Abstract
Association of antigenic peptides with newly synthesized major histocompatibility complex (MHC) class I molecules occurs in the endoplasmic reticulum and is a critical early step for the initiation of cytotoxic T lymphocyte (CTL)-mediated immune defenses. Pathogen-derived peptides compete with a plethora of endogenous peptides for MHC class I grooves. We find that two H2-Kd–restricted peptides, which derive from the Listeria monocytogenes p60 antigen, accumulate in infected cells with different kinetics. Although competition assays suggest that both epitopes are bound with equivalent affinity, they dissociate from MHC class I molecules at markedly different rates. p60 217-225 forms complexes with H2-Kd with a half-life >6 h, while p60 449-457 dissociates from H2-Kd with a half-life of ∼1 h. We find that p60 449-457–H2-Kd complexes retained intracellularly with brefeldin A have a half-life of 30 min, and thus are less stable than surface complexes. While peptide dissociation from retained MHC class I molecules is enhanced, retained H2-Kd molecules maintain a remarkable capacity to bind new T cell epitopes. We find that intracellular H2-Kd molecules can bind new CTL epitopes for up to 3 h after their synthesis. Our studies provide a glimpse of peptide interaction with MHC class I molecules in the endoplasmic reticulum/proximal Golgi complex of intact, infected cells. We propose that the increased intracellular lability of peptide–MHC class I complexes may function to optimize the spectrum of peptides presented to T lymphocytes during cellular infection.
The MHC class I antigen–processing pathway is a constitutive surveillance system that allows CTLs to monitor intracellular proteins (1–3). While the principal purpose of MHC class I antigen presentation is in the defense against viral infection, MHC class I molecules also present peptides derived from bacterial and protozoal pathogens (4). The essential aspects of the MHC class I antigen–processing pathway have been defined. First, endogenous and pathogen-derived proteins in the cytosol are degraded into peptides by the multicatalytic protease complex, or proteasome (5). The precise source of antigenic peptides is somewhat controversial since, in the case of antigens synthesized by the host cell, proteasomes may degrade either the mature protein or defective products generated during translation (6). Peptide fragments are then moved in an ATP-dependent process into the lumen of the endoplasmic reticulum (ER)1 by the transporter associated with antigen processing (TAP, reference 7). While peptides are being generated in the cytosol, MHC class I molecules are being synthesized and inserted into the ER membrane, where they first associate with β2-microglobulin and then TAP in a process that involves calnexin, calreticulin, and a newly described protein called tapasin (8). MHC molecules remain associated with TAP until they bind a peptide, at which time they dissociate from TAP and traffic, as an MHC class I–β2-m–peptide heterotrimer, to the cell surface (9). Although it is clear that antigenic peptides associate with MHC class I molecules in the ER (10), it is unclear how MHC class I molecules manage to selectively capture high affinity peptides. While the function of tapasin in MHC peptide loading is unclear (8), there is evidence that association of MHC class I with TAP may induce a conformational change that makes MHC class I molecules more peptide receptive (11).
In the absence of infection, peptides generated by proteasome-mediated degradation of endogenous proteins are the major substrates transported by TAP and bound by MHC class I H chains (12). Because intracellular pathogens constitute only a very small fraction of the total protein of an infected cell, even in heavily infected cells the majority of peptides associated with MHC class I H chains are derived from the host cell. Although it would be advantageous to the infected host to provide foreign peptides with open access to H chain grooves, it is unclear how accomodating the MHC class I surveillance system is to pathogen derived peptides.
We have used the intracellular pathogen Listeria monocytogenes to investigate the MHC class I antigen–processing pathway. L. monocytogenes is a gram-positive bacterium that causes severe disease in pregnant and immunocompromised individuals (13). Upon phagocytosis by macrophages, L. monocytogenes enters the cytosol by secreting listeriolysin O (LLO), a pore-forming hemolysin which destroys the phagolysosomal membrane (14). Two bacterially secreted proteins, LLO and the murein hydrolase p60, are degraded by proteasomes in the host cell cytosol to form three nonamer peptides: LLO 91-99, p60 217-225, and p60 449-457 (15– 18). These three Listeria-derived peptides are presented to CTL by H2-Kd MHC class I molecules. The number of secreted antigen molecules that the host cell degrades to generate a single H2-Kd-associated epitope ranges from 3–11 (for LLO 91-99 and p60 449-457) to 35 (for p60 217-225) (17, 19, 20). While the majority of antigen molecules that are degraded by the host cell do not yield an antigenic peptide, it is unclear where the losses occur. Each step of the MHC class I antigen–processing pathway, from proteasomemediated antigen degradation to peptide transport by TAP and association with H-2Kd grooves, may exact a substantial loss of epitopes (21–24).
In this report, we investigate the kinetics of CTL epitope generation and loss in L. monocytogenes–infected cells. Because intracellular L. monocytogenes and the host cell are metabolically distinct, it is possible to selectively manipulate antigen production and host cell antigen processing in intact cells with specific inhibitors. We exploit this ability to show that two peptides that are bound with high affinity dissociate from H2-Kd molecules at remarkably different rates. We find that one of the epitopes, p60 217-225, forms a stable complex with H2-Kd while the other epitope, p60 449457 forms a more labile complex with H2-Kd. p60 449457 dissociation occurs more rapidly when H2-Kd molecules are retained within the cell with brefeldin A (BFA) than when they transit to the cell surface, which suggests that intracellular conditions decrease the strength of MHC class I–peptide interactions. Although synthesis of new MHC class I molecules is essential for the generation of CTL epitopes, we find that intracellularly retained MHC molecules continue to bind new L. monocytogenes peptides for up to 3 h. Our findings demonstrate the tremendous capacity of class I molecules to bind pathogen-derived peptides upon cellular infection. Furthermore, enhanced peptide dissociation from MHC class I molecules in the ER/Golgi may enhance cell surface presentation of long-lived epitopes.
Materials and Methods
Bacteria and Cell Lines.
L. monocytogenes strain 43251 was obtained from the American Type Culture Collection (Rockville, MD) and grown in brain hearth infusion. H2-Kd-transfected RMA-S cells (25) were provided by M. Bevan (University of Washington, Seattle, WA) and grown in HL-1 medium (BioWhittaker, Inc., Walkersville, MD) with 400 μg/ml G418 (GIBCO BRL, Gaithersburg, MD). Macrophage cell line J774 (BALB/c, H-2d) and the mastocytoma cell line P815 (DBA/2, H-2d) (both from American Type Culture Collection) were grown in RPMI 1640 (GIBCO BRL) with 10% FCS, 2 mM l-glutamine, 20 mM Hepes (pH 7.5), 50 μM 2-mercaptoethanol, penicillin, streptomycin, and gentamicin (RP10 media). CTL clones L12.3, L9.6, and WP11.12 were derived from L. monocytogenes–primed C57BL/6 × BALB/c (H-2bxd) mice and maintained as described (16). L12.3 is specific for LLO 91-99, L9.6 is specific for p60 217-225, and WP11.12 is specific for p60 449-457, each in the context of the H2-Kd molecule.
Peptides and Reagents.
Synthetic LLO 91-99 (GYKDGNEYI), p60 217-225 (KYGVSVQDI), and p60 449-457 (IYVGNGQMI) peptides were purchased from Research Genetics, Inc. (Huntsville, AL). Peptides were HPLC purified and quantified by amino acid analysis and mass spectrometry. BFA was purchased from Epicentre Technologies (Madison, WI) and cycloheximide (CHX) and anisomycin (ANM) from Sigma Chemical Co. (St. Louis, MO).
CTL Assays.
CTL assays were performed as previously described (16). In short, 104 [51Cr]sodium chromate P815 target cells were added to HPLC-fractionated peptides or synthetic peptides dissolved in PBS. CTL were added at an E/T ratio of 1–3:1 and the assays were incubated for 4–5 h at 37°C. Supernatants were assayed for 51Cr-release in a gamma counter. The percentage of specific lysis was determined as described previously (15).
Peptide Competition Assays.
To estimate the affinity of H2-Kd for p60 217-225 and p60 449-457, P815 target cells were incubated in the presence of 10−10 M LLO 91-99 and a range of either p60 217-225 or p60 449-457 concentrations. Target cells were incubated with peptides for 30 min before the addition of CTL clone L12.3 (specific for LLO 91-99) at an effector to target ratio of 2:1. The percent specific lysis was determined 4 h after the addition of CTL.
Epitope Extraction and HPLC Fractionation.
J774 cells were grown to confluence in 27-ml RP10 media without antibiotics (RP10−) in 150-mm tissue-culture plates. Cells were infected with 3 ml of a log phase culture of L. monocytogenes (OD600 of 0.1) for 30 min. The media was then replaced with RP10− containing 5 μg/ml gentamicin. Infected J774 macrophages were harvested by scraping with PBS, pelleted by centrifugation, and stored at −80°C. To extract peptides, pellets were resuspended in 10 ml 0.1% TFA, dounce homogenized, sonicated, and centrifuged, at 100,000 g for 35 min at 4°C. Supernatants were lyophilized, pellets were resuspended in 0.1% TFA, spun through a Centricon-10 membrane, and fractionated by reverse-phase HPLC using a 0–60% acetonitrile gradient. Separated peptide fractions were lyophilized and used for CTL assays (16).
Epitope Quantification.
Lyophilized HPLC fractions were dissolved in 200 μl PBS. Samples of 5–50 μl were loaded on P815 target cells and tested for the presence of LLO 91-99, p60 217225, or p60 449-457 with CTL clones L12.3, L9.6, and WP11.12, respectively. Peptide-containing fractions were titrated and assayed in triplicate along with standard concentrations of synthetic peptides. Molar amounts of peptide were determined and epitope numbers were calculated as described (16).
H2-Kd Stability.
RMA-S Kd cells were seeded in the wells of 24-well plates at a concentration of 106 cells/ml (1 ml/well) in serum-free HL-1 medium with 400 μg/ml G418. Cells were incubated with 30 μM synthetic LLO 91-99, p60 217-225, or p60 449-457, or without peptide at 37°C overnight (o/n). Cells were harvested, washed three times with PBS, and chased at 37°C in the absence of peptide. Samples were taken after 0, 2, 4, and 6 h, washed with ice-cold PBS with 0.5% bovine serum albumin and 0.02% NaN3 (PBS buffer) and stained for H2-Kd class I expression.
Flow Cytometry.
4 × 105 cells were incubated with an excess amount of SF1-1.1.1 (a conformation-dependent mAb specific for the H2-Kd α3 domain) or without antibody for 30 min on ice. Cells were washed with PBS buffer and then incubated with FITCconjugated goat anti–mouse Ig for 30 min, washed twice, and then fixed in 1% paraformaldehyde in PBS. Fluorescence was measured with a FACScan® flow cytometer and data were analyzed using the CELLQuest application programs (Becton Dickinson, Mountain View, CA).
H2-Kd Trafficking.
H2-Kd was metabolically labeled and immunoprecipitated as previously described (26). In short, 2–4 × 106 J774 cells were seeded in 25-cm2 tissue-culture flasks in 10 ml RP10 media without antibiotics (RP10−) and incubated at 37°C o/n. Cells were infected with 1 ml of log phase L. monocytogenes (OD600 of 0.1) for 30 min, washed with PBS, and placed in RP10− with 50 μg/ml gentamicin for 3 h.
Monolayers of uninfected or infected J774 cells were washed with PBS and placed in 1 ml of methionine-free medium (methionine-free DMEM with 3% dialyzed FCS, 2 mM l-glutamine, 20 mM Hepes, pH 7.5). After 30 min, 50 μCi [35S]methionine (translabel, ICN Biomedicals Inc., Irvine, CA) was added and cells were pulse labeled for 15 min. Chases were performed in 4 ml RP10−, and 20 μg/ml tetracycline was added to L. monocytogenes–infected cells. Cells were lysed on ice in 1 ml lysis buffer (1% Triton X-100 in 140 mM NaCl, 10 mM Tris-HCl, pH 7.4 [Tris-buffered saline] with 0.5 mM PMSF and 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone), and the detergent lysates were centrifuged (6 min, 14,000 rpm at 4°C) and precleared with 25 μl protein A–Sepharose (50% slurry) and 5 μl mouse IgG for 4 h at 4°C. H2-Kd was immunoprecipitated with 25 μl protein A–Sepharose and 5 μl SF11.1.1 (1.4 mg/ml) for 30 min at 4°C. Sepharose beads were washed with Tris-buffered saline/0.5% Triton X-100, resuspended in endoglycosidase H (endo H) buffer (0.1 M phosphate buffer, pH 6.5, 0.5% SDS), and boiled for 2 min. The samples were split; half was mock-digested, and the other half digested with 2 × 10−3 U endo H at 37°C o/n. Sample buffer was added and proteins were separated on 10% polyacrylamide gels under reducing conditions. Radioactive signals were enhanced by incubation with 0.5 M salicylate for 15 min and visualized by autoradiography.
Results
Kinetics of CTL Epitope Loss from L. monocytogenes–infected Cells.
In previous studies we have shown that p60 449457 is generated in L. monocytogenes infected J774 cells earlier and faster than p60 217-225 (17). During the first few hours of infection, ∼10 times as much p60 449-457 can be extracted from infected cells as p60 217-225. Since both peptides derive from the same antigen and bind to H2-Kd molecules, the difference in numbers is attributable to a greater efficiency of p60 449-457 generation (17). To determine if MHC–peptide complex stability plays a role in the kinetics of p60 217-225 and p60 449-457 accumulation in L. monocytogenes–infected cells, we treated cells with tetracycline, which rapidly inhibits bacterial protein synthesis. Epitopes were quantified at hourly intervals after the cessation of antigen synthesis. In agreement with previous findings (19), we found that p60 217-225 epitope numbers are stable in infected cells (Fig. 1 A). However, the number of p60 449-457 epitopes that can be extracted from infected cells after tetracycline treatment drops abruptly, from 9,000 epitopes/cell to only 1,000 epitopes/cell 3 h later (Fig. 1 B). In the absence of tetracycline, p60 217-225 levels continue to increase in number between 4 and 7 h of cellular infection. In contrast, the number of p60 449-457 epitopes per cell remains stable between 5 and 7 h after infection in the absence of tetracycline. Taken together, our results indicate that the plateau levels of p60 449-457 epitopes during the later stages of cellular infection reflect an equilibrium, with equal amounts of p60 449-457 being generated and lost within infected cells.
Figure 1.
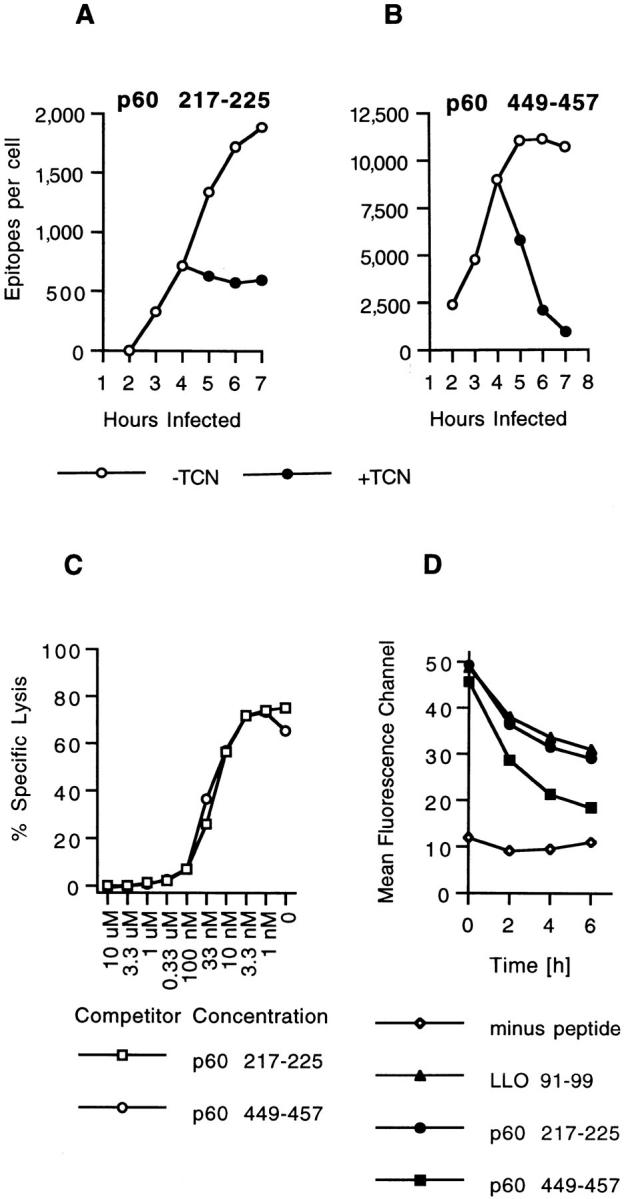
Two H2-Kd–binding p60 epitopes accumulate with different kinetics in infected cells. (A and B) J774 cells were infected with L. monocytogenes and p60 epitope accumulation followed in the absence and in the presence of 20 μg/ml tetracycline (TCN), a specific inhibitor of bacterial protein synthesis. TCN was added 4 h after infection. Cells were harvested at the indicated time periods, peptides were acid extracted, HPLC fractionated, and the amount of p60 217-225 and p60 449-457 was determined with CTL clone L9.6 (A) and WP11.12 (B). Epitope numbers were quantified and expressed as epitopes per infected cell. (C) Comparison of the binding affinities of p60 217-225 and p60 449-457 for H2-Kd molecules. P815 target cells were coated with 10−10 M LLO 91-99 in the presence of a range of p60 peptide concentrations and tested for recognition by LLO 91-99–specific CTL clone L12.3 as described in Materials and Methods. (D) Stability of H2-Kd-Listeria peptide complexes on the cell surface of RMA-S cells. Class I expression on RMA-S Kd cells was induced by incubation in the presence of 30 μM synthetic LLO 91-99, p60 217-225, or p60 449-457 at 37°C o/n. Control cells were incubated in the absence of peptide. Cells were chased at 37°C in the absence of peptide, and the stability of surface H2-Kd molecules was determined with H2-Kd–specific antibody SF1-1.1.1 and flow cytometry as described in Materials and Methods. Mean fluorecence channels of 10,000 gated events are depicted on the y-axis. Similar results were obtained in three independent experiments.
p60 217-225 and p60 449-457 Bind H2-Kd with High Affinity.
The different rates of p60 217-225 and p60 449-457 loss from L. monocytogenes–infected cells might be accounted for by distinct affinities of H2-Kd for the two peptides. The relative affinities of H2-Kd for p60 217-225 and p60 449457 were estimated by peptide competition assays against a third H2-Kd–restricted peptide, LLO 91-99. Using LLO 91-99 as the targeting peptide with CTL clone L12.3, which is specific for LLO 91-99, we found that p60 217225 and p60 449-457 were identical at competing for H2Kd binding (Fig. 1 C). These results confirm our previous findings (17) and indicate that the different kinetics of p60 217-225 and p60 449-457 loss in L. monocytogenes–infected cells cannot be attributed to different affinities, as determined by competition assays, of H2-Kd for the epitopes.
p60 449-457 Dissociates More Readily from H2-Kd than LLO 91-99 or p60 217-225.
The most likely explanation for the rapid loss of p60 449-457 from tetracyclinetreated cells is that p60 dissociates from H2-Kd and is destroyed by intra- and extracellular proteases. To determine if the two p60 epitopes and LLO 91-99 (which forms stable complexes with H2-Kd in infected cells; reference 20) differ in their dissociation from class I molecules, we incubated H2-Kd–transfected RMA-S cells with the three L. monocytogenes peptides o/n and then determined the stability of surface H2-Kd expression by flow cytometry. All three peptides stabilized surface H2-Kd after o/n incubation (Fig. 1 D). However, when cells were incubated at 37°C in the absence of peptides, surface H2-Kd complexes generated with p60 449-457 were markedly less stable than those generated with either LLO 91-99 or p60 217-225 (Fig. 1 D). These findings suggest that the loss of p60 449-457 in infected cells results from the enhanced dissociation of p60 449-457 relative to the other two epitopes. The decrease in surface expression of H2-Kd complexes with LLO 91-99 and p60 217-225 detected in this assay probably results from (a) the early loss of peptides that were only partially bound, (b) the more prolonged decrease attributable H2-Kd dilution caused by continued cell division, and (c) the intrinsic turnover rate of peptide-stabilized H2-Kd. The finding that H2-Kd complexes with p60 449-457 appear to be more stable in the RMA-S-Kd assay than p60 449-457 in infected cells, likely results from an overestimation of peptide-containing surface complexes on RMA-S cells, since MHC class I molecules may be detected by conformationspecific antibody for some time after peptide dissociation.
Enhanced Dissociation of p60 449-457 from H2-Kd Molecules in the ER/Golgi.
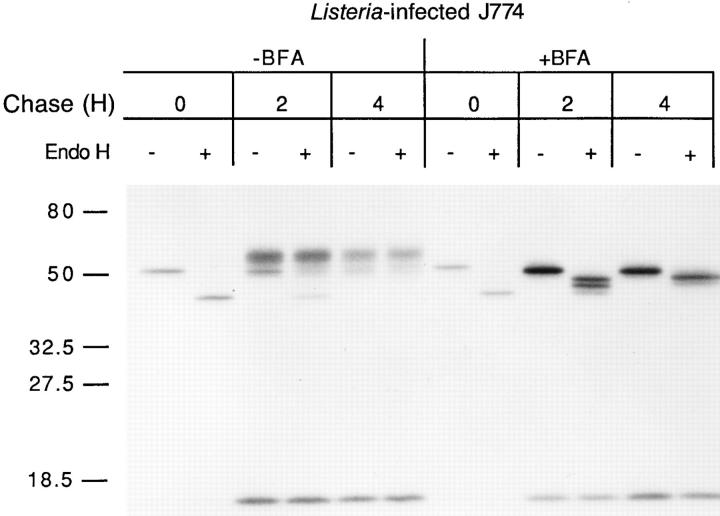
Since H2-Kd molecules travel to the cell surface rapidly in L. monocytogenes–infected cells (26), the majority of CTL epitopes extracted from tetracyclinetreated infected cells are likely to come from cell-surface H2-Kd. Thus, the 1 h half-life of p60 449-457 in infected cells principally reflects cell surface dissociation of p60 449457 from H2-Kd. We decided to determine if the stability of intracellular MHC peptide complexes is similar to surface complexes. BFA is an antibiotic that disrupts the Golgi complex, thereby preventing the movement of ER contents to the cell surface (27). BFA does not detectably affect L. monocytogenes multiplication, protein synthesis, or protein secretion (results not shown). We investigated the trafficking of metabolically labeled H2-Kd in L. monocytogenes– infected J774 cells in the absence and presence of BFA. In the absence of BFA, the great majority of pulse labeled H2Kd becomes endo H resistant after a 2 h chase, indicating that the complexes have trafficked through the Golgi complex (Fig. 2). In the presence of BFA, however, immunoprecipitated H2-Kd molecules remain endo H–sensitive, indicating that they have not traversed the Golgi complex (Fig. 2). It is noteworthy that retained H2-Kd molecules are stable, with minimal loss during the 4-h chase period.
Figure 2.
BFA treatment induces intracellular retention of H2-Kd molecules in infected J774 cells. J774 cells were infected with L. monocytogenes for 3 h and treated with 5 μg/ml BFA or left untreated. Cells were then pulse labeled with [35S]methionine and chased for the indicated time periods in the presence or absence of BFA. H2-Kd molecules were immunoprecipitated and either endo H–digested (+) or mock-digested (−) and subjected to SDS-PAGE. Molecular weight markers are shown on the left.
We next investigated the effect of BFA treatment on the generation of p60 217-225 and p60 449-457 epitopes in L. monocytogenes–infected J774 cells. The amount of p60 217225 that was extracted from infected cells was not affected by BFA (Fig. 3 A). This result confirms the finding that BFA does not inhibit the generation of CTL epitopes (10). In contrast, ∼60% more p60 449-457 epitopes were extracted from BFA-treated cells 6–7 h after infection than from untreated cells. This finding may be attributable to unusually high concentrations of H2-Kd and β2-microglobulin in the ER (28). To determine the stability of p60 217-225 and p60 449-457 when H2-Kd is retained intracellularly, we inhibited L. monocytogenes protein synthesis in infected, BFA treated cells and quantified epitope numbers. Surprisingly, although the amount of p60 217-225 decreased only minimally (Fig. 3 C), the loss of p60 449-457 in BFAtreated cells was very rapid (Fig. 3 D). The half-life of p60 449-457 associated with retained and normally trafficking H2-Kd molecules is shown in Table 1. In three separate experiments, the half-life of p60 449-457 in BFA-treated cells was reduced by ∼50%. Thus, in L. monocytogenes–infected cells, p60 449-457–H2-Kd complexes are less stable intracellularly than on the cell surface.
Figure 3.
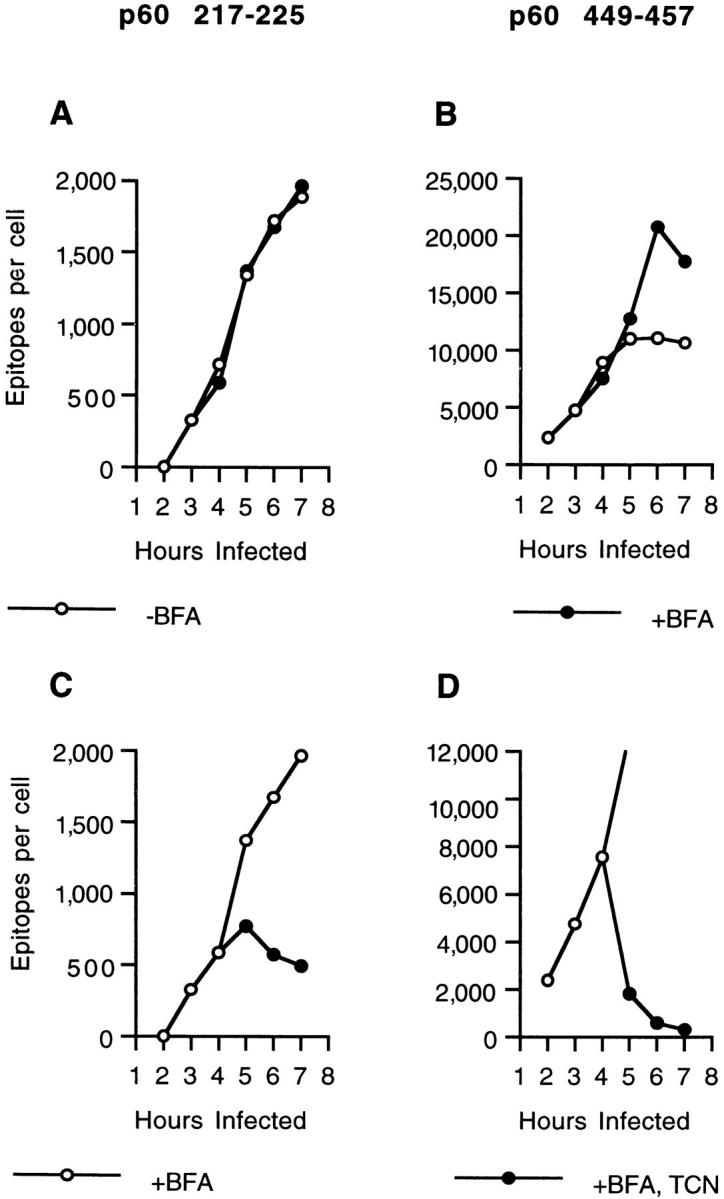
p60 449-457 dissociates at an enhanced rate from intracellularly-retained H2-Kd molecules. (A and B) J774 cells were infected with L. monocytogenes, and after 3 h, incubated in the absence or presence of BFA. p60 epitope accumulation was determined at the indicated times. CTL clone L9.6 was used to quantify p60 217-225 (A) and clone WP11.12 to quantify p60 449-457 (B). (C and D) p60 epitope accumulation and loss was also determined in infected, BFA-treated J774 cells in the presence and absence of TCN. BFA was added 3 h after infection and TCN was added 4 h after infection. CTL clone L9.6 was used to quantify p60 217-225 (C) and clone WP11.12 to quantify p60 449-457 (D).
Table 1.
Intracellular Retention of H2-Kd–p60 449-457 Complexes Shorten Their Half-life
| Experiment | Half-life of p60 449-457 | |||
|---|---|---|---|---|
| − BFA | + BFA | |||
| (min) | ||||
| 1 | 58 | 27 | ||
| 2 | 82 | 28 | ||
| 3 | 62 | 33 | ||
| Mean | 67 | 29 | ||
In three separate experiments, the loss of p60 449-457 epitopes from tetracycline-treated, L. monocytogenes–infected J774 cells was determined as described in Materials and Methods, and the half-life was determined.
H2-Kd Molecules Remain Peptide Receptive in the ER.
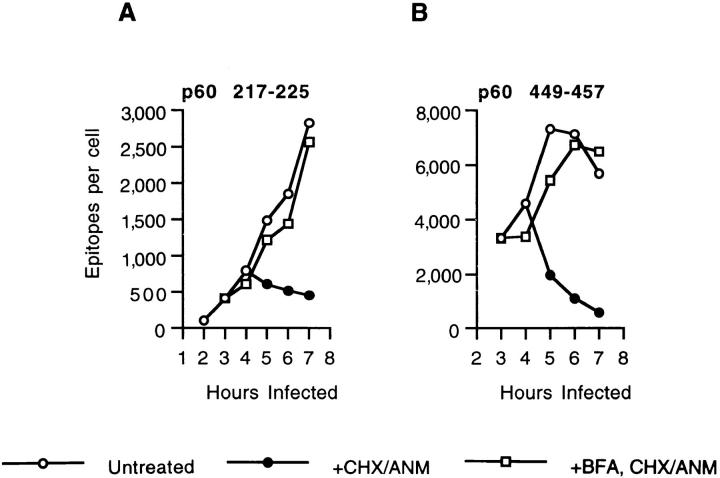
In the preceeding experiment, trafficking from the ER to the cell surface was inhibited with BFA, but host cell protein synthesis continued. Thus, new H2-Kd molecules were being synthesized and inserted into the ER membrane and then binding newly-generated p60 217-225 and p60 449-457. We next determined if p60 217-225 and p60 449-457 associate with newly synthesized H2-Kd, or if H2-Kd molecules that had been retained intracellularly for several hours could also bind epitopes. CHX and ANM, specific inhibitors of eukaryotic but not prokaryotic protein synthesis, abolish H2Kd synthesis in J774 cells (results not shown). J774 cells were infected with L. monocytogenes and then treated with CHX–ANM. The number of p60 217-225 epitopes stops increasing when host cell protein synthesis is inhibited since H2-Kd molecules, by leaving the ER, rapidly become unavailable for peptide binding (Fig. 4 A). The number of p60 449-457 epitopes per cell decreases after CHX–ANM treatment since this epitope dissociates at a rapid rate from H2-Kd (Fig. 4 B). To determine if H2-Kd molecules retained in the ER could continue to bind peptides, we treated cells with BFA and then stopped further host cell protein synthesis with CHX–ANM. As demonstrated in Fig. 4, H2-Kd molecules retained in the ER remained capable of binding new p60 217-225 and p60 449-457 epitopes for up to 3 h. Although the number of p60 449-457 epitopes does not increase in BFA-treated cells when host cell protein synthesis has been inhibited for 2–3 h (see Fig. 3 B, 6 and 7 h time points), new H2-Kd–p60 449-457 complexes are being generated since the number of p60 449-457 epitopes rapidly delines in the absence of antigen (see Fig. 3 D).
Figure 4.
Accumulation of p60 peptides in CHX–ANM-treated infected J774 cells. J774 cells were infected with L. monocytogenes for 3 h and then treated with BFA or left untreated. CHX and ANM were added 4 h after infection to inhibit cellular protein synthesis and p60 epitope production was determined. p60 217-225 and p60 449-457 epitopes were quantified with CTL clone L9.6 (A) and WP11.12 (B), respectively.
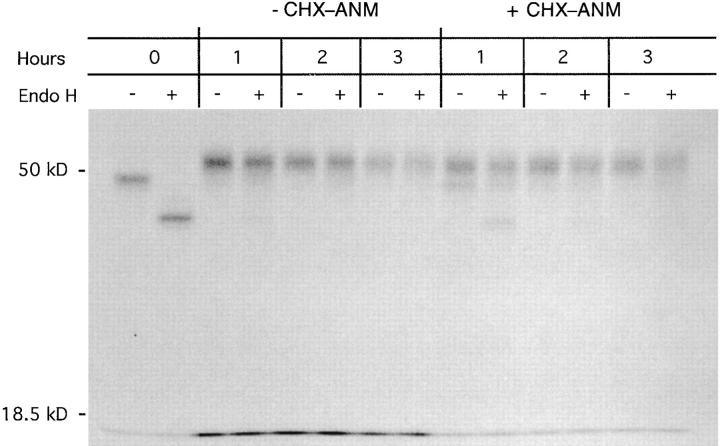
Most H2-Kd molecules exit the ER within 1 to 2 h after synthesis, indicating that they have bound a peptide during this time interval. Therefore, our finding that retained H2Kd molecules bind newly generated p60 217-225 and p60 449-457 for up to 3 h suggests that some of the H2-Kd molecules binding the L. monocytogenes epitopes previously bound other peptides. A potential problem with this interpretation is that protein synthesis inhibition, while preventing new H2-Kd synthesis, may also inhibit production of endogenous peptides from newly synthesized proteins. This would be particularly true if defective ribosomal products (6) are a major source of MHC class I–associated peptides. Thus, inhibiting protein synthesis may lead to a paucity of endogenous peptides and an abundance of empty H2-Kd molecules in the ER, thereby enhancing bacterial peptide capture. The importance of endogenous peptide production for normal H2-Kd trafficking to the cell surface can be readily demonstrated with proteasome inhibitors, which inhibit cytosolic peptide generation and retard H2-Kd trafficking to the cell surface (18). To address the issue of peptide production when host cell protein synthesis is inhibited, we pulse labeled BFA-treated J774 cells with [35S]methionine and chased for 1, 2, and 3 h in the presence of CHX– ANM. Despite inhibition of cellular protein synthesis, pulse-labeled H2-Kd molecules acquired endo H resistance upon BFA removal, indicating that they trafficked through the Golgi (Fig. 5). Thus, sufficient quantities of peptide are generated in the absence of host cell protein synthesis to enable H2-Kd transit with essentially normal kinetics. Importantly, all detectable H2-Kd molecules trafficked through the Golgi within 2 h after release of the BFA blockade, indicating that within this time period they had acquired an endogenous peptide. In contrast, H2-Kd molecules retained intracellularly with BFA continued to acquire L. monocytogenes epitopes 2–3 h after H2-Kd synthesis (Fig. 4). These findings suggest that H2-Kd molecules, if retained in a cellular compartment in which peptide loading occurs, are capable of binding peptides for a period of time that exceeds the time necessary for the acquisition of endogenous peptides.
Figure 5.
H2-Kd trafficking in CHX–ANM-treated J774 cells. J774 cells were pulse labeled in the presence of 5 μg/ml BFA to retain class I molecules, and then chased in the presence of CHX (50 μg/ml) and ANM (30 μg/ml). BFA was removed from the culture medium immediately after the pulse, and cells were either harvested immediately (0 h) or chased 1, 2, or 3 h in medium with or without CHX and ANM. H2-Kd molecules were immunoprecipitated, mock- or endo H–digested, and analyzed by SDS PAGE. Molecular weight markers are indicated on the left.
Discussion
Our studies of L. monocytogenes–infected cells have demonstrated several previously uncharacterized aspects of the MHC class I antigen–processing pathway. (a) Peptides with very similar affinities for MHC class I, as determined by competition assays, can have dramatically different dissociation rates, which will influence their surface representation. (b) Dissociation of peptides, particularly high off-rate peptides, occurs more rapidly intracellularly than on the cell surface. (c) Intracellular MHC class I molecules remain peptide receptive for a period of time that exceeds their usual residence in the ER. These findings have important implications for our understanding of T cell responses to intracellular pathogens.
The rate of antigenic peptide dissociation from MHC class I molecules determines the density of surface complexes available for T lymphocyte detection. Stable MHC class II– peptide complexes, for example, persist on the cell surface longer, giving them a greater opportunity to be of immunologic significance (29). In this report we demonstrate that epitope dissociation from naturally loaded MHC class I molecules can vary dramatically between peptides that appear, by competition assays, to have similar affinities for the class I molecule. The prevalence of p60 449-457 in infected cells is greater than either LLO 91-99 or p60 217225 throughout cellular infection because p60 449-457 is processed very efficiently (17). When p60 synthesis is made transient by tetracycline treatment of infected cells, the prevalence of p60 217-225 becomes greater than p60 449457 within several hours (Fig. 1). Thus, differences in peptide–MHC stability during continuous antigen production are less apparent than when antigen production is transient, as is typical of many viral infections. In the case of EpsteinBarr virus infection, the disparate amount of two EpsteinBarr virus epitopes has been attributed to different stabilities of their respective complexes with HLA-A11 (30).
Although it is unclear how important epitope density is in determining the magnitude of CTL responses after infection, it is very likely that differences in the half-lives of antigenic complexes on the cell surface will have implications for CTL responses. After infection with L. monocytogenes, H2-Kd–p60 217-225 will be present much longer than H2-Kd–p60 449-457 complexes. The importance of H2-Kd/peptide stability may be reflected by the finding that, after murine infection with L. monocytogenes, the number of CTL responding to p60 217-225 is 10 times greater than the number responding to p60 449-457 (31). A correlation between MHC–peptide stability and immunodominance has been suggested by an analysis of peptides that varied in their dissociation rate from MHC class I molecules (32).
While the selective presentation of epitopes that are stably bound by MHC class I molecules might enhance the T cell response to an intracellular pathogen, there is no known mechanism in the ER for differentiating high offrate from slow off-rate peptides. To increase the likelihood that an MHC class I molecule will bind a slow off-rate peptide, accelerated peptide dissociation from MHC class I molecules in the ER would be advantageous. Our finding that the half-life of intracellularly retained H2-Kd–p60 449457 is decreased suggests that such a mechanism may exist. How dissociation of p60 449-457 from H2-Kd is enhanced intracellularly is unknown. It is possible that physiologic conditions in the ER result in MHC class I conformations that loosen the association between the H chain groove and the antigenic peptide. Alternatively, ER resident proteins may enhance dissociation of fast off-rate peptides. In the MHC class II antigen processing pathway, the HLA-DM molecule catalyzes dissociation of invariant chain derived CLIP peptides from MHC class II molecules and facilitates loading with antigenic peptides (33–36). Recent studies indicate that HLA-DM may selectively enhance the dissociation of peptides with rapid off-rates (37). Whether the more rapid dissociation of low affinity peptides from class I molecules in the ER results from a similar interaction with a chaperone molecule, or perhaps tapasin, remains to be examined.
Current models of MHC class I antigen processing propose that newly synthesized MHC class I molecules form a complex with β2-microglobulin and associate with TAP molecules in the ER. When the MHC class I–β2-microglobulin complex binds a peptide, the trimeric complex dissociates from TAP and travels to the cell surface. The length of time from H chain synthesis to emigration from the ER varies with the different allotypes of MHC class I, but generally occurs within 30–60 min. In the context of the rapid association of MHC class I molecules with endogenous peptides, our finding that intracellularly retained H2-Kd molecules can bind additional p60 217-225 and p60 449-457 for up to 3 h after their synthesis is remarkable (Fig. 4). Since the length of time that retained H2-Kd molecules can bind new CTL epitopes significantly exceeds the residence time of H2-Kd in the ER in the absence of BFA, our findings suggest that MHC class I molecules may bind different peptides sequentially. This suggests that enhanced dissociation of fast off-rate peptides in the ER may liberate MHC class I molecules for subsequent interactions with better, slower off-rate peptides.
Based on our data, we propose the following model for MHC class I antigen presentation. J774 cells constitutively synthesize H2-Kd molecules at a high rate that exceeds the rate of high affinity, slow off-rate endogenous peptide production. As a consequence, newly synthesized H2-Kd molecules bind TAP-translocated peptides of varying affinities and off-rates. In the absence of higher affinity or slower off-rate peptides, H-2Kd molecules with fast off-rate peptides travel to the cell surface where their more rapid turnover is inconsequential. Upon cellular infection, however, new peptides are generated and offered to MHC class I molecules. The best peptides to present will be those that form the most stable complex on the cell surface with MHC class I molecules. We suggest that the ER provides a microenvironment in which fast off-rate peptides dissociate more readily, giving slow off-rate peptides a selective advantage to reach the cell surface.
Acknowledgments
This work was supported by grants from the Infectious Diseases Society of America, Arthritis Foundation, the Pew Charitable Trust, and the United States Public Health Service (grant AI33143) to E.G. Pamer, and by a fellowship from the Dutch Cancer Society to A.J.A.M. Sijts.
We would like to thank Peter Cresswell and members of his laboratory for helpful discussions during the course of this work.
Footnotes
1 Abbreviations used in this paper: ANM, anisomycin; BFA, brefeldin A; CHX, cycloheximide; endo H, endoglycosidase H; ER, endoplasmic reticulum; LLO, listeriolysin O; o/n, overnight; TAP, transporter associated with antigen processing.
References
- 1.Braciale TJ, Braciale VL. Viral antigen presentation and MHC assembly. Sem Immunol. 1992;4:81–84. [PubMed] [Google Scholar]
- 2.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 3.Yewdell JW, Bennink JR. Cell biology of antigen processing and presentation to major histocompatibility complex class I molecule–restricted T lymphocytes. Adv Immunol. 1992;52:1–123. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 4.Kaufmann SHE. CD8+ T lymphocytes in intracellular microbial infections. Immunol Today. 1990;11:129–136. doi: 10.1016/0167-5699(88)91292-3. [DOI] [PubMed] [Google Scholar]
- 5.Goettrup M, Soza A, Kuckelkorn U, Kloetzel P-M. Peptide antigen production by the proteasome: complexity provides efficiency. Immunol Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 6.Yewdell JW, Anton LC, Bennink JR. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? . J Immunol. 1996;157:1823–1826. [PubMed] [Google Scholar]
- 7.Androlewicz MJ, Cresswell P. How selective is the transporter associated with antigen processing? . Immunity. 1996;5:1–5. doi: 10.1016/s1074-7613(00)80304-0. [DOI] [PubMed] [Google Scholar]
- 8.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 9.Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/β2-microglobulin complexes associate with TAP transporters before peptide binding. Nature (Lond) 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 10.Lapham CK, Bacik I, Yewdell JW, Kane KP, Bennink JR. Class I molecules retained in the endoplasmic reticulum bind antigenic peptides. J Exp Med. 1993;177:1633–1641. doi: 10.1084/jem.177.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day PM, Esquivel F, Lukszo J, Bennink JR, Yewdell JW. Effect of TAP on the generation and intracellular trafficking of peptide-receptive major histocompatibility complex class I molecules. Immunity. 1995;2:137–147. doi: 10.1016/s1074-7613(95)80014-x. [DOI] [PubMed] [Google Scholar]
- 12.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 13.Gellin BG, Broome CV. Listeriosis. JAMA (J Am Med Assoc) 1989;261:1313–1320. [PubMed] [Google Scholar]
- 14.Bielecki J, Youngman P, Connelly P, Portnoy DA. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenescan grow in mammalian cells. Nature (Lond) 1990;345:175–176. doi: 10.1038/345175a0. [DOI] [PubMed] [Google Scholar]
- 15.Pamer EG, Harty JT, Bevan MJ. Precise prediction of a dominant class I MHC–restricted epitope of Listeria monocytogenes. . Nature (Lond) 1991;353:852–855. doi: 10.1038/353852a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pamer EG. Direct sequence identification and kinetic analysis of an MHC class I–restricted Listeria monocytogenesCTL epitope. J Immunol. 1994;152:686–694. [PubMed] [Google Scholar]
- 17.Sijts AJAM, Neisig A, Neefjes J, Pamer EG. Two Listeria monocytogenesCTL epitopes are processed from the same antigen with different efficiencies. J Immunol. 1996;156:685–692. [PubMed] [Google Scholar]
- 18.Sijts AJAM, Villanueva MS, Pamer EG. CTL epitope generation is tightly linked to cellular proteolysis of a Listeria monocytogenesantigen. J Immunol. 1996;156:1497–1503. [PubMed] [Google Scholar]
- 19.Villanueva MS, Fisher P, Feen K, Pamer EG. Efficiency of MHC class I antigen processing: a quantitative analysis. Immunity. 1994;1:479–489. doi: 10.1016/1074-7613(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 20.Villanueva MS, Sijts AJAM, Pamer EG. Listeriolysin is processed efficiently into an MHC class I–associated epitope in Listeria monocytogenes–infected cells. J Immunol. 1995;155:5227–5233. [PubMed] [Google Scholar]
- 21.Niedermann G, Butz S, Ihlenfeldt HG, Grimm R, Lucchiari M, Hoschützky H, Jung G, Maier B, Eichmann K. Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity. 1995;2:289–299. doi: 10.1016/1074-7613(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 22.Ossendorp F, Eggers M, Neisig A, Ruppert T, Goettrup M, Sijts A, Mengedé E, Kloetzel P-M, Neefjes J, Koszinowski U, Melief C. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- 23.Gournier H, Pascolo S, Siegrist CA, Jehan J, Perarnau B, Garcia Z, Rose T, Neefjes J, Lemonnier TA. Restriction of self-antigen presentation to cytolytic T lymphocytes by mouse peptide pumps. Eur J Immunol. 1995;25:2019–2026. doi: 10.1002/eji.1830250733. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Khilko S, Fecondo J, Margulies DH, McClukey J. Determinant selection of major histocompatibility complex class I–restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J Exp Med. 1994;180:1471–1483. doi: 10.1084/jem.180.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ljunggren, H.G., C. Öhlen, P. Höglund, L. Franksson, and K. Kärre. 1991. The RMA-S lymphoma mutant: consequences of a peptide loading defect on immunological recognition and graft rejection. Int. J. Cancer. (Suppl.)6:38–44. [DOI] [PubMed]
- 26.Villanueva MS, Beckers CJM, Pamer EG. Infection with Listeria monocytogenesimpairs sialic acid addition to host cell glycoproteins. J Exp Med. 1994;180:2137–2145. doi: 10.1084/jem.180.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiertz EJHJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 29.Nelson CA, Petzold SJ, Unanue E. Peptides determine the lifespan of MHC class II molecules in the antigen-presenting cell. Nature (Lond) 1994;371:250–252. doi: 10.1038/371250a0. [DOI] [PubMed] [Google Scholar]
- 30.Levitsky V, Zhang Q-J, Levitskaya J, Masucci MG. The life span of major histocompatibility complex–peptide complexes influences the efficiency of presentation and immunogenicity of two class I–restricted cytotoxic T lymphocyte epitopes in the Epstein-Barr virus nuclear antigen 4. J Exp Med. 1996;183:915–926. doi: 10.1084/jem.183.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijh, S., and E.G. Pamer. 1997. Immunodominant and subdominant CTL responses to Listeria monocytogenes infection. J. Immunol. In press. [PubMed]
- 32.van der Burg SH, Visseren MJW, Brandt RMP, Kast WM, Melief CJM. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC–peptide complex stability. J Immunol. 1996;156:3308–3314. [PubMed] [Google Scholar]
- 33.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II αβ dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 34.Fung-Leung W-P, Surh CD, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H-2-M–deficient mice. Science (Wash DC) 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 35.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain–derived peptide. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 36.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature (Lond) 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 37.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR–bound peptides in the presence of HLA-DM. Science (Wash DC) 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]