Abstract
A new surface protein, named NspA, which is distinct from the previously described Neisseria meningitidis outer membrane proteins was identified. An NspA-specific mAb, named Me-1, reacted with 99% of the meningococcal strains tested indicating that the epitope recognized by this particular mAb is widely distributed and highly conserved. Western immunoblotting experiments indicated that mAb Me-1 is directed against a protein band with an approximate molecular mass of 22,000, but also recognized a minor protein band with an approximate molecular mass of 18,000. This mAb exhibited bactericidal activity against four meningococcal strains, two isolates of serogroup B, and one isolate from each serogroup A and C, and passively protected mice against an experimental infection. To further characterize the NspA protein and to evaluate the protective potential of recombinant NspA protein, the nspA gene was identified and cloned into a low copy expression vector. Nucleotide sequencing of the meningococcal insert revealed an ORF of 525 nucleotides coding for a polypeptide of 174 amino acid residues, with a predicted molecular weight of 18,404 and a isoelectric point of 9.93. Three injections of either 10 or 20 μg of the affinity-purified recombinant NspA protein efficiently protected 80% of the mice against a meningococcal deadly challenge comparatively to the 20% observed in the control groups. The fact that the NspA protein can elicit the production of bactericidal and protective antibodies emphasize its potential as a vaccine candidate.
N eisseria meningitidis causes both endemic and epidemic diseases, principally meningitidis and meningococcemia (1, 2). This pathogenic bacteria primarily affects young children between 6 mo and 2 yr of age, but often infects teenagers (1). The incidence per year of meningococcal diseases during endemic periods is normally ∼1–3 cases per 100,000 in developed countries, but it can be as high as 500 per 100,000 during epidemics (2, 3).
N. meningitidis is classified into 12 serogroups based on the immunological characteristics of the capsular polysaccharides found at their surface. Within serogroups, different serotypes, subtypes, and immunotypes can be identified based on the antigenic specificity of the major outer membrane (OM)1 proteins and LPS (4). Approximately 90% of all meningococcal diseases worldwide are caused by isolates of serogroups A, B, and C (5). Vaccines based on the capsular polysaccharides of serogroups A, C, W-135, and Y were developed and proved efficient to control outbreaks and epidemics of meningococcal diseases (6). However, these vaccines are poorly immunogenic in very young children. Moreover, they do not induce immunological memory and the duration of the protection they provide is relatively short (5, 7–11). Recently, it was demonstrated that conjugation of capsular polysaccharides of serogroups A and C to carrier proteins resulted in a better immunogenicity and a longer persistence of specific antibodies against isolates of these serogroups (12–16). Attempts to develop an efficient vaccine against serogroup B isolates, which are responsible for 50–70% of the meningococcal disease in the developed countries were unsuccessful because the group B capsular polysaccharide is not a good immunogen in human, inducing only a poor IgM response of low specificity which is not protective (17–19). Furthermore, the presence of closely similar, cross-reactive structures in the glycoproteins of neonatal human brain tissue might discourage attempts to improve the immunogenicity of serogroup B polysaccharide (10).
To develop a vaccine effective against meningococci of serogroup B several non-capsular surface structures are under investigation (6, 10). Importantly, the presence of bactericidal antibodies against N. meningitidis have been strongly correlated with human immunity and protection (20–22). For that reason, it is believed that non-capsular surface antigens shown to stimulate bactericidal antibodies should be considered as the prime vaccines candidates (6). Early studies using sera of immunized volunteers and convalescent patients indicated that certain meningococcal surface proteins such as the ones responsible for serotype specificity and LPS could induce bactericidal antibodies and be involved in protection (23, 24). mAbs were then used to clearly establish the protective potential of certain meningococcal major surface proteins such as the PorA (class 1), PorB (class 2/3), and Opc (class 5C) (25–28).
Different vaccines based on OM proteins were recently evaluated in clinical trials and efficiency between 50 and 80% were recorded (6, 10). These first generation OM proteins vaccines often induced protection against a limited number of strains. Thus, these vaccines could be used during meningococcal epidemics when the antigenic variation of the meningococci causing diseases is relatively low. The specificity of the bactericidal antibodies induced by these vaccines was determined to be directed mainly against PorA and Opc proteins (29, 30). However, the PorA-specific bactericidal antibodies were found to be directed against epitopes located in surface-exposed highly variable regions (31). Moreover, the Opc protein was shown to be produced by only 60% of strains of different serogroups (32), and by ∼20% of serogroup B isolates (33). To improve the protection conferred by the PorA protein, strategies such as multivalent PorA vaccines or the incorporation of additional epitopes on PorA protein are presently under study (34–36). Proteins induced by iron limitation such as FrpB and Tbp-2 are also likely vaccine candidates, but they also show type specificity with respect to the induction of bactericidal antibodies (37–40). Antigenically conserved proteins such as the Lip (or H.8) (41, 42) and the Rmp (or class 4) (43) proteins were identified in the meningococcal OM, but antibodies directed against these proteins were found to be nonbactericidal. Moreover, high concentrations of antibodies to the Rmp protein were also reported to block the bactericidal activity of antibodies directed against PorA protein and could prevent the efficient killing of meningococcal cells (43).
In the present report, we described a new highly conserved protein, called NspA for Neisserial surface protein A, which was shown to be present in the OM of all N. meningitidis strains tested. An mAb, named Me-1, which is directed against the NspA protein was found to be bactericidal and to passively protect BALB/c mice against experimental infections. To further characterize this protein and to clearly establish its protective potential, the nspA gene was identified, sequenced and cloned into a low copy expression vector to obtain large quantities of the recombinant protein. In addition, we present data that demonstrate that the injection of purified recombinant NspA protein efficiently protect BALB/c mice against a bacterial challenge with a lethal dose of a meningococcal strain of serogroup B.
Materials and Methods
Bacterial Strains and Plasmids.
A collection of 250 N. meningitidis strains was used in this study. The panel of strains included 22 isolates of serogroup A, 44 isolates of serogroup B, 56 isolates of serogroup C, 1 isolate of serogroup 29-E, 7 isolates of serogroup W-135, 1 isolate of serogroup X, 3 isolates of serogroup Y, 2 isolates of serogroup Z, and 114 isolates that were not serogrouped. The following Neisseria species were also tested: 43 N. gonorrhoeae, 4 N. cinerea, 7 N. lactamica, 1 N. flava, 1 N. flavescens, 3 N. mucosa, 4 N. perflava/sicca, 4 N. perflava, 1 N. sicca, 1 N. subflava and 5 Moraxella catarrhalis. The Neisseria strains were grown overnight on chocolate agar plates at 37°C in an atmosphere containing 5% CO2. Cultures were stored at −70°C in heart infusion broth (Difco Laboratories, Detroit, MI) with 20% (vol/vol) glycerol (Sigma Chemical Co., St. Louis, MO). These isolates were obtained from the Caribbean Epidemiology Centre (Port of Spain, Trinidad); Children's Hospital of Eastern Ontario (Ottawa, Canada); Department of Saskatchewan Health (Regina, Canada), Laboratoire de Santé Publique du Québec (Montréal, Canada); Max-Planck Institut fur Molekulare Genetik (Berlin, Germany); Montreal Children Hospital (Montreal, Canada), Victoria General Hospital (Halifax, Canada), Laboratory Centre for Disease Control (Ottawa, Ontario), and our own strains collection.
The following control bacterial species were also used: Alcaligenes feacalis (ATCC 8750; American Type Culture Collection, Rockville, MD), Bordetella pertussis 9340, Citrobacter freundii (ATCC 2080), Edwarsiella tarda (ATCC 15947), Enterobacter cloaca (ATCC 23355), Enterobacter aerogenes (ATCC 13048), Flavobacterium odoratum (LSPQ2135), Haemophilus influenzae type b (Eagan strain), Klebsiella pneumoniae (ATCC 13883), Proteus rettgeri (ATCC 25932), Proteus vulgaris (ATCC 13315), Pseudomonas aeruginosa (ATCC 9027), Salmonella typhimurium (ATCC 14028), Serratia marcescens (ATCC 8100), Shigella flexneri (ATCC 12022), Shigella sonnei (ATCC 9290) and Streptococcus pnenumoniae WU2.
Escherichia coli JM109 (endA1 recA1 syrA96 thi-1 hsdR17(rk− mk+) relA1 supE44 λ− Δ(lac-proAB) [F'traD36 proAB lacIqZ\xc6 M15]) (44), LE392 (F− hsdRS514(rk−,mk−) supE44 supF58 lacY1 or Δ(lacIZY)6 galK2 galT22 metB1 trpR55 λ−) (45), and BL21(DE3) (F− dcm ompT hsdS(rB− mbB−) gal λ(DE3) (46) were grown on LB agar or broth (GIBCO BRL, Gaithersburg, MD) at 37°C. Where appropriate E. coli strains or transformants were grown on media containing 25 μg of ampicillin (Sigma) per ml. The low copy plasmid pWKS30, which carries a gene for resistance to ampicillin, a multiple cloning region flanked by the T7 and T3 RNA polymerase promoters, the lacZα gene, and the bacteriophage f1 origin of replication for production of single stranded DNA was described in details elsewhere (47).
Antigen Preparation.
Lithium chloride extractions of OM preparations from one N. gonorrhoeae strain A1 and nine meningococcal strains two of serogroup A (604A and Z4063), one of serogroup B (608B [B:2a:P1.2:L3]), two of serogroup C (2241C and 59C), one of serogroup 29-E, one of serogroup W-135, one of serogroup Y (SLATY) and one of serogroup Z (SLATZ) were performed as described by Brodeur et al. (28). Protein concentrations were determined by the Lowry method adapted to membrane fractions (48).
Generation of mAb Me-1.
Mice were injected intraperitoneally with 20 μg of OM preparation extracted from the meningococcal strain 604A mixed with Freund's complete adjuvant (GIBCO BRL), and then 3 wk later were injected with 20 μg of OM preparation extracted from the meningococcal strain 2241C mixed with Freund's incomplete adjuvant. 3 d before the fusion procedure, the selected mouse received a final intravenous injection of 10 μg of OM preparation extracted from the meningococcal strain 2241C. The fusion protocol used to produce the hybridoma cell lines was described previously by Hamel et al. (49). Hybrid clone supernatants were tested for specific antibody production by ELISA, as previously described (49), using meningococcal OM preparations as coating antigens (7.5 μg of protein per ml). Specific hybrids were cloned by sequential limiting dilutions, expanded, and frozen in liquid nitrogen. The class, subclass and light-chain type of the mAb were determined by ELISA with commercially available reagents (Southern Biotechnology Associates Inc., Birmingham, AL). A dot enzyme immunoassay was used for the rapid screening of the mAb against the bacterial strains as described elsewhere (50).
SDS-PAGE, Western Immunoblotting, Plaque, and Colony Blot Assays.
Meningococcal OM preparations were resolved by electrophoresis by using the discontinuous buffer system of Laemmli (51) with 14% (wt/vol) gels, and Western blot analyses were performed as described previously with the following modifications (52). The proteins were transferred from the gels to the nitrocellulose membranes using a semi-dry apparatus (Bio-Rad Labs., Hercules, CA). A current of 60 mA per gel was applied for 20 min in the electroblot buffer consisting of 25 mM Tris-HCl, 192 mM glycine, and 20% (vol/vol) methanol, pH 8.3. The presence of LOS in the different antigen preparations was visualized by silver staining as described by Tsai and Frasch (53). Recombinant plaques were blotted onto nitrocellulose membranes using the following protocol. The plates were incubated 15 min at −20°C to harden the top agar, and nitrocellulose membranes were gently applied onto the surface of the plates for 30 min at 4°C to absorb the proteins produced by the recombinant viral clones. The membranes were then washed in PBS Tween 0.02% (vol/vol). The plaque blots were sequentially incubated with tissue culture supernatant of mAb Me-1, peroxidase-labeled goat anti–mouse immunoglobulin (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) and the o-dianisidine (Sigma) peroxidase substrate as described previously (52). Colony blots of E. coli transformants were performed as described by (45).
Cloning the Gene for the NspA Protein.
A λGEM-11 genomic DNA library from N. meningitidis strain 608B was constructed according to the manufacturer's recommendations (Promega, Madison, WI). The recombinant phages were used to infect E. coli strain LE392 which was then plated onto LB agar. The plaques were screened with mAb Me-1 and the reactive clones were plaque purified twice. The phage DNA from one positive clone was extracted and purified from 10 ml of phage lysate using the LambdaSorb Phage Adsorbent following the manufacturer's recommendations (Promega). The DNA was then digested with SacI, and the fragments were purified by phenol extraction from low melting agarose gels, and ligated into the SacI-digested plasmid pWKS30 and transformed into E. coli strain JM109 by electroporation according to the manufacturer's recommendations (Bio-Rad). To further reduce the size of the insert, the SacI fragment was digested with ClaI, and the fragments were purified by agarose gel electrophoresis and ligated into the ClaI-digested plasmid pWKS30 and the recombinant plasmid was used to transform E. coli strain JM109 by electroporation. The transformants were screened with mAb Me-1, and two plasmids recovered from two reactive clones were designated pN2202 and pN2203.
DNA Sequencing.
The Double Stranded Nested Deletion Kit from Pharmacia Biotech Inc. (Piscataway, NJ) was used according to the manufacturer's instructions to generate a series of nested deletions from the recombinant plasmids pN2202 and pN2203. The resulting truncated inserts were sequenced from the M13 forward primer with the Taq Dye Deoxy terminator Cycle Sequencing Kit using an Applied Biosystems Inc. (Foster City, CA) automated sequencer model 373A according to the manufacturer's recommendations. Both strands of the insert were sequenced. Alignment of the sequences was accomplished by using Geneworks Software (Intelligenetics, Inc., Mountain View, CA). Sequence homology searches in GenBank and Swissprot databases were accomplished using the Geneworks, BLAST, BLIZT, and FASTA software.
NH2-terminal Amino Acid Sequence Analysis of the Native Meningococcal NspA Protein.
The protein present in the OM of N. meningitidis strain 608B were resolved by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad) by the method of Matsudaira (54). The PVDF membrane was stained with Coomassie Blue, the NspA band was excised, and then the NH2-terminal amino acid sequence was analyzed with an automated protein sequencer by the Service de Séquence des Protéines de l'Est du Québec (Centre de Recherche du CHUL, Québec, Canada).
Production and Purification of the Recombinant NspA Protein.
The recombinant NspA protein was purified from the culture supernatant by affinity chromatography. To generate the affinity chromatography media, mAb Me-1 was first purified from ascitic fluid using protein A–Sepharose CL-4B (Pharmacia) and was then immobilized on CNBr-activated Sepharose 4B (Pharmacia) according to the manufacturer's instructions. To obtain the NspA recombinant protein, the plasmid pN2202 was used to transform E. coli strain BL21(DE3) by electroporation. An overnight culture of the transformed E. coli strain BL21(DE3) was inoculated in LB broth containing 25 μg/ml of ampicillin and was incubated 4–5 h at 37°C with agitation. No induction step was used since the expression of the NspA protein was under the control of the gene's own promoter. The bacterial cells were removed from the culture media by centrifugation at 12,000 g for 30 min at 4°C. The supernatant was filtered onto a 0.22-μm membrane and concentrated using an ultrafiltration apparatus (Amicon Inc., Beverly, MA) and a membrane with a molecular cut off of 10,000. To solubilize the membrane vesicles, Empigen BB (Calbiochem Novabiochem, La Jolla, CA) was added to the concentrated culture supernatant to a final concentration of 1% (vol/vol), incubated at room temperature for one h, and then dialyzed overnight against 10 mM Tris-HCl buffer, pH 7.3, containing 0.05% Empigen (vol/vol). The antigen preparation was added to the affinity gel and incubated overnight at 4°C with constant agitation. The gel was poured into a chromatography column and washed extensively with of 10 mM Tris-HCl buffer, pH 7.3, containing 0.05% Empigen (vol/vol). The NspA protein was eluted from the column with 1 M LiCl in Tris-HCl buffer, pH 7.3. The fractions containing the NspA protein were pooled and dialyzed extensively against 10 mM Tris-HCl buffer, pH 7.3, containing 0.05% Empigen (vol/vol).
Bactericidal Assay.
The bactericidal activity of purified mAb Me-1 was tested in vitro as described previously (55) with the following modifications. 10 μl of a meningococcal suspension adjusted to 3 × 105 CFU/ml was dispensed into the wells of sterile flat-bottom 96-well plates (GIBCO BRL) containing 70 μl of PBS with 0.15 mM CaCl2, 0.5 mM MgCl2, and 0.1% BSA, 10 μl of guinea pig serum as a source of complement, and 10 μl of different dilution of protein A purified mAb Me-1, or heat-inactivated mouse serum. Duplicate antigen/antibody mixtures were incubated either in the absence of complement or with heat-inactivated guinea pig serum. After incubation for 1 h at 37°C, 10 μl of each mixture were plated onto chocolate agar plates. The plates were incubated overnight at 37°C, after which time the bacterial colonies were counted.
Protection Experiments.
The mouse model of infection used for the passive and active protection experiments was described previously (28). In brief, the N. meningitidis strain 608B (B: 2a: P1.2:L3) was passaged twice in mice before the bacterial challenge. For inoculation of mice, meningococci were removed from the chocolate agar plates after ∼20 h of incubation and suspended in PBS. The bacterial suspension was adjusted to an optical density of 0.25 (λ = 490 nm) and diluted to obtain 1,000 CFU per ml in brain heart infusion broth (Difco) containing 4% mucin (from porcine stomach type II; Sigma) and 1.5% hemoglobin (Oxoid Ltd., Nepean, Canada). All inocula were verified by colony counts. The number of surviving mice were recorded 24 and 72 h after the bacterial challenge. The mice that survived a bacterial challenge were always monitored for an additional 2 wk to see if there was any sign of infection. To evaluate the protective potential of mAb Me-1, 600 μl of ascitic fluid per mouse was injected intraperitoneally 18 h before the challenge. The mice in the control groups received the same volume of ascitic fluid containing an unrelated mAb or PBS. For the active protection experiments, groups of mice were injected three times at 3-wk intervals with 10 or 20 μg of affinity-purified NspA recombinant protein and 25 μg of QuilA (CedarLane Laboratories, Hornby, Ontario, Canada) as the adjuvant. Control mice were injected with either 20 μg of BSA (Sigma), concentrated E. coli BL21(DE3) supernatant, or PBS. 2 wk after the third injection the mice were used for the protection experiments. Blood samples were collected via the periorbital sinus before each immunization and 10 d after the third injection for mice receiving 20 μg of purified NspA protein or E. coli concentrated supernatant. The specific titer of each of these sera was determined by ELISA against meningococcal OM preparation extracted from the meningococcal strain 608B. The serum dilution for which an absorbance reading of 0.1 (λ = 410/630 nm) was recorded after background subtraction was considered to be the titer of this serum. The bactericidal activity of these sera was evaluated as described previously and the serum dilution giving 50% killing of meningococci was considered to be the bactericidal titer.
Results
Identification of the Meningococcal NspA Protein.
Mice were immunized with different combinations of meningococcal OM preparations in order to obtain hybridoma clones secreting mAbs specific for highly conserved surface antigens. mAb Me-1 was derived from one of these fusion experiments. The mouse that was used for that particular fusion was successively immunized with OM preparations obtained from one serogroup A strain (604A) and one serogroup C strain (2241C). mAb Me-1 was selected because it reacted with all nine meningococcal OM preparations tested, but did not recognize the OM preparation extracted from N. gonorrhoeae strain A1. The hybridoma cell line secreting mAb Me-1 was subcloned twice by limiting dilution. The class, subclass, and light chain specificity of this mAb were determined to be IgG2aκ. A dot enzyme assay using whole cell preparations as the source of antigen was used to evaluate the specificity and the antigenic conservation of the epitope recognized by mAb Me-1. mAb Me-1 reacted with 248 out of the 250 meningococcal strains tested (99.2%). The panel of strains used for the dot assay is described in detail in Materials and Methods. Only one non-typable meningococcal strain and one serogroup B strain were not recognized by the mAb. This result clearly indicates that the epitope recognized by this particular mAb is widely distributed among meningococcal strains. It also suggests that the particular epitope recognized by mAb Me-1 is not restricted to serologically related isolates since the panel of meningococcal strains were chosen to represent the major disease causing groups of strains. Moreover, two other NspA-specific mAbs, called Me-2 and Me-7, which were generated from other fusion experiments, reacted with the two meningococcal strains that were not recognized by mAb Me-1 (56). This latter result indicates that the epitopes recognized by these NspA-specific mAbs are present in the OM of all meningococcal strains tested so far. The reactivity of the mAb Me-1 was also evaluated with other Neisseria species. Out of the 74 non-meningococcal strains tested, mAb Me-1 reacted only with one N. lactamica strain, but not with any other Neisseria species tested. This mAb did not recognize any other gram-negative and gram-positive bacterial strains tested.
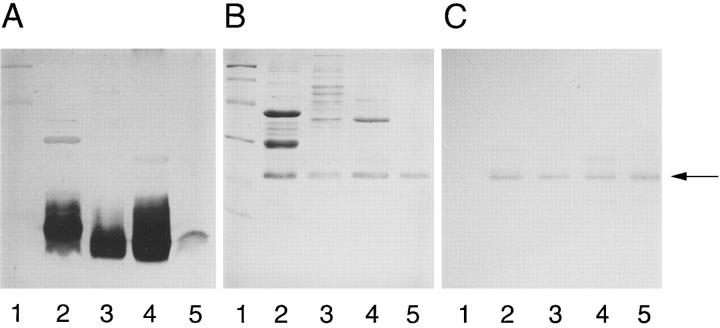
Western immunoblotting experiments indicated that mAb Me-1 reacted with a protein band with apparent molecular mass of 22,000 (Fig. 1 B). This mAb also reacted with a minor protein band with apparent molecular mass of 18,000. To observe the minor band, the total protein concentration of 5 μg that is normally applied to each well had to be increased to 7.5 or 10 μg. Analysis of SDS-PAGE and their corresponding Western immunoblots indicated that despite the variation normally observed in the OM protein migration profiles, the molecular mass of the 22,000 protein band is constant among meningococcal isolates. However, the amount of 22-kD protein band present in the different OM preparations varied from one meningococcal strain to another.
Figure 1.
Coomassie bluestained 14% SDS-PAGE gel (A) and Western immunoblots (B) showing the reactivity of mAb Me-1 with different meningococcal OM preparations (7.5 μg/lane): serogroup A strain Z4063 (lane 2); serogroup B strain 608B (lane 3); serogroup C strain 59 (lane 4); serogroup W-135 strain (lane 5); and serogroup Y strain SLATY (lane 6). The arrow indicates the location of the 22-kD NspA protein band. On the left, mol wt standard proteins are phosphorylase b (97,400), BSA (66,200), ovalbumin (45,000), carbonic anhydrase (31,000), soybean trypsin inhibitor (21,500), and lysozyme (14,400).
Cloning of the nspA Gene.
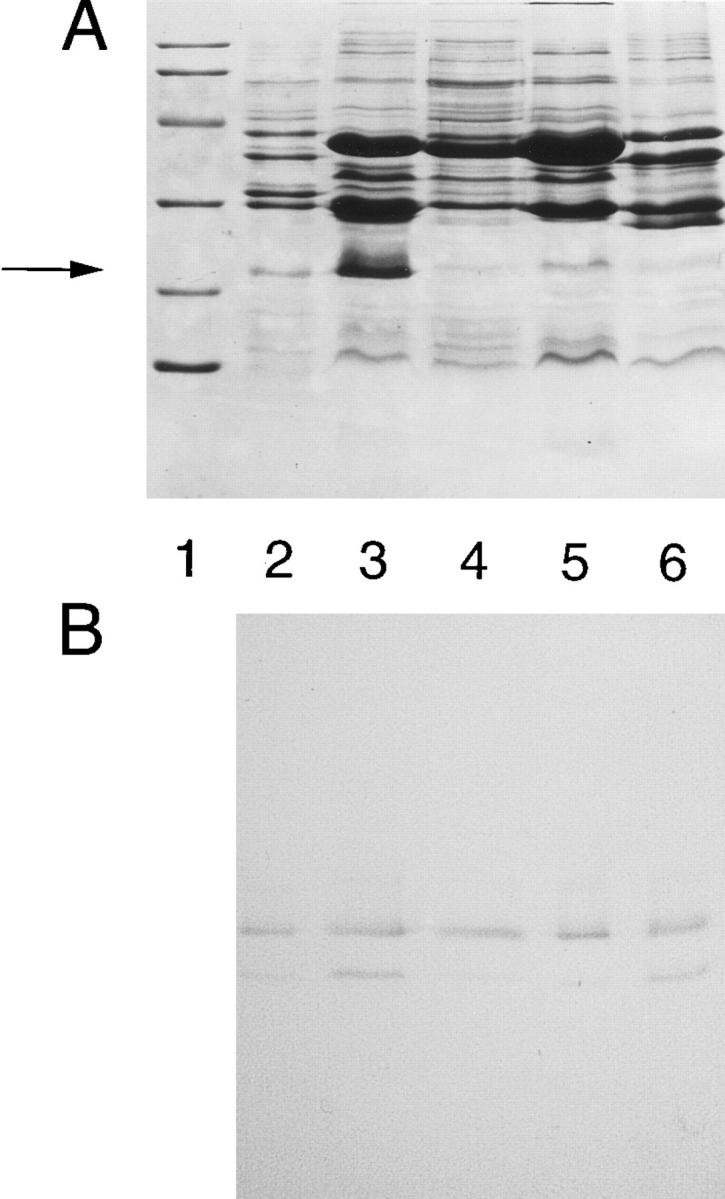
A meningococcal chromosomal library was constructed from N. meningitidis strain 608B in LambdaGEM-11. The genomic DNA was partially digested with Sau3AI and fragments ranging between 9 and 23 kb were purified and ligated to the BamHI sites of the LambdaGEM-11 bacteriophage arms. 19 positive plaques were identified after the immunoscreening of the library with mAb Me-1. After amplification and DNA purification, one viral clone which had a 13-kb insert was selected for the subcloning experiments. After digestion of this clone with SacI, two fragments of 5 and 8 kb were obtained, purified, and ligated into the SacI restriction site of the plasmid pWKS30. E. coli strain JM109 transformed with the recombinant plasmids were screened with mAb Me-1. Positive colonies were observed only when the bacteria were transformed with the plasmid carrying the 8-kb insert. Western blot analysis of the positive clones showed that the protein expressed by E. coli strain JM109 was complete and migrated on SDS-PAGE gel like the N. meningitidis NspA protein. A purified 8-kb fragment obtained from a positive clone was digested with ClaI to further reduce the size of the insert. The resulting 2.75-kb fragment was then ligated into the ClaI site of the pWKS30 plasmid. Western blot analysis of the resulting clones clearly indicated once again that the protein expressed by E. coli strain JM109 was complete and migrated on SDS-PAGE gel like the native N. meningitidis NspA protein (Fig. 2, B and C, lane 2). Both the 22,000 and the minor 18,000-protein band were produced by recombinant E. coli cells and were recognized by mAb Me-1. Restriction analysis indicated that two clones, designated pNP2202 and pNP2203, carried the 2.75-kb insert in opposite orientations.
Figure 2.
Silver-stained (A), Coomassie blue-stained (B) 14% SDSPAGE gels and corresponding Western immonoblots (C) showing the affinity-purified NspA protein that was injected to BALB/c mice used for the active protection experiments and the reactivity of mAb Me-1 with 5 μg of meningococcal OM preparation extracted from strain 608B (lane 2), 5 μg of OM protein preparation obtained from E. coli strain BL21(DE3) transformed with plasmid pN2202 (lane 3), concentrated culture supernatant obtained after 4 h of incubation of E. coli strain BL21(DE3) transformed with plasmid pN2202 (lane 4) and 2 μg of affinity-purified NspA protein (lane 5). The arrow indicates the location of the 22-kD NspA protein band. On the left, mol wt standard proteins are Phosphorylase b (97,400), BSA (66,200), ovalbumin (45,000), carbonic anhydrase (31,000), soybean trypsin inhibitor (21,500), and lysozyme (14,400).
Sequence Analysis of nspA.
The two plasmids pN2202 and pN2203 were selected to proceed with the sequencing of the nspA gene. Both expressed the NspA protein, indicating that the gene is being transcribed from a promoter located on the DNA insert. Nested sets of deletions were made for both clones by ExoIII digestion. The resulting truncated inserts were then sequenced from the M13 forward primer present on the plasmid pWKS30. The DNA sequence deduced from the DNA templates of the deletion subclones was used to design complementary oligonucleotide primers to complete the sequencing of the entire open reading frame (ORF) encoding the nspA gene. Fig. 3 shows the sequence of a portion of the 2.75-kb insert which contains the nspA gene. This sequence was submitted to GenBank and assigned the accesssion number U52066. The sequence analysis revealed an ORF of 525 nucleotides which start with an ATG codon at nucleotide 143 and ends with a TGA stop codon at nucleotide 667. A putative ribosome binding (AGGA) site was identified eight nucleotides upstream from the start codon and is boxed in the Fig. 3. This open reading frame codes for a 174–amino acid residues polypeptide with a predicted pI of 9.93 and a molecular mass of 18,404.
Figure 3.

Nucleotide and amino acid sequences of the gene encoding the N. meningitidis strain 608B NspA protein. The open reading frame of 525 bp extends from the start codon at base 143 to the stop codon at base 667. A 19–amino acid leader peptide is underlined, and a putative ribosome binding site is boxed. These sequence data are available from GenBank under accession number U52066.
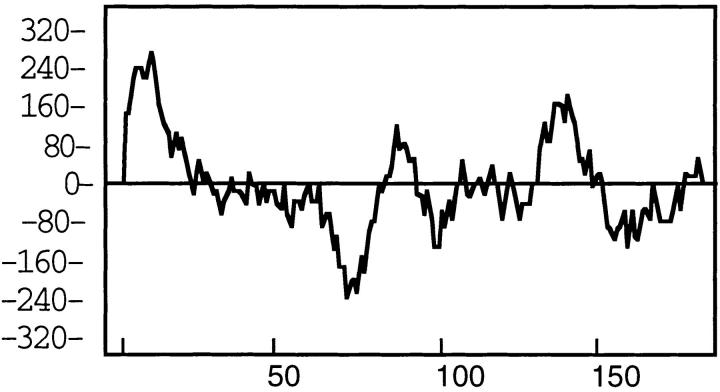
NH2-terminal amino acid analysis of the meningococcal 22-kD protein band present in the OM preparation of strain 608B indicated that the first 10 amino acid residues were E-G-A-S-G-F-Y-V-Q-A. Comparison of this sequence with the deduced amino sequence obtained after sequencing the nspA gene indicated the existence of a 19– amino acid residues leader peptide which is cleaved in the mature meningococcal protein (underlined in Fig. 3). Moreover, the NH2-terminal portion of the predicted polypeptide was found using Kyte-Doolittle analysis (Fig. 4) to be hydrophobic, which is typical of leader peptides of other known membrane proteins (57). The low amount of 18-kD protein band present in the meningococcal OM preparation after separation on SDS-PAGE prevented the same analysis. For that reason, affinity-purified recombinant protein was used to overload a gel in order to obtain enough of the minor protein band to resolve its NH2-terminal amino acid sequence. The first five amino acid residues of the minor 18-kD band were determined to be E-G-A-S-G-F, which corresponded exactly to the first amino acid residues of the mature meningococcal protein without its leader peptide. This result and the observed reactivity of mAb Me-1 with both protein bands suggest that the 22- and the 18-kD protein bands are related and possibly are the same protein.
Figure 4.
Schematic representation of a Kyte-Doolittle hydrophobicity plot of the N. meningitidis NspA protein.
No significant homology was found between the deduced amino acid sequence of the NspA protein and the sequences compiled in GenBank and Swissprot databases indicating that this protein has never been described previously. These searches indicated the existence of weak homology of the NspA protein with the Neisseria opacity (Opa) family of protein. Pairwise comparison of the different Opa protein sequences with the NspA sequence revealed identities between 20 and 28%. Closer analysis indicated that the observed homology with the Opa proteins seems to be clustered in two particular regions on the NspA protein. The first region that showed 62.5% homology with some of the Opa proteins is composed of 24 amino acid residues located between position 126 and 149. The second region which showed 60% homology with the carboxyl-terminal regions of Opa proteins, is made up of the last 10 amino acid residues at the carboxyl-terminal of the NspA protein.
Production and Purification of the Recombinant NspA Protein.
The purification protocol was based on the observation that the recombinant NspA protein produced by E. coli BL21(DE3) strain carrying the plasmid pNP2202 can be found in large amounts in the OM, but can also be obtained from the culture supernatant in which it is one of the most abundant proteins (Fig. 2 B). Preliminary experiments indicated that an induction step using isopropylβ-d-thiogalactopyranoside (IPTG) was not necessary since it did not significantly increase the level of NspA protein which was constitutively produced by E. coli BL21(DE3) strain transformed with plasmid pN2202. This result indicated that this neisserial promoter can efficiently control the expression of the nspA gene in E. coli. Therefore, the culture supernatant was the material used to purify the recombinant NspA protein using affinity chromatography. Silver-stained gel indicated that the amount of E. coli LPS present after affinity chromatography is greatly reduced compared to the amount initially present in the concentrated supernatant (Fig. 2 A). The recombinant NspA protein was still recognized by mAb Me-1 after solubilization and purification (Fig. 2 C). The purity of the NspA protein used to immunize mice was estimated by SDS-PAGE to be >90%.
Determination of the Bactericidal Activity and the Protective Potential of mAb Me-1.
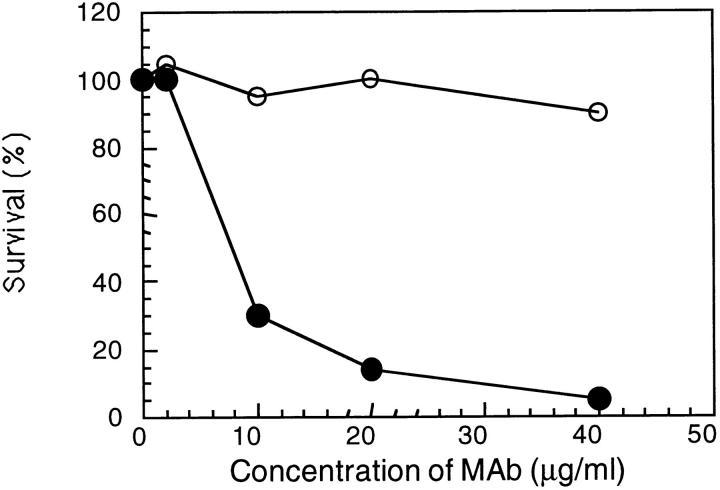
To demonstrate the bactericidal activity of mAb Me-1 an in vitro assay was performed using guinea pig serum and protein A–purified mAb. The bactericidal activity of mAb Me-1 against the meningococcal strain 608B is presented in Fig. 5. 7 μg/ml of purified mAb Me-1 were required to kill 50% of the meningococcal cells. Higher concentrations of purified mAb resulted in a sharp decrease in the recorded CFU ⩽100%. Heat-inactivation for 30 min at 56°C of the guinea pig serum completely abolished the bactericidal activity of mAb Me-1. 50% of killing were recorded for three other meningococcal strains, one of each serogroup A (strain Z4063), B ([B: 15:P1.7] strain 164), and C (strain 3), when 20 μg/ml of purified Me-1 was used in the bactericidal assay.
Figure 5.
Bactericidal activity of protein A–purified mAb Me-1 against N. meningitidis strain 608B. Guinea pig serum (closed circles) or heat inactivated guinea pig serum (open circles) were used as the source of complement to evaluate the bactericidal activity.
The ability of mAb Me-1 to passively protect mice against a lethal infection with the meningococcal strain 608B was evaluated and the combined results from three separate experiments are presented in Table 1. Intraperitoneal injection of 600 μl of ascitic fluid containing mAb Me-1 18 h before the bacterial challenge with a 1,000 CFU increased the survival rate of mice ⩽76% comparatively to 30% observed in the control groups receiving ascitic fluid containing either an unrelated mAb or PBS.
Table 1.
Passive Protection of Balb/c Mice Conferred by mAb Me-1 Against Infection with N. meningitidis Strain 608B
| Groups | Number of living mice after the bacterial challenge* | % of survival (range)‡ | ||||
|---|---|---|---|---|---|---|
| 24 h | 72 h | |||||
| Me-1–injected mice§ | 29/30 | 23/30 | 76 | |||
| (60–100) | ||||||
| Control mice‖ | 10/30 | 9/30 | 30 | |||
| (0–40) | ||||||
The mice were challenged with 1 ml of a suspension containing 1,000 CFU of N. meningitidis strain 608B, 4% mucin (Sigma), and 1.6% hemoglobin (Oxoid).
Combined results of three separate protection experiments. The range indicates the lowest and highest survival rates obtained.
600 μl of ascitic fluid containing mAb Me-1 were injected intraperitoneally 18 h before the bacterial challenge.
The mice in the control group received 600 μl of ascitic fluid containing unrelated mAb or phosphate buffered saline.
Immunization of BALB/c Mice with Affinity-purified Recombinant NspA Protein and Protection Experiments.
Affinity-purified recombinant meningococcal NspA protein was used to immunize BALB/c mice in order to determine its ability to induce a protective immune response against challenge with a lethal dose of N. meningitidis 608B strain (Table 2). 80% of the mice immunized with three injections of either 10 or 20 μg of purified recombinant meningococcal NspA protein survived the bacterial challenge comparatively to 0 to 20% in the control groups. Survivors at 72 h did not succumb during an additional two weeks of observation. The mice in the control group injected with concentrated E. coli culture supernatant were not protected against the bacterial challenge indicating that the components present in the culture media and other E. coli antigens that might be present in small amounts after purification do not contribute to the observed protection against N. meningitidis.
Table 2.
Immunization of Balb/c Mice with Affinity-purified Recombinant NspA Protein Confers Protection against Subsequent Challenge with N. meningitidis Strain 608B
| Number of living mice after the bacterial challenge* | ||||||
|---|---|---|---|---|---|---|
| Groups | 24 h | 72 h | % Survival | |||
| 10 μg of purified NspA protein | 20/20 | 16/20 | 80 | |||
| 20 μg of purified NspA protein | 9/10 | 8/10 | 80 | |||
| 20 μg of BSA | 6/10 | 2/10 | 20 | |||
| 20 μg of concentrated E. coli supernatant | 7/10 | 2/10 | 20 | |||
| Phosphate buffered saline | 8/10 | 0/10 | 0 | |||
The mice were challenged with 1 ml of a suspension containing 1,000 CFU of N. meningitidis strain 608B, 4% mucin (Sigma), and 1.6% hemoglobin (Oxoid).
Serum samples were obtained before immunization and after the third injection but before the bacterial challenge for mice immunized with 20 μg of affinity-purified protein or E. coli concentrated supernatant. The titer of these sera were determined by ELISA using meningococcal OM preparation as coating antigen. The reciprocal serum titers varied from 4,800 to 51,200 for the mice injected with 20 μg of protein, but were below 200 for the mice in the control group. Western immunoblotting experiments confirmed that the antibodies present in the sera obtained from the immunized mice recognized the recombinant protein, but also reacted with the native meningococcal NspA protein present in the OM preparation (data not shown). The reciprocal bactericidal titers of these sera varied from 10 to 90 when tested against the homologous strain 608B. Bactericidal titers between 10 and 20 were recorded when these sera were tested against the meningococcal strain 44 (B:4: P1.12). This latter result indicated that the injection of recombinant NspA protein to mice generate cross-reactive bactericidal antibodies, and correlated well with the ELISA titers. No bactericidal activity was recorded for the sera obtained from mice in the control group. Analysis of the sera obtained from the two mice who died following the bacterial challenge revealed that they had the lowest ELISA titers (4,800) as well as no bactericidal activity.
Discussion
In addition to the well known major OM proteins, it is believed that other antigenically stable surface proteins could play an important role in the protection against N. meningitidis and could become for that reason interesting vaccine candidates. Already, it was demonstrated that minor meningococcal antigens such as iron-regulated proteins could induce bactericidal antibodies that protected mice against experimental infection (37, 38). We have identified a minor antigen which is present in the OM of all meningococcal strains tested so far. To our knowledge this protein, which was called NspA for Neisserial surface protein A, was not described previously. Interestingly, comparison of the deduced amino acid sequence obtained after sequencing the nspA gene and the protein sequences compiled in the available databases revealed that two regions located between amino acid residues 126 and 149 and the last 10 residues at the carboxyl end showed ∼60% homology with members of the Neisserial opacity protein (Opa) family of proteins. These Opa proteins appear to be involved in cells adherence and invasion of epithelial or endothelial human cells (58). The observed similarity is clustered in two regions that were described to be highly conserved among Opa proteins (59, 60). The first conserved region is located on Opa proteins between two highly variable regions, named HV1 and HV2, which display considerable heterogeneity in sequence and in length, while the second region is found at the carboxyl end. These results suggest that the NspA protein could somehow be a smaller homologue of the Opa proteins and for that reason could be implicated in the meningococcal pathogenesis. However, beside the observed homology in the conserved regions, the NspA protein does not share two of the well-known characteristics of the Opa proteins such as the antigenic variability and the translational control of their expression during phase variation by multiple repeats of the nucleotide sequence (CTCTT) (59, 60). This homology can also be explained by the fact that the two regions of the NspA protein could be included in functional domains that are common to several OM proteins. Indeed, these two regions are hydrophobic as determined by a Kyte-Doolittle analysis (Fig. 4) and could be part of transmembrane domains which would normally be antigenically conserved and embedded in the meningococcal OM. More information about the physico-chemical properties and the mechanism which control the expression of the proteins is needed to clearly associate the NspA protein with the Opa proteins or to any other known meningococcal surface protein.
Even if the biological function of the NspA protein is not known at the moment, it is possible to determine using mAb Me-1 whether this protein can induce an immune response that can protect against meningococcal infection. Mab Me-1 was shown to be bactericidal against four meningococcal strains, one serogroup A, two serogroup B, and one serogroup C, indicating that the observed bactericidal activity is not restricted to one particular strain. It has been well documented that serum bactericidal activity is the major defense mechanism against N. meningitidis and that protection against invasion by the bacteria correlates with the presence in human sera of anti-meningococcal antibodies (20, 21). Conclusive evidence for the importance of bactericidal antibodies came from observations that individuals with intact opsonophagocytic capacity (61), but with deficiencies in one of the terminal complement components, have enhanced risk of contracting meningococcal infections (62). To evaluate whether the observed bactericidal activity of mAb Me-1 is not limited to a particular group of strains, we are presently evaluating its activity against a larger panel of serologically distinct meningococcal strains in the presence of human complement. Additional information about the localization of the NspA protein at the surface of the meningococcal cells can also be deduced from the bactericidal activity of mAb Me-1. Already, analysis of the migration profiles of meningococcal OM preparations have shown that the NspA protein is present in these preparations (Fig. 1). The bactericidal assay results indicate that the epitope recognized by mAb Me-1 is exposed at the surface of intact meningococcal cells where it is accessible to the antibodies. We are presently using a radioimmunobinding assay (56) and immunoelectron microscopy to further study the exposure of the NspA protein at the surface of intact meningococcal cells and its accessibility to specific antibodies. It is important to note that unlike the common occurrence for OM proteins to have their highly conserved regions embedded in the membrane while their variable regions are surface exposed (63, 64), the epitope on the NspA recognized by mAb Me-1 is accessible at the surface of intact bacteria. Furthermore, mAbs can be used with different animal models of infection to demonstrate the protective potential of a particular bacterial antigen. Using this approach, mAbs directed against PorA (25–27), PorB (25, 27, 28), Opc (27, 65), and LPS (27, 66) were shown to passively protect against meningococcal experimental infection. The injection of the bactericidal mAb Me-1 considerably improved the survival rate of the mice against a deadly infection with the homologous meningococcal strain 608B (Table 1). The combined results obtained using the bactericidal assay and the passive protection clearly demonstrated that antibodies that are specific for the NspA protein can efficiently protect against an experimental meningococcal infection. A new set of experiments will have to be conducted in order to determine the extend of protection conferred by mAb Me-1 when other serologically distinct strains are used to challenge the mice.
There are several advantages in favor of recombinant proteins for vaccine development: production of large amount of protein, lack of contaminating undesirable material, and the possibility to genetically modify the protein to increase its immunogenicity or to reduce its toxicity. For all these reasons we decided to use recombinant NspA protein instead of the native protein to clearly establish the protective potential of this meningococcal minor surface antigen. It is important to ascertain that the configuration of the recombinant protein is close enough to the native protein to generate functionally active antibodies. Indeed, Idänpään-Heikkilä et al. (67) reported the production of recombinant PorA protein in Bacillus subtilis as inclusion bodies. They observed that the native-like epitopes necessary to generate bactericidal and protective antibodies were present only when the recombinant PorA protein was allowed to refold in the presence of LPS or when it was incorporated into liposome. In our case, analysis of migration profiles by SDS-PAGE of different antigenic preparations extracted from E. coli strain BL21(DE3) transformed with the plasmid pN2202 indicated that the recombinant NspA protein is found in the OM as well as secreted in the culture supernatant (Fig. 3). Since there was no extensive solubilization and extraction steps necessary, we decided to purify the recombinant NspA protein from the culture supernatant by affinity chromatography. We hypothesized that the recombinant NspA protein, which was secreted in the culture supernatant through the bacterial OM, would be correctly folded and would generate functionally active antibodies. Data obtained from the protection experiments and the subsequent analysis of the immune sera confirmed that the recombinant protein shared enough immunological characteristics with the native protein to induce such antibodies. Indeed, 80% of the mice immunized with the recombinant NspA protein survived a deadly meningococcal challenge (Table 2). This result clearly linked the observed protection to the development of a specific immune response. Analysis of in vitro bactericidal results also indicated that cross-reactive bactericidal antibodies were present in the sera obtained from all the mice who survived the bacterial challenge.
In this report, we have presented results demonstrating that the recombinant NspA protein can induce an immune response that can protect against a lethal meningococcal infection. We also presented data indicating that this protein is present in the OM of all meningococcal strains where it is accessible to the antibodies at the surface of living cells. For all these reasons we believe that this protein possesses all the important characteristics to be considered as a potential vaccine candidate. We are presently constructing meningococcal mutant strains in order to study the function of the NspA protein and to determine its possible role in the meningococcal pathogenesis.
Footnotes
1 Abbreviations used in this paper: pI, isoelectric point; OM, major outer membrane; Opa, opacity protein; PVDF, polyvinylidene difluoride.
References
- 1.Peltola H. Meningococcal disease: still with us. Rev Infect Dis. 1983;5:71–91. doi: 10.1093/clinids/5.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz, B., P.S. Moore, and C.V. Broome. 1989. Global epidemiology of meningococcal disease. Clin. Microbiol. Rev. 2(Suppl.):S118–S124. [DOI] [PMC free article] [PubMed]
- 3.Whittle HC, Greenwood BM. Meningococcal meningitis in the northern savanna of Africa. Trop Doct. 1976;6:99–104. doi: 10.1177/004947557600600303. [DOI] [PubMed] [Google Scholar]
- 4.Frasch CE, Zollinger WD, Poolman JT. Serotype antigens of Neisseria meningitidisand a proposal scheme for designation of serotypes. Rev Infect Dis. 1988;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 5.Frasch, C.E. 1989. Vaccines for prevention of meningococcal disease. Clin. Microbiol. Rev. 2(Suppl.):S134–S138. [DOI] [PMC free article] [PubMed]
- 6.Frasch, C.E. 1995. Meningococcal vaccines: past, present and future. In Meningococcal Disease. K. Cartwright, editor. John Wiley & Sons Ltd, New York, NY. 245–283.
- 7.Artenstein MS, Gold R, Zimmerly JG, Wyle FA, Schneider H, Harkins C. Prevention of meningococcal disease by group C polysaccharide vaccine. N Engl J Med. 1970;282:417–420. doi: 10.1056/NEJM197002192820803. [DOI] [PubMed] [Google Scholar]
- 8.Peltola H, Mäkelä PH, Käyhty H, Jousimies H, Hera E, Hällström K, Sivonen A, Renkonen O-V, Pettay O, Karanko V, et al. Clinical efficacy of meningococcus group A capsular polysaccharide vaccine in children three months to five years of age. N Engl J Med. 1977;297:686–691. doi: 10.1056/NEJM197709292971302. [DOI] [PubMed] [Google Scholar]
- 9.Reingold AL, Broome CV, Hightower AW, Ajello GW, Bolan GA, Adamsbaum C, Jones EE, Phillips C, Tiendrebeogo H, Yada A. Age specific difference in duration of clinical protection after vaccination with meningococcal polysaccharide A vaccine. Lancet. 1985;2:114–118. doi: 10.1016/s0140-6736(85)90224-7. [DOI] [PubMed] [Google Scholar]
- 10.Poolman JT. Development of a meningococcal vaccine. Infect Agents Dis. 1995;4:13–28. [PubMed] [Google Scholar]
- 11.Lepow ML, Goldschneider I, Gold R, Randolph M, Gotschlich EC. Persistence of antibody following immunization of children with groups A and C meningococcal polysaccharide vaccines. Pediatrics. 1977;60:673–680. [PubMed] [Google Scholar]
- 12.Costantino P, Viti S, Podda A, Velmonte MA, Nencioni L, Rappuoli R. Development and phase 1 clinical testing of a conjugate vaccine against meningococcus A and C. Vaccine. 1992;10:691–698. doi: 10.1016/0264-410x(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 13.Jennings HJ, Lugowski C. Immunochemistry of groups A, B, and C meningococcal polysaccharide-tetanus toxoid conjugates. J Immunol. 1981;127:1011–1018. [PubMed] [Google Scholar]
- 14.Beuvery EC, Miedema F, van Delft R, Haverkamp K. Preparation and immunochemical characterization of meningococcal serogroup C polysaccharide-tetanus toxoid conjugates as a new generation of vaccines. Infect Immun. 1983;40:39–45. doi: 10.1128/iai.40.1.39-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beuvery EC, Kaaden A, Kanhai V, Leussink AB. Physiochemical and immunochemical characterization of meningococcal serogroup A polysaccharide-tetanus toxoid conjugates prepared by two methods. Vaccine. 1983;1:31–36. doi: 10.1016/0264-410x(83)90010-5. [DOI] [PubMed] [Google Scholar]
- 16.Anderson EL, Bowers T, Mink CM, Kennedy DJ, Belshe RB, Harakeh H, Pais L, Holder P, Carlone GM. Safety and immunogenicity of meningococcal A and C polysaccharide conjugate vaccine in adults. Infect Immun. 1994;62:3391–3395. doi: 10.1128/iai.62.8.3391-3395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zollinger, W.D., and E. Moran. 1991. Meningococcal vaccines—present and future. Trans. Roy. Soc. Trop. Med. Hyg. 85(Suppl.):37–43. [DOI] [PubMed]
- 18.Mandrell RE, Zollinger WD. Measurement of antibodies to meningococcal serogroup B polysaccharide: Low avidity binding and equilibrium binding constants. J Immunol. 1982;129:2172–2177. [PubMed] [Google Scholar]
- 19.Zollinger WD, Mandrell RE. Importance of complement source in bactericidal activity of human antibody and murine monoclonal antibody to meningococcal group B polysaccharide. Infect Immun. 1983;40:257–264. doi: 10.1128/iai.40.1.257-264.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of antibodies. J Exp Med. 1969;129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. II. Development of natural immunity. J Exp Med. 1969;129:1327–1378. doi: 10.1084/jem.129.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotschlich EC, Goldschneider I, Artenstein MS. Human immunity to the meningococcus. IV. Immunogenicity of serogroup A and serogroup C polysaccharides in human volunteers. J Exp Med. 1969;129:1367–1384. doi: 10.1084/jem.129.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frasch CE, Chapman SS. Classification of Neisseria meningitidisserogroup b into distinct serotypes. III. Application of a new bactericidal inhibition technique to distribution of serotypes among cases and carriers. J Infect Dis. 1973;127:149–154. doi: 10.1093/infdis/127.2.149. [DOI] [PubMed] [Google Scholar]
- 24.Jones DM, Eldridge J. Development of antibodies to meningococcal protein and lipopolysaccharide antigens in healthy carriers. J Med Microbiol. 1979;12:107–111. doi: 10.1099/00222615-12-1-107. [DOI] [PubMed] [Google Scholar]
- 25.Saukkonen K, Abdillahi H, Poolman JT, Leinonen M. Protective efficacy of monoclonal antibodies to class 1 and class 3 outer membrane proteins of Neisseria meningitidisB:15:P1.16 in infant rat infection model: new prospects for vaccine development. Microb Pathog. 1987;3:261–267. doi: 10.1016/0882-4010(87)90059-3. [DOI] [PubMed] [Google Scholar]
- 26.Saukkonen K, Leinonen M, Abdillahi H, Poolman JT. Comparative evaluation of potential components for group B meningococcal vaccine by passive protection in the infant rat model and in vitro bactericidal assay. Vaccine. 1989;7:325–328. doi: 10.1016/0264-410x(89)90194-1. [DOI] [PubMed] [Google Scholar]
- 27.Frasch CE, Tsai C-M, Mocca LF. Outer membrane proteins of Neisseria meningitidis: structure and importance in meningococcal disease. Clin Invest Med. 1986;9:101–107. [PubMed] [Google Scholar]
- 28.Brodeur BR, Larose Y, Tsang P, Hamel J, Ashton F, Ryan A. Protection against infection with Neisseria meningitidisgroup B serotype 2b by passive immunization with serotype-specific monoclonal antibody. Infect Immun. 1985;50:510–516. doi: 10.1128/iai.50.2.510-516.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Høiby EA, Rosenqvist EA, Frøholm LO, Bjune G, Feiring B, Nøkleby H, Rønnild E. Bactericidal antibodies after vaccination with the Norwegian meningococcal serogroup B outer membrane vesicle vaccine: a brief survey. NIPH (Natl Inst Public Health) Ann (OSLO) 1991;14:147–56. [PubMed] [Google Scholar]
- 30.Zollinger WD, Boslego J, Moran E, Garcia J, Cruz C, Ruiz S, Brandt B, Martinez M, Arther J, Underwood P, et al. Meningococcal serogroup B vaccine protection trial and follow-up studies in Chile. NIPH (Natl Inst Public Health) Ann (OSLO) 1991;14:211–213. [PubMed] [Google Scholar]
- 31.van der Ley PA, Heckels JE, Virji M, Hoogerhout P, Poolman JT. Topology of outer membrane porins in pathogenic Neisseriaspp. Infect Immun. 1991;59:2963–2971. doi: 10.1128/iai.59.9.2963-2971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olyhoek AJM, Sarkari J, Bopp M, Morelli G, Achtman M. Cloning and expression in Escherichia coli of Opc, the gene for an unusual class 5 outer membrane protein from Neisseria meningitidis . Microbiol Pathog. 1991;11:249–257. doi: 10.1016/0882-4010(91)90029-a. [DOI] [PubMed] [Google Scholar]
- 33.Rosenqvist E, Høiby EA, Wedege E, Kusecek B, Achtman M. The 5c protein of Neisseria meningitidisis highly immunogenic in humans and induces bactericidal antibodies. J Infect Dis. 1993;167:1065–1073. doi: 10.1093/infdis/167.5.1065. [DOI] [PubMed] [Google Scholar]
- 34.van der Ley PA, van der Biezen J, Peeters CCAM, Poolman JT. Use of transformation to construct antigenic hybrids of the class 1 outer membrane protein in Neisseria meningitidis . Infect Immun. 1993;61:4724–4733. doi: 10.1128/iai.61.10.4217-4224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Ley PA, van der Biezen J, Poolman JT. Construction of Neisseria meningitidis strains carrying multiple chromosomal copies of the porA gene for use in the production of a multivalent outer membrane vesicle vaccine. Vaccine. 1995;13:401–407. doi: 10.1016/0264-410x(95)98264-b. [DOI] [PubMed] [Google Scholar]
- 36.van der Ley PA, Poolman JT. Construction of multivalent class 1 OMP expressing meningococcal vaccine strain. Infect Immun. 1992;60:3156–3161. doi: 10.1128/iai.60.8.3156-3161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danve B, Lissolo L, Mignon M, Dumas P, Colombani PP, Colombani S, Schryvers AB, Quentin-Millet M-J. Transferrin-binding proteins isolated from Neisseria meningitidiselicit protective and bactericidal antibodies in laboratory animals. Vaccine. 1993;11:1214–1220. doi: 10.1016/0264-410x(93)90045-y. [DOI] [PubMed] [Google Scholar]
- 38.Pettersson A, Kuipers AJ, Pelzer M, Verhagen EPM, Tiesjema RH, Tommassen J, Poolman JT. Monoclonal antibodies against the 70-kilodalton iron-regulated protein of Neisseria meningitidisare bactericidal and strains-specific. Infect Immun. 1990;58:3036–3041. doi: 10.1128/iai.58.9.3036-3041.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatnagar NB, Frasch CE. Expression of Neisseria meningitidisiron-regulated outer membrane proteins including a 70-Kilodalton transferrin receptor, and their potential for use as vaccines. Infect Immun. 1990;58:2875–2881. doi: 10.1128/iai.58.9.2875-2881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ala'Aldeen DA, Borriello SP. The meningococcal transferrin-binding proteins 1 and 2 are both surface exposed and generate bactericidal antibodies capable of killing homologous and heterologous strains. Vaccine. 1996;14:49–53. doi: 10.1016/0264-410x(95)00136-o. [DOI] [PubMed] [Google Scholar]
- 41.Cannon JG, Black WJ, Nachamkin I, Stewart PW. Monoclonal antibody that recognizes an outer membrane antigen common to the pathogenic Neisseriaspecies. Infect Immun. 1984;43:994–999. doi: 10.1128/iai.43.3.994-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharjee AK, Moran EE, Zollinger WD. Antibodies to meningococcal H.8 (Lip) antigen fail to show bactericidal activity. Can J Microbiol. 1990;36:117–122. doi: 10.1139/m90-021. [DOI] [PubMed] [Google Scholar]
- 43.Munkley A, Tinsley CR, Virji M, Heckels JE. Blocking of bactericidal killing of Neisseria meningitidisby antibodies directed against class 4 outer membrane proteins. Microbiol Pathol. 1991;11:447–452. doi: 10.1016/0882-4010(91)90041-8. [DOI] [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Viera J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene (Amst) 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Plainview, NY. 12.21–12.23.
- 46.Studier, F.W., A.H. Rosenberg, J.J. Dunn, and J.W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. In Methods Enzymol. J.N. Abelson and M.I. Simon, editors. Academic Press, Inc., San Diego, CA. 185: 60–89. [DOI] [PubMed]
- 47.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli . Gene (Amst) 1991;100:195–199. [PubMed] [Google Scholar]
- 48.Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 49.Hamel J, Brodeur BR, Larose Y, Tsang PS, Belmaaza A, Montplaisir S. A monoclonal antibody directed against a serotype-specific, outer-membrane protein of Haemophilus influenzaetype b. J Med Microbiol. 1987;23:163–170. doi: 10.1099/00222615-23-2-163. [DOI] [PubMed] [Google Scholar]
- 50.Lussier M, Brodeur BR, Winston S. Detection of Neisseria gonorrhoeaeby dot enzyme immunoassay using monoclonal antibodies. J Immunoassay. 1989;10:373–394. doi: 10.1080/01971528908053248. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Martin D, Larose Y, Hamel J, Lagacé J, Brodeur BR. Heterohybridomas secreting human monoclonal antibodies against Haemophilus influenzaetype b. Eur J Immunol. 1988;18:601–606. doi: 10.1002/eji.1830180417. [DOI] [PubMed] [Google Scholar]
- 53.Tsai C-M, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Annal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 54.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 55.Martin D, Peppler MS, Brodeur BR. Immunological characterization of the lipooligosaccharide B band of Bordetella pertussis . Infect Immun. 1992;60:2718–2725. doi: 10.1128/iai.60.7.2718-2725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin, D., N. Cadieux, J. Hamel, and B.R. Brodeur. 1996. Identification of a highly conserved outer membrane protein of Neisseria meningitidis Am. Soc. Microbiol. 187(Abstr.):187.
- 57.Michaelis S, Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli . Ann Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- 58.Virji M, Makepeace K, Ferguson DJP, Achtman M, Moxon ER. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol Microbiol. 1993;10:499–510. doi: 10.1111/j.1365-2958.1993.tb00922.x. [DOI] [PubMed] [Google Scholar]
- 59.Aho EL, Dempsey JA, Hobbs MM, Klapper DG, Cannon JG. Characterization of the opa (class 5) gene family of Neisseria meningitidis. . Mol Microbiol. 1991;5:1429–1437. doi: 10.1111/j.1365-2958.1991.tb00789.x. [DOI] [PubMed] [Google Scholar]
- 60.Hobbs MM, Seiler A, Achtman M, Cannon JG. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis . Mol Microbiol. 1994;12:171–180. doi: 10.1111/j.1365-2958.1994.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 61.Ross SC, Rosenthal PJ, Berberic HM, Densen P. Killing of Neisseria meningitidisby human neutrophils: implications for normal and complement deficient individuals. J Infect Dis. 1987;155:1266–1275. doi: 10.1093/infdis/155.6.1266. [DOI] [PubMed] [Google Scholar]
- 62.Densen, P. 1989. Interaction of complement with Neisseria meningitidis and Neisseria gonorrhoeae Clin. Microbiol. Rev. 2(Suppl.): S11–S17. [DOI] [PMC free article] [PubMed]
- 63.Martin D, Munson R, Jr, Grass S, Chong P, Hamel J, Zobrist G, Klein M, Brodeur BR. Mapping of B-cell epitopes on the outer membrane P2 porin protein of Haemophilus influenzaeby using recombinant proteins and synthetic peptides. Infect Immun. 1991;59:1457–1464. doi: 10.1128/iai.59.4.1457-1464.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McGuinness BT, Lambden PR, Heckels JE. Class 1 outer membrane protein of Neisseria meningitidis: epitope analysis of the antigenic diversity between strains, implications for subtype definition and molecular epidemiology. Mol Microbiol. 1993;7:505–514. doi: 10.1111/j.1365-2958.1993.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez de Cossio ME, Ohlin M, Llano M, Selander B, Cruz S, Del Valle J, Borrebaeck CA. Human monoclonal antibodies against an epitope on the class 5c outer membrane protein common to many pathogenic strains of Neisseria meningitidis . J Infect Dis. 1992;166:1322–1328. doi: 10.1093/infdis/166.6.1322. [DOI] [PubMed] [Google Scholar]
- 66.Verheul AF, Kuipers AJ, Braat AK, Dekker HA, Peeters CC, Snippe H, Poolman JT. Development, characterization, and biological properties of meningococcal immunotype L3,7,(8),9-specific monoclonal antibodies. Clin Diagn Lab Immunol. 1994;1:729–736. doi: 10.1128/cdli.1.6.729-736.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Idänpään-Heikkilä I, Muttilainen S, Wahlström E, Saarinen L, Leinonen M, Sarvas M, Mäkelä PH. The antibody response to a prototype liposome vaccine containing Neisseria meningitidis outer membrane protein P1 produced in Bacillus subtilis . Vaccine. 1995;13:1501–1508. doi: 10.1016/0264-410x(95)00101-6. [DOI] [PubMed] [Google Scholar]