Abstract
A hammerhead ribozyme has been localized to the yeast nucleolus by using the U3 small nucleolar RNA as a carrier. The hybrid small nucleolar RNA:ribozyme, designated a “snorbozyme,” is metabolically stable and cleaves a target U3 RNA with nearly 100% efficiency in vivo. This is the most efficient in vivo cleavage reported for a trans-acting ribozyme. A key advantage of the model substrate featured is that a stable, trimmed cleavage product accumulates. This property allows accurate kinetic measurements of authentic cleavage in vivo. The system offers new avenues for developing effective ribozymes for research and therapeutic applications.
Keywords: yeast, nucleolus
Ribozymes have enormous potential in basic research and biotechnology with special promise for applications in human medicine (1, 2). In this context, a catalytic RNA is designed to specifically recognize, interact with, and alter a target molecule. The best-characterized ribozymes are hydrolytic nucleases that perform site-specific cleavage of a target RNA (3, 4). The hammerhead ribozyme has received most attention thus far because of its early discovery, small size, and potential to cleave almost any RNA molecule (5–7). Naturally occurring hammerhead ribozymes cleave target sequences in the same molecule, i.e., in cis (ref. 8; see also ref. 9). It is possible however, to engineer artificial ribozymes, which cleave RNA molecules in trans. In principle, this strategy can be used to alter the level of any RNA in a living organism, and achieving this goal has been a major focus of ribozyme research (see refs. 10 and 11). Despite much effort, progress has been slow in attaining highly efficient cleavage of target molecules in living cells (12–17). In no case known to us has complete or nearly complete cleavage been observed in vivo for any trans-acting hammerhead ribozyme. Overcoming this limitation is critical if the full potential for modulating gene expression with ribozymes is to be realized.
To achieve efficient cleavage in vivo, three key conditions must be satisfied: (i) an intracellular concentration of ribozyme that is not limiting, which requires a robust transcription rate and/or good metabolic stability of the ribozyme; (ii) a high degree of colocalization of the ribozyme to the same cellular compartment(s) as the target molecule (13, 16); and (iii) highly productive ribozyme–target interaction, i.e., good accessibility of the relevant RNA sequences in the catalytic and substrate molecules (18). In this last regard, either or both RNAs may be large and complex and bound to proteins; thus, the potential for interference effects can be high. Lack of detailed information about the in vivo structures and dynamics of the interacting RNAs continues to be a major problem. In the present study we sought to satisfy the requirements for effective in vivo cleavage by using small nucleolar RNAs (snoRNAs) as both catalytic and target molecules and to demonstrate that complete or near-complete cleavage is indeed possible with a trans-acting ribozyme.
EXPERIMENTAL PROCEDURES
Yeast Strains and Manipulations.
In vivo studies with cis-acting snorbozymes were performed with a haploid yeast strain transformed with appropriate plasmids. The host strain in these experiments was YSD2 (MATa ura3 ade2 his3 leu2 lys1 trp1) prepared by sporulating the isogenic MH2 strain (kindly provided by Molly Fitzgerald-Hayes, Univ. of Massachusetts, Amherst, MA). In vivo studies with the trans-acting snorbozymes were conducted with diploid yeast strains containing appropriate plasmid pairs. The latter strains were prepared as follows. First, two isogenic haploid strains, YSD2 and YSD1, obtained after sporulation of diploid MH2 and differing only in mating type (a vs. α), were transformed with plasmids expressing either target- (YSD2) or ribozyme (YSD1)-containing molecules. The transformants obtained were then crossed to yield diploids expressing the desired pairs of RNA molecules. Plasmids were introduced into yeast cells by using a lithium acetate transformation procedure (19).
Analysis of Ribozyme Activity.
The RNA molecules used in the in vitro ribozyme studies were prepared by transcribing DNA templates with T7 RNA polymerase. The templates were prepared by PCR amplification of the plasmids used in the in vivo ribozyme studies (see below). The cis reaction occurred during the transcription incubation (1 h) with almost 100% efficiency. In the trans ribozyme reactions, mutant and wild-type ribozyme and substrate transcripts were quantified spectrophotometrically or estimated from [32P]UTP incorporation measurements. A constant amount of substrate (2 nM) was incubated with 10, 20, 40, 60, 80, and 100 nM ribozyme in 20 mM Tris⋅HCl (pH 7.9)/100 mM NaCl/10 mM MgCl2/2 mM spermidine. Ribozyme and substrate were first combined in the above conditions, but without Mg2+, heated at 80°C for 1 min, and cooled on ice. Reactions were started by adding Mg2+ and incubated for 1 h at 37°C. Under these conditions, ≈40% of the substrate was cleaved at the highest ribozyme concentration (data not shown). The efficiency of ribozyme cleavage in vivo was assessed by Northern blot analysis.
RNA isolation and Northern blot analyses were performed as described (20). Total RNA for Northern blots was prepared from cells grown overnight in selective liquid YNB medium (0.67% yeast nitrogen base). Radiolabeled oligonucleotides used to probe the Northern blots were C106, which recognizes yeast U14 snoRNA (internal control); SD13*, which hybridizes to all hybrid molecules used in the cis ribozyme studies as well as substrate and product molecules featured in the trans ribozyme studies (this probe specifically recognizes a unique hairpin sequence in a modified U3*; see Fig. 1); and SD90, which recognizes specifically ribozyme-carrying molecules used in the trans ribozyme studies (this probe hybridizes to a core sequence in the hammerhead ribozyme, on the 5′ side of the nucleotide used to prepare inactive ribozyme variants).
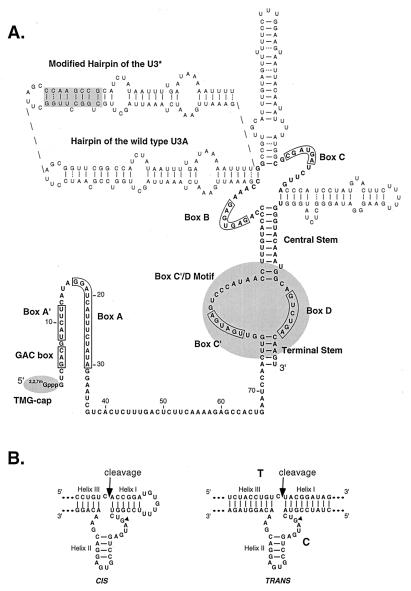
Figure 1.
Structure of the U3 snoRNA-hammerhead ribozyme (snorbozyme) chimeras featured in the present study. (A) Map of S. cerevisiae U3 (22). The yeast U3 molecule consists of (i) a 5′ region with little or no secondary folding; (ii) a single-stranded hinge region, and (iii) a highly structured 3′ domain. The 5′ and 3′ ends are protected from exonucleolytic damage by a trimethyl guanosine (TMG) cap and one or more proteins complexed with the box C′/D motif. Key elements incorporated into the design of the snorbozyme system are emphasized. The 5′ domain contains highly conserved box elements GAC, A′, and A, which are implicated in direct interaction with pre-rRNA. The box C′/D motif is a nucleolar localization signal and provides metabolic stability. The in vivo studies were carried out with cells expressing wild-type U3, which is essential for growth, and one or two U3 variants containing the experimental sequences. One variant (U3*) contains a unique hybridization tag created by modifying the sequence of one of the helical stems (upper left). The second is a functional “mini-U3” molecule (U3del), in which all hairpin domains are absent. The ribozyme and target sequences were introduced into the 5′ region of the U3 host molecules in place of conserved boxes A′ and A and a portion of the hinge segment. (B) Structures of the hammerhead ribozyme elements featured, in cis and trans configurations (8, 25). A G nucleotide essential for ribozyme cleavage is marked with a filled triangle. Inactive snorbozymes containing a C at this position were used in control experiments. The target and catalytic sequences in the trans configuration are labeled T and C, respectively.
In situ hybridization and probe synthesis were performed as described (21). The probes used were RNA molecules conjugated to either CY3 or Oregon green 488. These RNAs were obtained by using in vitro transcription of plasmid templates containing either the ribozyme or the U3* hybridization tag sequences.
DNA Constructs.
The RNA molecules used in the in vitro studies were prepared by transcribing DNA templates with T7 RNA polymerase according to a protocol provided by the manufacturer (Promega). The DNA templates were prepared by PCR amplification of the plasmids used in the in vivo studies (see below) and are as follows. Active cis-snorbozyme, plasmid pRS:T/C(+) with primers SD101 and SD102; inactive cis-snorbozyme, plasmid pRS:T/C(−) with primers SD101 and SD102; active trans-snorbozyme, plasmid pRS:C(+) with primers SD99 and SD101; inactive trans-snorbozyme, plasmid pRS:C(−) with primers SD99 and SD101; substrate molecule (trans ribozyme analyses), plasmid pRS:T with primers SD100 and SD101. Before T7 transcription, the DNA templates were treated with HpaI to generate RNA molecules with uniform 3′ ends. Plasmids for the in vivo snorbozyme studies were prepared by substituting major portions of the 5′ segment of the U3-coding region with the appropriate experimental sequences, in plasmids pRU3*-313, pRU3(del)-316 or YEp-U3(del) plasmids (see below). Replacements were performed by using PCR mutagenesis strategies described (22). The DNA templates and essential PCR primers were: pRU3*-313 and SD75, for pRS:T/C(+); pRU3*-313 and SD76, for pRS:T/C(−); pRU3(del)-313 and SD88, for pRS:C(+) and YEp:C(+); pRU3(del)-313 and SD89, for pRS:C(−) and YEp:C(−); pRU3*-313 and SD86, for pRS:T or pRS:T(9:9); pRS:T and SD97, for pRS:T(6:6); pRS:T and SD95, for pRS:T(6:9); pRS:T and SD96, for pRS:T(9:6); pRU3*-313 and SD87, for pRS:T(12:12); and pRS:T and SD98, for pRS:T(15:15). Plasmids pRU3*-313, pRU3(del)-316 and YEp-U3(del) were prepared by inserting a 1.3-kb HindIII fragment encoding U3* or U3(del) variants of the U3A gene (22) into cloning vectors pRS313 (encoding U3*, into SalI site), pRS316 [encoding U3(del), into SalI site], and YEp24 m [encoding U3(del), into ClaI site]. Vectors pRS313 and pRS316 are described by Sikorski & Hieter (23).
Oligonucleotides.
Sequences of the oligonucleotides used in this study are as follows: for SD13,* 5′-GCGGCTTAGGCTAAGCTAAGGCCAGC-3′; SD75, 5′-TGACTCTGTCGACGTACCTGTCACCGGATGTGTTTTCCGGTCTGATGAGTCCGTGAGGACGAAACAGGAATCCAACTTGGTTGATGAG-3′; SD76, 5′-TGACTCTGTCGACGTACCTGTCACCGGATGTGTTTTCCGGTCTCATGAGTCCGTGAGGACGAAACAGGAATCCAACTTGGTTGATGAG-3′; SD86, 5′-GACTCTGTCGACTCTACCTGTCTACGGATAGCCACTGAATCCAACTTGGTTG-3′; SD87, 5′-GACTCTGTCGACACGTCTACCTGTCTACGGATAGTGACCACTGAATCCAACTTGGTT-3′; SD88, 5′-GACTCTGTCGACTCACTATCCGTACTGATGAGTCCGTGAGGACGAAACAGGTAGACGTCCACTGAATCCAACTTGGTTG-3′; SD89, 5′-GACTCTGTCGACTCACTATCCGTACTCATGAGTCCGTGAGGACGAAACAGGTAGACGTCCACTGAATCCAACTTGGTTG-3′; SD90, 5′-ACGTCTACCTGTTTCGTCCTCACGGACTCAT-3′; SD95, 5′-ATCTGTGTCGACCGAACCTGTCTAC-3′; SD96, 5′-ATCTGTGTCGACTCTACCTGTCTACGGAATCCCACTGAATC-3′; SD97, 5′-ATCTGTGTCGACCGAACCTGTCTACGGAATCCCACTGAATC-3′ and for SD98, 5′-ATCTGTGTCGACTGGACGTCTACCTGTCTACGGATAGTGAGTCCCACTGAATCCAACTTG-3′; SD99, 5′-CTCTACGTAATACGACTCACTATAGGGTCGACTCACTATCCGTAC-3′; SD100, 5′-CTCTACGTAATACGACTCACTATAGGGTCGACTCTACCTGTCTAC-3′; SD101, 5′-AAGTGGTTAACTTGTCAGACTGCC-3′; SD102, 5′-CTCTACGTAATACGACTCACTATAGGGTCGACGTACCTGTCACCG-3′.
RESULTS
Recent advances in snoRNA research suggested to us that a snoRNA molecule could be used as a ribozyme carrier to target nucleolar RNAs. This approach was initially prompted by identification of a common snoRNA structure motif, known as the box C/D motif, which confers metabolic stability on the snoRNA and also causes the RNA to be localized to the nucleolus (ref. 21, and refs. therein). Other key advances were generation of a detailed structure–function map for the U3 box C/D snoRNA from Saccharomyces cerevisiae, and development of valuable information about the structure of the natural U3 snoRNP complex in this organism (refs. 22 and 24, and refs. therein).
Snorbozyme Experimental System.
The U3 snoRNA is conserved among all eukaryotes and is required for processing of rRNA. The structure of U3 from S. cerevisiae is shown in Fig. 1. Major features include (i) a trimethyl guanosine cap, which protects the RNA from 5′ → 3′ exonucleolytic degradation; (ii) a 5′ segment, which is involved in direct pre-rRNA binding and is thought to be largely or completely single stranded; and (iii) a highly folded 3′ region, which contains the box C′/D motif (an analog of the canonical box C/D motif in other snoRNAs; see also above). Because the U3 segment involved in pre-rRNA binding is known to be exposed in the mature snoRNP particle (24), we reasoned that a ribozyme sequence placed in this segment would be able to interact with a target molecule. By the same reasoning, we predicted that a target sequence embedded in the same region in a second U3 molecule would be accessible to the ribozyme. In addition to achieving good accessibility, targeting cleavage to this particular segment of U3 also was expected to yield a stable byproduct, which would facilitate characterization of the system. In this regard, the initial cleavage products are expected to be altered by endogenous exonucleases (see diagrams in Figs. 2 and 4). The short 5′ segment would likely be degraded completely; however, the larger 3′ product would be protected from complete degradation by proteins bound to the box C′/D motif. This final byproduct would be both stable and smaller than the initial 3′ cleavage product. These properties would make measurements of authentic in vivo cleavage both simple and reliable. We call the hybrid snoRNA:ribozyme molecules snorbozymes.
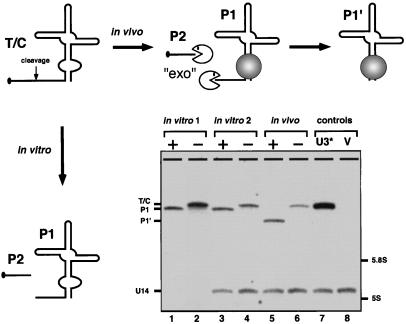
Figure 2.
Cleavage activity in the cis configuration. A cis-acting snorbozyme was constructed in which a hammerhead sequence (52 nt) was inserted into the full-size U3* molecule in place of nucleotides 10–67. The Northern blot identifies RNAs expressed in yeast (in vivo) or with T7 RNA polymerase in vitro (in vitro 1). To verify that cleavage and subsequent trimming of the in vivo-expressed snorbozyme did not occur during RNA isolation, the in vitro-transcribed molecules were added to intact yeast cells and passed through the same RNA isolation procedure (in vitro 2). + refers to the active ribozyme; − refers to the defective ribozyme (see Fig. 1B). Additional control samples correspond to total RNA isolated from cells expressing the U3* or empty host vector (V). The biochemical events leading to the final products obtained in vivo and in vitro are shown schematically. The P2 product is not identified in the Northern blot (in vitro lanes) because of its small size (14 nt).
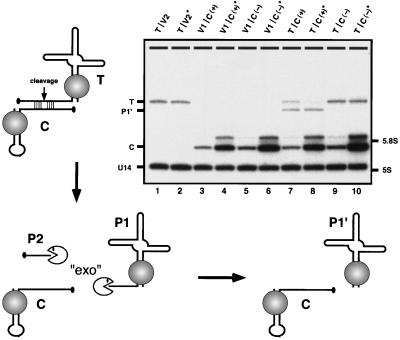
Figure 4.
Cleavage in trans. The in vivo cleavage efficiency in trans was assessed with snorbozyme genes that encode the catalytic and target elements in separate transcripts. The catalytic portion of the ribozyme (46 nt) was embedded in the mini-U3 variant (U3del) in place of nucleotides 6–60; C(+) refers to the active ribozyme, and C(−) refers to the defective ribozyme. The target sequence (19 nt) was placed into U3*, replacing the same wild-type segment; the target RNA is designated T. U3 snoRNA derivatives containing substrate and catalytic sequences were coexpressed in yeast cells from two different low-copy plasmid vectors (V1 and V2). To determine whether the ribozyme level was limiting, the catalytic RNA was also expressed from a multicopy vector (V*). The diagram depicts the principal events predicted to occur, and the Northern blot identifies the RNA species observed. Bands marked on the Northern blot correspond to: target RNA (T), cleaved and trimmed byproduct (P1′), catalytic RNA (C), U14 snoRNA (≈130 nt) as a control for RNA loading, and the positions of 5.8S (≈160 nt) and 5S (≈120 nt) rRNAs. Samples are given two-part names, which refer to the vector pairs carrying the target and catalytic RNA genes, respectively. Combinations of empty vectors and experimental genes are also included as controls. The samples are arranged as follows: lanes 1 and 2, target molecule (T) only, produced in the presence of low- and high-copy empty vectors (V2 and V2*); lanes 3 and 4, active catalytic RNAs produced from low- and high-copy vectors, C(+) and C(+)*, respectively, in the presence of empty vector V1; lanes 5 and 6, inactive catalytic RNA produced from low- and high-copy vectors, C(−) and C(−)*, respectively, in the presence of empty vector V1; lanes 7 and 8, target molecules coexpressed with active catalytic RNA encoded by low- and high-copy vectors, C(+) and C(+)*, respectively; lanes 9 and 10, target molecules coexpressed with inactive catalytic RNA encoded by low- and high-copy vectors, C(−) and C(−)*, respectively. The minor bands observed just above the catalytic RNAs (C) correspond to molecules with extensions at the 3′ end (22). The cleavage efficiency for active ribozyme produced from low- and high-copy vectors is estimated at ≈60% and 90%, respectively (lanes 7 and 8; compare with lanes 9 and 10).
Snorbozyme Cleavage in Cis.
Our initial tests were carried out with a cis-acting snorbozyme (Fig. 2). To distinguish this molecule from natural U3, which is required for cell growth, the snorbozyme was constructed from a U3 variant that contains a unique hybridization tag (see Fig. 1). In our scheme, the hammerhead ribozyme sequence (52 nt) was substituted for a U3 segment of similar size (57 nt). Nearly complete cleavage was observed with the cis-acting snorbozyme, both in vitro (>95%) and in vivo (>90%). Control experiments with mutant catalytic sequences demonstrated that cleavage is ribozyme-dependent and the shorter, exonucleolytically trimmed 3′ byproduct is only formed in living cells and not in vitro or after cell disruption. These results show the hammerhead ribozyme is highly active in the context of the U3 snoRNA and that a stable byproduct accumulates as predicted.
Trans-Acting Snorbozyme.
We next examined cleavage efficiency in the trans configuration. Two structural variants of U3 were used, which allowed easy discrimination of experimental and endogenous U3 molecules. The target RNA contained the same hybridization tag used for the cis snorbozyme. The catalytic sequence was placed into a U3 mini-variant, which is about half the size of wild-type U3. The intracellular locations of the target- and ribozyme-containing molecules were examined by using fluorescent in situ hybridization microscopy (Fig. 3). The patterns obtained show that the ribozyme and target molecules colocalize precisely to the nucleolus, as expected. Cleavage efficiency was assessed by using Northern blotting, for transformants expressing catalytic and target RNAs in various combinations. These included expression of catalytic molecules from low- and high-copy vectors and inactive catalytic RNA as a control (Fig. 4). Coexpression of target and catalytic molecules from low-copy plasmids, which produce approximately equimolar RNA amounts, resulted in ≈60% cleavage of the target molecule (lane 7). The cleavage efficiency increased to ≈90% when the catalytic RNA was expressed from a high-copy vector, i.e., in ≈10-fold molar excess (lane 8). These results show that the trans-acting U3-hammerhead snorbozyme is a stable and powerful catalyst in a cellular environment. To our knowledge, this is the most potent trans-acting ribozyme activity yet observed in vivo. Interestingly, in parallel analyses done in vitro, only ≈40% of the target U3 RNA was cleaved at a saturating condition (data not shown). We presume that the lower activity in vitro reflects different and less well controlled folding properties of both the catalytic and substrate RNAs.
Figure 3.
The U3-based snorbozymes localize to the nucleolus. The intracellular location of the trans-acting ribozyme components was examined by fluorescent in situ hybridization microscopy. Catalytic and target RNA molecules were expressed in yeast cells from separate low copy plasmids (see text and Fig. 4). Yeast nucleoli were identified by using a probe specific for the box C/D U14 snoRNA. (A) Hybridization with a probe specific for the catalytic molecule. (B) Hybridization with a probe specific for an internal segment of the substrate RNA. This probe recognizes both the full-length target molecule and the cleaved and trimmed byproduct (see also Fig. 4). (C) Double hybridization with probes that recognize the catalytic molecule and the target and cleaved byproduct molecules. The labels in each image correspond to the RNA molecules identified.
Optimization of in Vivo Cleavage.
With a view to further optimizing the trans cleavage efficiency in vivo, we examined the effect of altering the complementarity between the catalytic and substrate RNAs (Fig. 5). In the previous experiments, the target–catalyst pair had the potential to form 18 bp, corresponding to 9 bp each in helices I and III (see also Fig. 1). In the new experiments, the total complementarity was varied from 12 to 30 bp by adjusting the sequences of the target RNA. Coexpression of the test pairs—the target RNA from the low-copy plasmid and catalytic RNA from the high-copy plasmid—showed that the best cleavage efficiency occurred with an intermediate and asymmetric complementarity of 15 bp [9 bp in helix III and 6 bp in helix I (lane 7)]. These results suggest that 9:6 bp complementarity provides optimal binding and cleavage in vivo, which is in good agreement with in vitro data. Lengthening helix I to more than 6 bp has been observed to slow the rate of cleavage in vitro, in some cases by as much as 100 fold. Shortening the total complementarity to less than 10–12 bp, on the other hand, also reduced the efficiency (27; see also ref. 28). Strikingly, the cleavage efficiency was nearly complete in the optimal case shown here.
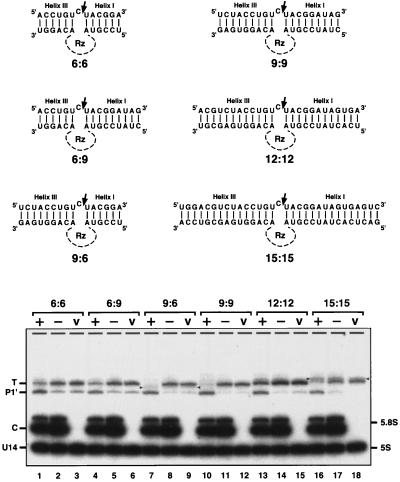
Figure 5.
Effect of flanking complementarity on trans-acting ribozyme cleavage activity. (A) A set of substrates able to form different numbers of base pairs with the initial ribozyme molecule was prepared. The relevant target region is on top, paired with the corresponding ribozyme moiety on the bottom (the catalytic core of the ribozyme Rz is omitted for simplicity). The trans-acting ribozyme complex examined earlier is the 9:9 pair. (B) Efficiency of cleavage was assessed by using Northern blot analysis. Each substrate RNA was coproduced with an active (+) or inactive (−) ribozyme molecule or in the presence of an empty host vector (V). Variable amounts of truncated substrates are evident in the negative control samples. The yield of these molecules reflects differences in the 5′ sequences of the substrates and the growth stage of the cells used in the analysis (more is present at later stages of growth). These products are presumed to arise from random degradation of the substrate 5′ end. The origin of the truncated substrate variant in lane 17 is not clear. Accumulation of this product might reflect a nonspecific cleavage process that depends on the inactive ribozyme. Alternatively, cleavage might be mediated by the “inactive” ribozyme, which may have low residual activity that is magnified in this particular, high-affinity context. The RNA species marked with triangles (lanes 7–9 and 16–18) most likely originate from alternative folding of target molecules. Similar effects have been observed previously with internally modified U2 and U3 RNAs (22, 26). Interestingly, these target molecules are completely resistant to ribozyme cleavage.
Kinetics of the in Vivo Snorbozyme Cleavage.
An important advantage of the new in vivo system is that the ribozyme, substrate and cleavage molecules can all be quantified, as predicted. Product accumulation is unusual in ribozyme studies, and this property makes accurate in vivo kinetic studies possible for the present system. Although detailed measurements are beyond the scope of the present report, several preliminary observations can be made from the results obtained, for both the cis- and trans-acting ribozymes. The efficiency of the cis-acting snorbozyme in vivo is ≈90%. If the half-life of the U3 derivatives is similar to that of natural U3, estimated at 90 min (29), the rate of the ribozyme reaction is deduced to be 0.08 min−1 in vivo (see Appendix). In trans, the best cleavage efficiency observed was better yet, for a ribozyme–substrate complementarity of 9:6 bp and expression of the ribozyme from a high-copy vector (≈10-fold excess over the target). Assuming an efficiency of >95%, the rate of reaction, kobs, is estimated at >0.15 min−1 (see Appendix). These values are consistent with the lower range of activities observed in vitro with trans-acting hammerhead ribozymes (30), indicating that the maximal reaction rate, i.e., the rate of the cleavage step, may be roughly comparable in vivo and in vitro. When the ribozyme and the substrate have 9:9 bp of complementarity, the reaction rate, kobs, increased from 0.01 min−1 (60% cleavage), when the ribozyme was expressed from a low-copy vector, to 0.08 min−1 (90% cleavage), when the ribozyme was expressed from a high-copy vector. This result suggests that for the ribozyme levels evaluated, the rate of ribozyme–substrate association is limiting in vivo. We estimate the ribozyme concentration to be in the range of 3 μM (low-copy vector) to 30 μM (high-copy vector) in the nucleolus, significantly more than the concentration required to saturate the reaction in vitro (see Appendix). Although preliminary in nature, this is direct evidence that ribozyme–substrate binding is much slower in vivo than in vitro. Finally, it is known that the rate of product release affects the overall activity of a trans-acting ribozyme in vitro (31). However, in the present study, product release did not seem to be an issue in vivo. The fact that cleaved and nontrimmed product does not accumulate in the cells indicates that the initial cleavage product is rapidly degraded and is not associated with the ribozyme.
DISCUSSION
Taken together, the results of this study allow a number of important conclusions and predictions to be made. First, and most significant, the findings validate the long-standing prediction that gene expression in a living cell can, indeed, be effectively blocked with a powerful artificial ribozyme. It is clear now that a trans-acting hammerhead ribozyme can cleave a target RNA with near-perfect efficiency in vivo. It follows that all key reaction requirements have been satisfied, including ribozyme stability, colocalization of the catalytic and target RNAs, and accessibility of the relevant RNA regions.
In addition to showing that a trans-acting ribozyme can cleave RNA nearly completely, we have established an effective strategy for delivering a ribozyme to a specific subcellular site for targeting a specific type of RNA, in the present case the nucleolus and an artificial nucleolar RNA. This particular approach could be valuable for modulating levels of natural snoRNAs and ribosomal RNAs as well as other RNAs proposed to be associated with the nucleolus. The latter class includes various mRNAs, tRNAs and splicing snRNAs, and the RNA components of telomerase, the signal recognition particle, MRP RNase and RNase P (ref. 32, and refs. therein). Because artificial box C/D snoRNAs are also known to localize to coiled bodies in mammalian cells (ref. 21, and refs. therein), it may also be possible to target snorbozymes to RNAs in coiled bodies, such as splicing snRNAs and the U7 snRNA involved in histone mRNA maturation (33–38). By extension of the concept used here, it seems reasonable to predict that alternative artificial ribozyme–protein complexes that function in different cellular compartments can be designed by creative use of other stabilizing and localizing determinants.
Beyond establishing an effective approach to localization and cleavage, the U3 snorbozyme system described has the key advantage of yielding a stable cleavage byproduct that can be quantified. Formation of this stable and, importantly, endogenously trimmed product provides the twin benefits of (i) distinguishing authentic in vivo cleavage from cleavage that may occur during or after RNA purification and (ii) the ability to measure product formation quantitatively. These properties allow detailed and accurate kinetics studies to be performed in vivo. Our initial results indicate that the in vivo performance of the trans-acting U3-based snorbozymes compares favorably with that of small model ribozyme and substrate molecules analyzed in vitro. The rate of cleavage is quite comparable, whereas substrate binding requires high snorbozyme concentrations.
The present study also demonstrates the feasibility of using yeast for in vivo ribozyme studies. Earlier results with this organism were much less striking, with only 20–50% reduction of target mRNA levels observed for a trans-acting ribozyme (39, 40). The success experienced here indicates that the powerful genetic advantages of yeast can now be exploited to characterize and optimize ribozyme performance in vivo.
Acknowledgments
We thank Dr. Molly Fitzgerald-Hayes for providing yeast strain MH2 and Drs. John J. Rossi, John M. Burke, and Thomas L. Mason for critical reading of the manuscript. The research was supported by National Institutes of Health Grant GM 19351 and National Science Foundation Grant MCB-9419007 (D.A.S. and M.J.F), a grant from the Medical Research Council of Canada (G.F. and R.C.), and National Institutes of Health Grants GM 54887 and GM 57071 (E.B. and R.H.S.). This paper is dedicated to the memory of one of the coauthors, Dr. Robert Cedergren, a wonderful scientist, mentor, and friend.
ABBREVIATION
- snoRNA
small nucleolar RNA
Appendix
Parameters Used in the in Vivo Kinetics Calculations.
The efficiency of the cis-acting snorbozyme in vivo is ≈90%. If the half-life of the U3 derivatives is similar to that of natural U3, estimated at 90 min (29), the rate of the ribozyme reaction is deduced to be 0.08 min−1 in vivo (for the formulas used in calculations see ref. 41). In trans, the best cleavage efficiency observed was better yet, for a ribozyme–substrate complementarity of 9:6 bp and expression of the ribozyme from a high-copy vector (≈10-fold excess over the target). Assuming an efficiency of >95%, the rate of reaction, kobs, is estimated at >0.15 min−1 (see below). When the ribozyme and the substrate have 9:9 bp of complementarity, the reaction rate, kobs, increased from 0.01 min−1 (60% cleavage) when the ribozyme was expressed from a low-copy vector, to 0.08 min−1 (90% cleavage) when the ribozyme was expressed from a high-copy vector. This effect suggests that for the ribozyme levels evaluated, the rate of ribozyme-substrate association is limiting in vivo. The ribozyme concentration is estimated to be in the range of 3 μM (low-copy vector) to 30 μM (high-copy vector) in the nucleolus, significantly higher than the concentration required to saturate the reaction in vitro (30). The volume of the yeast nucleolus is roughly 0.5 × 10−15 liters (1 μm × 1 μm × 0.5 μm; E.B., unpublished data). Assuming ribozyme expressed from a low-copy vector (1–3 copies per cell) occurs at a concentration approximating that of natural U3 (normally from two gene copies), i.e., about 1,000 molecules per cell, as reported by Hughes et al. (42), the ribozyme concentration is estimated to be 3 μM.
In Vivo Trans-Cleavage Model.
The ribozyme cleavage of an RNA target in trans and in vivo can be described by the following kinetic model: ![]()
where E is the ribozyme; S is the substrate; P1 and P2 are the two cleavage products; P1′ is exo-trimmed P1; and & signifies a complex. kdeg is the degradation rate of natural U3 and is assumed to be the same for all U3 derivatives involved here. Based on estimation of the U3 half-life as 90 min (29), kdeg should be ≈0.008 min−1. k2 is the rate of the cleavage step and is assumed to be much faster than kdeg. In agreement, k2 is estimated to be 0.08 min−1 in the cis-acting ribozyme experiment. k3 is the rate of conversion of P1 into P1′ and is also assumed to be much faster than kdeg. Indeed, k3 is much faster than k2, because P1 cannot be detected in vivo. The equilibrium attained in vivo implies that the concentration of each molecular species is constant over time. If we define the overall reaction rate, kobs, as the fraction of substrate which is cleaved per unit time, then kobs = k2 × [E&S]/[S0], where [S0] is the total concentration of substrate. From the equilibrium equations, it follows that in vivo, kobs = kdeg × [P1′]/[S0]. The value for kobs can be calculated experimentally, because kdeg is known, and [P1′]/[S0] ratio can be estimated from the Northern blots.
References
- 1.Rossi J J. Trends Biotechnol. 1995;13:301–306. doi: 10.1016/S0167-7799(00)88969-6. [DOI] [PubMed] [Google Scholar]
- 2.Couture L A, Stinchcomb D T. Trends Genet. 1996;12:510–515. doi: 10.1016/s0168-9525(97)81398-4. [DOI] [PubMed] [Google Scholar]
- 3.Symons R H. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- 4.Pyle A M. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 5.Bratty J, Chartrand P, Ferbeyre G, Cedergren R. Biochim Biophys Acta. 1993;1216:345–359. doi: 10.1016/0167-4781(93)90001-t. [DOI] [PubMed] [Google Scholar]
- 6.McKay D B. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- 7.Birikh K R, Heaton P A, Eckstein F. Eur J Biochem. 1997;245:1–16. doi: 10.1111/j.1432-1033.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- 8.Forster A C, Symons R H. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 9.Symons R H. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haseloff J, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 11.Usman N, Stinchcomb D T. In: Catalytic RNA. Eckstein F, Lilley D M J, editors. Berlin: Springer; 1996. pp. 243–264. [Google Scholar]
- 12.Sarver N, Cantin E M, Chang P S, Zaia J A, Ladne P A, Stephens D A, Rossi J J. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 13.Sullenger B A, Cech T R. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 14.Scanlon K J, Ishida H, Kashani-Sabet M. Proc Natl Acad Sci USA. 1994;91:11123–11127. doi: 10.1073/pnas.91.23.11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck J, Nassal M. Nucleic Acids Res. 1995;23:4954–4962. doi: 10.1093/nar/23.24.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke J M. Nat Biotechnol. 1997;15:414–415. doi: 10.1038/nbt0597-414. [DOI] [PubMed] [Google Scholar]
- 17.Montgomery R A, Dietz H C. Hum Mol Genet. 1997;6:519–525. doi: 10.1093/hmg/6.4.519. [DOI] [PubMed] [Google Scholar]
- 18.Lieber A, Strauss M. Mol Cell Biol. 1995;15:540–551. doi: 10.1128/mcb.15.1.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose M D, Winston F, Heiter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 20.Samarsky D A, Balakin A G, Fournier M J. Nucleic Acids Res. 1995;23:2548–2554. doi: 10.1093/nar/23.13.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samarsky D A, Fournier M J, Singer R H, Bertrand E. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samarsky D A, Fournier M J. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 25.Clouet-d’Orval B, Uhlenbeck O C. Biochemistry. 1997;36:9087–9092. doi: 10.1021/bi9710941. [DOI] [PubMed] [Google Scholar]
- 26.Shuster E O, Guthrie C. Cell. 1988;55:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- 27.Hendry P, McCall M. Nucleic Acids Res. 1996;24:2679–2684. doi: 10.1093/nar/24.14.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stage-Zimmermann T K, Uhlenbeck O C. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes J M X, Ares M., Jr EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson J B, Tucsl T, Eckstein F. In: Catalytic RNA. Eckstein F, Lilley D M J, editors. Berlin: Springer; 1996. pp. 173–196. [Google Scholar]
- 31.Hertel K J, Herschlag D, Uhlenbeck O C. Biochemistry. 1994;33:3374–3385. doi: 10.1021/bi00177a031. [DOI] [PubMed] [Google Scholar]
- 32.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brasch K, Ochs R L. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- 34.Carmo-Fonseca M, Pepperkok R, Carvalho M T, Lamond A I. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matera A G, Ward D C. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frey M R, Matera A G. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gall J G, Tsvetkov A, Wu Z, Murphy C. Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- 38.Wu C H, Murphy C, Gall J G. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- 39.Ferbeyre G, Bratty J, Chen H, Cedergren R. Gene. 1995;155:45–50. doi: 10.1016/0378-1119(94)00891-u. [DOI] [PubMed] [Google Scholar]
- 40.Ferbeyre G, Bratty J, Chen H, Cedergren R. J Biol Chem. 1996;271:19318–19323. doi: 10.1074/jbc.271.32.19318. [DOI] [PubMed] [Google Scholar]
- 41.Donahue C P, Fedor M J. RNA. 1997;3:961–973. [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes J M X, Konings D A M, Cesareni G. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]