yeast continue to exhibit nuclear cNLS–GFP localization compared with 18% of wild-type yeast (Fig. 5 B, 0 min). Thus, nup60Δ yeast display a small, yet reproducible, defect in diffusion of cNLS–GFP protein out of the nucleus during the poisoning phase. Although NPCs lacking Nup60p do not exhibit gross abnormalities in composition or distribution of Nups (Fig. 2 B), they may have altered permeability for diffusion of small proteins. Other Nups required for normal NPC permeability include Nup170p and Nup188p (Shulga et al., 2000). At 2 min into the recovery phase, a smaller fraction of nup60Δ yeast exhibits nuclear cNLS-GFP fluorescence compared with wild-type (Fig. 5 B; P < 0.01). The relative cNLS import rate during the first 2 min of recovery was always slower in nup60Δ yeast (13% recovered cells/min) compared with wild-type yeast (25% recovered cells/min), indicating a slower initial rate of import in nup60Δ yeast. This import deficiency may be caused by the loss of a Kap95p–Kap60p docking site in the NPC (i.e., no Nup60p) or by the mislocalization of Nup2p, since it also functions in cNLS-mediated nuclear import (Solsbacher et al., 2000). Similar experiments are being conducted to determine if Nup60p also functions in Kap123p-dependent transport.
The Nup60p–Nup2p complex promotes efficient export of Kap60p from the yeast nucleus
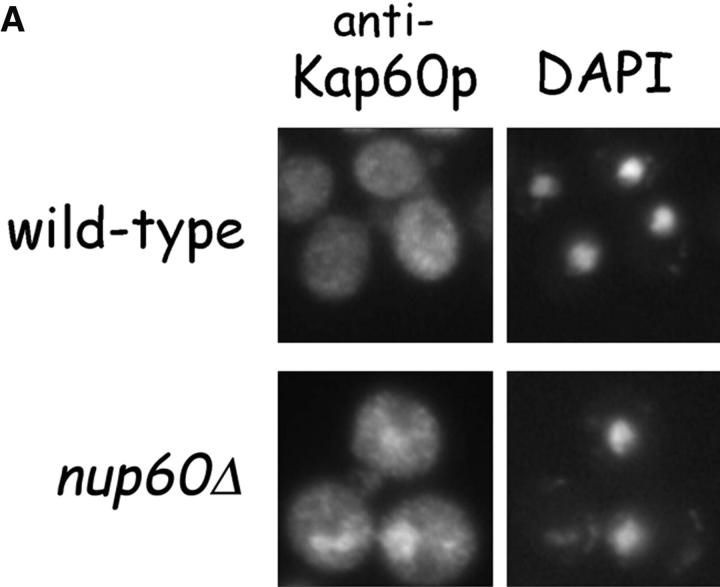
Previous studies described a role for Nup2p in the Cse1p-dependent export of Kap60p from the nucleus (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000). As Nup60p tethers Nup2p to the NPC in vivo (Fig. 2 A), it may also influence the Cse1p-mediated export of Kap60p. The subcellular location of Kap60p in wild-type and nup60Δ yeast was examined by indirect immunofluorescence using specific anti-Kap60p antibodies. We find that Kap60p partially accumulates in the nucleoplasm of nup60Δ yeast, but not wild-type yeast (Fig. 7 A). Thus, yeast lacking Nup60p exhibit a mild defect in Kap60p export, likely caused by the loss of Nup2p from the NPC.
Figure 7.
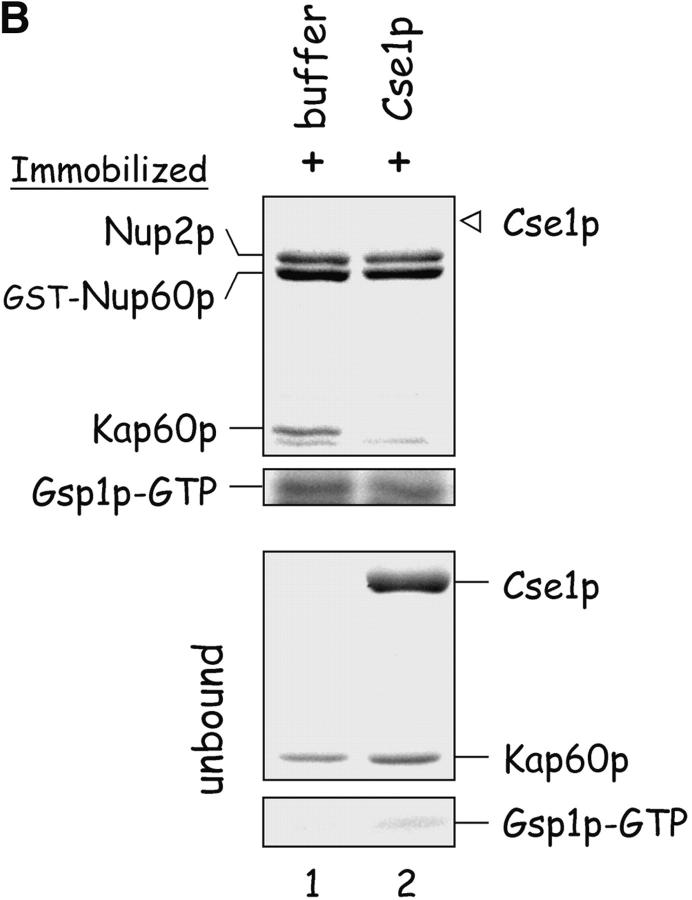
Nup60p plays a role in the nuclear export of Kap60p. (A) The location of Kap60p in wild-type and nup60Δ yeast was detected by indirect immunofluorescence using affinity-purified anti-Kap60p antibodies. Yeast grown to early log phase at 30°C in rich media were fixed in 3.7% formaldehyde for 1 h and processed for immunofluorescence microscopy (left). Note the moderate accumulation of Kap60p in nuclei of nup60Δ yeast compared with wild-type. (B) Cse1p accepts Kap60p and Gsp1p–GTP from a donor Nup2p–Gsp1p–GTP–Nup2p–Kap60p complex in vitro. GST–Nup60p (1 μg) was immobilized on beads and incubated with Nup2p (2 μg), Gsp1p–GTP (Q71L) (2 μg), and Kap60p (2 μg) for 1 h at 4°C to form the Nup2p–Gsp1p–GTP–Nup2p–Kap60p complex. After washing the beads to remove unbound proteins, the quaternary complex was mixed with buffer of Cse1p (1 μg). After 1 h at 4°C, unbound and bound proteins were collected, resolved by SDS-PAGE, and stained with Coomassie blue. Note that when Cse1p is present, all of the Kap60p and some Gsp1p are lost from the immobilized Nup60p. Also note that Cse1p does not bind to Nup60p–Nup2p complexes.
It was shown previously that Cse1p and Gsp1p–GTP cooperate to acquire Kap60p from the NH2 terminus of Nup2p (Booth et al., 1999). As Nup60p binds Gsp1p–GTP and Nup2p–Kap60p heterodimers simultaneously (Fig. 3 B), it was important to test whether the Nup60p–Gsp1p–Nup2p–Kap60p complex can donate Kap60p and Gsp1p–GTP to Cse1p. Here, the tetrameric complex was first reconstituted with purified proteins and then challenged with Cse1p or buffer alone (Fig. 7 B). Cse1p does not bind to the immobilized Nup60p–Nup2p complex; however, addition of Cse1p results in the removal all Kap60p and some Gsp1p–GTP from the complex (Fig. 7 B, lane 2). Thus, Nup60p can donate Kap60p and Gsp1p–GTP to Cse1p, indicating that Nup60p could function as a departure platform for Kap60p–Cse1p–Gsp1p–GTP complexes from the nucleus. Notably, Kap60p dissociates from the Nup60p–Gsp1p–Nup2p–Kap60p complex more readily than from Nup2p alone (data not shown), implying that Nup60p weakens the interaction between Nup2p and Kap60p upon binding Nup2p. This may contribute to the efficient transfer of Kap60p to its exportin Cse1p. We conclude that the Kap60p export defect observed in nup60Δ yeast (Fig. 7 A) is due to Nup2p mislocalization and to an increased stability of Nup2p–Kap60p heterodimers in the absence of Nup60p.
Discussion
Here we describe in detail a molecular mechanism for the dynamic assembly and disassembly of Nup60p–Nup2p complexes in response to the concentration of Gsp1p–GTP, and show that Nup60p contributes towards efficient import of cNLS-containing cargoes and nuclear export of Kap60p in vivo. Our biochemical reconstitution data suggests that Nup2p detaches from Nup60p in vivo and becomes a mobile component of the NPC when the intranuclear concentration of Gsp1p–GTP is low. This is consistent with the recent finding that Nup2p is a nucleocytoplasmic shuttling protein that changes localization upon cellular depletion of Gsp1p–GTP (Dilworth et al., 2001).
Three lines of evidence support the conclusion that Nup60p functions as an anchoring site for Nup2p at the NPC. First, Nup60p binds to Nup2p in the absence of additional proteins (KD ∼396 nM) (Fig. 3, A and D) and in crude yeast extracts in the presence of thousands of competitor proteins (Figs. 1 B and 3 C). Second, yeast lacking Nup60p show significant reduction in the amounts of Nup2p at the nuclear envelope, and consequently show accumulation of Nup2p throughout the nucleoplasm and cytoplasm (Fig. 2 A). This effect was specific for Nup2p, as neighboring Nups were unaffected (Fig. 2, A and B). Third, the function of Nup2p in the nuclear export of Kap60p (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000) seems compromised in yeast lacking Nup60p (Fig. 7 A). Nup2p may use its interaction with Nup60p to function more efficiently in Kap60p export. Despite the fact that Nup2p and Nup1p exhibit genetic interactions with each other and perform similar functions in Kap60p export and Kap95p–Kap60p import, there is no evidence of physical interaction between them.
Additional evidence suggests that the association between Nup2p and Nup60p at the NPC is dynamic. First, Nup2p fails to copurify with other Nups during the isolation of NPCs (Rout et al., 2000), implying that Nup2p dissociates readily from the NPC. Second, Nup2p–GFP accumulates in the nucleoplasm and cytoplasm of yeast lacking Nup60p (Fig. 2 A, middle), raising the possibility that Nup2p shuttles in and out of the nucleus and uses Nup60p as a rest-stop. Third, Nup2p can be tethered (indirectly) to a subset of FG Nups (Nup42p, Nup49p, Nup57p, Nup100p, and Nup116p) via the Kap95p–Kap60p heterodimer (Allen et al., 2001 and data not shown), suggesting that Nup2p moves across the NPC in a Kap95p–Kap60p-dependent manner.
The dynamic interaction between Nup60p and Nup2p is controlled by Gsp1p–GTP and Kap60p. Nup60p and Nup2p bind each other directly in the absence of additional proteins (KD ∼396 nM) (Figs. 3, A and C, and Fig. 6) and colocalize to the nuclear basket structure of the yeast NPC (Hood et al., 2000; Rout et al., 2000; Solsbacher et al., 2000). When Kap60p binds the NH2 terminus of Nup2p (KD ∼0.2 nM; data not shown) it prevents formation of the Nup60p–Nup2p complex (Fig. 3 A, lane 2). Similarly, when Kap95p binds Nup60p it prevents formation of the Nup60p–Nup2p complex (data not shown). Physiological concentrations of Gsp1p–GTP (i.e., 3 μM) enhance the affinity of the Nup60p–Nup2p complex 10-fold (to KD ∼38 nM; Fig. 3 D) and restore binding of Nup60p and Nup2p in the presence of Kap60p (Fig. 3 B, lane 3). As Gsp1p–GTP binds independently to Nup60p and Nup2p (Fig. 4 B), we suggest that Gsp1p bridges the interaction between Nup60p and Nup2p to form Nup60p–Gsp1p–GTP–Nup2p–Kap60p complexes (Figs. 3 B and 7 B, lane 1). These complexes could form readily in the nuclear basket structure of the yeast NPC, especially if Prp20p bound to Nup60p generates Gsp1p–GTP locally (see below).
What is the purpose of a regulated interaction between Nup60p and Nup2p at the NPC? The dynamic interaction between Nup2p and Nup60p may help maintain cell homeostasis by favoring different types of import or export reactions in response to the local concentration of Gsp1p–GTP. Our data suggests that when the concentration of Gsp1p–GTP is low at the nuclear basket structure, Nup2p detaches from Nup60p and becomes mobile via interaction with Kap60p–Kap95p heterodimers. The release of Nup2p from Nup60p may accomplish various tasks. For example, it may improve the efficiency of Kap95p–Kap60p-dependent import reactions, because a detachable docking site for Kap95p–Kap60p heterodimers could alleviate congestion of incoming Kap95p–Kap60p–cargo complexes that are not yet dissociated by Gsp1p–GTP. Alternatively, dissociation of Nup2p from the NPC could modulate the permeability barrier of the NPC. It is also possible that Nup2p moves within the NPC to perform an undetermined role in tRNA export (Hellmuth et al., 1998). We favor the explanation that Nup2p becomes mobile to help increase the free concentration of Gsp1p–GTP at the NPC. Nup2p may accomplish this task in two ways. First, Nup2p may accelerate the Kap60p-dependent release of Gsp1p–GTP from Kap95p (as Nup1p does) (Floer et al., 1997) at locations other than the nuclear basket structure (i.e., in the nucleoplasm). Second, Prp20p may piggy-back its way out of the nucleus when bound to Nup2p (Prp20p binds Nup2p; data not shown) and catalyze the conversion of Gsp1–GDP to Gsp1p–GTP at locations of the NPC other than the basket structure, such as the central transporter region or cytoplasmic fibrils, or even throughout the cytoplasm.
Nup60p is a novel Gsp1p binding protein. The observed binding of Gsp1p–GTP to Nup60p (Fig. 4) was unexpected, as the amino acid sequence of Nup60p does not predict a Gsp1p binding domain similar to those of Yrb1p, other Nups, or karyopherin βs (Dingwall et al., 1995; Gorlich et al. 1997). We determined that Nup60p is not a Gsp1p GEF or GAP, because it does not promote nucleotide exchange or stimulate the GTPase activity of Gsp1p in vitro (data not shown). Also, Nup60p did not protect Gsp1p–GTP from Rna1p action (data not shown) as Kap95p does (Floer et al., 1997).
Gsp1p–GTP may function as a physical bridge between Nup60p and Nup2p. Nup60p and Nup2p bind independently to Gsp1p–GTP (Fig. 4 B), so it is possible that Gsp1p–GTP contacts Nup60p and Nup2p simultaneously in the observed Nup60p–Gsp1p–Nup2p complexes (Figs. 3 B and 7 A). Affinity measurements demonstrate that binding of Nup60p, Nup2p, and Gsp1p–GTP is cooperative; for example, Gsp1p–GTP binds weakly to Nup60p (KD ∼5 μM) and Nup2p (KD ∼3.6 μM), but binds fivefold better to the Nup60p–Nup2p complex (KD ∼0.8 μM; Fig. 4 B). Similarly, Nup60p and Nup2p interact with 10-fold higher affinity in the presence of Gsp1p–GTP (KD ∼38 nM) compared with its absence (KD ∼396 nM; Fig. 3 D). A structural study of the Nup60p–Gsp1p–Nup2p complex will be needed to determine conclusively the contact sites between them.
Nup60p reduces (but does not abolish) Prp20p activity and thus functions as a mild Gsp1p GDI (Fig. 4 C). Since Nup60p binds directly (and independently) to Prp20p and Gsp1p (Fig. 6 B), it is unclear which interaction is responsible for reducing Prp20p activity (i.e., Nup60p–Prp20p or Nup60p–Gsp1p–GTP). Nevertheless, we note that Nup2p increases the affinity of Gsp1p–GTP towards Nup60p fivefold (Fig. 4 B), yet does not increase the inhibitory effect of Nup60p on Prp20p activity (Fig. 4 C). This suggests that Nup60p modulates the activity Prp20p via direct binding, rather than by sequestering Gsp1p–GTP away from Prp20p. In vivo, recruitment of Prp20p to the nuclear basket structure followed by reduction of its exchange activity could serve to create a unique local concentration of Gsp1p–GTP at the nuclear basket structure. A gradient of Prp20p activity could exist in the nucleus, whereby Prp20p activity would be high when bound to chromatin (Nemergut et al., 2001), intermediate when free in the nucleoplasm, and low when bound to Nup60p in the nuclear basket structure. A gradient of Prp20p activity would in turn generate a gradient of Gsp1p–GTP across the nucleoplasm. Such a gradient could guide the movement of importins and exportins towards (or away from) the NPC, or could serve as a positional marker for the release of cargo at distinct nuclear locations, assuming that karyopherins “sense” the concentration of Gsp1p–GTP differently when bound by different cargoes. We also observed that Prp20p binds directly to Nup2p, but this binding interaction does not affect the activity of Prp20p (Fig. 4 C and data not shown).
Nup60p functions as a docking site for Kap95p. Nup60p binds Kap95p monomers and Kap95p–Kap60p heterodimers (Figs. 5 A and 6 B), but only when the concentration of Gsp1p–GTP is low (Fig. 5 A; Allen et al., 2001). Kap95p monomers bind to the NH2 terminus and middle regions of Nup60p, which are devoid of FXF peptide repeats. However, when Kap95p is part of Kap95p–Kap60p heterodimers it no longer binds to the NH2 terminus of Nup60p, instead it binds to the COOH terminus which contains four FXF repeats (Fig. 6). At present, it is unclear why Kap95p chooses to bind different regions of Nup60p when Kap60p is present. Yeast lacking Nup60p import cNLS–GFP (the Kap95p–Kap60p dependent import cargo) into nuclei at a slower rate than wild-type yeast (Fig. 5 B). This may be due to the absence of Kap95p docking sites provided by Nup60p, or to a reduction in the levels of Gsp1p–GTP at the nuclear basket structure resulting from the loss of Prp20p binding sites provided by Nup60p. A mild nuclear import phenotype is consistent with the observation that nup60Δ yeast are healthy and grow as well as wild-type yeast. Indeed, the role of Nup60p in Kap95p-dependent import is likely redundant as each NPC contains many docking sites for Kap95p (Allen et al., 2001). We conclude that Nup60p functions as a docking site for Kap95p–Kap60p heterodimers when the concentration of Gsp1p–GTP is low, and as a tethering site for Nup2p–Kap60p heterodimers when the concentration of Gsp1p–GTP is high (Fig. 3, B–D). Kap123p is also among the proteins captured by Nup60p in yeast extracts (Fig. 1 B); it binds the NH2 terminus of Nup60p (Fig. 6). This suggests that Kap123p also uses Nup60p as a docking site in a manner similar to Kap95p. The interaction between Nup60p and Kap123p, and the interaction between Nup60p and Pab1p, will be explored in future experiments.
Nup60p functions as a platform for the assembly (and nuclear departure) of Kap60p–Cse1p–Gsp1p–GTP export complexes. Recruitment of Nup2p by Nup60p at the NPC may serve to enhance the efficiency of nuclear export of Kap60p. This is supported by the fact that absence of Nup60p from the NPC hinders Kap60p export (Fig. 7 A). It is likely that physical proximity between Kap60p and Gsp1p–GTP in the Nup60p–Gsp1p–GTP–Nup2p–Kap60p complex facilitates assembly of Kap60p–Cse1p–Gsp1p–GTP complexes in preparation for export. In favor of this model we find that Nup60p weakens the interaction between Kap60p and Nup2p (data not shown), and that the Nup60p–Gsp1p–Nup2p–Kap60p complex transfers Kap60p and Gsp1p–GTP readily to the exportin Cse1p (Fig. 7 B).
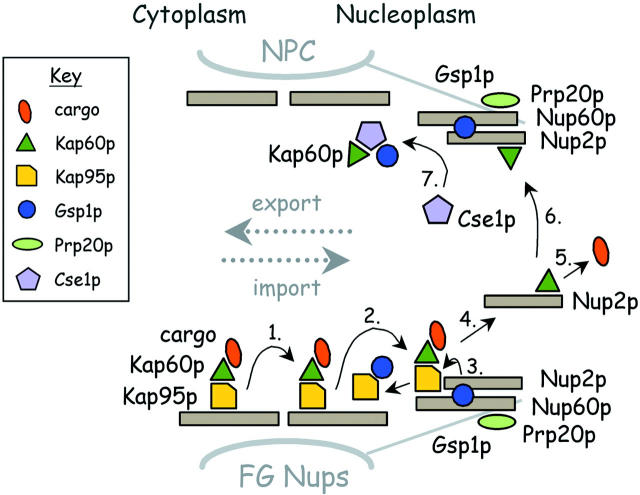
The cartoon in Fig. 8 depicts the proposed role of Nup60p in nuclear import and export reactions. It highlights the Nup60p–Gsp1p–Nup2p complex as an arrival terminal for incoming Kap95p–Kap60p cargo complexes (bottom half), and highlights the Nup60p–Gsp1p–Nup2p–Kap60p complex as a departure terminal for Kap60p–Cse1p–Gsp1p–GTP complexes (top half). Bottom half: an incoming Kap95p–Kap60p cargo complex binds to an FG Nup facing the nucleoplasmic side of the NPC (step 1); it then hops to a Nup60p–Gsp1p–GTP–Nup2p complex in the nuclear basket structure (step 2), where docking can occur via Kap95p–Kap60p binding to Nup60p or Nup2p. Prp20p bound to Nup60p generates Gsp1p–GTP locally and Gsp1p–GTP binds to Kap95p (step 3) to induce the release of the Kap60p–cargo complex from Kap95p (step 4; Rexach and Blobel, 1995). Upon dissociation from Kap95p, Kap60p lowers its affinity for cargo and binds Nup2p (step 5). In the absence of Gsp1p–GTP, Kap60p binding to Nup2p causes its dissociation from Nup60p (step 4), and the resulting Kap60p–Nup2p complex is free to diffuse in the nucleoplasm. Top half: Prp20p bound to Nup60p generates Gsp1p–GTP locally to promote binding of the Nup2p–Kap60p complexes to Nup60p, forming a Nup60p–Gsp1p–Nup2p–Kap60p complex (step 6). This complex functions as a platform for the assembly and departure of Kap60p–Cse1p–Gsp1p–GTP complexes from the nucleus (step 7). After donating Kap60p and Gsp1p–GTP to Cse1p, the resulting Nup60p–Nup2p complex becomes available for another round of Kap95p–Kap60p import and Kap60p export.
Figure 8.
A cartoon depicting the NPC and the proposed role of Nup60p in nuclear import and export reactions.
Materials and methods
Yeast strains
Table I contains a list of yeast strains used. Strains BY4741 and YD00407 were purchased from Research Genetics. Genomic integration of eGFP at the 3′-end of NUP2 was achieved by homologous recombination (Wach et al., 1997; Longtime et al., 1998) in strains BY4741 and YD00407, yielding strains MRY113 and MRY114. Expression of Nup2p–GFP was verified by Western blot analysis with anti-Nup2p and anti-GFP antibodies. Strains MRY115, MRY116, and MRY117 were obtained by mating MRY113 with YD13105, YD14906, and YD14917, respectively, followed by selection of His+ kanR sporulants. MRY118 and MRY119 were obtained by transforming BY4741 and YD00407 with pGAD-NLS-GFP (R. Wozniak, University of Alberta, Edmonton, Canada).
Table I. Yeast strains.
| Name | Genotype | Source |
|---|---|---|
| GPY60 | MATα his4-579 leu2-3,112 trp1-289 ura3-52 prb1 pep4::URA3 gal2 | Baker et al., 1988 |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Brachmann et al., 1998 |
| YD00407 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δnup60::kanMX4 | Winzeler et al., 1999 |
| YD13105 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Δnup170::kanMX4 | Winzeler et al., 1999 |
| YD14906 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Δnup120::kanMX4 | Winzeler et al., 1999 |
| MRY113 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 NUP2-GFP:HIS3 | This study |
| MRY114 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δnup60::kanMX4 NUP2-GFP:HIS3 | This study |
| MRY115 | MAT? his3Δ1 leu2Δ0 lys2Δ0? met 15Δ0? ura3Δ0 Δnup170::kanMX4 NUP2-GFP:HIS3 | This study |
| MRY116 | MAT? his3Δ1 leu2Δ0 lys2Δ0? met 15Δ0? ura3Δ0 Δnup120::kanMX4 NUP2-GFP:HIS3 | This study |
| MRY117 | MAT? his3Δ1 leu2Δ0 lys2Δ0? met 15Δ0? ura3Δ0 Δnup100::kanMX4 NUP2-GFP:HIS3 | This study |
| MRY118 | BY4741, plus pGAD-NLS-GFP | This study |
| MRY119 | YD00407, plus pGAD-NLS-GFP | This study |
Construction of recombinant proteins
Recombinant proteins used (except His-Gsp1p Q71L and His-Gsp1p) were expressed as GST fusions using vector pGEX-2TK (Amersham Pharmacia Biotech), which incorporates a thrombin cleavage site at the fusion junction as well as a specific kinase site that remains with the NH2 terminus of the fusion partner after cleavage. The NUP2, NUP60, KAP60, CSE1, KAP95, YRB1, and PRP20 genes and portions of NUP2 and NUP60 were amplified from yeast genomic DNA (Promega) using PCR. The PCR products were ligated into vector pGEX-2TK and transformed into the Escherichia coli strain BLR (Novagen). The His-Gsp1p Q71L and His-Gsp1p expression plasmids were obtained from K. Weis (University of California, Berkeley, CA).
Purification of recombinant proteins from E. coli
Yeast were grown in 1 liter of 2× YT medium with ampicillin (0.1 mg/ml) and 2% glucose at 37°C to OD600 = 1.0. Protein production was induced with IPTG for 15 min to 2 h at 24°C. Yeast were resuspended with 30 ml of chilled lysis buffer with protease inhibitors (0.1 mg/ml PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, and 2 μg/ml leupeptin) and lysed in a French Press cell (SLM Instruments). Cell debris was removed by centrifugation at 30,000 g for 15 min at 4°C, and Tween-20 (0.1%) was added to the supernatant. Recombinant proteins were purified on glutathione-Sepharose beads or nickel-Sepharose beads according to the manufacturer's instructions (Amersham Pharmacia Biotech and QIAGEN). When needed, the GST moiety was cleaved off by incubation with thrombin (Calbiochem) at 25°C (1 NIH unit thrombin/100 μg GST fusion) for controlled times. A molar excess of hirudin (Calbiochem) was added to neutralize thrombin, and free GST was removed using glutathione-Sepharose beads. Proteins were further purified in FPLC sizing columns equilibrated with 20 mM Hepes, pH 6.8, 150 mM KOAc, 2 mM Mg(OAc)2. Gsp1p was charged with GTP or GDP as described previously (Rexach and Blobel, 1995).
Preparation of S. cerevisiae extracts
GPY60 yeast was grown in 1 liter of YPD medium at 30°C to an OD600 = 2.0. Yeast were harvested by sedimentation at 5,000 g for 10 min and resuspended in 20 mM Hepes, pH 6.8, 150 mM KOAc, 250 mM sorbitol, 2 mM Mg(OAc)2, and protease inhibitors to a final volume of 40 ml. Yeast were lysed in a French Press cell and cell debris was removed by sedimentation at 30,000 g for 30 min at 4°C. The supernatant was desalted in a Sephadex G-25 fine column (Amersham Pharmacia Biotech) pre-equilibrated in 20 mM Hepes, pH 6.8, 150 mM KOAc, and 2 mM Mg(OAc)2. Pooled fractions were supplemented with 0.1% Tween, 2 mM DTT, and protease inhibitors.
Solution binding assays
Assays were performed in binding buffer (20 mM Hepes, pH 6.8, 150 mM KOAc, 2 mM Mg(OAc)2, 1 mM DTT, 0.1% Tween-20). For each experiment, glutathione-Sepharose beads and an E. coli extract containing the desired GST fusion were incubated for 15 min at 4°C. Beads were collected at 2,000 g for 30 s and washed repeatedly by resuspension and sedimentation. Equal aliquots of beads were then incubated with purified proteins or yeast extract for various times at 4°C. Beads were sedimented, the unbound material was collected, and the beads were washed repeatedly as before. In some cases, a second incubation with purified proteins was conducted. Bound proteins were eluted with 250 mM MgCl2 or Laemmli sample buffer, resolved by SDS-PAGE, and visualized with Coomassie blue. Gels were digitized in a scanner and assembled into figures using Adobe Photoshop® and Microsoft Powerpoint® software.
Gsp1p–GTP exchange assay
His-Gsp1p–[γ-32P]GTP was immobilized on Ni2+-NTA agarose beads (QIAGEN) at a final concentration of 15 nM within the beads. The immobilized His-Gsp1p–[γ-32P]GTP was incubated for 10 min at room temperature in the presence of 0.9 nM Prp20p, 1 mM GDP, and either Yrb1p, Nup2p, GST-Nup60p (aa 188–539), or GST (all at 4 μM). Prp20p activity was quenched by the addition of ice-cold buffer. After washes, the remaining [γ-32P]GTP bound to beads was quantified by scintillation counting (Beckman Coulter). Results were analyzed and graphed using GraphPad Prism™ (Biosoft).
Protein affinity measurements
Affinities were calculated using purified radiolabeled proteins in a bead-based solution binding assay. Purified Nup2p was phosphorylated at its NH2 terminus using bovine heart kinase and [γ-32P]ATP (NEN Life Science Products) as described in the Amersham Pharmacia Biotech GST handbook. His-tagged Gsp1p was loaded with [γ-32P]GTP (NEN Life Science Products) as described for GTP (Rexach and Blobel, 1995), and an unincorporated nucleotide was removed in G-25 desalting spin columns (Bio-Rad Laboratories). GST–Nup60p and GST–Nup2p were immobilized separately on beads and incubated with increasing amounts of radiolabeled Nup2p or Gsp1p–GTP at 4°C or room temperature in 20 mM Hepes, pH 6.8, 150 mM KOAc, 2 mM Mg(OAc)2, 1 mM DTT, 0.1% Tween-20, protease inhibitors, and 10 mg/ml BSA. After incubation, beads were isolated on filters using a vacuum manifold and specifically bound radiolabeled proteins were eluted with 1% SDS and quantified by scintillation counting. Binding curves were fit to the data using GraphPad Prism™ software (Biosoft).
Direct GFP fluorescence microscopy
Yeast-expressing GFP fusions were grown to midlog phase at 30°C in YP media with 2% glucose or galactose. Yeast were allowed to adhere to multiwell slides (Erie Scientific) coated with 0.1% (wt/vol) poly-l-lysine solution (Sigma-Aldrich). Nonadherent yeast were washed away and cover slips were coated in antifade solution (PBS, 90% glycerol, 250 ng/ml DAPI, 10 μg/ml p-phenylendiamine). GFP fusions were visualized live with a ZEISS Axiovert 35 epifluorescence microscope. Images were captured and adjusted using Metamorph and Photoshop 4.0 software.
Indirect immunofluorescence microscopy
Yeast were grown to early log phase at 30°C in YPD media and were fixed with 3.7% formaldehyde for 10 or 60 min for visualization of Nups or Kap60p, respectively. Yeast were collected by centrifugation, washed in 100 mM KH2PO4, pH 6.5, 500 μM MgCl2, and resuspended in the same buffer with 1.2 M sorbitol. Cell walls were digested with zymolase for 1 h at 25°C, and the yeast spheroplasts were adhered to poly-l-lysine–treated slides. The spheroplasts were postfixed for 6 min in dry ice–cold methanol followed by 30 s in acetone. Fixed yeast were washed with blocking solution and incubated with affinity-purified antibodies for 10 h at 4°C. After washes, yeast were incubated for 1 h at 4°C with FITC-conjugated anti–rabbit IgG (Jackson ImmunoResearch Laboratories) at a 1:1,000 dilution. After washing, cells were mounted for microscopic analysis as described above.
In vivo cNLS–GFP import assay
The in vivo nuclear import assay was performed as described in Shulga et al. (1996). In brief, MRY118 and MRY119 strains carrying the pGAD-cNLS–GFP plasmid (encoding the GFP with the cNLS of large T antigen) were grown at 30°C to an OD600 = 0.2. After induction of the cNLS–GFP fusion protein with 2% galactose for 2 h at 30°C, yeast were sedimented and washed with ice-cold water. Yeast were resuspended in 1 ml of growth medium containing 20 mM sodium azide and 20 mM 2-deoxyglucose and incubated for 1 h at 4°C to deplete cellular levels of ATP and block nucleocytoplasmic transport. Yeast were sedimented and resuspended in 50–100 μl of medium containing 2% dextrose and incubated at 4°C until assayed for recovery of nuclear import (judged by reimport of the cNLS–GFP protein). Nuclear import was initiated by transferring 2 μl of cell suspension onto a microscope slide at room temperature. Localization of the cNLS–GFP fusion protein was determined via fluorescence microscopy. Cells were scored as exhibiting diffuse cNLS–GFP fluorescence or predominantly nuclear cNLS–GFP, relying on standards described by Shulga et al. (1996). At least 50 cells were counted in 2-min intervals.
Antibodies
Anti-Nup2p, -Nup1p, -Nup100p, and -Kap60p antibodies were prepared by immunizing rabbits with GST-Nup2p (aa 562–720), GST-Nup1p (aa 963–1076), GST-Nup100p (aa 1–640), or GST-Kap60p (full length). Specific antibodies were affinity-purified from serum using the GST fusions as affinity resins. The affinity-purified anti-Nup100p antibodies crossreact with Nup116p (a homologue of Nup100p) and were therefore designated as anti-Nup100p/Nup116p. Anti-Kap60p antibodies were used at a 1:2,000 dilution; and anti-Nup2p, -Nup1p, and -Nup100p/Nup116p were used at a 1:5,000 dilution.
Quantitative Western blots
Yeast proteins bound to GST–Nup60p-coated beads in the presence or absence of additional proteins were eluted with SDS and resolved by SDS-PAGE. After transfer to Immobulon-P filter paper (Millipore), the Western blots were probed with affinity-purified anti-Nup2p or anti-GST antibodies (1:1,000 dilution) for 1 h in Tris-buffered saline containing 3% milk and 0.1% Tween-20. After washing, blots were probed with 125I-protein A (0.5 μCi; Amersham Pharmacia Biotech) for 1 h. 125I-labeled blots were exposed to Phosphor screens for 12 h and then digitized and quantified using ImageQuantTM v1.2 software (Molecular Dynamics).
Mass spectrometry
Proteins were excised from gels, destained in 25 mM ammonium bicarbonate/50% acetonitrile, and dried. Gel slices were resuspended in equal volume of 50 ng/ml trypsin (Promega) and digested for 24 h at 37°C. Peptides were extracted from gel slices using 50% acetonitrile/5% formic acid, and sample volumes were concentrated to 5 μl and readjusted to 20 μl with 0.1% formic acid. After the samples were cleaned in C18 ZipTips (Millipore) and resuspended in 50% acetonitrile/0.1% formic acid, portions were mixed with 2,5-dihydroxy benzoic acid (Aldridge) and analyzed by MALDI-TOF mass spectrometry (Voyager-DE STR; PerSeptive Biosystems). External and internal calibrations were done for all samples. Peptide masses were used to search the ProFound™ (Rockefeller University) and MS-Fit™ (University of California at San Francisco) databases for protein candidates.
Acknowledgments
We thank K. Weis for providing the HIS-Gsp1p expression plasmids.
This work was supported by the Searle Scholars Program (M. Rexach), the National Science Foundation and Stanford Graduate Fellowships (D. Denning), and the National Institutes of Health (B. Mykytka, N. Allen, L. Huang, and A. Burlingame).
Footnotes
Abbreviations used in this paper: aa, amino acids; cNLS, classic nuclear localization signal; GDI, guanine nucleotide dissociation inhibitor; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GST, glutathione S-transferase; NPC, nuclear pore complex; Nup, nucleoporin.
References
- Allen, N., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268–29274. [DOI] [PubMed] [Google Scholar]
- Baker, D., L. Hicke, M. Rexach, M. Schleyer, and R. Schekman. 1988. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 54:335–344. [DOI] [PubMed] [Google Scholar]
- Belanger, K.D., M.A. Kenna, S. Wei, and L.I. Davis. 1994. Genetic and physical interactions between Srp1p and nuclear pore complex proteins Nup1p and Nup2p. J. Cell Biol. 126:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, J.W., K.D. Belanger, M.I. Sannella, and L.I. Davis. 1999. The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J. Biol. Chem. 274:32360–32367. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Davis, L.I., and G.R. Fink. 1990. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell. 61:965–978. [DOI] [PubMed] [Google Scholar]
- Dilworth, D.J., A. Suprapto, J.C. Padovan, B.T. Chait, R.W. Wozniack, M.P. Rout, and J.D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall, C., S. Kandels-Lewis, and B. Seraphin. 1995. A family of Ran binding proteins that includes nucleoporins. Proc. Natl. Acad. Sci. USA. 92:7525–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel, C., G. Blobel, and M. Rexach. 1995. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 270:16499–16502. [DOI] [PubMed] [Google Scholar]
- Floer, M., G. Blobel, and M. Rexach. 1997. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J. Biol. Chem. 272:19538–19546. [DOI] [PubMed] [Google Scholar]
- Fontoura, B.M., G. Blobel, and N.R. Yaseen. 2000. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin β 2-mediated nuclear import. J. Biol. Chem. 275:31289–31296. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607–660. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., M. Dabrowski, F.R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi, S., Y. Rochon, B. Franza, and R. Abersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth, K., D.M. Lau, F.R. Bischoff, M. Kunzler, E. Hurt, and G. Simos. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18:6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, J.K., J.M. Casolari, and P.A. Silver. 2000. Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J. Cell Sci. 113:1471–1480. [DOI] [PubMed] [Google Scholar]
- Kehlenbach, R.H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna, M.A., J.G. Petranka, J.L. Reilly, and L.I. Davis. 1996. Yeast N1e3p/Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol. Cell. Biol. 16:2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, O., C.V. Heath, D.C. Amberg, T.C. Dockendorff, C.S. Copeland, M. Snyder, and C.N. Cole. 1995. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell. 6:401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J.D., L.I. Davis, and G.R. Fink. 1993. NUP2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol. Biol. Cell. 4:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtime, M., A. McKenzie, D. Demarini, N. Shah, A. Wach, A. Brachat, P. Philippsen, and J. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in S. cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell. 99:677–690. [DOI] [PubMed] [Google Scholar]
- Nemergut, M., C. Mizzen, T. Stukenberg, C. Allis, and I. Macara. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 292:1540–1543. [DOI] [PubMed] [Google Scholar]
- Pemberton, L.F., M.P. Rout, and G. Blobel. 1995. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 92:1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu, A., M.S. Moore, and G. Blobel. 1995. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 81:215–222. [DOI] [PubMed] [Google Scholar]
- Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 83:683–692. [DOI] [PubMed] [Google Scholar]
- Rout, M., J. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., and S.R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361–371. [DOI] [PubMed] [Google Scholar]
- Shah, S., and D.J. Forbes. 1998. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 8:1376–1386. [DOI] [PubMed] [Google Scholar]
- Shulga, N., P. Roberts, Z. Gu, L. Spitz, M.M. Tabb, M. Nomura, and D.S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga, N., N. Mosammaparast, R. Wozniak, and D.S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher, J., P. Maurer, F. Vogel, and G. Schlenstedt. 2000. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol. Cell. Biol. 20:8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman, K.S., S. Shah, M.A. Powers, and D.J. Forbes. 1999. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell. 10:649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, C. Alberti-Seguei, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS marker and GFP reporter molecules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., D.D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J.D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. [DOI] [PubMed] [Google Scholar]
- Yang, Q., M.P. Rout, and C.W. Akey. 1998. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell. 1:223–234. [DOI] [PubMed] [Google Scholar]
- Yaseen, N.R., and G. Blobel. 1999. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin nup358. J. Biol. Chem. 274:26493–26502. [DOI] [PubMed] [Google Scholar]